Abstract
The ability of salmonellae to become internalized and to survive and replicate in amoebae was evaluated by using three separate serovars of Salmonella enterica and five different isolates of axenic Acanthamoeba spp. In gentamicin protection assays, Salmonella enterica serovar Dublin was internalized more efficiently than Salmonella enterica serovar Enteritidis or Salmonella enterica serovar Typhimurium in all of the amoeba isolates tested. The bacteria appeared to be most efficiently internalized by Acanthamoeba rhysodes. Variations in bacterial growth conditions affected internalization efficiency, but this effect was not altered by inactivation of hilA, a key regulator in the expression of the invasion-associated Salmonella pathogenicity island 1. Microscopy of infected A. rhysodes revealed that S. enterica resided within vacuoles. Prolonged incubation resulted in a loss of intracellular bacteria associated with morphological changes and loss of amoebae. In part, these alterations were associated with hilA and the Salmonella virulence plasmid. The data show that Acanthamoeba spp. can differentiate between different serovars of salmonellae and that internalization is associated with cytotoxic effects mediated by defined Salmonella virulence loci.
Free-living amoebae and bacteria are involved in complex interactions, and it is known that amoebae act as environmental hosts of several intracellular pathogens, such as Legionella, Chlamydia, Mycobacterium, and Listeria spp. (3, 28, 32, 42, 47). Interestingly, gene functions required by Legionella spp. for infection of protozoa are also required for infection of mammalian cells (7, 13, 16, 39). It has even been suggested that growth in an amoebic intracellular environment might assist bacteria in their adaptation to mammalian phagocytic cells (17, 18). Moreover, incorporation of bacteria into amoebic cysts has been shown to confer resistance to adverse environmental conditions such as exposure to biocidal agents (4).
Salmonellae are a group of closely related gram-negative enteric bacteria, many of which act as facultatively intracellular pathogens (37). Salmonellae can also be isolated from the environment, where they share the habitat with a variety of other bacteria, plants, and protozoa. Members of the genus Salmonella infect an impressive spectrum of host organisms, either causing symptomatic infection after ingestion of contaminated food or water or establishing persistent carrier states. After colonizing the intestine, they may penetrate into the intestinal epithelium and Peyer's patches, spread into the draining mesenteric lymph nodes, and, via the bloodstream, access the spleen and liver, where they replicate in macrophages of the reticuloendothelial system (24, 25).
In model infections the pathogenicity of Salmonella is dependent on entry into, and proliferation inside, mammalian host cells. Notably, functions encoded by pathogenicity islands orchestrate these processes through the activities of several bacterial virulence proteins. Salmonella pathogenicity island 1 (SPI1) codes for a type III secretion system, required for invasion of epithelial cells, that translocates bacterial virulence proteins into the host cell (21, 22). In mice, macrophages appear to be a central host cell type for bacterial intracellular proliferation during later phases of infection (15, 36). Intramacrophage survival and proliferation is in part mediated by genes located on a second pathogenicity island, SPI2, which codes for another type III secretion system (8, 19, 36), and on a virulence-associated plasmid carrying the highly conserved spv (salmonella plasmid virulence) gene cluster (15).
Free-living amoebae, such as Acanthamoeba spp., are commonly found in natural aquatic systems and in soil (30), and even within the intestines of humans (46, 48) and reptiles (40); hence, they are expected to encounter and ingest salmonellae. Indeed, King et al. (26) reported the survival of Salmonella enterica serovar Typhimurium within Acanthamoeba castellanii during chlorination, suggesting a protective intracellular habitat for the bacteria. Yet, while one finds reports on recovery of Salmonella and Acanthamoeba species from soil and water at the same environmental locations (38), no data on the interaction between salmonellae and free-living amoebae have been provided. In this study we describe the establishment of conditions for the growth and replication of salmonellae in an amoebic intracellular environment, and we determine whether bacterial growth conditions affect the survival, replication, and cytotoxicity of salmonellae within Acanthamoeba rhysodes. In addition, the effects of the Salmonella virulence plasmid, the spv genes, and the SPI1 transcriptional activator of invasion gene hilA on bacterial survival, replication, and cytotoxicity for A. rhysodes were evaluated.
MATERIALS AND METHODS
Bacterial strains.
Salmonella serovar Typhimurium 14028 was obtained from the American Type Culture Collection. Salmonella enterica serovar Enteritidis, isolated from a Swedish patient, was obtained from the Swedish Institute for Infectious Disease Control. Salmonella enterica serovar Dublin strain SH9325 was obtained from Alistair J. Lax (43). Strains containing insertion elements in spvR, spvB, or hilA were derivatives of Salmonella serovar Dublin strain 2229. The insertion elements zzx-2558::Tn5 and zzx-2556::Tn5, disrupting the spvR and spvB genes (44), respectively, were introduced by transduction with phage P22 int (41). Insertion of the hilA::kan334 element (10), inactivating the hilA gene, generated an invasion-deficient strain. An isogenic derivative of Salmonella serovar Dublin 2229 cured of the virulence plasmid was obtained from Alistair J. Lax and was designated M173c (35). Strains were grown in Luria broth (LB) or on Luria agar (LA) with antibiotics. For plasmid segregation experiments, strains were transformed with plasmid pPir (34).
Cell lines and culture conditions.
The five isolates of amoebae used in these experiments were A. castellanii, A. rhysodes, Acanthamoeba sp. strain Ac896, isolated from a human keratitis case, and Acanthamoeba sp. strains I4 and V38, isolated from a geyser and a contaminated cell culture, respectively. These were obtained as axenic strains and were maintained as adherent cells in an axenic culture medium, peptone-yeast extract-glucose (PYG) broth, in 25-cm3 tissue culture flasks incubated at 30°C, until near-confluence was reached. Amoebic suspension was examined by bright-field microscopy before use, and the number of cells was determined by cell counting in a Burker chamber.
Epithelial Madin-Darby canine kidney (MDCK) cells from the American Type Culture Collection were maintained in RPMI 1640 medium supplemented with l-glutamine (final concentration, 2 mM), HEPES (final concentration, 10 mM), fetal bovine serum (final concentration, 10% [vol/vol]), and gentamicin sulfate (final concentration, 10 μg/ml).
Preparation of invasive and noninvasive bacterial inocula.
Bacterial strains were grown either in LB or on LA with appropriate antibiotics. Salmonellae grown to the late-logarithmic-growth phase in LB show clear induction of SPI1 invasion genes and invasion ability, whereas bacteria grown on LA show diminished invasiveness (10). Cocultivation of amoebae and bacteria was performed in PYG. To prepare noninvasive bacteria, salmonellae were collected from LA and suspended in phosphate-buffered saline (PBS). The bacteria were then diluted in PYG to give a concentration of 100 bacteria/amoeba. Invasion-competent bacterial cultures were prepared by diluting stationary-phase bacteria 1/10 in LB medium and subsequently incubating the culture for 1 h, 45 min, on a roller under microanaerobic conditions. The bacteria were then diluted in PYG to yield a concentration of 20 bacteria/amoeba.
Intracellular bacterial growth in acanthamoebae.
Amoebae were grown in 5 ml of PYG medium. The tissue culture flask was gently shaken, and the PYG containing nonadherent amoebae was removed. New PYG was added, and the amoebae were taken off by incubation on ice for 30 min. The suspension was centrifuged for 5 min at 200 × g, and the pellet was washed with PBS and resuspended in PYG. The suspension was added to each well of a 6-well plate (106 amoebae/well). Amoebae were then incubated for 24 h at 30°C to allow them to adhere. The number of amoebae/well was calculated once more before infection.
To infect amoebae, invasive or noninvasive bacterial inocula were diluted in PYG to give a final concentration of 20 to 100 bacteria per amoeba. The plate was gently centrifuged (for 5 min at 500 × g) in order to promote contact between bacteria and amoebae and was then incubated for 60 min at 37°C. After the incubation, the medium was changed to PYG supplemented with gentamicin sulfate (final concentration, 50 μg/ml) in order to kill extracellular bacteria. After incubation for 60 min at 37°C, the medium was changed to maintenance medium, PYG supplemented with gentamicin at a final concentration of 10 μg/ml, for continuing incubation. The cells were washed twice with PBS and lysed in 0.5% sodium deoxycholate (1), and the number of CFU was determined by plating samples on LA plates.
For plasmid segregation experiments, strains harboring pPir were grown overnight in the presence of 100 μg of ampicillin sodium salt (Sigma)/ml at 30°C as described by Benjamin et al. (5). Infection was performed as described above.
The sensitivity of intracellular SH9325 bacteria (devoid of pPir) to ampicillin (100 μg/ml) was determined as outlined by Abshire and Neidhardt (2).
Viability assay.
The viability of host cells was determined at 16 h postinfection by using trypan blue, followed by quantification of stained cells and total cells by use of a Burker chamber.
Immunofluorescence.
A. rhysodes was grown on Lab-Tek chamber slides with coverslips and infected as described above. The cells were washed twice with PBS and fixed with acetone for 20 min at −20°C. Cells were later stained with fluorescein isothiocyanate-phalloidin, tetramethyl rhodamine isothiocyanate-conjugated anti-rabbit immunoglobulin G, or a rabbit antiserum against S. enterica O-antigen 9 (Reagensia, Stockholm, Sweden).
Transmission electron microscopy.
A. rhysodes was infected with invasive Salmonella serovar Dublin as described above. At 16 h postinfection, the cells were washed twice with PBS and fixed with 50% sodium cacodylate (pH 7.4), 1% glutaraldehyde, and 3% paraformaldehyde for 1 h at 4°C. Cells were harvested by pipetting up and down, pelleted for 5 min at 600 × g and 4°C, and resuspended in fixation buffer diluted 1:10. Transmission electron microscopy was performed using standard procedures.
RESULTS
Internalization of invasive and noninvasive S. enterica into acanthamoebae.
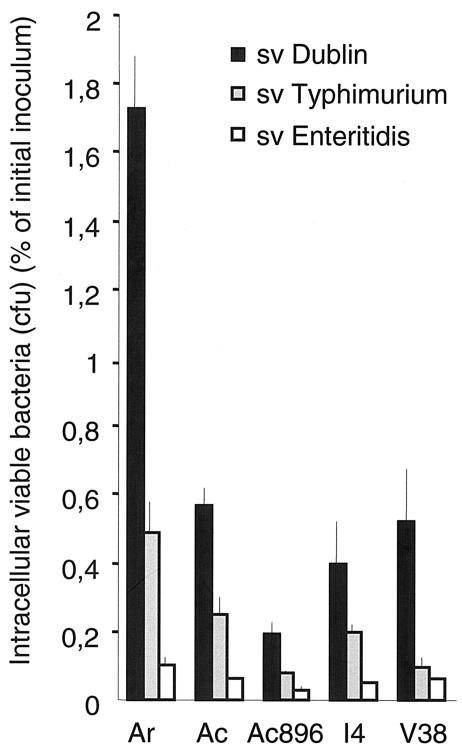
We initiated our experiments by infecting five different Acanthamoeba isolates (A. rhysodes, A. castellanii, A. castellanii 896, and isolates I4 and V38) with invasive (LB-grown) cultures of Salmonella serovar Dublin, Salmonella serovar Enteritidis, and Salmonella serovar Typhimurium. Two hours after infection, the number of viable intracellular bacteria was determined as a percentage of the initial inoculum (for details, see Materials and Methods). Salmonella serovar Dublin was recovered in higher numbers than Salmonella serovar Enteritidis or Salmonella serovar Typhimurium from all Acanthamoeba lines tested, whereas all serovars were most efficiently internalized by A. rhysodes (Fig. 1).
FIG. 1.
Internalization of invasive S. enterica into five Acanthamoeba isolates in gentamicin protection assays. Results are presented as viable intracellular bacteria recovered 2 h after challenge, given as a percentage of the initial inoculum. Values are means from three independent experiments, each carried out in duplicate wells. Error bars, standard deviations. Ar, A. rhysodes; Ac, A. castellanii; Ac896, Acanthamoeba sp. strain Ac896.
When noninvasive (LA-grown) cultures of Salmonella serovar Dublin or Salmonella serovar Typhimurium were used, we observed a significant decrease in the numbers of viable bacteria recovered from infected A. rhysodes (0.008 and 0.002%, respectively). Still, compared to invasive S. enterica cultures, the Escherichia coli laboratory strain TG1 was recovered in diminished amounts when either LB- or LA-grown cultures were used (0.002 and 0.007%, respectively).
Role of invasion functions for bacterial uptake.
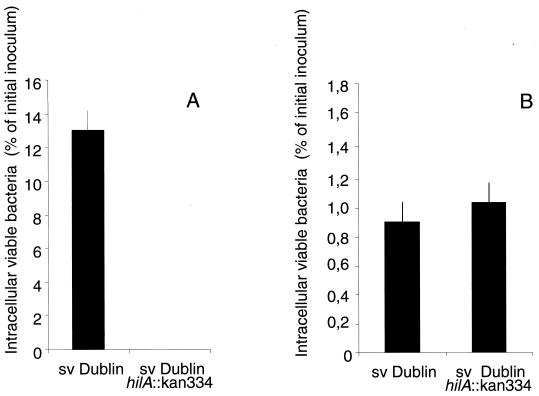
Because invasion of epithelial cells is mediated by a type III secretion system encoded on Salmonella pathogenicity island 1 (SPI1), we next studied the effect of inactivating the hilA gene in Salmonella serovar Dublin. The hilA gene codes for the SPI1 activator protein HilA, and hilA mutants are known to have a strongly reduced ability to enter and cross the intestinal epithelium (12, 29). In order to study the role of SPI1 in entry, epithelial MDCK cells and A. rhysodes were infected with LB-grown cultures of Salmonella serovar Dublin or with an isogenic hilA mutant. Two hours after infection, the number of viable intracellular bacteria was determined as a percentage of the initial inoculum (Fig. 2). MDCK cells infected with the Salmonella serovar Dublin hilA mutant showed a strongly reduced uptake of bacteria compared to cells infected with wild-type Salmonella serovar Dublin (hilA+) (Fig. 2A). In contrast, A. rhysodes infected with Salmonella serovar Dublin did not distinguish the hilA mutant from the hilA+ strain in terms of bacterial uptake (Fig. 2B). These observations indicate that hilA plays different roles in the entry of Salmonella into MDCK and A. rhysodes cells. Apparently, still, the bacterial growth condition had a significant effect on the mechanism of entry into amoebae.
FIG. 2.
Uptake of invasive wild-type Salmonella serovar Dublin (hilA+) and the Salmonella serovar Dublin hilA mutant into epithelial MDCK cells (A) and A. rhysodes (B).
Detachment of amoebae upon prolonged infection with S. enterica.
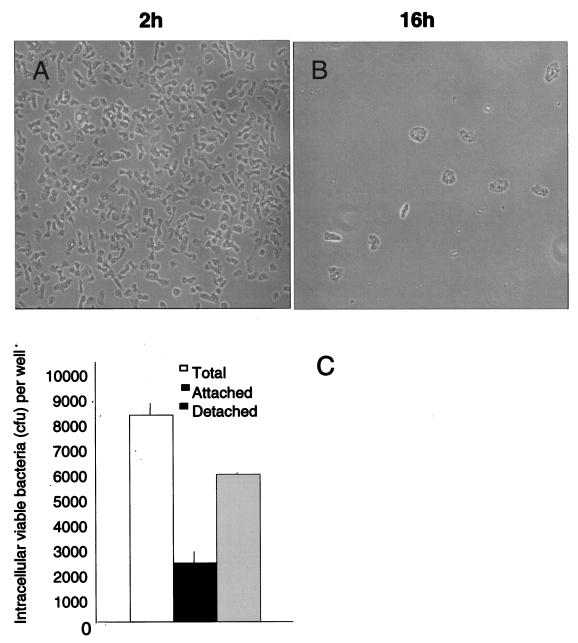
Under phase-contrast microscopy, A. rhysodes infected with invasive wild-type S. enterica serovar Dublin displayed a significant loss of adherent cells over 16 h of infection (Fig. 3A and B). A similar phenomenon is observed when human monocytic cells are infected with salmonellae: infected cells become detached, whereupon most of the viable intracellular bacteria can be recovered from detached cells (27). Therefore, the numbers of viable bacteria recovered from attached and detached A. rhysodes at 16 h postinfection were determined (Fig. 3C) by using LB-grown cultures of the bacteria. The results show that the number of viable Salmonella serovar Dublin bacteria recovered from the detached amoebae was higher than that recovered from the attached fraction.
FIG. 3.
(A and B) Phase-contrast images of A. rhysodes infected with invasive Salmonella serovar Dublin. Images were taken at 2 h (A) and 16 h (B) postinfection. (C) Proportion of live intracellular bacteria recovered from detached and attached A. rhysodes cells 16 h after infection with invasive Salmonella serovar Dublin. Values are means from three independent experiments, each carried out in duplicate wells. Error bars, standard deviations.
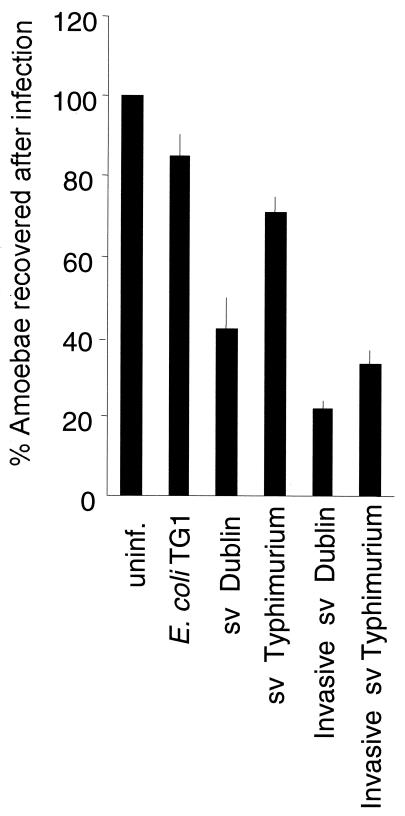
We then reasoned that detachment of A. rhysodes after infection with Salmonella serovar Dublin could be the result of a cytotoxic effect mediated by the infecting bacteria. A. rhysodes was infected with invasive (LB-grown) or noninvasive (LA-grown) bacteria at a multiplicity of infection of 20 or 100, respectively, and the cells were analyzed more carefully at 16 h postinfection (Fig. 4). When A. rhysodes was infected with invasive Salmonella serovar Dublin, 20% of the amoebae were recovered at 16 h postinfection relative to the recovery of uninfected controls (taken as 100%), whereas with noninvasive bacteria, the recovery of amoebae rose to 40% (Fig. 4). A similar trend was observed with invasive and noninvasive Salmonella serovar Typhimurium (Fig. 4). When the control strain E. coli TG1 was used, 90% of the infected amoebae were recovered (Fig. 4), implying that ingestion of bacteria per se did not induce detachment.
FIG. 4.
Proportion of A. rhysodes cells recovered after infection with invasive (LB-grown) or noninvasive (LA-grown) bacteria at a multiplicity of infection of 20 or 100, respectively, at 16 h postinfection. Values are means from three independent experiments, each carried out in duplicate wells. Error bars, standard deviations. uninf., uninfected controls.
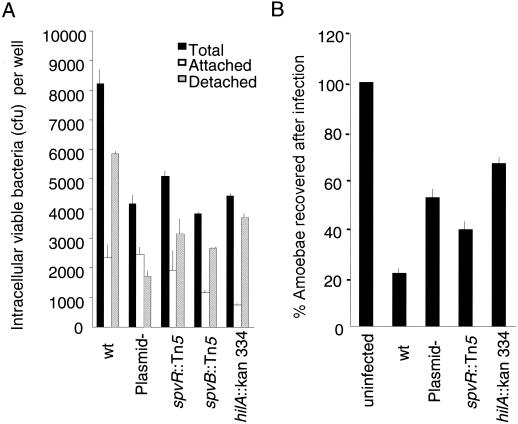
Next, hilA, spvB, and spvR mutants of Salmonella serovar Dublin and an isogenic virulence plasmid-cured strain were tested for intracellular fitness and cytotoxicity in A. rhysodes. Again, the numbers of viable bacteria recovered in detached and attached fractions were determined (Fig. 5A). Altogether, the number of total bacteria recovered was higher for the wild-type strain than for any of the mutant strains tested or the plasmid-cured strain. With the plasmid-cured strain, there was apparently a relative loss of viable bacteria recovered from the detached fraction, whereas the hilA mutant showed a more significant decrease in the number of bacteria in the attached population of amoebae.
FIG. 5.
Role of bacterial virulence determinants for bacteria intracellular growth yields and recovery of amoebae. (A) Amounts of viable bacteria recovered from detached and attached A. rhysodes cells 16 h after infection. wt, wild type; Plasmid-, virulence plasmid cured. (B) Proportion of A. rhysodes cells recovered after infection with invasive (LB-grown) bacteria at the same time point. Values are means from three independent experiments, each carried out in duplicate wells. Error bars, standard deviations.
In parallel, in A. rhysodes cultures infected with the hilA mutant of Salmonella serovar Dublin, the fraction of amoebae recovered over a 16-h incubation increased (to more than 60%) relative to that for cultures infected with the wild-type parental strain (Fig. 5B). Also, infection with the plasmid-cured strain resulted in an increased proportion of amoebae in long-term-infected cultures (Fig. 5B). Thus, it appears that both SPI1 and the virulence plasmid contribute to the interaction between salmonellae and A. rhysodes, although hilA is dispensable for uptake.
A. rhysodes-associated salmonellae are intracellular.
Because the prolonged cocultivation experiments presented above were conducted in the presence of gentamicin, the bacteria recovered from the amoebae are likely to derive from the inside of the host cell. In order to verify whether the salmonellae associated with A. rhysodes were indeed intracellular, cultures infected for 2 and 16 h were examined by immunofluorescence and transmission electron microscopy (Fig. 6).
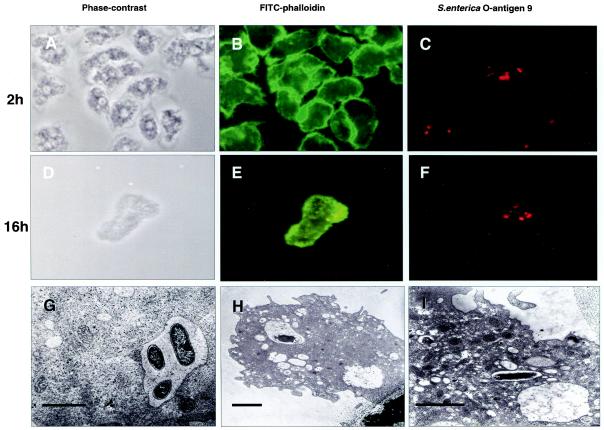
FIG. 6.
(A through F) Microscopy of A. rhysodes infected with invasive Salmonella serovar Dublin at 2 h (A through C) and 16 h (D through F) postinfection. (A and D) Phase-contrast images; (B and E) staining of cells with fluorescein isothiocyanate (FITC)-phalloidin; (C and F) staining of bacteria with a rabbit antiserum against S. enterica O-antigen 9 and rhodamine-conjugated anti-rabbit immunoglobulin G. (G through I) Transmission electron microscopy shows intravacuolar Salmonella serovar Typhimurium bacteria at 2 h postinfection (G) and Salmonella serovar Dublin at 16 h postinfection (H and I). (G) Magnification, ×5,000; bar, 3.4 μm. At this magnification, bacterial membranes appear intact. (H) Magnification, ×5,000; bar, 2.2 μm. (I) Magnification, ×8,000; bar, 2.3 μm.
Fluorescence microscopy showed dense layers of amoebae at 2 h postinfection, with the cells staining brightly for filamentous actin (Fig. 6A to C). The bacteria resided within the borders of the cells, consistent with an intracellular localization. At 16 h postinfection, the bacteria were similarly localized, but infected cells were less dense, were rounded, and showed a somewhat more diffuse granular staining for filamentous actin (Fig. 6D to F).
An actual intracellular localization was confirmed by transmission electron microscopy, which also showed that bacteria were contained within membrane-bound vacuoles in A. rhysodes (Fig. 6G to I).
Intracellular salmonellae are replicating.
Unlike normal macrophage phagosomes, in which bacteria are degraded, the Salmonella-containing-vacuole (SCV) appears to represent a unique type of intracellular vacuole that is permissive for bacterial replication (20). A general trend during the cocultivation experiments with A. rhysodes was the loss of viable bacteria recovered between the 2- and 16-h observation points. This could be due to efficient killing of the bacteria by A. rhysodes. Alternatively, or in parallel, the bacterium-mediated cytotoxicity may have caused bacteria to be released into the gentamicin-containing medium.
In order to demonstrate or exclude intracellular bacterial replication, A. rhysodes cells were infected with Salmonella serovar Dublin carrying a plasmid with a temperature-sensitive replicon and an ampicillin resistance marker. Because the plasmid applied segregates as a function of bacterial cell division at 37°C, loss of the antibiotic resistance marker among bacterial progenitors can be used as a measure of replication. This approach has been used in a number of other studies to determine Salmonella proliferation in infected organs (5, 14) as well as in cultured cells (23).
Thus, the percentages of viable ampicillin-resistant bacteria in the inoculum and among intracellular bacteria recovered at 2 and 16 h after challenge were determined. The results showed that the plasmid segregated from the bacterial cultures as a function of the infection of A. rhysodes (Fig. 7), implying the presence of intracellularly replicating bacteria. This interpretation was also corroborated by infecting A. rhysodes with ampicillin-sensitive Salmonella serovar Dublin and monitoring the decrease in viable counts in the presence and absence of ampicillin as described by Abshire and Neidhardt (2). In this experiment, the decline in the number of intracellular viable bacteria was much more pronounced in the presence of ampicillin (data not shown), a finding consistent with the presence of replicating bacteria inside the amoebae.
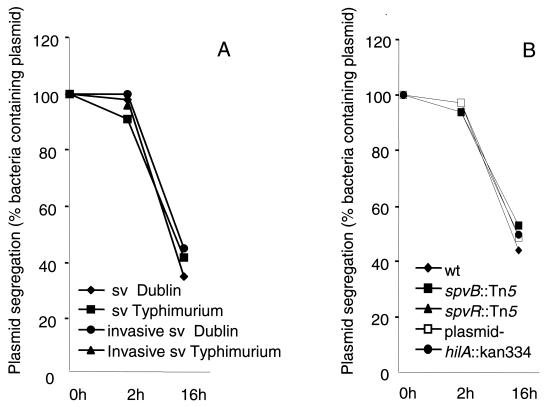
FIG. 7.
Demonstration of intracellular replication of Salmonella serovar Dublin within A. rhysodes. The percentage of viable ampicillin-resistant bacteria was determined for the inoculum and for bacteria recovered 2 and 16 h after challenge, and segregation of the ampicillin resistance plasmid was used as a readout for the efficiency of replication. (A) Replication of invasive and noninvasive Salmonella serovar Dublin and Salmonella serovar Typhimurium. (B) Replication of the hilA, spvR, and spvB mutants and of the plasmid-cured strain. wt, wild type; plasmid-, plasmid cured. Values are means from three independent experiments, each carried out in duplicate wells.
No significant differences could be observed between invasive and noninvasive Salmonella infection in terms of segregation of the ampicillin resistance marker (Fig. 7A). Nor could we show any effect of the hilA, spvR, or spvB mutant, or of the plasmid-cured strain, on replication (Fig. 7B). Combined, these data suggest that Salmonella serovar Dublin uses selected, defined virulence functions to mediate cytotoxicity but that these gene functions do not affect replication efficiency under the experimental conditions used.
DISCUSSION
Free-living amoebae are gaining increasing attention as ubiquitous eukaryotes influencing our perception of human bacterial pathogens in the environment. For example, the facultatively intracellular mammalian pathogens Legionella pneumophila and mycobacteria are also known to prosper within free-living amoebae, implying that such bacteria are adapted to an intracellular environment in a more general sense (32, 42). Furthermore, L. pneumophila is known to use the same sets of genes for multiplying in human macrophages and A. castellanii (17).
Salmonellae are rather promiscuous in their ability to invade and replicate in mammalian host cells. In contrast, the interaction between salmonellae and Acanthamoeba species has not been described in any detail. In the present study we observed a preferential uptake of Salmonella serovar Dublin over that of serovar Enteritidis or serovar Typhimurium in all isolates of Acanthamoeba tested (Fig. 1). Since by definition the bacterial serovars express different types of sugar entities in the surface O-antigen polysaccharide, one may suggest that this results in differential recognition by the amoebae and hence in different efficiencies of uptake mediated by the lectin-like receptors on the surfaces of the amoebae. We also found that bacterial growth conditions had significant effects on the efficiency of entry into amoebae. Broth (LB) cultures were more efficiently taken up by A. rhysodes than were bacteria propagated on solid (LA) medium. It is possible that this significant difference in uptake can be related to the different entry mechanisms used. Entry of salmonellae into mammalian host cells, especially epithelial cells of the gut, depends on virulence factors that are encoded by SPI1 and are induced under LB growth conditions (12, 29). However, the data reported here indicate that the SPI1 activator HilA was not required for the entry of Salmonella serovar Dublin into A. rhysodes (Fig. 2B).
Prolonged incubation of the bacterial-amoebic cultures resulted in a gradual change in morphology and eventually in the disappearance of the host cells (Fig. 3A). A related phenomenon has been reported by Ly and Müller (28) for cocultures of Listeria monocytogenes and Acanthamoeba species. Listeriae were taken up by the amoebae, and the bacteria replicated intracellularly. However, longer incubations led to release of bacteria and encystment, with the intracellular listeriae losing viability in cysts.
A portion of the observations implied that the loss of salmonellae in A. rhysodes was not simply a reflection of intracellular killing of the bacteria. Salmonella spp., like several other bacterial pathogens, including Mycobacterium, Legionella, and Brucella spp., replicate within membrane-bound vacuoles inside macrophages. While we did not attempt to define the nature of intracellular vesicles, electron microscopy of Salmonella-infected A. rhysodes showed that the bacteria were localized within membrane-bound vacuoles (Fig. 6G through I), and some bacteria apparently were replicating (Fig. 6G). When we used Salmonella serovar Dublin carrying a plasmid with a temperature-sensitive replicon in the intracellular replication experiment, the plasmid segregated as a function of time, implying that a least a portion of the bacteria were actually replicating inside the acanthamoebae (Fig. 7).
Finally, since we recovered more bacteria from detached host cells than from cells attached to the culture chamber (Fig. 3B), it appears that engulfment of Salmonella serovar Dublin resulted in cytotoxicity, leading to detachment and disintegration of the amoebae, which subsequently resulted in exposure of the bacteria to gentamicin. This resembles the scenario seen with human monocytic cells and MDCK epithelial cells infected with S. enterica in that the cells are intoxicated by the infecting bacteria (6, 25, 33, 45).
SPI1 is responsible for inducing a very rapid apoptotic response in mononuclear phagocytes (31). Another, later phase of cytotoxicity is dependent on SPI2 and the spv genes (6, 11, 33) and involves apoptosis and interference with the normal dynamics of the actin cytoskeleton. Concomitantly, the loss of infected Acanthamoeba cells and the apparent growth yields of the bacteria were found to be partly dependent on hilA and the S. enterica spv-carrying virulence plasmid (Fig. 5). However, it remains to be shown whether the effect of SPI1 and the spv genes is that of inducing programmed cell death in amoebae, or whether loss of cells is due to other effects of the corresponding virulence proteins.
Free-living amoebae can harbor bacteria inside their cysts, giving them a microhabitat protecting them from environmental hazards. Furthermore, a study by King et al. (26) showed that the bacterium-protozoan association provides increased numbers of bacteria with increased resistance to free chlorine residuals, which can lead to the persistence of bacteria in chlorine-treated water. It has also been reported that amoeba-grown L. pneumophila displays increased intracellular survival and replication in macrophages (9) and that intracellular growth in A. castellanii affects the monocyte entry mechanism and enhances the virulence of L. pneumophila.
Although the present study included only a restricted set of salmonellae and acanthamoebae, and therefore formally may not reflect all possible forms of interactions, our results show that A. rhysodes was able to ingest salmonellae and that subsequent events included intracellular bacterial replication. We also detected a bacterium-mediated cytotoxicity that appeared to be dependent on documented virulence genes, implying that genetic determinants of salmonellae used for invasion and intracellular proliferation in mammals could also be operative in the environment. It thus remains possible that free-living amoebae function as environmental hosts for salmonellae and that such amoebae participate in the transmission of salmonellosis.
REFERENCES
- 1.Abd, H., T. Johansson, I. Golovliov, G. Sandstrom, and M. Forsman. 2003. Survival and growth of Francisella tularensis in Acanthamoeba castellanii. Appl. Environ. Microbiol. 69:600-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abshire, K. Z., and F. C. Neidhardt. 1993. Growth rate paradox of Salmonella typhimurium within host macrophages. J. Bacteriol. 175:3744-3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann, R., N. Springer, W. Schonhuber, W. Ludwig, E. N. Schmid, K. D. Muller, and R. Michel. 1997. Obligate intracellular bacterial parasites of acanthamoebae related to Chlamydia spp. Appl. Environ. Microbiol. 63:115-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barker, J., and M. R. Brown. 1994. Trojan horses of the microbial world: protozoa and the survival of bacterial pathogens in the environment. Microbiology 140:1253-1259. [DOI] [PubMed] [Google Scholar]
- 5.Benjamin, W. H., Jr., P. Hall, S. J. Roberts, and D. E. Briles. 1990. The primary effect of the Ity locus is on the rate of growth of Salmonella typhimurium that are relatively protected from killing. J. Immunol. 144:3143-3151. [PubMed] [Google Scholar]
- 6.Browne, S. H., M. L. Lesnick, and D. G. Guiney. 2002. Genetic requirements for Salmonella-induced cytopathology in human monocyte-derived macrophages. Infect. Immun. 70:7126-7135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cianciotto, N. P., and B. S. Fields. 1992. Legionella pneumophila mip gene potentiates intracellular infection of protozoa and human macrophages. Proc. Natl. Acad. Sci. USA 89:5188-5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cirillo, D. M., R. H. Valdivia, D. M. Monack, and S. Falkow. 1998. Macrophage dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol. Microbiol. 30:175-188. [DOI] [PubMed] [Google Scholar]
- 9.Cirillo, J. D., S. L. G. Cirillo, L. Yan, L. E. Bermudez, S. Falkow, and L. S. Tompkins. 1999. Intracellular growth in Acanthamoeba castellanii affects monocyte entry mechanisms and enhances virulence of Legionella pneumophila. Infect. Immun. 67:4427-4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eriksson, S., J. Björkman, S. Borg, A. Syk, S. Pettersson, D. I. Andersson, and M. Rhen. 2000. Salmonella typhimurium mutants down regulating phagocyte nitric oxide production. Cell. Microbiol. 2:239-250. [DOI] [PubMed] [Google Scholar]
- 11.Fierer, J., and D. G. Guiney. 2001. Diverse virulence traits underlying different clinical outcomes of Salmonella infection. J. Clin. Investig. 107:775-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galán, J. E. 1996. Molecular genetic bases of Salmonella entry into host cells. Mol. Microbiol. 20:263-271. [DOI] [PubMed] [Google Scholar]
- 13.Gao, L. Y., O. S. Harb, and Y. Abu Kwaik. 1997. Utilization of similar mechanisms by Legionella pneumophila to parasitize two evolutionarily distant host cells, mammalian macrophages and protozoa. Infect. Immun. 65:4738-4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gulig, P. A., T. J. Doyle, M. J. Clare-Salzler, R. L. Maiese, and H. Matsui. 1997. Systemic infection of mice by wild-type but not Spv− Salmonella typhimurium is enhanced by neutralization of gamma interferon and tumor necrosis factor alpha. Infect. Immun. 65:5191-5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gulig, P. A., T. J. Doyle, and H. Matsui. 1998. Analysis of host cells associated with the Spv-mediated increased intracellular growth rate of Salmonella typhimurium in mice. Infect. Immun. 66:2471-2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hales, L. M., and H. A. Shuman. 1999. The Legionella pneumophila rpoS gene is required for growth within Acanthamoeba castellanii. J. Bacteriol. 181:4879-4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harb, O. S., and Y. Abu Kwaik. 2000. Interaction of Legionella pneumophila with protozoa provides lessons. ASM News 66:609-616. [Google Scholar]
- 18.Harb, O. S., L. Y. Gao, and Y. Abu Kwaik. 2000. From protozoa to mammalian cells: a new paradigm in the life cycle of intracellular bacterial pathogens. Environ. Microbiol. 2:251-265. [DOI] [PubMed] [Google Scholar]
- 19.Hensel, M., J. E. Shea, S. R. Waterman, R. Mundy, T. Nikolaus, G. Banks, A. Vazquez-Torres, C. Gleeson, F. C. Fang, and D. W. Holden. 1998. Genes encoding putative effector proteins of the type III secretion system of Salmonella pathogenicity island 2 are required for bacterial virulence and proliferation in macrophages. Mol. Microbiol. 30:163-174. [DOI] [PubMed] [Google Scholar]
- 20.Holden, D. W. 2002. Trafficking of the Salmonella vacuole in macrophages. Traffic 3:161-169. [DOI] [PubMed] [Google Scholar]
- 21.Hueck, C. J., M. J. Hantman, V. Bajaj, C. Johnston, C. A. Lee, and S. I Miller. 1995. Salmonella typhimurium secreted invasion determinants are homologous to Shigella Ipa proteins. Mol. Microbiol. 18:479-490. [DOI] [PubMed] [Google Scholar]
- 22.Hueck, C. J. 1998. Type III secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jantsch, J., C. Cheminay, D. Chakravortty, T. Lindig, J. Hein, and M. Hensel. 2003. Intracellular activities of Salmonella enterica in murine dendritic cells. Cell. Microbiol. 5:933-945. [DOI] [PubMed] [Google Scholar]
- 24.Jensen, V. B., J. T. Hartyand, and B. D. Jones. 1998. Interaction of the invasive pathogens Salmonella typhimurium, Listeria monocytogenes, and Shigella flexneri with M cells and murine Peyer's patches. Infect. Immun. 66:3758-3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones, B. D., N. Ghori, and S. Falkow. 1994. Salmonella typhimurium initiates murine infection by penetrating and destroying the specialized epithelial M cells of the Peyer's patches. J. Exp. Med. 180:15-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.King, C. H., E. B. Shotts, Jr., R. E. Wooley, and K. G. Porter. 1988. Survival of coliforms and bacterial pathogens within protozoa during chlorination. Appl. Environ. Microbiol. 54:3023-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Libby, S. J., M. Lesnick, P. Hasegawa, E. Weidenhammer, and D. G. Guiney. 2000. The Salmonella virulence plasmid spv genes are required for cytopathology in human monocyte-derived macrophages. Cell. Microbiol. 2:49-58. [DOI] [PubMed] [Google Scholar]
- 28.Ly, T. M. C., and H. E. Müller. 1990. Ingested Listeria monocytogenes survive and multiply in protozoa. J. Med. Microbiol. 33:51-54. [DOI] [PubMed] [Google Scholar]
- 29.Marcus, S. L., J. H. Brumell, C. G. Pfeifer, and B. B. Finlay. 2000. Salmonella pathogenicity islands: big virulence in small packages. Microbes Infect. 2:145-156. [DOI] [PubMed] [Google Scholar]
- 30.Martinez, A. J., and G. S. Visvesvara. 1997. Free-living, amphizoic and opportunistic amebas. Brain Pathol. 7:583-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Monack, D. M., B. Raupach, A. E. Hromockyj, and S. Falkow. 1996. Salmonella typhimurium invasion induces apoptosis in infected macrophages. Proc. Natl. Acad. Sci. USA 93:9833-9838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neumeister, B., M. Schoniger, M. Faigle, M. Eichner, and K. Dietz. 1997. Multiplication of different Legionella species in Mono Mac 6 cells and in Acanthamoeba castellanii. Appl. Environ. Microbiol. 63:1219-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paesold, G., D. G. Guiney, L. Eckmann, and M. F. Kagnoff. 2002. Genes in the Salmonella pathogenicity island 2 and the Salmonella virulence plasmid are essential for Salmonella-induced apoptosis in intestinal epithelial cells. Cell. Microbiol. 4:771-781. [DOI] [PubMed] [Google Scholar]
- 34.Pósfai, G., M. D. Koob, H. A. Kirkpatrick, and F. R. Blattner. 1997. Versatile insertion plasmids for targeted genome manipulations in bacteria: isolation, deletion, and rescue of the pathogenicity island LEE of the Escherichia coli O157:H7 genome. J. Bacteriol. 179:4426-4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pullinger, G. D., and A. J. Lax. 1992. A Salmonella dublin virulence plasmid locus that affects bacterial growth under nutrient-limited conditions. Mol. Microbiol. 6:1631-1643. [DOI] [PubMed] [Google Scholar]
- 36.Richter-Dahlfors, A., A. M. J. Buchan, and B. B. Finlay. 1997. Murine salmonellosis studied by confocal microscopy: Salmonella typhimurium resides intracellularly inside macrophages and exerts a cytotoxic effect on phagocytes in vivo. J. Exp. Med. 186:569-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodriguez, M., I. de Diego, and M. C. Mendoza. 1998. Extraintestinal salmonellosis in a general hospital (1991 to 1996): relationships between Salmonella genomic groups and clinical presentations. J. Clin. Microbiol. 36:3291-3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rude, R. A., G. J. Jackson, J. W. Bier, T. K. Sawyer, and N. G. Risty. 1984. Survey of fresh vegetables for nematodes, amoebae, and Salmonella. J. Assoc. Off. Anal. Chem. 67:613-615. [PubMed] [Google Scholar]
- 39.Segal, G., and H. A. Shuman. 1999. Legionella pneumophila utilizes the same genes to multiply within Acanthamoeba castellanii and human macrophages. Infect. Immun. 67:2117-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sesma, M. J., and L. Z. Ramos. 1989. Isolation of free-living amoebas from the intestinal contents of reptiles. J. Parasitol. 75:322-324. [PubMed] [Google Scholar]
- 41.Schieger, H. 1972. Phage P22 with increased or decreased transduction abilities. Mol. Gen. Genet. 119:75-88. [DOI] [PubMed] [Google Scholar]
- 42.Steinert, M., K. Birkness, E. White, B. Fields, and F. Quinn. 1998. Mycobacterium avium bacilli grow saprozoically in coculture with Acanthamoeba polyphaga and survive within cyst walls. Appl. Environ. Microbiol. 64:2256-2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sukopolvi, S., P. Riikonen, S. Taira, H. Saarilahti, and M. Rhen. 1992. Plasmid-mediated serum resistance in Salmonella enterica. Microb. Pathog. 12:219-225. [DOI] [PubMed] [Google Scholar]
- 44.Taira, S., and M. Rhen. 1989. Identification and genetic analysis of mkaA—a gene of the Salmonella typhimurium virulence plasmid necessary for intracellular growth. Microb. Pathog. 7:165-173. [DOI] [PubMed] [Google Scholar]
- 45.Tezcan-Merdol, D., T., Nyman, U. Lindberg, F. Haag, F. Koch-Nolte, and M. Rhen. 2001. Actin is ADP-ribosylated by the Salmonella enterica virulence-associated protein SpvB. Mol. Microbiol. 39:606-619. [DOI] [PubMed] [Google Scholar]
- 46.Thamprasert, K., S. Khunamornpong, and N. Morakote. 1993. Acanthamoeba infection of peptic ulcer. Ann. Trop. Med. Parasitol. 87:403-405. [DOI] [PubMed] [Google Scholar]
- 47.Winiecka-Krusnell, J., and E. Linder. 2001. Bacterial infections of free-living amoebae. Res. Microbiol. 152:613-619. [DOI] [PubMed] [Google Scholar]
- 48.Zaman, V., M. Zaki, and M. Manzoor. 1999. Acanthamoeba in human faeces from Karachi. Ann. Trop. Med. Parasitol. 93:189-191. [DOI] [PubMed] [Google Scholar]