Abstract
Altered expression of specific microRNAs (miRNAs) has been observed in human cervical cancer. However, the biological functions of many of these miRNAs are yet to be discovered. We previously showed that miR-944 is significantly more abundant in cervical cancer tissues than their normal counterparts. In this study, we investigated the functions and targets of miR-944 in human cervical cancer cells. MiR-944 is located in the intron of the tumor protein p63 (TP63) gene, which is frequently overexpressed in cervical carcinomas. Using gain- and loss-of-function experiments in vitro, we demonstrate that miR-944 promotes cell proliferation, migration and invasion, but has no effect on apoptosis, in human cervical cancer cells. To identify the targets of miR-944, we performed photoactivatable-ribonucleoside-enhanced crosslinking and immunoprecipitation followed by deep sequencing. Among the candidate targets, we validated HECW2 (HECT domain ligase W2) and S100PBP (S100P binding protein) as direct targets of miR-944 using luciferase reporter assays and western blot analysis. Our findings reveal novel functions and targets of miR-944 in human cervical cancer cells, which may provide new insights of its role in cervical carcinogenesis.
What's new?
While miR-944 has been shown to be associated with tumor development and progression in several tumor types, its functions and targets remain undetermined. This study stands out as the first report of miR-944 functions and targets in human cancer. The authors demonstrate that miR-944 functions as an oncogene in human cervical cancer cells by promoting cell proliferation, migration, and invasion. In addition, they identified and validated HECW2 and S100PBP as direct targets of miR-944 in human cervical cancer cells. The findings provide new insights into the biological roles of miR-944 in human cervical cancer cells.
Keywords: miR-944, PAR-CLIP, cervical cancer, proliferation, migration, invasion
MicroRNAs (miRNAs), small noncoding RNAs of ∼22 nucleotides in length, generally act as negative regulators of gene expression at post-transcriptional level through mRNA degradation and/or translation repression. These molecules play important roles in many important biological processes, including cell growth, apoptosis, viral infection and cancer development.
Previous studies from our group and others have shown that miRNAs are deregulated in human cervical cancer.1–7 Several of these differentially expressed miRNAs have been consistently observed in different studies, suggesting that these miRNAs (or their targets) are important for cervical cancer development and progression. Yet, only few miRNAs have been functionally characterized in human cervical cancer cells.
In this study, we investigated the functions and targets of miR-944, which is one of the most significant deregulated miRNAs in human cervical cancer identified in our previous study.6 MiR-944 was first identified in human cervical cells using a small RNA cloning approach.2 This miRNA is located in the intron of tumor protein p63 (TP63) gene and mapped to human 3q28, a region frequently amplified in cervical cancer.8,9 To date, nothing is known about the function of this miRNA and its targets remain to be identified.
Here, we used the PAR-CLIP (Photoactivatable-Ribonucleoside-Enhanced Crosslinking and Immunoprecipitation) sequencing approach to identify the targets of miR-944. This approach has been previously applied to identify RNA binding sites of miRNAs and RNA binding proteins.10 The method is based on the site-specific incorporation of photoreactive ribonucleoside analogs [e.g., 4-thiouridine (4-SU) and 6-thioguanosine (6-SG)] into RNA transcripts by living cells, followed by crosslinking of photoreactive nucleoside-labeled cellular RNAs to interacting RNA binding proteins by ultraviolet (UV) irradiation. This method provides more efficient UV crosslinking and immunoprecipitation and allows identification of the precise position of crosslinking by mutations residing in the sequenced cDNA; which makes it possible to be separated from the background sequences derived from abundant cellular RNAs.
Herein, we describe the functions and targets of miR-944 in human cervical cancer cells. Our data suggest that miR-944 plays an oncogenic role in cervical cancer cells by promoting cell proliferation, migration and invasion. Using the PAR-CLIP sequencing approach, we identified a set of miR-944 targets and two of them were further validated as direct targets of miR-944 by luciferase reporter assays and western blot analysis.
Material and Methods
Cervical cancer tissue samples and cell lines
Twenty-seven pairs of frozen cervical tumors and matched normal tissues were provided by the Gynecologic Oncology Group Tissue Bank (Columbus, OH). All samples were included in our previous sequencing-based small RNA profiling study.6 The study was approved by the local ethical committee. Seven human cervical cancer cell lines (CaSki, HeLa, SW756, ME-180, SiHa, C4I and C33A) were purchased from the American Type Culture Collection and the culture conditions were described previously.11 In brief, CaSki and ME-180 cells were cultured in RPMI 1640 and the other cell lines were grown in DMEM medium, supplemented with 10% FBS. Authentications of HeLa and CaSki cells were recently verified by short tandem repeats profiling, as performed by Bio-Synthesis (Lewisville, TX).
RNA extraction
mirVana miRNA isolation kit (Applied Biosystems/Ambion, Austin, TX) was used to extract RNA from tissue samples and cell lines. For tissue samples, extractions of small RNAs (<200-nt) and large RNAs (≥200-nt) were performed according to the manufacturer's protocol. For cell lines, total RNA isolation protocol was performed. RNA concentrations were measured using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE) and stored at −80°C for further application.
TaqMan reverse transcription quantitative PCR (RT-qPCR)
MiR-944 and TP63 expressions were determined by RT-qPCR using the StepOnePlus™ Real-Time PCR system or 7900HT Real-Time PCR System (Life technologies, Carlsbad, CA). Predesigned TaqMan assays for miR-944 (ID 002189), TP63 (ID Hs00978340_m1), RNU6B (ID 001093) and 18S (ID Hs99999901_s1) were purchased from Applied Biosystems.
For mature miRNA detection, cDNA was synthesized from 120 ng of total RNAs (cell lines) or 30 ng small RNAs (clinical samples) using TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems). For mRNA expression detection, cDNA was synthesized from 200 ng large RNAs using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). All reactions were performed in triplicate. The relative expression levels of miR-944 and TP63 were normalized by RNU6B and 18S, respectively, and reported as 2−ΔCT.
miR-944 overexpression and inhibition
All the miRNA mimics and inhibitors used in this study were purchased from Applied Biosystems/Ambion. For gain-of-function experiments, HeLa, CaSki and SW756 cells were transfected with 10 nM Pre-miR™ miR-944 precursor (ID PM12272) or Pre-miR Negative control #1 (ID AM17110). For loss-of-function experiments, CaSki cells were transfected with 50 nM of Anti-miR™ miR-944 inhibitor (ID AM12272) or Anti-miR Negative control #1 (ID AM17010) in parallel. All cells were transfected using siPORT NeoFX transfection agent (Applied Biosystems/Ambion) following the manufacturer's instruction.
Cell growth
Cell growth was assessed by WST-1 colorimetric assay (Roche Applied Science, Mannheim, Germany) and trypan blue exclusion assay. For WST-1 assay, a total of 2.5 × 103 HeLa or 5 × 103 CaSki cells per well in 100 μL culture medium were seeded into 96-well plate. At different time points (0, 24, 48, 72 and 96 hr post-transfection), 10 μL of WST-1 reagent was added into each well and incubated for 3 hr at 37°C. Absorbances at 450 nm (detection) and 650 nm (reference) were determined by VERSAmax microplate reader (Molecular Devices, Sunnyvale, CA) and analyzed with SoftMax Pro 5 software (Molecular Devices). Each experimental group consisted of eight replicate wells for each time point. Relative cell growth rate was evaluated by subtracting the background absorbance individually and normalized to 0 hr time point. All experiments were performed three times independently.
In trypan blue exclusion assay, 8 × 104 cells per well were seeded into 24-well plate. At different time points (0, 24, 48, 72 and 96 hr post-transfection), cells were collected and stained with 0.4% Trypan Blue Stain (Invitrogen, Camarillo, CA) and analyzed using the TC10™ automated cell counter (Bio-Rad, Hercules, CA). The relative cell growth rate was determined by calculating the total living cells and normalized to 0 hr time point. All experiments were performed three times independently.
Wound healing scratch assay
A total of 3.5 × 105 cells (in 2.5 mL/well) were transfected and seeded on a 6-well plate. After 48 hr of transfection, a scratch wound was made on the confluent monolayer cells and fresh culture medium with or without 5 mM hydroxyurea was added to the cells. The hydroxyurea concentration was determined by screening the effects of different concentrations on cell proliferation and viability in HeLa and CaSki cells, which indicated that 5 mM hydroxyurea was sufficient to inhibit cell growth with minimal effects on cell viability for both cell lines (data not shown). The scratch was imaged at three different time points (0, 12 and 18 hr) using an inverted microscope (Leica DM IL LED, Leica Microsystems GmbH, Wetzlar, Germany) equipped with ProgRes® MF camera (Jenoptik GmbH, Jena, Germany). Image J software version 1.43u (http://rsbweb.nih.gov/ij/) was used to process all images for quantification purposes. The percentage of wound closure (cell migration) was compared with the wound area at 0 hr. The relative wound closure in Pre-miR-944 or Anti-miR-944 treated group was normalized by the respective negative control. All experiments were performed independently in triplicate.
Transwell cell migration and invasion assays
For cell migration, polyethylene terephthalate (PET) cell culture inserts with 8.0 μm pores (BD Biosciences, Franklin lakes, NJ) were placed in 24-well plate and equilibration for 30 min with 750 μL of culture medium and 20% FBS in the lower chamber. Cells were harvested after 48 hr of transfection, washed with PBS three times and resuspended in serum-free medium at 5 × 105 cells/mL. Cells (100 μL) were added to the upper chamber and incubated for 24 hr (HeLa and SW756 cells) or 48 hr (CaSki cells) at 37°C with 5% CO2.
For cell invasion, the PET inserts were coated with 100 μL freshly diluted Basement Membrane Matrix (cat. no. 354234, BD Biosciences; 1:25 dilution with chilled 10 mM Tris/ 0.7% NaCl, pH 8.0), placed in a 24-well plate and incubated at 37°C for 3 hr. A total of 750 μL culture medium with 20% FBS was added into the lower chamber as chemoattractant. After 48 hr of transfection, cells were harvested, washed three times with PBS and resuspended in serum-free medium at 2 × 105 cells/mL. Five-hundred microliters of cells were added to the upper chamber and incubated for another 48 hr at 37°C with 5% CO2.
Cotton swabs were used to remove the cells or mixture of matrix and cells on the top surface of membrane. Migrated/invaded cells were fixed with 4% paraformaldehyde solution (USB Corporation, Cleveland, OH) for 10 min, washed with PBS and stained with 0.5% crystal violet (prepared in 20% ethanol) for 10 min. The inserts were rinsed with water and air-dried at room temperature. An inverted microscope (Leica DM IL LED) equipped with ProgRes® MF camera (Jenoptik GmbH) was used to take images. For quantification, the dyes obtained by migrated/invaded cells were dissolved in de-staining solution (50% ethanol/0.1% acetic acid) by gently shaking for 15 min at room temperature, followed by determining the absorbance reading at 570 nm (detection) and 630 nm (reference). Relative cell migration or invasion was normalized by the respective negative controls. All the experiments were carried out independently in triplicate.
Cell proliferation and migration analysis using the xCELLigence system
xCELLigence Real-Time Cell Analyzer DP instrument (Roche Applied Science) was used to monitor cell proliferation and migration in real-time. Microelectronic plates E-plate 16 and CIM-plate 16 were used for cell proliferation and cell migration measurements, respectively. For cell proliferation measurement, the E-plate was filled with 100 μL culture medium per well and put in the cradle of the instrument for equilibration and background measurement for 30 min. A total of 1 × 104 cells (in 100 μL culture medium, collected at 48 hr post-transfection) were seeded in each well. The E-plate was placed in the RTCA DP instrument equilibrated in a CO2 incubator. Proliferation of the cells was measured over a time period of up to 72 hr.
For cell migration measurement, 160 μL prewarmed culture medium supplemented with 20% FBS were added into the lower chamber of the CIM-plate and the upper chamber was filled with 30 μL of serum-free medium to immense the PET membrane surface. The CIM-plate was loaded onto the RTCA DP analyzer inside the CO2 incubator and incubated for 2 hr to equilibrate the membrane surface with the medium. Cells were collected after 48 hr of transfection, washed with PBS and resuspended in serum-free medium at 5 × 105 cells/mL. Cell suspension (100 μL) was added into each well. Migration of the cells was measured over a time period of up to 27 hr. At least four replicate wells were performed in each experimental group and all experiments were performed at least twice independently. Data were analyzed by RTCA software version 1.2.1 (Roche Applied Science).
Apoptosis assays
Apoptosis was performed using the ApoTarget Annexin-V FITC apoptosis kit (Invitrogen) and/or caspase-3 colorimetric assay (BioVision, Mountain View, CA). For Annexin-V assay, cells were collected after 66 hr of transfection, washed with PBS twice, followed by 1× Binding Buffer once and resuspended in 1× Binding Buffer at 2 × 106 cells/mL. Five microliters Annexin-V-FITC was added to 100 μL cell suspension, gently mixed and incubated for 15 min in the dark followed by addition of 400 μL 1× Binding Buffer and 5 μL propidium iodide (PI). The cells were analyzed by flow cytometry (Cytomics FC 500; Beckman Coulter, Brea, CA) and quantified by FlowJo software version 7.6.2 (http://www.flowjo.com/index.php).
Caspase-3 colorimetric assay was performed as described previously.11 In brief, 3 × 106 transfected cells were harvested after 72 hr of transfection and lysed in chilled cell lysis buffer. One-hundred micrograms of protein lysate (in 50 μL) was mixed with equal volume of 2× Reaction Buffer and 5 μL of 4 mM caspase-3 substrate (DEVD-pNA) and incubated for 1 hr at 37°C. Detection of the chromophore p-nitroaniline (pNA) was measured at 405 nm using a VERSAmax microplate reader (Molecular Devices) and analyzed with SoftMax Pro 5 software (Molecular Devices). As for positive control, cells were treated with 100 μM camptothecin (Sigma-Aldrich) for 15 hr before apoptosis assay was performed. All experiments were performed independently in triplicate.
Cell cycle analysis
One million cells were harvested after 48 hr of transfection, washed with PBS and then fixed in 50% ethanol. Cells were treated with 2.5 μL RNase A (20 mg/mL; Sigma-Aldrich) for 1 hr at 37°C. Ten microliters of PI (1 mg/mL; Sigma-Aldrich) was added into cell suspension and mixed gently. PI staining was determined by flow cytometry (Cytomics FC 500) and further analyzed by FlowJo software version 7.6.2. All experiments were performed independently in triplicate.
Photoactivatable-ribonucleoside-enhanced crosslinking and immunoprecipitation (PAR-CLIP)
PAR-CLIP was performed as previously described,10 with slight modifications. Approximately 2 × 107 HeLa cells were transfected with 10 nM Pre-miR-944 and seeded in ten pieces of 10-cm tissue culture plates. Mock transfection was used as a negative control. After 48 hr of transfection, 4-thiouridine (4-SU; 100 μM; Sigma-Aldrich) was added and incubated for 14–18 hr. The cells were then crosslinked with 365 nm UV light (UVGL-25; UVP, Upland, CA) for 30 sec. Cells were lysed in an equal volume (w/v) of NP40 lysis buffer (Invitrogen), supplemented with 1 mM phenylmethanesulfonyl fluoride (PMSF; Sigma-Aldrich), 1 mM dithiothreitol (DTT; Invitrogen), 1% Protease Inhibitor Cocktail (Sigma-Aldrich) and 200 U/mL RNase inhibitor (Applied Biosystems). The lysates were cleared by centrifugation at 14,000 rpm for 30 min at 4°C. To prepare antibody-coated beads, 120 μL of Protein G Sepharose 4 Fast Flow bead slurry (GE Healthcare, Little Chalfont, UK) was washed three times with 1 mL of NT2 buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM MgCl2 and 0.5% NP40) and incubated with 5 μg of mouse anti-human Ago2/eIF2C2 antibody (ab57113; Abcam, Cambridge, UK) on a rotator at 4°C for 6 hr. The beads were then washed with cold NT2 buffer three times to remove the unbound antibodies. For immunoprecipitation, the cleared lysates were incubated with the antibody-coated Sepharose beads (in NT2 buffer supplemented with 1 mM DTT, 200 U/mL RNase inhibitor and 20 mM EDTA) overnight at 4°C on a rotator. The beads were washed three times with cold NT2 buffer for 10 min each at 4°C, followed by RNase T1 treatment (100 U/μL; Fermentas, St. Leon-Rot, Germany) for 10 min at 22°C and cooled on ice for 5 min. Subsequently, the beads were washed three times with NT2 buffer and incubated with proteinase K (10 mg/mL) for 1 hr at 55°C. The RNA was extracted with TRIzol reagent (Invitrogen) and stored at −80oC for further use.
Construction of small RNA sequencing (sRNA-seq) libraries
The RNAs obtained by Ago2 IP (∼200 ng in each sample) from four replicate experiments (Pre-miR-944 versus mock control in HeLa cells) were used for sRNA-seq preparation following protocols as described previously,6 with minor modifications. In brief, the RNAs were incubated with alkaline phosphatase (Takara Bio Inc, Otsu, Japan) at a final concentration of 0.8 U/μL for 30 min at 37°C. After phenol-chloroform extraction and ethanol precipitation, the RNAs were incubated with 100 μM preadenylated 3′-adaptor oligonucleotide (Linker-1, IDT Inc; Coralville, Iowa), 5% DMSO (Sigma-Aldrich) and 200 units T4 RNA ligase 2 (truncated K227Q; New England Biolabs, Ipswich, MA) at 16°C for overnight. Next, the ligated products were purified on a 12% denaturing polyacrylamide gel, followed by incubation with T4 polynucleotide kinase (New England Biolabs) at 37°C for 30 min and a second ligation reaction with a 5′-adaptor oligonucleotide, 5′-ACG CTC TTC CGA TCTv−3′ (uppercase, DNA; v represents barcode with triple RNA molecules, aaa, ggg, ccc or uuu; IDT) at 37oC for 1 hr. The doubly ligated RNA molecules were reverse transcribed using 150 units Superscript II (Invitrogen) and RT primer, 5′-ATT GAT GGT GCC TAC AG-3′. The cDNA was amplified with 20 PCR cycles, using a forward primer 5′-GAT ACG GCG ACC ACC GAG ATC TAC ACT CTT TCC CTA CAC GAC GCT CTT CCG ATC T-3′ and a reverse primer 5′-CAA GCA GAA GAC GGC ATA CGA GCT CTT CCG ATC TAT TGA TGG TGC CTA CAG-3′ to produce sequencing libraries that were subjected to Solexa/Illumina sequencing platform.
PAR-CLIP sequencing data analysis
After removal of barcodes, adaptors and linker-linker ligated sequences, we compressed the identical sequences as one unique sequence read by recording read counts. The collapsed sequence reads were aligned to UCSC human genome (hg19) using Bowtie (v2.0.0) and then processed by PARalyzer v1.1 (http://www.genome.duke.edu/labs/ohler/research/PARalyzer) using the default settings that allow a minimum sequence length of 13 nucleotides, at most two conversions (T to C) per read, a sequence read count must occur at least five times, extension of overlapped region by five nucleotides on each side and reads without T to C conversions were discarded.12 Reads mapped to repeat elements were filtered and the remaining reads that overlapped by at least a single nucleotide were grouped together as a cluster. The clusters were annotated based on human gene annotation from UCSC genome browser (hg19). The candidate targets were compared with the targets predicted by TargetScan 6.2 (http://www.targetscan.org/).
Luciferase reporter assay
The pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega Corporation, Madison, WI) was used to construct all the reporter vectors. The wild-type and mutated-type of the putative miR-944 binding sites of HECT domain ligase W2 (HECW2) and S100PBP were cloned into 3′-UTR downstream of luc2 firefly luciferase gene at PmeI and XbaI sites following the manufacturer's instruction. All constructs were confirmed by sequencing at KIGene core facility using pmirGLO R primer (5′-CAC TGC ATT CTA GTT GTG GT-3′). All plasmids were purified using Qiagen Plasmid Mini Kit (Qiagen, Hildane, Germany).
The pmirGLO reporter plasmids (60 ng) and Pre-miR-944 or Pre-miR-NC#1 (5 pmol) were cotransfected into HeLa cells (2 × 104 cells/well in a 96-well plate) using Lipofectamine 2000 Transfection Reagent (Invitrogen). After 24 hr of transfection, Dual-GLO Luciferase Assay System (Promega Corporation) was used to determine the firefly and Renilla luciferase luminescence signals using a microplate luminometer (Centro LB 960, Berthold Technologies, Bad Wildbad, Germany). Data were analyzed by dividing firefly luciferase activity with Renilla luciferase activity for each transfectant and further normalized to the empty pmirGLO vector transfectant. All experiments were performed independently three times.
Western blot analysis
After 48 hr of transfection, cells were collected and lysed for immunoblot analysis as described previously.11 In brief, 200 μg whole cell lysate was separated in 4–12% Bis-Tris gel (Invitrogen) and transferred to nitrocellulose membrane (Invitrogen). Rabbit polyclonal HECW2/NEDL2 antibody (1:1000; ab154888, Abcam, Cambridge, UK) and mouse polyclonal S100PBP antibody (1:500; ab68733, Abcam) were used to determine HECW2 and S100PBP expressions, respectively. GAPDH (1:10000; sc-47724; Santa Cruz Biotechnology) was used as normalization. Signals were detected by LAS-1000 Image Analyzer (Fujifilm, Tokyo, Japan) and protein expressions were quantified using Image Gauge v4.0 (Fujifilm).
Statistical analysis
All analyses were performed using Statistica 7.0 (StatSoft, Tulsa, OK) or MS Office Excel 2007. Student t-test with equal variance was performed to compare mean relative changes between the tested and control samples from three independent experiments. All analyses were 2-tailed and p-values <0.05 were considered statistically significant.
Results
miR-944 and TP63 expressions in human cervical cancer samples and cell lines
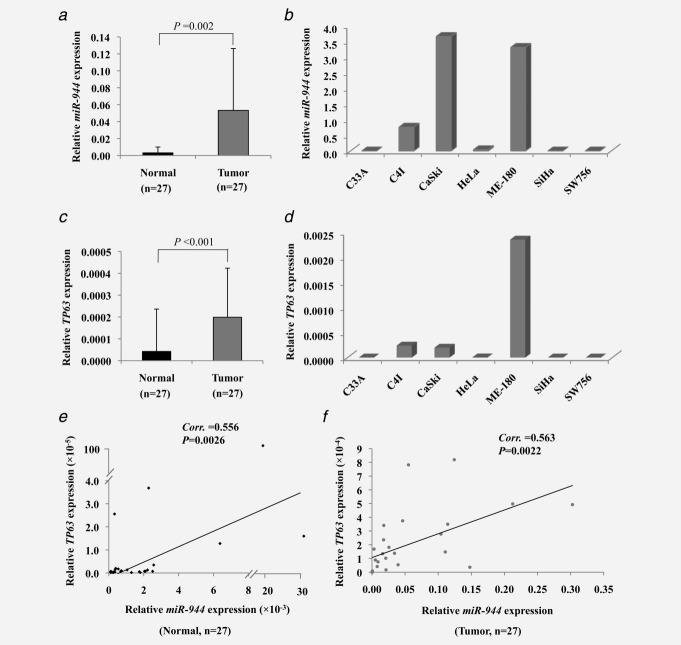
We investigated the expressions of miR-944 and its host gene TP63 in a series of 27 cervical tumor-normal pairs and seven cervical cancer cell lines by RT-qPCR. miR-944 expression was significantly higher in the cancer samples compared with matched normal tissues (p = 0.002; Fig. 1a and Supporting Information Fig. S1a). Similarly, we also observed significantly higher expression of TP63 in the cancer samples than their normal counterparts (p < 0.001; Fig. 1c and Supporting Information Fig. S1b). The expression of miR-944 was significantly correlated with the expression of TP63 (Corr. = 0.556, p = 0.0026 in normal tissues; Corr. = 0.563, p = 0.0022 in tumors; Figs. 1e and 1f); supporting that miR-944 is likely co-transcribed with its host gene TP63. Among the cervical cancer cell lines analyzed, miR-944 and TP63 expressions were relatively higher in CaSki, ME-180 and C4I cells, while their levels were very low in C33A, HeLa, SiHa and SW756 (Figs. 1b and 1d).
Figure 1.
miR-944 and TP63 expressions in human cervical tumor-normal pairs and cancer cell lines quantified by RT-qPCR. Relative miR-944 (a,b) and TP63 (c,d) expressions in normal cervices, cervical tumors and cervical cancer cell lines as normalized to RNU6B and 18S, respectively. The correlation of miR-944 and TP63 expressions in normal cervices (e) and cervical tumors (f) were evaluated by Pearson's correlation analysis. p < 0.05 was considered statistically significant.
For functional studies, we performed miR-944 overexpression in HeLa cells, which expressed very low level of miR-944. In addition, we also performed both gain- and loss-of-function of miR-944 in CaSki cells to complement each other's effects. To assess the efficiency of miR-944 overexpression and inhibition, we quantified the changes of miR-944 expression by RT-qPCR after 48 hr of transfection. As shown in Supporting Information Figure S2a and b, miR-944 expression was significantly increased in both HeLa and CaSki cells transfected with Pre-miR-944, or significantly decreased in CaSki cells transfected with Anti-miR-944, as compared with their respective negative controls.
miR-944 expression promotes cell proliferation
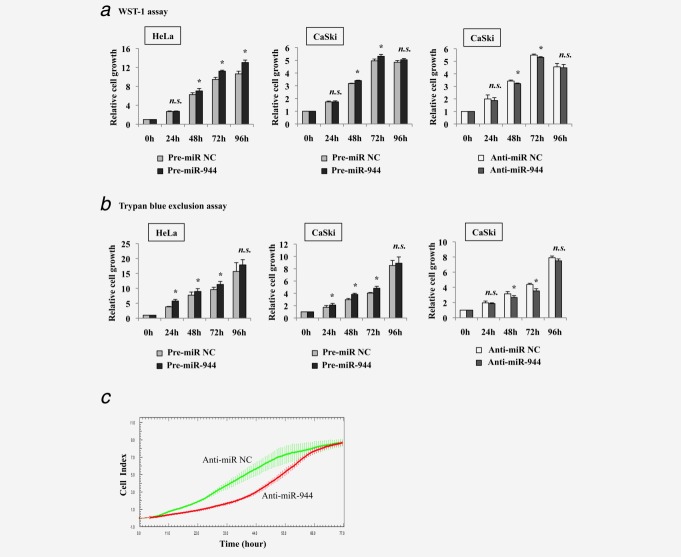
Assessment from using both WST-1 assay and trypan blue exclusion assay, we observed that overexpression of miR-944 in HeLa and CaSki cells resulted in significant increases of cell proliferation (Figs. 2a and 2b). In complement to the gain-of-function experiments, inhibition of miR-944 expression in CaSki cells led to a significant decrease in cell growth (Figs. 2a and 2b).
Figure 2.
Regulation of cell proliferation by miR-944 expression in cervical cancer cells. Relative cell growth rates were evaluated in miR-944 overexpressing HeLa cells and CaSki cells and miR-944 inhibition in CaSki cells using (a) WST-1 assay and (b) trypan blue exclusion assay at different time points and normalized to 0 hr. Data represent mean of three independent experiments. Error bars represent standard deviations from the mean. All comparisons were evaluated using t-test. *p < 0.05; n.s.: not significant. (c) xCELLigence system was used to monitor dynamic cell proliferation in real-time. CaSki cells transfected with Anti-miR-944 or Anti-miR NC (negative control) were seeded on E-plate of the xCELLigene RTCA instrument after 48 hr of transfection and monitored for another 72 hr. Cell index refers to a relative change in electrical impedance representing the number of cells detected on the microelectrodes on the bottom of the plate.
In addition, we also measured the effect of miR-944 deregulation on cell proliferation in real-time using the xCELLigence system. We observed decrease of cell proliferation in CaSki cells upon miR-944 inhibition from ∼12 hr and the effect was maintained for ∼60 hr after seeding (i.e., five days after transfection; Fig. 2c).
miR-944 expression promotes cell migration and invasion
We evaluated the effect of miR-944 regulation on cell migration using three different methods: real-time monitoring cell migration using the xCELLigence system, transwell cell migration assay and wound healing scratch assay. Using xCELLigence system in HeLa cells, we started to observe different migration rates between cells with and without miR-944 overexpression at ∼8 hr (Fig. 3a). To determine the cell migration rates, we compared the slopes of the curves between 8 and 27 hr (which represent the rate of change of the cell index) from cells transfected with Pre-miR-944 and its negative control. A significantly higher migration rate was observed in the Pre-miR-944 treated cells compared with its negative control (p = 0.006; Fig. 3b).
Figure 3.
Effect of cell migration and invasion by miR-944 expression in cervical cancer cells. (a,b) Cell migration in HeLa cells transfected with Pre-miR-944 or Pre-miR NC was measured by the xCELLigence RTCA DP instrument. After 48 hr of transfection, cells were seeded on CIM-plate and monitored for additional 27 hr. (a) Cell index represents relative number of migrated cells detected by the microelectrodes on the bottom of the PET membrane. (b) The migration rate was calculated from the slopes of the curves between 8 and 27 hr. (c) Cell migration and (d) cell invasion were evaluated using Transwell assays. Relative cell migration and invasion in miR-944 overexpression or inhibition were normalized to their respective control treated cells. Data presented represent mean of three independent experiments. Error bars represent standard deviations from the mean. All comparisons were evaluated using t-test. **p < 0.01; n.s.: not significant.
Similar results were observed using the transwell cell migration assay, in which cell migration was significantly enhanced in miR-944 overexpressing HeLa (∼24%; p = 0.005; Fig. 3c and Supporting Information Fig. S3a) and SW756 cells (∼29%; p = 0.05; Supporting Information Fig. S3b). However, we did not observe any significant changes of cell migration upon miR-944 overexpression or inhibition in CaSki cells (Fig. 3c and Supporting Information Fig. S3a).
In wound healing scratch assay, we revealed that miR-944 overexpression in both HeLa and CaSki cells significantly enhanced the ability to close the wound areas as compared with the respective negative controls at 18 hr (p = 0.01 and p = 0.003, respectively; Supporting Information Fig. S4a). However, miR-944 suppression in CaSki cells did not show any significant difference on wound closure rate compared with its negative control (Supporting Information Fig. S4a). To distinguish the contributions of cell proliferation and migration to wound closure, we re-evaluated the wound healing assay in the presence of cell cycle blocker hydroxyurea. We did not observe any significant effect in HeLa cells upon miR-944 overexpression or in CaSki cells upon miR-944 overexpression or suppression (Supporting Information Fig. S4b); suggesting that the wound closure effect by miR-944 is likely contributed by cell proliferation, instead of cell migration.
For cell invasion, we applied the transwell invasion assays to evaluate cell invasion capacity of HeLa and CaSki cells upon modulation of miR-944 expression levels. In HeLa cells, we showed that cell invasion was significantly increased upon miR-944 overexpression (p = 0.002). However, we did not observe any significant changes of cell invasion in CaSki cells either with miR-944 overexpression or suppression (Fig. 3d and Supporting Information Fig. S3c).
Altered miR-944 expression has no effect on cell apoptosis and cell cycle
To determine the effect on cell apoptosis, we used Annexin-V/PI double staining and caspase-3 assays in miR-944 overexpression of HeLa and CaSki cells, as well as miR-944 suppression of CaSki cells. In Annexin-V assay, we did not observe any significant changes of cell apoptosis (both early and late events) neither in HeLa nor CaSki cells comparing with their respective negative controls (Supporting Information Figs. S5a and S5b). Both miR-944 overexpression and inhibition had no significant effects on cell apoptosis. In concordance with the Annexin-V results, we did not observe any significant changes of caspase-3 activity in Pre-miR-944 treated HeLa cells as compared with the negative control (Supporting Information Fig. S5c).
To determine whether miR-944 plays any role in cell cycle, we analyzed cell cycle distribution in HeLa, CaSki and SW756 cells transfected with Pre-miR-944, as well as in CaSki cells transfected with Anti-miR-944 using PI staining. In all three cell lines, we did not observe any cell cycle arrest neither at G0/G1 nor G2/M phase (Supporting Information Fig. S5d).
Identification of miR-944 targets by PAR-CLIP sequencing
To further understand the biological roles of miR-944, we performed PAR-CLIP approach to identify its targets. In total, we obtained 1,324,451 reads from eight libraries (four replicates from each miR-944 overexpressing and mock control HeLa cells). Among them, 1,118,875 reads were obtained from Pre-miR-944 treated cells and the remaining 205,576 reads were obtained from the control groups. Due to the sequence reads in each library were not sufficient for quantitative analysis, we compared the presence of transcripts between the Pre-miR-944 treated and control groups. We found 58 transcripts (in 64 clusters) present in the miR-944 overexpressing cells and only two of them (i.e., CRYAB and RRBP1) were also present in the control group. Among the unique transcripts present in the miR-944 overexpressing cells, 25 were aligned to 3′UTR, 23 to coding region and four to 5′UTR of target mRNAs and the remaining six transcripts were aligned to noncoding RNAs (Supporting Information Table S1).
Interestingly, 19 of the 58 transcripts from the PAR-CLIP data are also predicted targets of miR-944 by TargetScan 6.2. Of these 19 candidate targets, two of them, HECW2 and S100PBP, have conserved miR-944 binding sites and the remaining 17 targets have poorly conserved sites (Table1). All transcripts present in the miR-944 overexpressing and the control groups are listed in Supporting Information Tables S1 and S2, respectively.
Table 1.
Candidate targets of miR-944 found in both PAR-CLIP sequencing and TargetScan 6.2
| TargetScan 6.2 |
||||||
|---|---|---|---|---|---|---|
| Gene symbol | Description | Read count | Transcript location | Binding site(s) | Type of seed match | |
| HECW2 | HECT, C2 and WW domain containing E3 ubiquitio protein ligase 2 | 13 | 3′UTR | 11 | 7mer-m8 | |
| KLHDC10 | Kelch domain containing 10 | 11 | 3′UTR | 1 | 7mer-m8 | |
| SEC22B | SEC22 vesicle trafficking protein homolog B (S. cerevisiae) | 8 | 3′UTR | 1 | 7mer-1A | |
| RPP30 | Ribonuclease P/MRP 30kDa subunit | 7 | 3′UTR | 1 | 7mer-m8 | |
| BVES | Blood vessel epicardial substance | 7 | 3′UTR | 1 | 7mer-1A | |
| SHE | Src homolog 2 domain containing E | 6 | 3′UTR | 1 | 7mer-m8 | |
| FGD4 | FYVE, RhoGEF and PH domain containing 4 | 6 | 3′UTR | 3 | 7mer-m8, 7mer-1A | |
| S100PBP | S100P binding protein | 5 | 3′UTR | 11 | 7mer-m8 | |
| DDHD1 | DDHD domain containing 1 | 5 | 3′UTR | 1 | 7mer-m8 | |
| ACTR2 | ARP2 actin-related protein 2 homolog (yeast) | 5 | 3′UTR | 2 | 8mer | |
| ACSL3 | Acyl-CoA synthetase long-chain family member 3 | 5 | 3′UTR | 1 | 7mer-1A | |
| LRBA | LPS-responsive vesicle trafficking, beach and anchor containing | 5 | 3′UTR | 1 | 7mer-m8 | |
| PLEKHA2 | Pleckstrin homology domain containing, family A, member 2 | 5 | 3′UTR | 1 | 7mer-m8 | |
| PRPS2 | Phosphoribosyl pyrophosphate synthetase 2 | 5 | 3′UTR | 1 | 7mer-1A | |
| MCHR2 | Melanin-concentrating hormone receptor 2 | 7 | CDS | 1 | 7mer-1A | |
| ABCC2 | ATP-binding cassette, subfamily C (CFTR/MRP), member 2 | 5 | CDS | 2 | 7mer-m8, 7mer-1A | |
| RAB11A | RAB11A, member RAS oncogene family | 6 | CDS/intron | 3 | 7mer-m8, 7mer-1A | |
| MACF1 | Microtubule-actin crosslinking factor 1 | 5 | CDS/intron | 1 | 7mer-m8 | |
| MAP4K5 | Mitogen-activated protein kinase kinase kinase kinase 5 | 8 | 5′UTR | 1 | 7mer-1A | |
Conserved binding sites.
Abbreviations: 7mer-1A: an exact match to positions 2-7 of the mature miRNA followed by an “A”; 7mer-m8: an exact match to positions 2-8 of the mature miRNA; 8mer: an exact match to positions 2-8 of the mature miRNA followed by an “A”; CDS: coding sequence; UTR: untranslated region.
Validation of HECW2 and S100PBP as miR-944 target genes
We chose the two target genes, HECW2 and S100PBP, with conserved miR-944 binding sites for further validations using luciferase reporter assay and western blot analysis. We constructed the wild-type and mutated (four mismatches in the seed region) putative miR-944 binding sites in the 3′UTR of HECW2 and S100PBP (Figs. 4a and 4b, upper panel) into pmirGLO Dual-luciferase miRNA target expression vector. As shown in Figures 4a and 4b (lower panel), the luciferase activities were significantly decreased when the wild-type constructs of HECW2 and S100PBP were co-transfected with Pre-miR-944 as compared with the Pre-miR negative control (p = 0.0212 and p = 0.0037, respectively). As expected, the seed mutant constructs rescued the suppression of luciferase activities by miR-944.
Figure 4.
Verification of HECW2 and S100PBP as direct targets of miR-944. (a,b) The upper panels show the sequences alignment of miR-944 and the wild-type (wt) and the mutated (mut; underlined) target sequences of HECW2 (a) and S100PBP (b). The seed sequence of miR-944 is indicated in bold. The lower panels show the effect of miR-944 on luciferase activity using the luciferase reporter gene containing the wild-type and mutant of HECW2 or S100PBP 3'UTR, as evaluated 24 hr after co-transfection Pre-miR-944 or Pre-miR NC with the wild-type and mutant reporter constructs in HeLa cells. Data represent mean of three independent experiments. (c,d) Western blot analysis of HECW2 (c) and S100PBP (d) expressions upon miR-944 overexpression or inhibition in HeLa and CaSki cells. Top: Representative western blots show decreased HECW2 and S100PBP expressions in miR-944 overexpressing HeLa and CaSki cells and increased HECW2 and S100PBP expressions in miR-944 inhibiting CaSki cells, as compared with their respective negative controls. Bottom: Quantification of HECW2 and S100PBP protein levels in three independent experiments. GAPDH was used as a loading control. Error bars indicate standard deviations. All comparisons were evaluated using t-test. * p < 0.05; ** p < 0.01; n.s.: not significant.
We further analyzed HECW2 and S100PBP expressions upon miR-944 overexpression in HeLa and miR-944 overexpression or suppression in CaSki cells by western blot analysis. As shown in Figures 4c and 4d, HECW2 and S100PBP expressions were significantly decreased in miR-944 overexpressing HeLa and CaSki cells, while their expressions were significantly increased in CaSki cells upon miR-944 inhibition.
Discussion
MiR-944 expression has been detected in several cancer types, including cervical,2,6 melanoma,13 colorectal14 and bladder cancers.15 In cervical cancer and melanoma, miR-944 is more abundant in tumor samples than in their normal counterparts.6,13 High expression of miR-944 is also associated with tumor recurrence in colorectal cancer,14 and poor chemotherapy response and survival in bladder cancer.15 These findings suggest that miR-944 may play an important role in tumorigenesis and progression of several cancer types. Despite its expression pattern, nothing is known about its functions and targets. In this study, we sought to investigate the functions and targets of miR-944 in human cervical cancer cells.
miR-944 plays an oncogenic role in cervical cancer cell lines
Here, we show that miR-944 promotes cell proliferation, migration and invasion in HeLa cells. Similarly, significant effect on cell proliferation was also observed in CaSki cells. Despite that the effect of miR-944 on cell growth was subtle, significant effects were consistently observed in different employed methods. An explanation of the subtle phenotypic effect could be due to the target gene, which is involved in cell growth, is targeted by multiple miRNAs. Modulation of miR-944 expression itself is not sufficient to yield strong phenotypic effect.
Unlike HeLa cells, we did not observe significant effects on cell migration and invasion in CaSki cells neither with miR-944 overexpression nor inhibition. The apparent lack of cell migration and invasion phenotypes observed in CaSki cells could probably due to higher level of endogenous miR-944 in CaSki cells that would affect the functional consequences. Since the Anti-miR-944 treatment in CaSki cells did not completely knock down the expression of miR-944, the minute residual miR-944 expression could be sufficient to retain the phenotypes. Given that HeLa cell was derived from cervical adenocarcinoma and CaSki cell was established from cervical squamous cell carcinoma, the different effect observed between the two cell lines may suggest that miR-944 may have different functions in different histological subtypes. In line with this notion, the host gene encoding miR-944, i.e. TP63, is a squamous differentiation marker that can distinguish squamous cell carcinoma from adenocarcinoma.16 Further investigations are warranted to determine whether miR-944 expression can distinguish between these two tumor subtypes and its specific functions in squamous cell carcinoma and adenocarcinoma.
Evaluations of PAR-CLIP sequencing data
Despite that we obtained millions of raw reads per library, a large proportion of the reads were mapped to linker-linker ligation products. This is due to the minute amount of Ago2-immunoprecipitated RNA for RNA-sequencing library preparation. Given that the sequencing depth was insufficient for enrichment quantification, we combined all replicates in each experimental conditions and compared the transcripts present in miR-944 overexpressing and control cells. Nonetheless, our PAR-CLIP sequencing revealed several Ago2 bound transcripts and miRNA targets.
Our PAR-CLIP data revealed that, among the mRNA transcripts, 48% corresponded to 3′UTR, 44% to CDS and 8% to 5′UTR, which is concordant with the findings reported by Hafner et al.10 In addition to mRNAs, we also found six noncoding RNAs (ncRNAs) in our PAR-CLIP data. Similarly, several classes of ncRNAs, including pseudogenes, long non-coding RNAs, transfer RNAs, ribosomal RNAs and small nuclear RNAs, have been found directly interacting with miRNA using CLASH approach (crosslinking, ligation and sequencing of hybrids).17 These findings suggest the possibility of miRNA-ncRNA interactions that have yet to be explored. However, a much higher sequencing depth is needed for complete identification of miR-944 targets.
HECW2 and S100PBP as direct targets of miR-944 in human cervical cancer cells
Using luciferase reporter assays, we verified the direct targeting of 3′UTR of HECW2 and S100PBP by miR-944. HECW2 is a poorly characterized HECT domain ubiquitin ligase.18 This protein has been shown to stabilize tumor suppressor p73 and enhance its transcriptional activity.18 Recently, Muralikrishna et al. reported that HECW2 is involved in degradation of ATR kinase in HeLa cells with lamin mutants or lamin silenced HeLa cells.19 Interestingly, another HECT-type ubiquitin ligase Itch has been shown to promote degradation of p63 through ubiquitination.20 It is tempting to speculate that HECW2 may also regulate protein stability of p63 and miR-944 suppresses HECW2 expression to protect the stability of p63. Further studies are warranted to determine the functional role of HECW2 in cervical cancer and p63-miR-944 regulatory network.
Another miR-944 target, S100PBP interacts with the S100 calcium-binding protein P21 and its increased or decreased expression has been observed in different cancer types.22 Low or undetectable level of S100PBP was found in lung squamous cell carcinoma, prostate and breast adenocarcinoma compared with their normal counterparts. However, its expression was higher in thyroid, liver and ovarian cancers than their respective normal tissues. Functionally, S100PBP expression has been shown to suppress cell invasion and adhesion in pancreatic cancer cell lines.22 Given that we observed an increase of cell invasion upon miR-944 overexpression in HeLa cells, it will be interesting to determine whether S100PBP is involved in cell invasion in cervical cancer cells.
In summary, we show that miR-944 plays an oncogenic role in human cervical cancer cells by promoting cell proliferation, migration and invasion. In addition, we identified a set of novel targets of miR-944 by using the PAR-CLIP sequencing approach and validated HECW2 and S100PBP as bona fide direct targets of miR-944 in human cervical cancer cells. Our findings provide new insights into the biological roles of miR-944 in human cervical cancer cells.
Acknowledgments
The authors thank LinJing Zhu and Zhangsen Huang (Karolinska Institutet) for technical support of flow cytometry, Deniz M. Özata (Karolinska Institutet) for helps with Illumina sequencing, Iryna Kolosenko (Karolinska Institutet) for technical support of bioluminescence detection and all other members from Medical Genetics research group for valuable discussions and suggestions.
Supporting Information
Additional Supporting Information may be found in the online version of this article.
Supplementary Information
Supplementary Information
Supplementary Information
Supplementary Information
Supplementary Information
Supplementary Information
Supplementary Information
References
- 1.Lee JW, Choi CH, Choi JJ, et al. Altered MicroRNA expression in cervical carcinomas. Clin Cancer Res. 2008;14:2535–2542. doi: 10.1158/1078-0432.CCR-07-1231. [DOI] [PubMed] [Google Scholar]
- 2.Lui WO, Pourmand N, Patterson BK, et al. Patterns of known and novel small RNAs in human cervical cancer. Cancer Res. 2007;67:6031–6043. doi: 10.1158/0008-5472.CAN-06-0561. [DOI] [PubMed] [Google Scholar]
- 3.Martinez I, Gardiner AS, Board KF, et al. Human papillomavirus type 16 reduces the expression of microRNA-218 in cervical carcinoma cells. Oncogene. 2008;27:2575–2582. doi: 10.1038/sj.onc.1210919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pereira PM, Marques JP, Soares AR, et al. MicroRNA expression variability in human cervical tissues. PLoS One. 2010;5:e11780. doi: 10.1371/journal.pone.0011780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang X, Tang S, Le SY, et al. Aberrant expression of oncogenic and tumor-suppressive microRNAs in cervical cancer is required for cancer cell growth. PLoS One. 2008;3:e2557. doi: 10.1371/journal.pone.0002557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Witten D, Tibshirani R, Gu SG, et al. Ultra-high throughput sequencing-based small RNA discovery and discrete statistical biomarker analysis in a collection of cervical tumours and matched controls. BMC Biol. 2010;8:58. doi: 10.1186/1741-7007-8-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yao Q, Xu H, Zhang QQ, et al. MicroRNA-21 promotes cell proliferation and down-regulates the expression of programmed cell death 4 (PDCD4) in HeLa cervical carcinoma cells. Biochem Biophys Res Commun. 2009;388:539–542. doi: 10.1016/j.bbrc.2009.08.044. [DOI] [PubMed] [Google Scholar]
- 8.Yang YC, Shyong WY, Chang MS, et al. Frequent gain of copy number on the long arm of chromosome 3 in human cervical adenocarcinoma. Cancer Genet Cytogenet. 2001;131:48–53. doi: 10.1016/s0165-4608(01)00510-6. [DOI] [PubMed] [Google Scholar]
- 9.Schmitz M, Driesch C, Jansen L, et al. Non-random integration of the HPV genome in cervical cancer. PLoS One. 2012;7:e39632. doi: 10.1371/journal.pone.0039632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hafner M, Landthaler M, Burger L, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xie H, Zhao Y, Caramuta S, et al. miR-205 expression promotes cell proliferation and migration of human cervical cancer cells. PLoS One. 2012;7:e46990. doi: 10.1371/journal.pone.0046990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corcoran DL, Georgiev S, Mukherjee N, et al. PARalyzer: definition of RNA binding sites from PAR-CLIP short-read sequence data. Genome Biol. 2011;12:R79. doi: 10.1186/gb-2011-12-8-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stark MS, Tyagi S, Nancarrow DJ, et al. Characterization of the Melanoma miRNAome by Deep Sequencing. PLoS One. 2010;5:e9685. doi: 10.1371/journal.pone.0009685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christensen LL, Tobiasen H, Holm A, et al. MiRNA-362-3p induces cell cycle arrest through targeting of E2F1, USF2 and PTPN1 and is associated with recurrence of colorectal cancer. Int J Cancer. 2013;133:67–78. doi: 10.1002/ijc.28010. [DOI] [PubMed] [Google Scholar]
- 15.Nordentoft I, Birkenkamp-Demtroder K, Agerbaek M, et al. miRNAs associated with chemo-sensitivity in cell lines and in advanced bladder cancer. BMC Med Genomics. 2012;5:40. doi: 10.1186/1755-8794-5-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang TY, Chen BF, Yang YC, et al. Histologic and immunophenotypic classification of cervical carcinomas by expression of the p53 homologue p63: a study of 250 cases. Hum Pathol. 2001;32:479–486. doi: 10.1053/hupa.2001.24324. [DOI] [PubMed] [Google Scholar]
- 17.Helwak A, Kudla G, Dudnakova T, et al. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell. 2013;153:654–665. doi: 10.1016/j.cell.2013.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyazaki K, Ozaki T, Kato C, et al. A novel HECT-type E3 ubiquitin ligase, NEDL2, stabilizes p73 and enhances its transcriptional activity. Biochem Biophys Res Commun. 2003;308:106–113. doi: 10.1016/s0006-291x(03)01347-0. [DOI] [PubMed] [Google Scholar]
- 19.Muralikrishna B, Chaturvedi P, Sinha K, et al. Lamin misexpression upregulates three distinct ubiquitin ligase systems that degrade ATR kinase in HeLa cells. Mol Cell Biochem. 2012;365:323–332. doi: 10.1007/s11010-012-1272-4. [DOI] [PubMed] [Google Scholar]
- 20.Rossi M, Aqeilan RI, Neale M, et al. The E3 ubiquitin ligase Itch controls the protein stability of p63. Proc Natl Acad Sci USA. 2006;103:12753–12758. doi: 10.1073/pnas.0603449103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dowen SE, Crnogorac-Jurcevic T, Gangeswaran R, et al. Expression of S100P and its novel binding partner S100PBPR in early pancreatic cancer. Am J Pathol. 2005;166:81–92. doi: 10.1016/S0002-9440(10)62234-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lines KE, Chelala C, Dmitrovic B, et al. S100P-binding protein, S100PBP, mediates adhesion through regulation of cathepsin Z in pancreatic cancer cells. Am J Pathol. 2012;180:1485–1494. doi: 10.1016/j.ajpath.2011.12.031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Supplementary Information
Supplementary Information
Supplementary Information
Supplementary Information
Supplementary Information
Supplementary Information