Abstract
Although diverse natural products have been isolated from the benthic, filamentous cyanobacterium Lyngbya majuscula, it is unclear whether this chemical variation can be used to establish taxonomic relationships among disparate collections. We compared morphological characteristics, secondary-metabolite compositions, and partial 16S ribosomal DNA (rDNA) sequences among several collections of L. majuscula Gomont, Lyngbya spp., and Symploca spp. from Guam and the Republic of Palau. The morphological characteristics examined were cell length, cell width, and the presence or absence of a calyptra. Secondary metabolites were analyzed by two-dimensional thin-layer chromatography. Each collection possessed a distinct cellular morphology that readily distinguished Lyngbya spp. from Symploca spp. Each collection yielded a unique chemotype, but common chemical characteristics were shared among four collections of L. majuscula. A phylogeny based on secondary-metabolite composition supported the reciprocal monophyly of Lyngbya and Symploca but yielded a basal polytomy for Lyngbya. Pairwise sequence divergence among species ranged from 10 to 14% across 605 bp of 16S rDNA, while collections of L. majuscula showed 0 to 1.3% divergence. Although the phylogeny of 16S rDNA sequences strongly supported the reciprocal monophyly of Lyngbya and Symploca as well as the monophyly of Lyngbya bouillonii and L. majuscula, genetic divergence was not correlated with chemical and morphological differences. These data suggest that 16S rDNA sequence analyses do not predict chemical variability among Lyngbya species. Other mechanisms, including higher rates of evolution for biosynthetic genes, horizontal gene transfer, and interactions between different genotypes and environmental conditions, may play important roles in generating qualitative and quantitative chemical variation within and among Lyngbya species.
The benthic, filamentous cyanobacterium Lyngbya majuscula Gomont is distributed throughout the tropics in reef and lagoonal habitats (18, 61, 62), often forming dense mats that carpet benthic substrates. L. majuscula can compete with macroalgae (59) and is unpalatable to fish, crabs, urchins, and other macroherbivores (43, 50, 57). However, specialized mesoherbivores, such as the sea hare Stylocheilus striatus, may preferentially consume L. majuscula (8, 41, 51). Secondary metabolites produced by L. majuscula are responsible for both the deterrence of feeding by macroherbivores and the stimulation of feeding by mesoherbivores (41, 51, 57).
Over 100 novel secondary metabolites have been isolated from collections of L. majuscula. Although these collections have been globally distributed, there is little evidence that any given types of secondary metabolites are associated with specific geographic regions (10). Indeed, collections within limited geographic areas are often extremely diverse. On Guam, L. majuscula collections have yielded indanone metabolites (45), lyngbyastatins (16, 27, 64), malyngamides (4, 5, 38), malyngolide (7), majusculamides (35), pitiamide (42), and pitipeptolides (25). These compounds have been implicated in cases of swimmer's itch, human poisonings, and fish kills and are of interest to pharmaceutical and biochemical researchers due to their selective cytotoxicity (6, 39, 40, 44). Collections of the filamentous cyanobacterium Symploca hydnoides (Harvey) Kützing on Guam have yielded additional cytotoxic compounds, including symplostatins (14, 15).
Traditional taxonomy within the cyanobacterial family Oscillatoriaceae is largely based on morphological measurements, including cell length and cell width, of axenic cultures (9, 65). However, many cyanobacteria cannot be grown on artificial media or undergo morphological changes when grown under different culture conditions (23, 46, 65). Advances in molecular systematics have yielded cyanobacterial phylogenies based on several genes, with 16S ribosomal DNA (rDNA), nifH, and phycocyanin sequences being the most prevalent (1, 33, 47, 48, 49, 67, 68). These phylogenies have revealed inconsistencies in the morphological classification of several cyanobacterial taxa. For example, although Oscillatoria and Microcoleus are traditionally described as morphologically distinct genera, differences in morphology are not reflected in analyses of their 16S rDNA (66, 67). Gugger et al. (11, 12) reported that cyanobacteria in the genera Anabaena and Aphanizomenon appear morphologically distinct but show a high degree of similarity in 16S rDNA (11, 33) and ribulose-1,5-bisphosphate carboxylase/oxygenase sequences (11) and have similar cellular fatty acid profiles (12).
The diverse secondary metabolites produced by L. majuscula may provide chemotaxonomic markers that are correlated with both morphological and genetic variations. Most studies of cyanobacterial chemotaxonomy have focused on primary metabolites and include studies of cellular fatty acid composition (12, 20, 22), carotenoids (17, 67), and aromatic amino acid biochemical pathways (13). Recent investigations have examined variation in secondary-metabolite production, using DNA sequences to differentiate between toxin-producing and nontoxic strains of Anabaena, Aphanizomenon, and Microcystis (2, 11, 21, 33). These studies have often found that the traditional morphological taxonomy of cyanobacteria is neither supported by phylogenetic analyses nor correlated with chemical variation.
In this study, we compared morphological and chemical characteristics among several collections of L. majuscula, Lyngbya spp., and Symploca spp. from Guam and the Republic of Palau. We also amplified and sequenced 16S rDNA from these same collections. We examined the ability of morphological and chemical characteristics to establish species relationships among these taxa and compared these relationships to a phylogeny constructed from 16S rDNA sequences.
MATERIALS AND METHODS
Sample collection.
Specimens of L. majuscula were collected from three locations on Guam: Piti Bomb Holes, Cocos Lagoon, and Pago Bay. Three replicate specimens, separated by at least 5 m, were collected from each location. Other species of cyanobacteria were collected from long-term monitoring transects on Guam (58). These included Lyngbya sp. 1 aff. majuscula Gomont from Piti Bomb Holes (42), Lyngbya sp. 2 aff. semiplena (C. Agardh) J. Agardh from Tumon Bay (45), L. confervoides C. Agardh from Piti Bomb Holes, Lyngbya sp. 3 aff. polychroa (Meneghini) Rabenhorst from Double Reef, and L. bouillonii L. Hoffmann et V. Demoulin from Piti Bomb Holes. Specimens of S. hydnoides were collected from Pago Bay and Piti Bomb Holes. The Pago Bay collection formed small upright tufts on the reef flat and was epiphytic on forereef macroalgae, while the Piti Bomb Holes collection was found underneath soft corals (Sinularia spp.) in long, rope-like strands. Additional specimens were collected from several locations in the Republic of Palau, including Ulong Channel (Lyngbya sp. 4 aff. polychroa and L. bouillonii), Mecherchar Cove (L. majuscula), and Short Drop-Off (Symploca sp. aff. hydnoides). When samples could not be processed immediately, they were stored in 80% ethanol. Voucher specimens are stored in 10% formalin at the University of Guam Marine Laboratory.
Morphological characteristics.
The cell lengths and cell widths of 10 filaments from each replicated sample were measured at ×500 and ×1,250 magnifications. These two variables were used to generate a scatter plot of the mean values for each collection. The presence or absence of a calyptra on the terminal cells of filaments was also noted. Species names were assigned following the criteria of Desikachary (9), Hoffmann (18), and Littler and Littler (24). The identification of L. bouillonii was based on descriptions by Hoffmann and Demoulin (19).
Chemical characteristics.
Fresh collections of cyanobacteria were extracted in CH2Cl2-methanol (MeOH) (1:1, vol/vol). Solvents were removed by rotary evaporation. Two-dimensional thin-layer chromatography (TLC) analyses followed the protocol of Nagle and Paul (43). Each crude extract was dissolved in CH2Cl2-MeOH (1:1, vol/vol) and applied to form a single spot in a corner of an aluminum-backed silica gel TLC sheet (10 by 10 cm). The TLC sheets were developed in CH2Cl2-MeOH (9:1) and removed after the solvent front traveled 8 cm. The TLC sheets were dried and then placed into another solvent chamber (ethyl acetate-hexanes, 1:1) so that the developed extract was placed horizontally above the second solvent. The TLC sheets were removed after the second solvent front traveled 8 cm. After the sheets were dried, they were marked with brackets to indicate pigmented compounds. The sheets were placed in a UV (254-nm-wavelength) viewing cabinet, and locations of UV-fluorescing compounds were recorded on the sheets with circles. The sheets were coated with H2SO4 in ethanol (1:19) and heated with a hot-air gun. Acid-charring compounds were marked on the sheets with arrows. Comparisons of TLC sheets among extracts allowed the presence or absence of particular compounds to be noted for each cyanobacterium. Pigments were not included in these analyses.
16S rDNA sequencing.
Genomic DNA was isolated from collections of cyanobacteria by using a G-NOME DNA isolation kit (Bio 101), following the manufacturer's suggested protocol. PCR amplification with cyanobacterium-specific primers followed the protocol of Nübel et al. (49), with forward primer CYA106F (5′-CGGACGGGTGAGTAACGCGTGA-3′) and an equimolar mixture of CYA781Ra (5′-GACTACTGGGGTATCTAATCCCATT-3′) and CYA781Rb (5′-GACTACAGGGGTATCTAATCCCTTT-3′) as reverse primers. Preliminary analyses of GenBank sequences indicated that this approximately 660-bp region of 16S rDNA accounts for over half of the variable bases reported for Oscillatoriales. PCR products were cleaned with preparatory columns (Wizard PCR Preps; Promega) and then directly sequenced at a commercial facility (Davis Sequencing, Davis, Calif.). Sequences were aligned manually by using the Se-Al sequence alignment program (A. Rambaut, University of Oxford, Oxford, England). We included the sequence reported for Lyngbya strain PCC7419 (GenBank accession number AJ000714) (49) in our analyses as a reference.
Phylogenetic analyses.
All phylogenetic analyses were conducted using PAUP* [Phylogenetic Analysis Using Parsimony (*and other methods), version 4.0b3a, 1999; D. L. Swofford, Sinauer, Sunderland, Mass.]. For each sample, the presence or absence of 21 chemical compounds was coded into a PAUP data matrix by using binary, unordered characters. A phylogeny based on these chemical data was constructed with a branch-and-bound search and a maximum parsimony optimality criterion. Support for this phylogeny was examined by using 1,000 bootstrap replicates of the branch-and-bound search.
Phylogenetic trees were constructed from DNA sequence data by using both maximum parsimony and maximum likelihood optimality criteria. The maximum parsimony tree was obtained through a heuristic search with 10 random stepwise addition sequences. Support for each node was evaluated with 500 bootstrap replicates of a fast heuristic search with a single random stepwise addition sequence for each replicate. Likelihood ratio tests comparing hierarchical models of DNA substitution were evaluated by Modeltest (52). The maximum likelihood tree was generated from the likelihood settings calculated by Modeltest in a heuristic search with 10 random stepwise addition sequences. Support for each node was evaluated with 100 bootstrap replicates of a heuristic search with 10 random stepwise addition sequences for each replicate.
RESULTS
Morphological variation.
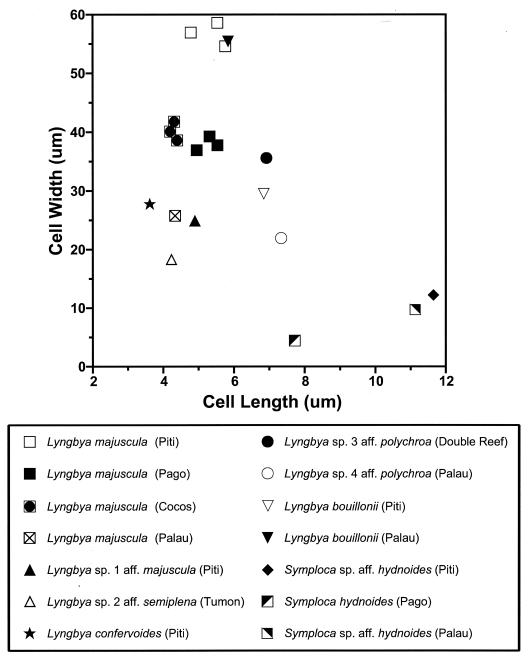
Morphological characteristics clearly distinguished Lyngbya spp. from Symploca spp., as Lyngbya specimens typically contained wider, shorter cells and Symploca specimens contained narrower, longer cells (Fig. 1). The morphology of each species varied considerably among sampling locations, with a large amount of overlapping variation among the five species of Lyngbya (Fig. 1). Only L. semiplena possessed a calyptra at the end of each filament. Within L. majuscula, samples collected from Piti Bomb Holes had coarser filaments than samples collected from Cocos Lagoon and Pago Bay. The Cocos Lagoon samples had slightly shorter cells than samples from Piti Bomb Holes and Pago Bay. L. majuscula specimens from Palau had much narrower cells than specimens from Guam.
FIG. 1.
Variations in cell length (x axis) and cell width (y axis), measured in micrometers, among cyanobacterial collections. Standard error bars for both measurements are smaller than the size of each symbol. Each species of Lyngbya and Symploca has a distinct morphology at each location.
Chemical variation.
Each species also showed chemical variation among locations, with a unique chemotype associated with each collection location. Some compounds were specific to certain genera (e.g., compound 7 was found in all Lyngbya spp. but not in Symploca spp.), while others were specific to certain collection locations (e.g., compound 15 was found only in L. majuscula from Piti Bomb Holes). Additional variation was found among samples of L. majuscula collected from Pago Bay and Cocos Lagoon, as individual samples contained unique combinations of compounds. Compounds that matched known standards included compound 6 (which matched malyngamide A), compound 4 (malyngamide B), compound 5 (majusculamides A and B), and compound 14 (lyngbyastatin 1).
Genetic variation.
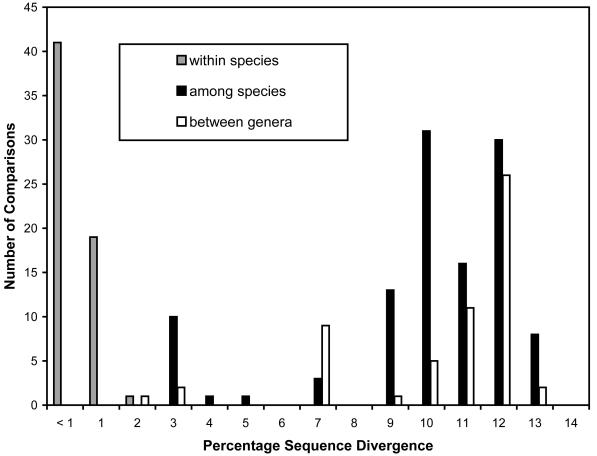
An approximately 660-bp fragment was amplified from each collection. Sequences of these fragments have been deposited in GenBank under accession numbers AF510963 to AF510983. Of the 605 aligned positions, 131 were variable and 105 were parsimony informative. Base frequencies averaged 27.0% for A, 19.4% for T, 32.2% for G, and 21.4% for C, with a transition-to-transversion ratio of 1.15. The sequence determined for S. hydnoides (Pago Bay) was identical to the sequence of Symploca strain VP377 collected from Pago Bay and reported by Hoffmann et al. (GenBank accession number AF306497). The sequences obtained for L. bouillonii from Guam (Piti Bomb Holes 2) and for L. bouillonii from Palau (Ulong Channel) were 99.8% similar to those for Lyngbya strain VP417 (from Apra Harbor, Guam; GenBank accession number AY049751) (28) and Lyngbya strain NIH309 (from Short Drop-Off, Palau; GenBank accession number AY049752) (28), respectively. Pairwise comparisons of sequence divergence percentages among specimens of L. majuscula showed 0 to 1.3% divergence (Fig. 2). Sequence divergence among species of Lyngbya was as high or higher than between Lyngbya and Symploca (Fig. 2). Comparisons among samples of L. bouillonii showed 0.2 to 1.4% divergence, and comparisons among samples of S. hydnoides showed 0.3 to 0.7% divergence.
FIG. 2.
Distribution of pairwise sequence divergence across an alignment of 605 bp of 16S rDNA, indicating divergence within species of Lyngbya and Symploca (gray bars), among Lyngbya species (black bars), and between the genera Lyngbya and Symploca (white bars). Pairwise comparisons are adjusted for missing data and are uncorrected for multiple substitutions.
Phylogenetic analyses.
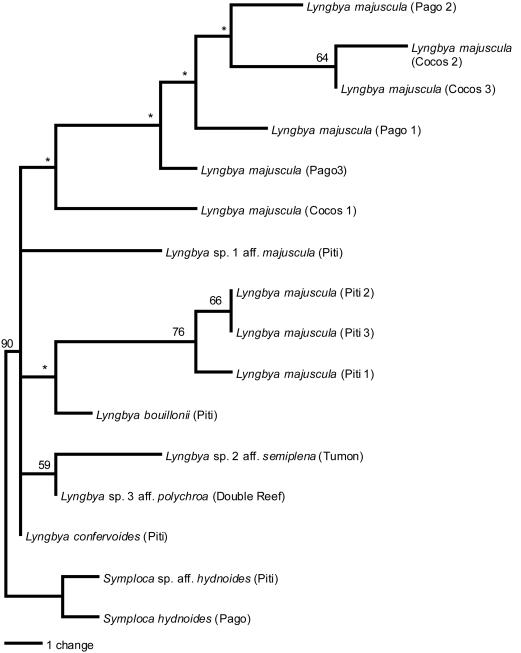
Phylogenetic analyses based on chemical characteristics yielded a single most-parsimonious tree with a length of 33 and a consistency index of 0.636 (Fig. 3). Although bootstrap analyses strongly supported the reciprocal monophyly of Lyngbya and Symploca, nodes within the genus Lyngbya were weakly supported, suggesting a basal polytomy for the genus Lyngbya. Within L. majuscula, three collections from Piti Bomb Holes formed a distinct clade, while two collections from Cocos Lagoon formed a separate clade, illustrating the substantial chemical variation observed among these populations.
FIG. 3.
Maximum parsimony phylogeny based on the presence of secondary metabolites in collections of Lyngbya and Symploca species, as analyzed by two-dimensional TLC. Numbers above branches indicate bootstrap values (percentages) from 1,000 replicates of a branch-and-bound search executed using PAUP* version 4.0b3a. Asterisks indicate bootstrap support of less than 50%.
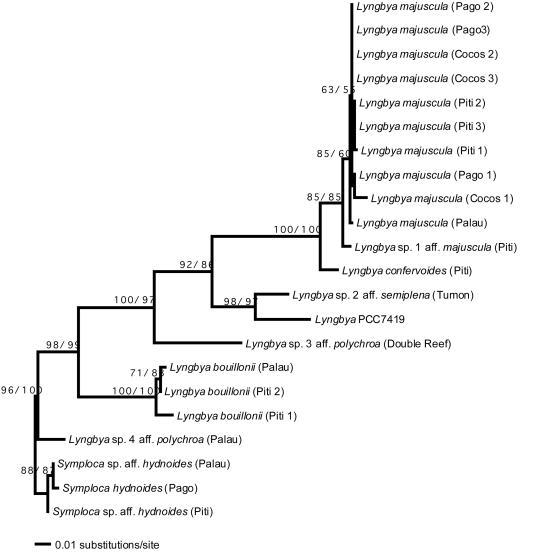
Hierarchical models of DNA substitution indicated that a general time-reversible model (53) that included both the proportion of invariable sites and heterogeneous rates of substitution among variable sites best fit our data. Model parameters estimated using Modeltest (52) included the substitution rate matrix (A→C = 1.245, A→G = 1.551, A→T = 1.335, C→G = 0.325, C→T = 3.289, and G→T = 1.000), the proportion of invariable sites (I = 0.500), and the gamma distribution shape parameter (G = 0.486). This model generated a maximum likelihood tree with a likelihood score of 2048.686; bootstrap analyses supported all nodes except those within the monophyletic clade of L. majuscula collected on Guam (Fig. 4). Maximum parsimony analyses constructed 12 equally parsimonious trees with a length of 235 and a consistency index of 0.753. The maximum parsimony bootstrap consensus tree shared the same topology as the maximum likelihood tree, again with no support for nodes within the clade of L. majuscula collected on Guam. The phylogenies based on 16S rDNA sequences (Fig. 4) strongly supported the reciprocal monophyly of Lyngbya and Symploca. Both L. bouillonii and L. majuscula were well supported as monophyletic clades.
FIG. 4.
Maximum likelihood phylogram of collections of Lyngbya and Symploca spp., constructed from an alignment of 605 bp of 16S rDNA. The maximum parsimony bootstrap consensus tree shares the same topology. Numbers above branches indicate likelihood/parsimony bootstrap values (percentages). The maximum likelihood analysis was based on a general time-reversible model that included both the proportion of invariable sites and heterogeneous rates of substitution among variable sites, with model parameters estimated with Modeltest (52) and trees evaluated with PAUP* version 4.0b3a.
DISCUSSION
Recent reviews of secondary metabolites produced by Lyngbya species and other filamentous marine cyanobacteria have stressed the tremendous variation in metabolite quantity and quality among collections (10, 39, 43). Since many of these compounds have promising pharmaceutical applications, including curacin A (from L. majuscula) and symplostatin 1 (from S. hydnoides), there is growing interest in determining the ecological and evolutionary mechanisms that generate this variation (10, 50). We examined the morphology, secondary-metabolite composition, and 16S rDNA sequences of 21 collections of Lyngbya and Symploca species from Guam and the Republic of Palau. Cell length and cell width are the morphological characteristics traditionally used to differentiate species of Lyngbya (9, 19, 24). In our study, these characteristics distinguished Lyngbya from Symploca but did not clearly separate species of Lyngbya. A phylogeny based on secondary-metabolite characteristics also differentiated Lyngbya from Symploca but did not resolve species relationships within Lyngbya. For Lyngbya species, each collection location yielded samples with unique cellular morphologies and chemical profiles, providing very little phylogenetic signal and indicating that these characteristics are not suitable for a phylogenetically based classification of Lyngbya species.
Sequence analysis of the 16S rDNA subunit can provide a much higher degree of resolution among cyanobacterial taxa than either morphological or chemical traits (67). The majority of comparisons of sequence divergence among Lyngbya spp. demonstrated 10 to 14% divergence (86 to 90% similarity), as did comparisons between Lyngbya and Symploca spp. These values are similar to the range of average cyanobacterial similarities (83.7 to 88.7%) reported by Wilmotte (67). Large differences within genera may reflect the ancient lineages of cyanobacteria, while the transition-to-transversion ratio of 1.15 may indicate increasing saturation of these sequences. However, partial 16S rDNA sequences strongly supported the reciprocal monophyly of the genera Lyngbya and Symploca, as well as the monophyly of L. bouillonii and L. majuscula, with high bootstrap support for nodes separating species. Although all collections of L. majuscula were grouped into a monophyletic clade, these sequences did not resolve phylogenetic relationships among populations of L. majuscula with a high degree of certainty.
Although many collections of L. majuscula possessed both a unique chemotype and a unique cellular morphology, this variation was not correlated with variability in 16S rDNA sequences. For example, while L. majuscula collections Cocos 2 and Cocos 3 were readily distinguished from collections Pago 2 and Pago 3 on the basis of chemical and morphological characteristics, these four collections possessed identical 16S rDNA sequences. Given this conservation of 16S rDNA sequences, the high degree of plasticity observed for both morphology and secondary-metabolite composition may be due to three nonexclusive biological processes: (i) genes responsible for secondary-metabolite biosynthesis and morphology may evolve at higher rates than the 16S ribosomal gene; (ii) horizontal gene transfer may enhance variability in secondary-metabolite biosynthesis; and (iii) cyanobacteria may display plastic morphological and chemical responses to the environmental conditions found at each collection location.
Divergence in chemosynthetic genes may not be reflected in 16S rDNA sequences if the ribosomal sequences are relatively more conserved. Toxins produced by cyanobacteria can be associated with genetic differences among strains, but differences in toxin production have also been reported among genetically similar strains (2). For example, although hepatotoxic strains of Anabaena are genetically distinct from neurotoxic strains of Anabaena and nontoxic Aphanizomenon (11, 33), neurotoxic Anabaena and nontoxic Aphanizomenon strains show 99.9 to 100% similarity in their 16S rDNA sequences. Higher rates of evolution in toxin-encoding genes than in ribosomal genes have also been reported for Microcystis strains (36, 60). Future phylogenetic analyses of genes responsible for the biosynthesis of Lyngbya metabolites, e.g., polyketide synthases and nonribosomal peptide synthetases (21, 37), analyses of the internally transcribed spacer between the 16S and the 23S rDNA subunits (56), and repetitive extragenic palindromic fingerprinting (33) may reveal greater amounts of within-species genetic variation than observed in the 16S rDNA subunit.
Analyses of the substitution patterns found within Lyngbya biosynthetic genes may provide evidence for horizontal gene transfer (54, 55). Horizontal gene transfer among cyanobacterial strains has been demonstrated for genes encoding both primary metabolites (3, 34, 54, 55) and secondary metabolites, including the Microcystis mcy operon (36, 60). Horizontal gene transfer may be more frequent among closely related strains (54), providing a potential explanation for the diversity of metabolites isolated from neighboring Lyngbya populations.
Environmental variation among collection locations could also influence the observed variation in chemical and morphological characteristics. For example, the Cocos Lagoon samples of L. majuscula were collected at a depth of 3 to 5 m in relatively calm waters, while the Pago Bay samples were collected at a depth of less than 1 m on a wave-swept reef flat. Additional laboratory and field experiments, including reciprocal transplants, are needed to determine whether variation in morphological and chemical characteristics reflects the responses of different genotypes to changing environmental conditions or whether this variation reflects genetic differences that are expressed regardless of environmental conditions.
Taxonomic difficulties may confound any current analyses of the geographic distributions of cyanobacterial compounds. The 16S rDNA phylogeny indicates that L. majuscula and L. bouillonii are genetically distinct, with greater than 10% average sequence divergence. L. bouillonii can be recognized in the field by its dark red coloration and thick, net-like mats, but it shows an extremely wide variation in cell shape, cell size, and trichome width (19). However, several compounds that originate from L. bouillonii have been attributed to L. majuscula, including apratoxins A (30), B, and C (28), lyngbyabellins A, B, and D (31, 32, 63), lyngbyapeptin A (32), ulongamides (26), and other alkaloids (29). Future investigations of cyanobacteria that yield pharmaceutically active compounds should rely on a combination of both morphological and molecular taxonomy for identifications, as an enhanced chemical and molecular database may clarify patterns in the evolution and biogeography of these natural products.
Acknowledgments
We thank the Division of Marine Resources, Republic of Palau, and the Koror State Government for providing marine research permits. In addition, we thank the Guam Division of Aquatic and Wildlife Resources for permits to collect cyanobacteria at the Piti Bomb Holes and Tumon Bay marine reserves. D. Nagle provided assistance with the chemical analyses, while U. Nübel and G. Muyzer provided helpful advice on the amplification of cyanobacterial rDNA sequences. We are grateful to two anonymous reviewers for their comments and suggestions on the manuscript.
The U.S. ECOHAB program is sponsored by NOAA, NSF, EPA, NASA, and ONR. Additional support for this research was provided by the University of Alabama at Birmingham.
Although the research described in this article was funded in part by the U.S. EPA through grant R82-6220, it has not been subjected to the Agency's required peer and policy review and therefore does not necessarily reflect the views of the Agency; no official endorsement should be inferred.
Footnotes
Contribution 93 of the U.S. ECOHAB program, contribution 513 of the University of Guam Marine Laboratory, and contribution 580 of the Smithsonian Marine Station at Fort Pierce.
REFERENCES
- 1.Abed, R., S. Golubic, F. Garcia-Pichel, G. F. Camoin, and S. Sprachta. 2003. Characterization of microbialite-forming cyanobacteria in a tropical lagoon: Tikehau Atoll, Tuamotu, French Polynesia. J. Phycol. 39:862-873. [Google Scholar]
- 2.Baker, J. A., B. Entsch, B. A. Neilan, and D. B. McKay. 2002. Monitoring changing toxigenicity of a cyanobacterial bloom by molecular methods. Appl. Environ. Microbiol. 68:6070-6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barker, G. L. A., B. A. Handley, P. Vacharapiyasophon, J. R. Stevens, and P. K. Hayes. 2000. Allele-specific PCR shows that genetic exchange occurs among genetically diverse Nodularia (cyanobacteria) filaments in the Baltic Sea. Microbiology 146:2865-2875. [DOI] [PubMed] [Google Scholar]
- 4.Cardellina, J. H., II, D. Dalietos, F.-J. Marner, J. S. Mynderse, and R. E. Moore. 1978. (−)-Trans-7(S)-methoxytetradec-4-enoic acid and related amides from the marine cyanophyte Lyngbya majuscula. Phytochemistry 17:2091-2095. [Google Scholar]
- 5.Cardellina, J. H., II, F.-J. Marner, and R. E. Moore. 1979. Malyngamide A, a novel chlorinated metabolite of the marine cyanophyte Lyngbya majuscula. J. Am. Chem. Soc. 101:240-241. [Google Scholar]
- 6.Cardellina, J. H., II, F.-J. Marner, and R. E. Moore. 1979. Seaweed dermatitis: structure of lyngbyatoxin A. Science 204:193-195. [DOI] [PubMed] [Google Scholar]
- 7.Cardellina, J. H., II, R. E. Moore, E. V. Arnold, and J. Clardy. 1979. Structure and absolute configuration of malyngolide, an antibiotic from the marine blue-green alga Lyngbya majuscula Gomont. J. Org. Chem. 44:4039-4042. [Google Scholar]
- 8.Cruz-Rivera, E., and V. J. Paul. 2002. Coral reef benthic cyanobacteria as food and refuge: diversity, chemistry, and complex interactions, p. 515-520. In M. K. Kasim Moosa, S. Soemodihardjo, A. Nontji, A. Soegiarto, K. Rominohtarto, Sukarno, and Suharsono (ed.), Proceedings of the Ninth International Coral Reef Symposium. Indonesian Institute of Sciences and State Ministry for Environment, Jakarta, Republic of Indonesia.
- 9.Desikachary, T. V. 1959. Cyanophyta. Indian Council of Cultural Research, New Delhi, India.
- 10.Gerwick, W. H., L. T. Tan, and N. Sitachitta. 2001. Nitrogen-containing metabolites from marine cyanobacteria. Alkaloids Chem. Biol. 57:75-184. [DOI] [PubMed] [Google Scholar]
- 11.Gugger, M., C. Lyra, P. Henriksen, A. Couté, J.-F. Humbert, and K. Sivonen. 2002. Phylogenetic comparison of the cyanobacterial genera Anabaena and Aphanizomenon. Int. J. Syst. Evol. Microbiol. 52:1867-1880. [DOI] [PubMed] [Google Scholar]
- 12.Gugger, M., C. Lyra, I. Suominen, I. Tsitko, J.-F. Humbert, M. S. Salkinoja-Salonen, and K. Sivonen. 2002. Cellular fatty acids as chemotaxonomic markers of the genera Anabaena, Aphanizomenon, Microcystis, Nostoc, and Planktothrix (cyanobacteria). Int. J. Syst. Evol. Microbiol. 52:1007-1015. [DOI] [PubMed] [Google Scholar]
- 13.Hall, G. C., M. B. Flick, R. L. Gherna, and R. A. Jensen. 1982. Biochemical diversity for biosynthesis of aromatic amino acids among the cyanobacteria. J. Bacteriol. 149:65-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harrigan, G. G., H. Luesch, W. Y. Yoshida, R. E. Moore, D. G. Nagle, V. J. Paul, S. L. Mooberry, T. H. Corbett, and F. A. Valeriote. 1998. Symplostatin 1: a dolastatin 10 analogue from the marine cyanobacterium Symploca hydnoides. J. Nat. Prod. 61:1075-1077. [DOI] [PubMed] [Google Scholar]
- 15.Harrigan, G. G., H. Luesch, W. Y. Yoshida, R. E. Moore, D. G. Nagle, and V. J. Paul. 1999. Symplostatin 2: a dolastatin 13 analogue from the marine cyanobacterium Symploca hydnoides. J. Nat. Prod. 62:655-658. [DOI] [PubMed] [Google Scholar]
- 16.Harrigan, G. G., W. Y. Yoshida, R. E. Moore, D. G. Nagle, P. U. Park, J. Biggs, V. J. Paul, S. L. Mooberry, T. H. Corbett, and F. A. Valeriote. 1998. Isolation, structure determination, and biological activity of dolastatin 12 and lyngbyastatin 1 from Lyngbya majuscula/Schizothrix calcicola cyanobacterial assemblages. J. Nat. Prod. 61:1221-1225. [DOI] [PubMed] [Google Scholar]
- 17.Hertzberg, S., S. Liaaen-Jensen, and H. W. Siegelman. 1971. The carotenoids of blue-green algae. Phytochemistry 10:3121-3127. [Google Scholar]
- 18.Hoffmann, L. 1994. Marine Cyanophyceae of Papua New Guinea. VI. The genus Lyngbya S.L. Belg. J. Bot. 127:79-86. [Google Scholar]
- 19.Hoffmann, L., and V. Demoulin. 1991. Marine Cyanophyceae of Papua New Guinea. II. Lyngbya bouillonii sp. nov., a remarkable tropical reef-inhabiting blue-green alga. Belg. J. Bot. 124:82-88. [Google Scholar]
- 20.Holton, R. W., H. H. Blecker, and T. S. Stevens. 1968. Fatty acids in blue-green algae: possible relation to phylogenetic position. Science 160:545-547. [DOI] [PubMed] [Google Scholar]
- 21.Kaebernick, M., and B. A. Neilan. 2001. Ecological and molecular investigations of cyanotoxin production. FEMS Microbiol. Ecol. 35:1-9. [DOI] [PubMed] [Google Scholar]
- 22.Kenyon, C. N., and R. Y. Stanier. 1970. Possible evolutionary significance of polyunsaturated fatty acids in blue-green algae. Nature 227:1164-1166. [DOI] [PubMed] [Google Scholar]
- 23.Laamanen, M. J., L. Forsström, and K. Sivonen. 2002. Diversity of Aphanizomenon flos-aquae (Cyanobacterium) populations along a Baltic Sea salinity gradient. Appl. Environ. Microbiol. 68:5296-5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Littler, D. S., and M. M. Littler. 2000. Caribbean reef plants. Offshore Graphics, Inc., Washington, D.C.
- 25.Luesch, H., R. Pangilinan, W. Y. Yoshida, R. E. Moore, and V. J. Paul. 2001. Pitipeptolides A and B, new cyclodepsipeptides from the marine cyanobacterium Lyngbya majuscula. J. Nat. Prod. 64:304-307. [DOI] [PubMed] [Google Scholar]
- 26.Luesch, H., P. G. Williams, W. Y. Yoshida, R. E. Moore, and V. J. Paul. 2002. Ulongamides A-F, new β-amino acid containing cyclodepsipeptides from Palauan collections of the marine cyanobacterium Lyngbya sp. J. Nat. Prod. 65:996-1000. [DOI] [PubMed] [Google Scholar]
- 27.Luesch, H., W. Y. Yoshida, R. E. Moore, and V. J. Paul. 1999. Lyngbyastatin 2 and norlyngbyastatin 2, analogues of dolastatin G and nordolastatin G from the marine cyanobacterium Lyngbya majuscula. J. Nat. Prod. 62:1702-1706. [DOI] [PubMed] [Google Scholar]
- 28.Luesch, H., W. Y. Yoshida, R. E. Moore, and V. J. Paul. 2002. New apratoxins of marine cyanobacterial origin from Guam and Palau. Bioorg. Med. Chem. 10:1973-1978. [DOI] [PubMed] [Google Scholar]
- 29.Luesch, H., W. Y. Yoshida, R. E. Moore, and V. J. Paul. 2002. Structurally diverse new alkaloids from Palauan collections of the apratoxin-producing marine cyanobacterium Lyngbya sp. Tetrahedron 58:7959-7966. [Google Scholar]
- 30.Luesch, H., W. Y. Yoshida, R. E. Moore, V. J. Paul, and T. H. Corbett. 2001. Total structure determination of apratoxin A, a potent novel cytotoxin from the marine cyanobacterium Lyngbya majuscula. J. Am. Chem. Soc. 123:5418-5423. [DOI] [PubMed] [Google Scholar]
- 31.Luesch, H., W. Y. Yoshida, R. E. Moore, V. J. Paul, and S. L. Mooberry. 2000. Isolation, structure determination, and biological activity of lyngbyabellin A from the marine cyanobacterium Lyngbya majuscula. J. Nat. Prod. 63:611-615. [DOI] [PubMed] [Google Scholar]
- 32.Luesch, H., W. Y. Yoshida, R. E. Moore, V. J. Paul, and S. L. Mooberry. 2000. Isolation and structure of the cytotoxin lyngbyabellin B and absolute configuration of lyngbyapeptin A from the marine cyanobacterium Lyngbya majuscula. J. Nat. Prod. 63:1437-1439. [DOI] [PubMed] [Google Scholar]
- 33.Lyra, C., S. Suomalainen, M. Gugger, C. Vezie, P. Sundman, L. Paulin, and K. Sivonen. 2001. Molecular characterization of planktic cyanobacteria of Anabaena, Aphanizomenon, Microcystis, and Planktothrix genera. Int. J. Syst. Evol. Microbiol. 51:513-526. [DOI] [PubMed] [Google Scholar]
- 34.Manen, J. F., and J. Falquet. 2002. The cpcB-cpcA locus as a tool for the genetic characterization of the genus Arthrospira (Cyanobacteria): evidence for horizontal transfer. Int. J. Syst. Evol. Microbiol. 52:861-867. [DOI] [PubMed] [Google Scholar]
- 35.Marner, F.-J., and R. E. Moore. 1977. Majusculamides A and B, two epimeric lipodipeptides from Lyngbya majuscula Gomont. J. Org. Chem. 42:2815-2819. [Google Scholar]
- 36.Mikalsen, B., G. Boison, O. M. Skulberg, J. Fastner, W. Davies, T. M. Gabrielsen, K. Rudi, and K. S. Jakobsen. 2003. Natural variation in the microcystin synthetase operon mcyABC and impact on microcystin production in Microcystis strains. J. Bacteriol. 185:2774-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moore, B. S., and C. Hertweck. 2002. Biosynthesis and attachment of novel bacterial polyketide synthase starter units. Nat. Prod. Rep. 19:70-99. [DOI] [PubMed] [Google Scholar]
- 38.Moore, R. E. 1981. Constituents of blue-green algae, p. 1-52. In P. J. Scheuer (ed.), Marine natural products, vol. 4. Academic Press, New York, N.Y. [Google Scholar]
- 39.Moore, R. E. 1996. Cyclic peptides and depsipeptides from cyanobacteria: a review. J. Ind. Microbiol. 16:134-143. [DOI] [PubMed] [Google Scholar]
- 40.Nagai, H., T. Yasumoto, and Y. Hokama. 1996. Aplysiatoxin and debromoaplysiatoxin as the causative agents of a red alga Gracilaria coronopifolia poisoning in Hawaii. Toxicon 37:753-761. [DOI] [PubMed] [Google Scholar]
- 41.Nagle, D. G., F. T. Camacho, and V. J. Paul. 1998. Dietary preferences of the opisthobranch mollusc Stylocheilus longicauda for secondary metabolites produced by the tropical cyanobacterium Lyngbya majuscula. Mar. Biol. 132:267-273. [Google Scholar]
- 42.Nagle, D. G., P. U. Park, and V. J. Paul. 1997. Pitiamide A, a new chlorinated lipid from a mixed marine cyanobacterial assemblage. Tetrahedron Lett. 38:6969-6972. [Google Scholar]
- 43.Nagle, D. G., and V. J. Paul. 1999. Production of secondary metabolites by filamentous tropical marine cyanobacteria: ecological functions of the compounds. J. Phycol. 35:1412-1421. [Google Scholar]
- 44.Nagle, D. G., V. J. Paul, and M. A. Roberts. 1996. Ypaoamide, a new broadly acting feeding deterrent from the marine cyanobacterium Lyngbya majuscula. Tetrahedron Lett. 37:6263-6266. [Google Scholar]
- 45.Nagle, D. G., Y-D. Zhou, P. U. Park, V. J. Paul, I. Rajbhandari, C. J. G. Duncan, and D. S. Pasco. 2000. A new indanone from the marine cyanobacterium Lyngbya majuscula that inhibits hypoxia-induced activation of the VEGF promoter in Hep3B cells. J. Nat. Prod. 63:1431-1433. [DOI] [PubMed] [Google Scholar]
- 46.Neilan, B. A., B. P. Burns, D. A. Relman, and D. R. Lowe. 2002. Molecular identification of cyanobacteria associated with stromatolites from distinct geographic locations. Astrobiology 2:271-280. [DOI] [PubMed] [Google Scholar]
- 47.Neilan, B. A., D. Jacobs, and A. E. Goodman. 1995. Genetic diversity and phylogeny of toxic cyanobacteria determined by DNA polymorphisms within the phycocyanin locus. Appl. Environ. Microbiol. 61:3875-3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nelissen, B., R. DeBaere, A. Wilmotte, and R. DeWachter. 1996. Phylogenetic relationships of nonaxenic filamentous cyanobacterial strains based on 16S rRNA sequence analysis. J. Mol. Evol. 42:194-200. [DOI] [PubMed] [Google Scholar]
- 49.Nübel, U., F. Garcia-Pichel, and G. Muyzer. 1997. PCR primers to amplify 16S rRNA genes from cyanobacteria. Appl. Environ. Microbiol. 63:3327-3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paul, V. J., R. W. Thacker, and E. Cruz-Rivera. 2001. Chemical mediation of seaweed-herbivore interactions: ecological and evolutionary perspectives, p. 227-265. In J. McClintock and B. Baker (ed.), Marine chemical ecology. CRC Press, New York, N.Y.
- 51.Pennings, S. C., A. M. Weiss, and V. J. Paul. 1996. Secondary metabolites of the cyanobacterium Microcoleus lyngbyaceus and the sea hare Stylocheilus longicauda: palatability and toxicity. Mar. Biol. 126:735-743. [Google Scholar]
- 52.Posada, D., and K. A. Crandall. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 53.Rodríguez, F., J. L. Oliver, A. Marín, and J. R. Medina. 1990. The general stochastic model of nucleotide substitution. J. Theor. Biol. 142:485-501. [DOI] [PubMed] [Google Scholar]
- 54.Rudi, K., O. M. Skulberg, and K. S. Jakobsen. 1998. Evolution of cyanobacteria by exchange of genetic material among phyletically related strains. J. Bacteriol. 180:3453-3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rudi, K., T. Fossheim, and K. S. Jakobsen. 2002. Nested evolution of a tRNALeu(UAA) group I intron by both horizontal intron transfer and recombination of the entire tRNA locus. J. Bacteriol. 184:666-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scheldeman, P., D. Baurain, R. Bouhy, M. Scott, M. Mühling, B. A. Whitton, A. Belay, and A. Wilmotte. 1999. Arthrospira (′Spirulina') strains from four continents are resolved into only two clusters, based on amplified ribosomal DNA restriction analysis of the internally transcribed spacer. FEMS Microbiol. Lett. 172:213-222. [DOI] [PubMed] [Google Scholar]
- 57.Thacker, R. W., D. G. Nagle, and V. J. Paul. 1997. Effects of repeated exposures to marine cyanobacterial secondary metabolites on feeding by juvenile rabbitfish and parrotfish. Mar. Ecol. Prog. Ser. 167:21-29. [Google Scholar]
- 58.Thacker, R. W., and V. J. Paul. 2001. Are benthic cyanobacteria indicators of nutrient enrichment? Relationships between cyanobacterial abundance and environmental factors on the reef flats of Guam. Bull. Mar. Sci. 69:497-508. [Google Scholar]
- 59.Thacker, R. W., D. W. Ginsburg, and V. J. Paul. 2001. Effects of herbivore exclusion and nutrient enrichment on coral reef macroalgae and cyanobacteria. Coral Reefs 19:318-329. [Google Scholar]
- 60.Tillett, D., D. L. Parker, and B. A. Neilan. 2001. Detection of toxigenicity by a probe for the microcystin synthetase A gene (mcyA) of the cyanobacterial genus Microcystis: comparison of toxicities with 16S rRNA and phycocyanin operon (phycocyanin intergenic spacer) phylogenies. Appl. Environ. Microbiol. 67:2810-2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Whitton, B. A., and M. Potts. 1982. Marine littoral, p. 515-542. In N. G. Carr and B. A. Whitton (ed.), The biology of cyanobacteria. Blackwell Science, Oxford, England.
- 62.Whitton, B. A., and M. Potts. 2000. The ecology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 63.Williams, P. G., H. Luesch, W. Y. Yoshida, R. E. Moore, and V. J. Paul. 2003. Continuing studies on the cyanobacterium Lyngbya sp.: isolation and structure determination of 15-norlyngbyapeptin A and lyngbyabellin D. J. Nat. Prod. 66:595-598. [DOI] [PubMed] [Google Scholar]
- 64.Williams, P. G., R. E. Moore, and V. J. Paul. 2003. Isolation and structure determination of lyngbyastatin 3, a lyngbyastatin 1 homologue from the marine cyanobacterium Lyngbya majuscula. Determination of the configuration of the 4-amino-2,2-dimethyl-3-oxopentanoic acid unit in majusculamide C, dolastatin 12, lyngbyastatin 1, and lyngbyastatin 3 from cyanobacteria. J. Nat. Prod. 66:1356-1363. [DOI] [PubMed] [Google Scholar]
- 65.Wilmotte, A. 1991. Taxonomic study of marine oscillatoriacean strains (Cyanophyceae, Cyanobacteria) with narrow trichomes. I. Morphological variability and autecological features. Algol. Stud. 64:215-248. [Google Scholar]
- 66.Wilmotte, A. 1992. Taxonomic study of marine oscillatoriacean strains (Cyanobacteria) with narrow trichomes. II. Nucleotide sequence analysis of the 16S ribosomal RNA. J. Phycol. 28:828-838. [Google Scholar]
- 67.Wilmotte, A. 1994. Molecular evolution and taxonomy of the Cyanobacteria, p. 1-25. In D. A. Bryant (ed.), The molecular biology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 68.Zehr, J. P., M. T. Mellon, and W. D. Hiorns. 1997. Phylogeny of cyanobacterial nifH genes: evolutionary implications and potential applications to natural assemblages. Microbiology 143:1443-1450. [DOI] [PubMed] [Google Scholar]