Abstract
Heat shock proteins (HSPs) consist of a large group of chaperones whose expression is induced by high temperature, hypoxia, infection and a number of other stresses. Among all the HSPs, Hsp40 is the largest HSP family, which bind to Hsp70 ATPase domain in assisting protein folding. In this study, we identified 57 hsp40s in channel catfish (Ictalurus punctatus) through in silico analysis using RNA-Seq and genome databases. These genes can be classified into three different types, Type I, II and III, based on their structural similarities. Phylogenetic and syntenic analyses provided strong evidence in supporting the orthologies of these HSPs. Meta-analyses of RNA-Seq datasets were conducted to analyze expression profile of Hsp40s following bacterial infection. Twenty seven hsp40s were found to be significantly up- or down-regulated in the liver after infection with E. ictaluri; 19 hsp40s were found to be significantly regulated in the intestine after infection with E. ictaluri; and 19 hsp40s were found to be significantly regulated in the gill following infection with F. columnare. Altogether, a total of 42 Hsp40 genes were regulated under disease situations involving three tissues and two bacterial infections. The significant regulated expression of Hsp40 genes after bacterial infection suggested their involvement in disease defenses in catfish.
Introduction
Molecular chaperones include a large and diverse group of proteins that assist in folding or unfolding of other proteins or macromolecule structures in cells [1]. Among these, heat shock proteins (HSPs) constitute the largest groups of chaperones. They were found to be induced under various stresses such as elevated temperature, hypoxia [2] and diseases [3], [4]. HSPs are classified based on their molecular weight to include Hsp105/110, Hsp90, Hsp70, Hsp60, Hsp40, Hsp10, and small HSPs (sHSPs) [5].
Hsp40 proteins, also known as DnaJ proteins, constitute one of the largest families among heat shock proteins. They regulate the ATPase activity of Hsp70 proteins whose function is reversibly binding to partially denatured protein substrates to avoid the aggregation of themselves or with other molecules [6]–[8]. In the Hsp70-Hsp40 co-chaperone system, the association between Hsp70 proteins and substrates requires an ATP binding to ATPase domain and then being hydrolyzed to change conformation of the binding domain. Thus, various Hsp70's substrates could specifically bind to its least conserved C-terminal at a higher affinity. However, because the ATPase activity of Hsp70s is extremely weak, The J-domain of Hsp40s is needed for activating the ATPase domain of Hsp70s [8], [9].
Hsp40s share a 70-amino acids conserved J-domain, similar to the 73-amino acid-domain of prototypical DnaJ protein in Escherichia coli [10]. The DnaJ of E. coli typically consists of four regions: N-terminus J-domain, glycine/phenylalanine-rich region, Cysteine repeats and variable C-terminus domain (CTD) [10]. According to the homology of the DnaJ protein of E.coli, Hsp40 proteins were classified into three types: Type I DnaJ proteins (DnaJA) possess all four parts of DnaJ protein in E. coli; Type II DnaJ proteins (DnaJB) possess the N-terminus J-domain and the glycine/phenylalanine-rich region; Type III DnaJ proteins (DnaJC) only have a J-domain, which is not necessarily located at N-terminus of the protein [11], [12]. Recently, type IV DnaJ protein family was added, which differs from the other three types of DnaJ proteins in that it owns a ‘J-like’ domain [6], [13], [14] containing various mutations in a highly conserved histidine, proline, and aspartic acid–HPD motif located between helices II and III in DnaJ domain [6], [15]–[17]. However, Peter Walsh et al. (2004) proposed that the term J-proteins should be used more strictly to describe only J proteins with well-conserved J-domain in the HPD motif, while structurally less-conserved proteins should be referred to as J-like proteins [6].
Although heat shock proteins are traditionally regarded as being induced by heat and stresses, recent studies suggested HSPs may actually play important roles in immune responses [18]. For example, HSPs are considered to mediate humoral and cellular innate immune responses [19]; HSPs in extracellular environment serve as a danger signal to activate innate immune cells such as dendritic cells and macrophages [20]–[22]. Several cytokines can be induced by HSPs, including TNF-α, IL-1β, IL-12, nitric oxide and some chemokines [23]; HSPs can also stimulate adaptive immune responses as potent antigen carriers. Hsp60, Hsp70, Hsp90 have been reported to interact with immune cells as a ligand for a variety of cell-surface receptors such as Toll-like receptors [25], [26] and a number of CDs such as CD14 and CD91 [24]–[27].
Due to Hsp40 and Hsp70 worked together as a co-chaperone, the immune function especially the expression fold change trend of this two proteins should be taken into consideration at the same time. In teleost, hsp70 genes have been found to be involved in bacterial kidney disease in coho salmon [28] and vibriosis in rainbow trout [29]. In olive flounder, Hsp40 proteins were found to be up-regulated in flounder embryonic cells (FEC) after viral infection and a flounder hsp70 gene was also expressed in heat-shocked and virus treated FEC cells [30], indicating hsp40 and hsp70 functioned as co-chaperone in antiviral immune responses. In the kidney of olive flounder, dnaja4, dnajb6 and dnajb11 were found to be expressed after being infected by Streptococcus parauberis [31]. However only limited studies have been done on the roles of Hsp40s in disease resistance.
RNA-Seq-based expression analysis has become a robust method to assess transcriptional profile to different challenge experiments [32]. In our recent RNA-Seq studies, we have successfully obtained comprehensive transcriptome assemblies from catfish intestine and liver after E. ictaluri infection and from catfish gill after F. Columnare infection [33]–[35]. The expression patterns of differentially expressed genes from these three studies were validated by quantitative real-time RT-PCR with average correlation coefficient around 0.9 (p<0.001).
Channel catfish (Ictalurus punctatus) is the leading aquaculture species in the United States. Its genomic resources have been well developed in recent years, particularly ESTs [36], transcriptome sequences generated by RNA-Seq [37], [38] and draft whole genome sequence (unpublished data). These resources make it feasible to conduct systematic analysis of hsp40 genes in channel catfish genome. The objective of this study was to determine the involvement of hsp40 genes in disease responses after bacterial infection in catfish. Here we report the genome-wide identification of a full set of 57 hsp40 genes, their phylogenetic and syntenic analyses, and their involvement in disease responses after bacterial infection with ESC and columnaris using RNA-Seq datasets.
Materials and Methods
Database mining and sequence analyses
In order to identify the full set of hsp40 genes in channel catfish, we collected all Hsp40 proteins from teleost fishes (zebrafish, three-spined stickleback, medaka, tilapia and fugu) and other species (human, mouse, platypus, chicken, turtle and frog) (S1 Table). These sequences were retrieved from NCBI (http://www.ncbi.nlm.nih.gov) and Ensembl (http://www.ensembl.org) databases and used as queries to search against channel catfish RNA-Seq datasets. The e-value was set at intermediately stringent level of e-10 for collecting as many as potential hsp40-related sequences for further analysis. The retrieved sequences were then translated using ORF finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). Further, the predicted ORFs were verified by BLASTP against NCBI non-redundant (Nr) protein sequence database. The simple modular architecture research tool (SMART) [39] was used to predict the conserved domains based on sequence homology and further confirmed by conserved domain prediction from BLAST. The predicted catfish Hsp40s proteins and all other query sequences were utilized to search against catfish genome database using TBLASTN program. The retrieved genome scaffolds were then predicted by FGENESH in SoftBerry (http://linux1.softberry.com/berry.phtml?topic=fgenesh&group=programs&subgroup=gfind).
Phylogenetic and conserved syntenic analyses
All the amino acids from channel catfish and other species were used to construct phylogenetic tree. Multiple protein sequences alignments were conducted using the Clustal W2 program [40] and MUSCLE 3.8 [41]. Three alignment methods: L-INS-i, E-INS-i and G-INS-i were applied from MAFFT 7.01 [42] and the best alignment with highest score was evaluated by program MUMSA [43]. JTT+I+G model [Jones-Taylor-Thornton (JTT) matrix incorporated a proportion of invariant sites (+I) and the gamma distribution for modeling rate heterogeneity (+G)] was selected as the best-fit model by ProtTest 3 program [44] according to the Bayesian information criterion. Maximum likelihood phylogenetic tree was constructed using MEGA5.2.2 [45] with bootstrap test of 1,000 replicates. Final phylogenetic tree was separated into three different phylogenetic trees according to classification of subfamilies due to the large size.
Conserved syntenic regions surrounding the relevant hsp40 genes were searched by examining the conserved co-localization of neighboring genes on scaffold of channel catfish and other species based on genome information from Ensembl (Release 74) and NCBI database. Neighbor genes of channel catfish Hsp40 genes were predicted by FGENESH [46] and BLASTP.
Meta-analysis of expression of hsp40 genes and bacterial challenge
The Illumina-based RNA-Seq reads were retrieved from bacterial challenge experiments in catfish: intestine samples challenged with Edwardsiella ictaluri (SRA accession number SRP009069) [34], liver samples challenged with E. ictaluri challenge (SRA number SRP028159) [33] and gill samples challenged with Flavobacterium Columnare (SRA number SRP012586) [35]. Trimmed high-quality reads were mapped onto the catfish Hsp40 genes using CLC Genomics Workbench software (version 5.5.2; CLC bio, Aarhus, Denmark). Mapping parameters were set as ≥95% of the reads in perfect allignment and ≤2 mismatches. The total mapped reads number for each transcript was determined and normalized to analyze RPKM (Reads Per Kilobase of exon model per Million mapped reads). The proportions-based Kal's test was performed to identify the differently expressed genes comparing with control sample and fold changes were calculated. Transcrirps with absolute fold change value ≥1.5, p-value ≤0.05 and total read number ≥5 were included in the analyses as significantly differently expressed genes.
Results
Identification of Hsp40 genes in catfish
A total of 57 Hsp40 genes were identified in channel catfish. Their classification, domain structures, and GenBank accession numbers are summarized in Table 1. Type I included 6 Hsp40 genes; type II included 16 Hsp40 genes; and type III included 35 subfamily C Hsp40 genes. Among all these genes, almost all sequences were identified in both transcriptome and genome databases with full-length except dnajb9 with partial sequences in both databases. These catfish hsp40 genes were named following Zebrafish Nomenclature Guidelines (https://wiki.zfin.org/display/general/ZFIN+Zebrafish+Nomenclature+Guidelines).
Table 1. Summary of 57 Hsp40 genes identified in the catfish genome.
| Name | Type | ORF | Domain Structure | Accession number |
| dnaja1 | I | complete | DnaJ-CXXCXGXG-DnaJ_C | JT413966 |
| dnaja2 | I | complete | DnaJ-CXXCXGXG-DnaJ_C | JT408916 |
| dnaja2l | I | complete | DnaJ-CXXCXGXG-DnaJ_C | JT412411 |
| dnaja3a | I | complete | DnaJ-CXXCXGXG-DnaJ_C | JT425696 |
| dnaja3b | I | complete | DnaJ-CXXCXGXG-DnaJ_C | JT410497 |
| dnaja4 | I | complete | DnaJ-CXXCXGXG-DnaJ_C | JT340875 |
| dnajb1a | II | complete | DnaJ-DnaJ_C | JT410595 |
| dnajb1b | II | complete | DnaJ-DnaJ_C | JT405623 |
| dnajb2 | II | complete | DnaJ-3*(UIM) | JT413231 |
| dnajb4 | II | complete | DnaJ-DnaJ_C | JT418024 |
| dnajb5 | II | complete | DnaJ-DnaJ_C | JT410284 |
| dnajb5L | II | complete | DnaJ-DnaJ_C | JT407526 |
| dnajb6a | II | complete | DnaJ | JT280415 |
| dnajb6b | II | complete | DnaJ | JT477624 |
| dnajb9L1 | II | complete | DnaJ | JT406977 |
| dnajb9L2 | II | complete | DnaJ | JT244156 |
| dnajb9 | II | Partial | DnaJ | JT383860 |
| dnajb11 | II | complete | DnaJ-DnaJ_C | JT410155 |
| dnajb12a | II | complete | DnaJ-DUF1977 | JT411397 |
| dnajb12b | II | complete | DnaJ-DUF1977 | JT348699 |
| dnajb13 | II | complete | DnaJ-DnaJ_C | JT413284 |
| dnajb14 | II | complete | DnaJ-DUF1977 | JT417082 |
| dnajc1 | III | complete | DnaJ-SANT | JT407463 |
| dnajc2 | III | complete | (DnaJ)-2*(SANT) | JT342108 |
| dnajc3(prkri) | III | complete | 7*(TPR)-DnaJ | JT407497 |
| dnajc3 | III | complete | 7*(TPR)-DnaJ | JT410821 |
| dnajc4 | III | complete | DnaJ | JT400171 |
| dnajc5ab | III | complete | DnaJ | JT412549 |
| dnajc5aa | III | complete | DnaJ | JT483994 |
| dnajc5gb | III | complete | DnaJ | JT406264 |
| dnajc6 | III | complete | (PTPc_DSPc)-DnaJ | JT410375 |
| dnajc7 | III | complete | 7*(TPR)-DnaJ | JT278330 |
| dnajc8 | III | complete | DnaJ | JT278586 |
| dnajc9 | III | complete | DnaJ | JT223126 |
| dnajc10 | III | complete | DnaJ-4*(Thioredoxin) | JT424718 |
| dnajc11 | III | complete | DnaJ-DUF3395 | JT406478 |
| dnajc12 | III | complete | DnaJ | JT411276 |
| dnajc13 | III | complete | DnaJ | JT411283 |
| dnajc14 | III | complete | DnaJ | JT340805 |
| dnajc15 | III | complete | DnaJ | JT417797 |
| dnajc16 | III | complete | DnaJ-Thioredoxin | JT405377 |
| dnajc16L | III | complete | DnaJ-Thioredoxin | JT413761 |
| dnajc17 | III | complete | DnaJ-RRM_1 | JT411855 |
| dnajc18 | III | complete | DnaJ-DUF1977 | JT414201 |
| dnajc19 | III | complete | DnaJ | JT348880 |
| dnajc20(hscb) | III | complete | DnaJ-HSCB_C | JT410908 |
| dnajc21 | III | complete | DnaJ-ZnF_U1- ZnF_C2H2 | JT414736 |
| dnajc22 | III | complete | TM2-4*(Transmembrane)- DnaJ | JT405460 |
| dnajc23(sec63) | III | complete | DnaJ-Sec63 | JT409443 |
| dnajc24 | III | complete | DnaJ-CSL zinc finger | JT407343 |
| dnajc25 | III | complete | DnaJ | JT406198 |
| dnajc26(gak) | III | complete | S_TKc-STYKc-PTPc_DSPc-PTEN_C2-STYKc-DnaJ | JT418500 |
| dnajc27 | III | complete | Small GTPase-DnaJ | JT464820 |
| dnajc28 | III | complete | DnaJ-DUF1992 | JT413325 |
| dnajc29(sacs) | III | complete | UBQ-2*(HATPase_c)-DnaJ-HEPN | JT399814/JT345237 |
| dnajc30a | III | complete | DnaJ | JT399756 |
| dnajc30b | III | complete | DnaJ | JT199100 |
Six type I genes were identified in the catfish genome including dnaJa1, dnaJa2, dnaJa2-like, dnaJa3a, dnaJa3b and dnaJa4. These Hsp40 genes are homologous to DnaJ of E.coli, whose structure is conservatively comprised of N-terminal J-domain, glycine/phenylalanine-rich region, cysteine repeats motif and variable C-terminus domain (CTD) (Fig. 1 and Table 1).
Figure 1. Schematic presentation of three sub-types of HSP40 family.
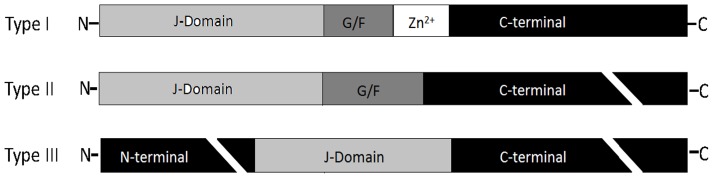
Type I DnaJ proteins have full domain conservation with Escherichia coli DnaJ, type II DnaJ proteins have a J domain and G/F (Gly/Phe-rich region) motif at N-terminus, type III DnaJ proteins only have a J domain anywhere in the protein.
Sixteen type II genes were identified in channel catfish including dnaJb1a, dnaJb1b, dnaJb2, dnaJb4, dnaJb5, dnaJb5-like, dnaJb6a, dnaJb6b, dnaJb9, dnaJb9-like1, dnaJb9-like2, dnaJb11, dnaJb12a, dnaJb12b, dnaJb13 .and dnaJb14., of which dnaJb1 was among the most highly heat-inducible HSPs in human [12].
A total of 35 type III hsp40 genes were identified from the catfish transcriptome and confirmed with the genome database including dnaJc1, dnajc2, dnajc3, dnajc3 (prkri), dnajc4, dnajc5aa, dnajc5ab, dnajc5gb, dnajc6, dnajc7, dnajc8, dnajc9, dnajc10, dnajc11, dnajc12, dnajc13, dnajc14, dnajc15, dnajc16, dnajc16-like, dnajc17, dnajc18, dnajc19, dnajc20 (hscb), dnajc21, dnajc22, dnajc23 (sec63), dnajc24, dnajc25, dnajc26 (gak), dnajc27, dnajc28, dnajc29 (sacs), dnajc30a and dnajc30b. Catfish type III Hsp40 proteins only have one J-domain in their structure, which is not necessarily located at N-terminus of the protein (Fig. 1 and Table 1). Orthologies were established for all the type III Hsp40s among human, zebrafish and catfish. However, several type III Hsp40s, i.e., Dnajc3 (Prkri), Dnajc20 (Hscb), Dnajc23 (Sec63), Dnajc26 (Gak), and Dnajc29 (Sacs) have not been annotated as DnaJC members. They are currently named according to aliases of human DNAJC proteins respectively [12].
Phylogenetic analysis of channel catfish Hsp40s
A total of 57 channel catfish Hsp40 genes have been phylogenetically analyzed. Each type of Hsp40 was subsequently analyzed separately (S1–S5 Figs.). Type III is divided into three parts due to its enormous size of the phylogenetic tree (S3–S5 Figs.). In a few cases where it was difficult to establish orthologies due to duplications (dnajb9, dnajb12 and dnajc30), syntenic analyses were also conducted (see below). We renamed some of the ambiguous names of hsp40 genes from other fish according to their relationship with the relevant zebrafish genes on the phylogenetic tree. The phylogenetic trees were then reconstructed after standardizing all the names. In the phylogenetic tree, all the members of catfish Hsp40 were well distributed into distinct clades and grouped with corresponding genes of zebrafish and other fishes, which were supported by strong bootstrap value (S1–S5 Figs.).
Syntenic analysis of channel catfish Hsp40s
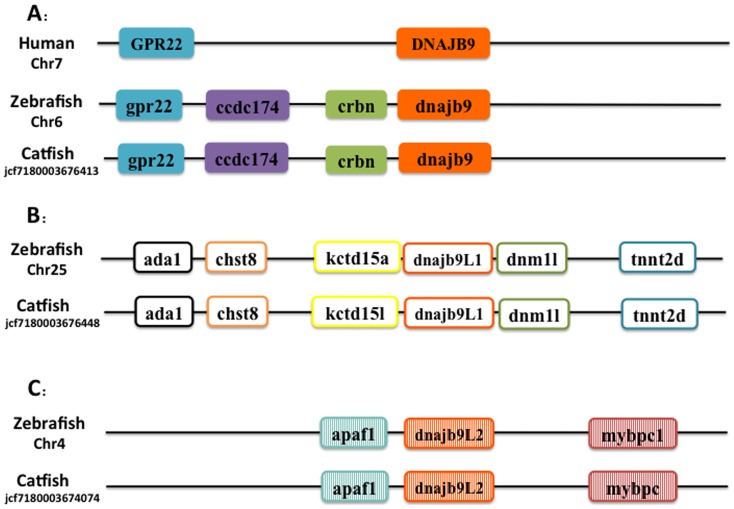
Though phylogenetic relationships provide strong support for the identities of most Hsp40 genes, syntenic analyses were required to provide additional evidence for orthologies or otherwise the paralogies for several hsp40 genes including the duplicated hsp40 genes such as dnajb9, dnajb12 and dnajc30. Positions of these catfish hsp40 genes and their neighbor genes were identified from the draft genome scaffolds. And the genes were also identified from the zebrafish genome. As shown in Fig. 2, three dnajb9-related genes were analyzed. The gene with the highest level of conservation in gene contents and gene orders as compared with the human DNAJB9 was named dnajb9 in zebrafish (accession number from ensembl: ENSDARP00000094644). Two other genes similar to dnajb9 in zebrafish with NCBI accession number: NP_001020355.1 and NP_001019564.1 were named dnajb9L1 and dnajb9L2 (L refers to “like”) respectively. All those genes of catfish, as well as those related genes from other fish species, were therefore named as dnajb9L1 and dnajb9L2 accordingly.
Figure 2. Schematic presentation of the conserved synteny blocks neighboring Dnajb9 gene (A) Dnajb9-like1 gene (B) and Dnajb9-like2 gene (C).
Note that in each case, the gene order and orientation was relatively conserved. Abbreviations: (A) GPR22, G Protein Receptor 22; Dnajb9, Dnaj (Hsp40) homolog, subfamily B, member 9; ccdc174, Coiled-Coil Domain Containing 174; crbn, cereblon. (B) ada1, Adenosine Deaminase; chst8, carbohydrate (N-acetylgalactosamine 4–0) sulfotransferase 8; kctd15a, potassium channel tetramerisation domain containing 15a; dnajb9L1 DnaJ (Hsp40) homolog, subfamily B, member 9 like 1; dnm1l, dynamin 1-like; tnnt2d, troponin T2d, cardiac. (C) apaf1, apoptotic protease activating factor 1; dnajb9l2 DnaJ (Hsp40) homolog, subfamily B, member 9 like 2; mybpc, myosin binding protein C.
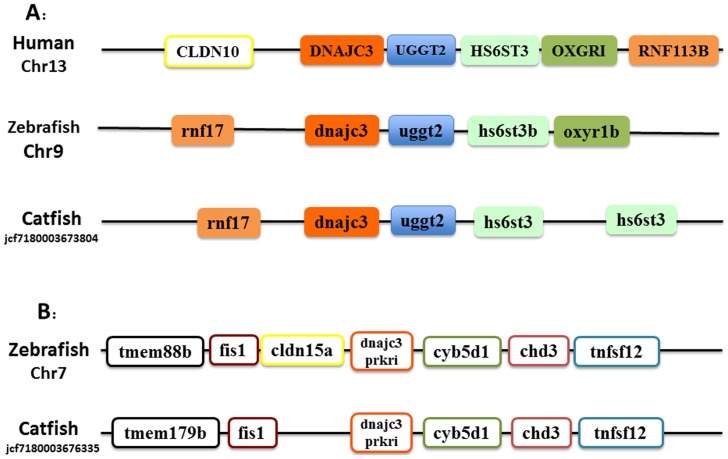
As shown in Fig. 3, of the two dnajc3-related genes, the neighboring genes surrounding the DNAJC3 gene is clearly well conserved among human, zebrafish and catfish (Fig. 3A), suggesting their orthologies. However, the genes surrounding the other dnajc3-related gene were quite different between the fish genes and the human genes (compare Fig. 3A and Fig. 3B), suggesting that they were not orthologous. However, the genes surrounding this gene are well conserved between zebrafish and catfish. We therefore annotated this genes as prkri as annotated in zebrafish.
Figure 3. Schematic presentation of the conserved synteny blocks neighboring DnaJc3 (A) and DnaJc3-prkri gene (B).
Note that in each case, the gene order and orientation was relatively conserved. Abbreviations: (A) CLDN10, claudin 10; rnf17, ring finger protein 17; DnaJc3, DnaJ (Hsp40) homolog, subfamily C, member 3; uggt2, UDP-glucose glycoprotein glucosyltransferase 2; hs6st3b, heparan sulfate 6-O-sulfotransferase 3b; oxyr1b, oxoglutarate (alpha-ketoglutarate) receptor 1b; RNF113B, ring finger protein 113B. (B) tmem88b, transmembrane protein 88 b; fis1, fission 1 (mitochondrial outer membrane) homolog (S. cerevisiae); cldn15a, claudin 15a; dnajc3 prkri, protein-kinase, interferon-inducible double stranded RNA dependent inhibitor; cyb5d1, cytochrome b5 domain containing 1; chd3, chromodomain helicase DNA binding protein 3; tnfsf12, tumor necrosis factor (ligand) superfamily, member 12.
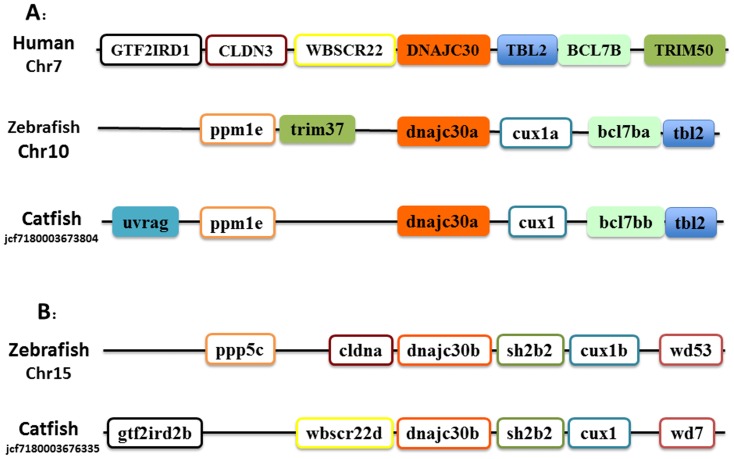
Two dnajc30-related genes were found in catfish and zebrafish. As shown in Fig. 4, these two genes were located on two different chromosomes of zebrafish, but genes surrounding the duplicated gene are well conserved among human, zebrafish and catfish, suggesting that they were derived from the whole genome duplication event. Therefore, they were annotated as dnajc30a and dnajc30b, following zebrafish gene nomenclature system.
Figure 4. Schematic presentation of the conserved synteny blocks neighboring DnaJc30a (A) and DnaJc30b gene (B).
Note that in each case, the gene order and orientation was relatively conserved.Abbreviations: (A) GTF2IRD1, GTF2I repeat domain containing 1; CLDN3, claudin 3; WBSCR22, Williams Beuren syndrome chromosome region 22; DnaJc30, DnaJ (Hsp40) homolog, subfamily C, member 30; TBL2, transducin (beta)-like 2; BCL7B, B-cell CLL/lymphoma 7B; TRIM50, tripartite motif containing 50; ppm1e, protein phosphatase 1E (PP2C domain containing); trim37, tripartite motif containing 37; cux1a, cut-like homeobox 1a; uvrag, UV radiation resistance associated. (B) ppp5c, protein phosphatase 5, catalytic subunit; cldna, claudin a; dnajc30b, DnaJ (Hsp40) homolog, subfamily C, member 30; sh2b2, fission 1 (mitochondrial outer membrane) homolog (S. cerevisiae); wd53, WD repeat domain 53; wd7, WD repeat domain 7; gtf2ird2b, GTF2I repeat domain containing 2b.
Hsp40s copy number variation among species
Catfish have almost all the orthologues of Hsp40s in human and zebrafish (Table 2), with exception of several genes existing in humans, but absent from teleost fishes including Dnajb3, Dnajb7, and Dnajb8. It is interesting that quite a few of the Dnaja genes and Dnajb genes have duplicated copies in various teleost species, however, most of the Dnajc genes have only a single copy in the teleost genomes (Table 2). Specifically, Dnaja2, Dnaja3, Dnajb1, Dnajb5, Dnajb6, Dnajb12, Dnajc3, Dnajc7, Dnajc11, Dnajc16 and Dnajc30 were found to have two duplicates, and Dnajb9 was found to have three copies in catfish and most of the teleost fish while only one copy was found in other species. Note that Dnajc5 have been found to have five copies in zebrafish, three copies in catfish and three copies in human as well. This is the only gene that has more than one copy in the human genome. Compared to zebrafish, channel catfish has fewer copies for Dnajb12, Dnajc5, Dnajc7 and Dnajc11 (Table 2).
Table 2. Comparison of copy numbers of HSP40 genes among selected vertebrate genomes.
| Gene | Human | Chicken | Frog | Zebrafish | Catfish | Medaka | Tilapia | Fugu | |
| Type I | Dnaja1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 |
| Dnaja2 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | |
| Dnaja3 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | |
| Dnaja4 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | |
| Subtotal | 4 | 4 | 5 | 6 | 6 | 6 | 6 | 5 | |
| Type II | Dnajb1 | 1 | 0 | 1 | 2 | 2 | 2 | 2 | 2 |
| Dnajb2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Dnajb3 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Dnajb4 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Dnajb5 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 3 | |
| Dnajb6 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | |
| Dnajb7 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Dnajb8 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Dnajb9 | 1 | 1 | 1 | 3 | 3 | 2 | 3 | 1 | |
| Dnajb11 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | |
| Dnajb12 | 1 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | |
| Dnajb13 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Dnajb14 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | |
| Subtotal | 13 | 10 | 11 | 17 | 16 | 17 | 16 | 15 | |
| Type III | Dnajc1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Dnajc2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Dnajc3 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | |
| Dnajc4 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Dnajc5 | 3 | 1 | 2 | 5 | 3 | 2 | 1 | 2 | |
| Dnajc6 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Dnajc7 | 1 | 1 | 1 | 2 | 1 | 2 | 2 | 2 | |
| Dnajc8 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Dnajc9 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Dnajc10 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Dnajc11 | 1 | 1 | 1 | 2 | 1 | 2 | 2 | 2 | |
| Dnajc12 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Dnajc13 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Dnajc14 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Dnajc15 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | |
| Dnajc16 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | |
| Dnajc17 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Dnajc18 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Dnajc19 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Hscb(Dnajc20) | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Dnajc21 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Dnajc22 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Dnajc23 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Dnajc24 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Dnajc25 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Dnajc26 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Dnajc27 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Dnajc28 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Dnajc29 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Dnajc30 | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 2 | |
| Subtotal | 32 | 29 | 31 | 39 | 35 | 35 | 34 | 35 | |
| Total | 49 | 43 | 47 | 62 | 57 | 58 | 56 | 55 |
Regulated expression of hsp40 genes in catfish after bacterial infection
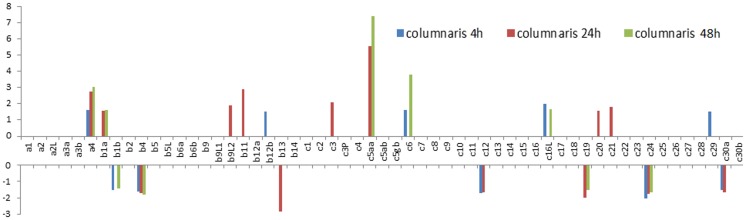
Using three bacterial challenged RNA-Seq datasets (intestine sample infected by E. ictaluri, liver sample infected by E. ictaluri, and intestine sample infected by F. columnare), the involvement of hsp40 genes after bacterial infection was determined. As shown in S2 Table, 42 out of a total of 57 hsp40s were involved in disease defense responses. Specifically, 19 hsp40s were found regulated in the gill following F. columnare infection. Among them, 12 genes were up-regulated (1.5× fold change cutoff) and 7 genes were down-regulated (1.5× fold change cutoff) after columnaris infection (Fig. 5). Among these regulated hsp40 genes, some of them are transiently up- or down-regulated while others are gradually induced or suppressed. For instance, Dnajb9L2, Dnajb11, Dnajb12b, Dnajc3, Dnajc20, Dnajc21, and Dnajc29 were up-regulated at only one time point (1.5× fold change cutoff); similarly, Dnajb13 was only down regulated at 24h after infection. In contrast, Dnaja4, Dnajb1a, Dnajc5aa, Dnajc6, and Dnajc16L were up-regulated in at least two time points after infection, suggesting their up-regulated expression was more lasting. Similar patterns were observed for down-regulated genes including Dnajb1b, Dnajb4, Dnajc12, Dnajc19, Dnajc24, and Dnajc30a (Fig. 5).
Figure 5. Column bar chart showing the fold change of Hsp40s expression in F. Columnare challenge experiments.
Vertical axis shows the value of fold change. Gene names are shown as clipped name without “dnaj”. For instance, a1 means “dnaja1”. “c3P” represents “dnajc3_prkri”.
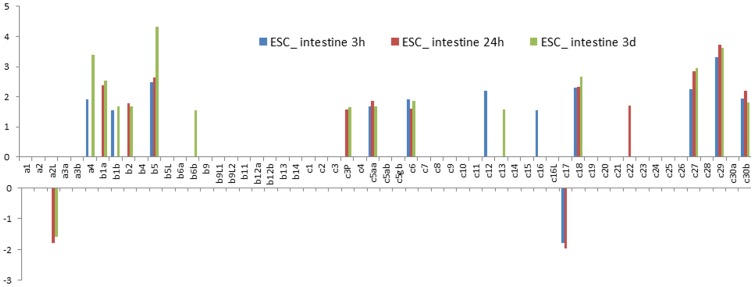
A total of 19 hsp40 genes were found to be regulated in the intestine after E. ictaluri infection. Among these, 17 were up-regulated while two were down-regulated. The upregulated hsp40 genes included Dnaja4, Dnajb1a, Dnajb1b, Dnajb2, Dnajb5, Dnajb6b, prkri, Dnajc5aa, Dnajc6, Dnajc12, Dnajc13, Dnajc16, Dnajc18, Dnajc22, Dnajc27, Dnajc29, and Dnajc30b. The two down-regulated hsp40 genes were Dnaja2L, and Dnajc17. Significantly regulated expression of most of these genes were observed in more than one time points with exception of Dnajb6b, Dnajc12, Dnajc13, Dnajc16, and Dnajc22 where the significantly regulated expression was observed at only one time point (Fig. 6).
Figure 6. Column bar chart shows the fold change of Hsp40s expression in intestine after E. ictaluri challenge experiments.
Vertical axis shows the value of fold change. Gene names are shown as clipped name without “dnaj”. For instance, a1 means “dnaja1”. “c3P” represents “dnajc3_prkri”.
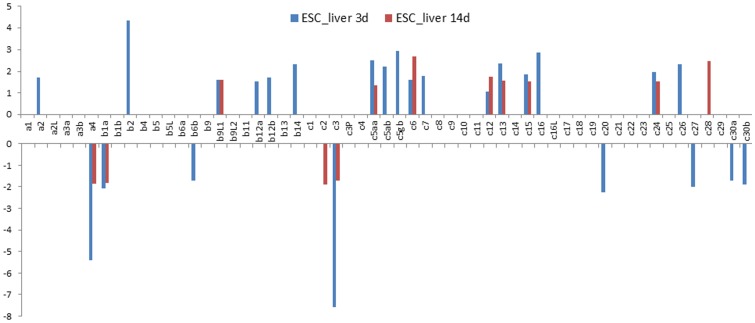
While RNA-Seq datasets for the gill tissue after columnaris infection and the intestine tissue after ESC infection were from early infection period of 4 h to 3days after infection, we analyzed RNA-Seq datasets from liver tissues collected at 3 days and 14 days after infection. As shown in Fig. 7, a total of 27 hsp40 genes were found to be regulated during this period in the liver. It is apparent that many of the hsp40 genes were induced at only one time points at 3 days or 14 days after infection, but not at both time points. For instance, Dnaja2, Dnajb2, Dnajb12a, Dnajb12b, Dnajb14, Dnajc5ab, Dnajc5gb, Dnajc7, Dnajc16, and Dnajc26 were up-regulated only at 3 days after infection, but not at 14 days after infection; similarly, Dnajc28 was up-regulated only at 14 days after infection, but not at 3 days after infection. Very similarly, of the 9 down-regulated genes, six were regulated at only one time point. Down-regulated expression was observed mostly at 3 days after infection as the up-regulated hsp40 genes, but not at 14 days after infection. Only Dnajc2 was found to be down-regulated 14 days after infection in the liver tissues (Fig. 7).
Figure 7. Column bar chart shows the fold change of Hsp40s expression in liver after E. ictaluri challenge experiments.
Vertical axis shows the value of fold change. Gene names are shown as clipped name without “dnaj”. For instance, a1 means “dnaja1”. “c3P” represents “dnajc3_prkri”.
Discussion
Hsp40 proteins play key roles in assisting protein folding by activating the ATPase domain of Hsp70. In spite of their importance, only a few Hsp40 genes were characterized from channel catfish [47]. Systematic analysis of channel catfish Hsp40 genes has been lacking. In this study, we identified a full set of 57 catfish Hsp40 genes in the catfish genome. This was achieved by thorough analysis of rich genomic and transcriptomic resources including several hundred thousands of ESTs [33]–[35], RNA-Seq transcriptome assemblies [37], [38], and the draft genome sequences (unpublished).
Phylogenetic and syntenic analyses allowed annotation of Hsp40 genes. It is apparent that catfish harbored most of hsp40 genes. Compared with the human genome, catfish lacked only three hsp40 genes, similar to the situation of other teleost fish species. However, a number of paralogues were also discovered in catfish due to duplication. The phylogenetic analysis strongly supported the nomenclature of catfish hsp40 genes in all three subfamilies. Most Hsp40s are conserved through evolution, while there are still special preferences made by teleost and catfish. First, the teleost tended to have more duplications than mammals, likely as a consequence of whole genome duplication. Among various fish species, DnaJa2, DnaJa3, DnaJb1, DnaJb5, DnaJb9, DnaJb12, DnaJc3, DnaJc7, DnaJc11, DnaJc16, and DnaJc30 were found to be duplicated while in mammals, birds and amphibians only one copy of these genes were found.
Meta-analysis of disease challenges revealed a gene fold change profile of catfish hsp40s involving in disease defense and stress protection. As mentioned in the introduction, although regulated expression of HSPs after infection have been reported in several fish species, systematic analysis of their involvement in diseases has not been conducted. This work, therefore, represents the first systematic analysis of Hsp40 involvement after bacterial infection among all species.
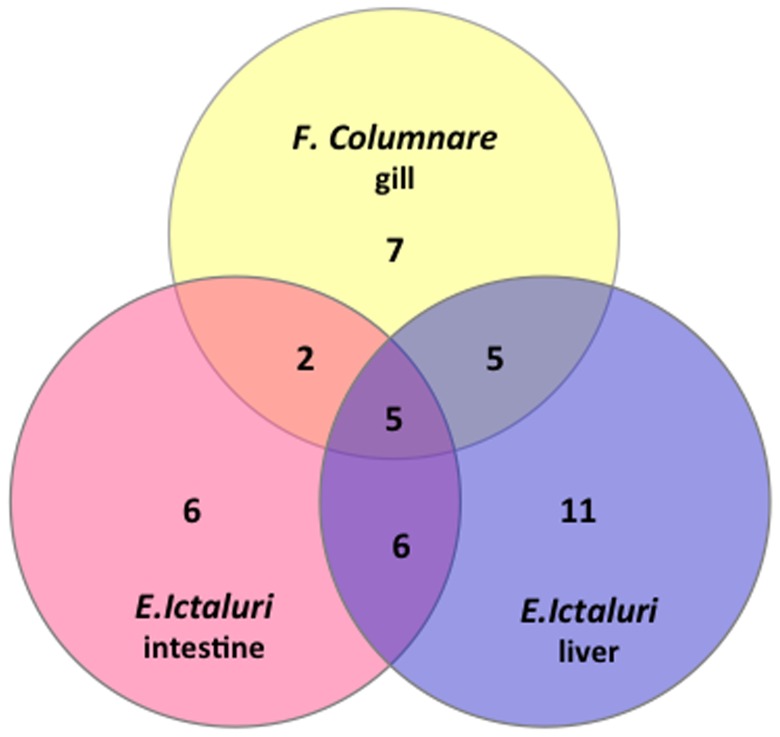
Surprisingly, a large number of hsp40 genes, 42 in total, were up- or down- regulated after bacterial infection, suggesting their extensive involvement in disease responses. Specifically, a total of 27 hsp40 genes were found up- or down- regulated in liver after E. ictaluri infection, 19 genes were found regulated in intestine after E. ictaluri infection, and 19 genes were found regulated in gill following F. columnare infection (Fig. 8). Of these 42 regulated hsp40 genes, 5 genes: dnaja4, dnajb1a, dnajc5aa, dnajc6 and dnajc12 were regulated in all three situations, suggesting their importance as general disease response hsps; 7 genes (dnajb1b, dnajc29, dnaja4, dnajb1a, dnajc5aa, dnajc6 and dnajc12) were commonly regulated with columnaris infection in the gill and ESC infection in the intestine; 10 genes (dnajb12b, dnajc3, dnajc20, dnajc24, dnajc30a, dnaja4, dnajb1a, dnajc5aa, dnajc6 and dnajc12) were commonly regulated with columnaris infection and ESC infection in the liver; and 11 genes (dnajb2, dnajb6b, dnajc13, dnajc16, dnajc27, dnajc30b, dnaja4, dnajb1a, dnajc5aa, dnajc6 and dnajc12) were commonly regulated with infection of ESC in the intestine and ESC infection in the liver (Fig. 8).
Figure 8. Venn diagram of catfish Hsp40 gene expression showing overlapped pattern of induced expression under various challenge conditions.
In spite of a set of commonly regulated hsps, disease- and tissue-specific genes in each challenge experiment were observed. For instance, after F. columnare infection, seven genes (dnajb4, dnajb9L2, dnajb11, dnajb13, dnajc16L, dnajc19 and dnajc21) were found to be specifically regulated in the gill. Similarly, six hsp40 genes (dnaja2L, dnajb5, prkri, dnajc17, dnajc18 and dnajc22) were found to be specifically regulated in the intestine after ESC infection; and 11 hsp40 genes (dnaja2, dnajb9L1, dnajb12a, dnajb14, dnajc2, dnajc5ab, dnajc5gb, dnajc7, dnajc15, dnajc26 and dnajc28) were found to be specifically regulated in the liver, suggesting their specific roles in a tissue- and time-dependent manner.
The vast majority of hsp40 genes were found to be significantly regulated soon after infection at 3h or 24 h after infection with both columnaris and ESC, suggesting their involvement in the early phases of disease response. Only dnajb6b and dnajc13 were not significantly up-regulated until 3 days after infection. However, in the liver at late stages of disease development, more hsp40 genes (nine) were found to be down-regulated. These findings could be explained by the co-chaperon system of Hsp40 and Hsp70 [48], [49]. Hsp70 regulates the intracellular function and fate of proteins through the formation of direct protein-protein interactions that occur largely through an EEVD-binding domain in its C terminus [50]–[52]. In innate immune system, Hsp70 can serve as the endogenous ligand of TLR2 and TLR4 and aid to recognize the bacteria [53]. Previous work demonstrated that TLRs were up-regulated at 1 h post challenge [54] by F. columnare and most up-regulated at 6 h and 24 h post challenge by E. ictaluri [55] in catfish. It is believed that since Hsp70 serves as a TLR4 agonist, there is a positive correlation between HSP70 and TLR4 in human. Therefore, the increased or decreased fold of Hsp40s after infection of bacteria can reflect the fold change of its co-chaperone Hsp70 and thus the associated TLRs [56], [57]. Furthermore, it was also reported that TLRs are down-regulated after 36 h. This could be the explanation to the large number of down-regulated in liver 3 days and 14 days post challenge from our meta-analysis. It is interesting to observe that seven genes were down-regulated with columnaris whereas only two genes were down-regulated with ESC at early stages of disease development after infection. This observation indicated that the immune response to the F. columare was faster than that to E. ictaluri. In general, hsp40 genes were up-regulated at early stages of diseases, and with time, they tended to be down regulated as the diseases progressed. In the same line of thoughts, columnaris disease may involve more rapid destructive processes than ESC diseases and therefore, more hsp40 genes were found to be down-regulated even at early stages of columnaris disease development (Fig. 5).
The level of regulated expression varied among the genes as well as tissues and time after infection. For most of the hsp40 genes, up- or down-regulation was less than five-fold as compared with the controls. Only three genes were regulated more than five times after bacterial infections. Dnajc5aa was up-regulated 6-8 times after columnaris infection (Fig. 5). No hsp40 genes were up- or down-regulated more than 5-fold in the intestine after ESC infection. However, in the liver, and Dnaja4 and Dnajc3 were down-regulated more than five-fold in the liver after infection with ESC.
Conclusions
We have identified and annotated a full set of 57 hsp40 genes. The vast majority (42 out of 57) of hsp40 genes were either up- or down-regulated after infection. Their patterns of regulation were tissue-, time- and disease-specific, but it appeared that at earliest stages of disease infections, the majority of hsp40 genes were up-regulated whereas with the progression of the diseases, more and more hsp40 genes became down-regulated.
Supporting Information
Phylogenetic tree of Hsp40s Type I. The phylogenetic tree was constructed by Mega5.2.2 using the Maximum Likelihood method based on the JTT matrix-based model of amino acid substitution as described in detail in Material and Method section. Numbers around the nodes correspond to bootstrap support values in percentages. A discrete Gamma distribution was used to model evolutionary rate differences among sites (5 categories (+G, parameter = 1.1981)). The rate variation model allowed for some sites to be evolutionarily invariable ([+I], 2.8567% sites). All positions with less than 95% site coverage were eliminated. That is, fewer than 5% alignment gaps, missing data, and ambiguous bases were allowed at any position. There were a total of 332 positions in the final dataset. Accession numbers for all protein sequences used in the analysis are provided in S1 Table. The black dots indicate catfish dnaja genes. Suffix “L” indicated “-like”, for instance, dnaja2L means dnaja2-like.
(TIF)
Phylogenetic tree of Hsp40s type II. The phylogenetic tree was constructed as in S1 Fig. Accession numbers for all sequences are provided in S1 Table. Suffix “L” indicated “-like”, for instance, dnajb5L means Dnajb5-like.
(TIF)
Phylogenetic tree of Hsp40s type III: Dnajc1 to Dnajc9. The phylogenetic tree was constructed as in S1 Fig. Accession numbers for all sequences are provided in S1 Table. The black dots indicate catfish Dnajc genes. Suffix “L” indicated “-like”, for instance, Dnajc7L means Dnajc7-like.
(TIF)
Phylogenetic tree of Hsp40s type III: Dnajc10 to Dnajc20. The phylogenetic tree was constructed as in S1 Fig. Accession numbers for all sequences are provided in S1 Table. The black dots indicate catfish Dnajc genes. Suffix “L” indicated “-like”.
(TIF)
Phylogenetic tree of Hsp40s type III: Dnajc21 to Dnajc30. The phylogenetic tree was constructed as in S1 Fig. Accession numbers for all sequences are provided in S1 Table. The black dots indicate catfish Dnajc genes. Suffix “L” indicated “-like”.
(TIF)
Gene names and accessions of reference Hsp40s used in this study.
(XLSX)
Fold changes of Hsp40s under each challenge. Bold font is the significant expression.
(XLSX)
Acknowledgments
The authors wish to thank Dr. Geoff Waldbieser for his collaborations in the catfish genome projects that led to the generation of genome sequences used in this study.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
This project was supported by Agriculture and Food Research Initiative Competitive Grants 2009-35205-05101 and 2010-65205-20356 from the USDA National Institute of Food and Agriculture (NIFA). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Ellis J (1987) Proteins as molecular chaperones. Nature 328:378–379. [DOI] [PubMed] [Google Scholar]
- 2. Welch WJ (1993) How cells respond to stress: during emergencies, cells produce stress proteins that repair damage, inquiry into how they work offers promise for coping with infection, autoimmune disease and even cancer. Scientific American 268:56–56. [DOI] [PubMed] [Google Scholar]
- 3. Pockley AG (2003) Heat shock proteins as regulators of the immune response. The lancet 362:469–476. [DOI] [PubMed] [Google Scholar]
- 4. Srivastava P (2002) Roles of heat-shock proteins in innate and adaptive immunity. Nature Reviews Immunology 2:185–194. [DOI] [PubMed] [Google Scholar]
- 5. Feder ME, Hofmann GE (1999) Heat-shock proteins, molecular chaperones, and the stress response: Evolutionary and Ecological Physiology. Annual Review of Physiology 61:243–282. [DOI] [PubMed] [Google Scholar]
- 6. Walsh P, Bursać D, Law Y, Cyr D, Lithgow T (2004) The J-protein family: modulating protein assembly, disassembly and translocation. European Molecular Biology Organization Report 5:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Beckmann RP, Mizzen L, Welch WJ (1990) Interaction of Hsp 70 with newly synthesized proteins: implications for protein folding and assembly. Science 248:850–854. [DOI] [PubMed] [Google Scholar]
- 8. Kelley WL (1998) The J-domain family and the recruitment of chaperone power. Trends in biochemical sciences 23:222–227. [DOI] [PubMed] [Google Scholar]
- 9. Qiu XB, Shao YM, Miao S, Wang L (2006) The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell Mol Life Sci 63:2560–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hennessy F, Nicoll WS, Zimmermann R, Cheetham ME, Blatch GL (2005) Not all J domains are created equal: implications for the specificity of Hsp40-Hsp70 interactions. Protein Sci 14:1697–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kampinga HH, Hageman J, Vos MJ, Kubota H, Tanguay RM, et al. (2009) Guidelines for the nomenclature of the human heat shock proteins. Cell Stress Chaperones 14:105–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cheetham ME, Caplan AJ (1998) Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell stress & chaperones 3:28–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Botha M, Pesce ER, Blatch GL (2007) The Hsp40 proteins of Plasmodium falciparum and other apicomplexa: regulating chaperone power in the parasite and the host. Int J Biochem Cell Biol 39:1781–1803. [DOI] [PubMed] [Google Scholar]
- 14. Morahan BJ, Strobel C, Hasan U, Czesny B, Mantel PY, et al. (2011) Functional analysis of the exported type IV HSP40 protein PfGECO in Plasmodium falciparum gametocytes. Eukaryot Cell 10:1492–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Douglas MG (1996) A Conserved HPD Sequence of the J-domain Is Necessary for YDJ1 Stimulation of Hsp70 ATPase Activity at a Site Distinct from Substrate Binding. Journal of Biological Chemistry 271:9347–9354. [DOI] [PubMed] [Google Scholar]
- 16. Mayer MP, Laufen T, Paal K, McCarty JS, Bukau B (1999) Investigation of the interaction between DnaK and DnaJ by surface plasmon resonance spectroscopy. J Mol Biol 289:1131–1144. [DOI] [PubMed] [Google Scholar]
- 17. Hennessy F, Cheetham ME, Dirr HW, Blatch GL (2000) Analysis of the levels of conservation of the J domain among the various types of DnaJ-like proteins. Cell stress & chaperones 5:347–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Roberts RJ, Agius C, Saliba C, Bossier P, Sung YY (2010) Heat shock proteins (chaperones) in fish and shellfish and their potential role in relation to fish health: a review. J Fish Dis 33:789–801. [DOI] [PubMed] [Google Scholar]
- 19. Sung Y, MacRae T (2011) Heat shock proteins and disease control in aquatic organisms. Journal of Aquaculture Research & Development S 2:006. [Google Scholar]
- 20. Singh-Jasuja H, Scherer HU, Hilf N, Arnold-Schild D, Rammensee HG, et al. (2000) The heat shock protein gp96 induces maturation of dendritic cells and down-regulation of its receptor. Eur J Immunol 30:2211–2215. [DOI] [PubMed] [Google Scholar]
- 21. Chen W, Syldath U, Bellmann K, Burkart V, Kolb H (1999) Human 60-kDa heat-shock protein: a danger signal to the innate immune system. J Immunol 162:3212–3219. [PubMed] [Google Scholar]
- 22. Kol A, Lichtman AH, Finberg RW, Libby P, Kurt-Jones EA (2000) Cutting edge: heat shock protein (HSP) 60 activates the innate immune response: CD14 is an essential receptor for HSP60 activation of mononuclear cells. The Journal of Immunology 164:13–17. [DOI] [PubMed] [Google Scholar]
- 23. Srivastava P (2002) Roles of heat-shock proteins in innate and adaptive immunity. Nat Rev Immunol 2:185–194. [DOI] [PubMed] [Google Scholar]
- 24. Basu S, Binder RJ, Ramalingam T, Srivastava PK (2001) CD91 is a common receptor for heat shock proteins gp96, hsp90, hsp70, and calreticulin. Immunity 14:303–313. [DOI] [PubMed] [Google Scholar]
- 25. Ohashi K, Burkart V, Flohe S, Kolb H (2000) Cutting edge: heat shock protein 60 is a putative endogenous ligand of the toll-like receptor-4 complex. J Immunol 164:558–561. [DOI] [PubMed] [Google Scholar]
- 26. Vabulas RM, Ahmad-Nejad P, da Costa C, Miethke T, Kirschning CJ, et al. (2001) Endocytosed HSP60s use toll-like receptor 2 (TLR2) and TLR4 to activate the toll/interleukin-1 receptor signaling pathway in innate immune cells. Journal of Biological Chemistry 276:31332–31339. [DOI] [PubMed] [Google Scholar]
- 27. Habich C, Baumgart K, Kolb H, Burkart V (2002) The receptor for heat shock protein 60 on macrophages is saturable, specific, and distinct from receptors for other heat shock proteins. The Journal of Immunology 168:569–576. [DOI] [PubMed] [Google Scholar]
- 28. Forsyth RB, Candido EPM, Babich SL, Iwama GK (1997) Stress Protein Expression in Coho Salmon with Bacterial Kidney Disease. Journal of Aquatic Animal Health 9:18–25. [Google Scholar]
- 29. Ackerman PA, Iwama GK (2001) Physiological and Cellular Stress Responses of Juvenile Rainbow Trout to Vibriosis. Journal of Aquatic Animal Health 13:173–180. [Google Scholar]
- 30. Dong CW, Zhang YB, Zhang QY, Gui JF (2006) Differential expression of three Paralichthys olivaceus Hsp40 genes in responses to virus infection and heat shock. Fish Shellfish Immunol 21:146–158. [DOI] [PubMed] [Google Scholar]
- 31. Cha IS, Kwon J, Park SB, Jang HB, Nho SW, et al. (2013) Heat shock protein profiles on the protein and gene expression levels in olive flounder kidney infected with Streptococcus parauberis . Fish Shellfish Immunol 34:1455–1462. [DOI] [PubMed] [Google Scholar]
- 32. Oshlack A, Robinson MD, Young MD (2010) From RNA-seq reads to differential expression results. Genome Biol 11:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang R, Sun L, Bao L, Zhang J, Jiang Y, et al. (2013) Bulk segregant RNA-seq reveals expression and positional candidate genes and allele-specific expression for disease resistance against enteric septicemia of catfish. BMC Genomics 14:929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li C, Zhang Y, Wang R, Lu J, Nandi S, et al. (2012) RNA-seq analysis of mucosal immune responses reveals signatures of intestinal barrier disruption and pathogen entry following Edwardsiella ictaluri infection in channel catfish, Ictalurus punctatus. Fish Shellfish Immunol 32:816–827. [DOI] [PubMed] [Google Scholar]
- 35. Sun F, Peatman E, Li C, Liu S, Jiang Y, et al. (2012) Transcriptomic signatures of attachment, NF-kappaB suppression and IFN stimulation in the catfish gill following columnaris bacterial infection. Dev Comp Immunol 38:169–180. [DOI] [PubMed] [Google Scholar]
- 36.Wang S, Liu Z (2011) SNP Discovery through EST Data Mining. Next Generation Sequencing and Whole Genome Selection in Aquaculture. Oxford, UK: Wiley-Blackwell.
- 37. Liu S, Zhou Z, Lu J, Sun F, Wang S, et al. (2011) Generation of genome-scale gene-associated SNPs in catfish for the construction of a high-density SNP array. BMC Genomics 12:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu S, Zhang Y, Zhou Z, Waldbieser G, Sun F, et al. (2012) Efficient assembly and annotation of the transcriptome of catfish by RNA-Seq analysis of a doubled haploid homozygote. BMC genomics 13:595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Letunic I, Doerks T, Bork P (2012) SMART 7: recent updates to the protein domain annotation resource. Nucleic acids research 40:D302–D305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. [DOI] [PubMed] [Google Scholar]
- 41. Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lassmann T, Sonnhammer EL (2006) Kalign, Kalignvu and Mumsa: web servers for multiple sequence alignment. Nucleic Acids Res 34:W596–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Darriba D, Taboada GL, Doallo R, Posada D (2011) ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27:1164–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Salamov AA, Solovyev VV (2000) Ab initio gene finding in Drosophila genomic DNA. Genome Res 10:516–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chen F, Lee Y, Jiang Y, Wang S, Peatman E, et al. (2010) Identification and Characterization of Full-Length cDNAs in Channel Catfish (Ictalurus punctatus) and Blue Catfish (Ictalurus furcatus). PLoS ONE 5:e11546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Freeman B, Myers M, Schumacher R, Morimoto R (1995) Identification of a regulatory motif in Hsp70 that affects ATPase activity, substrate binding and interaction with HDJ-1. The EMBO journal 14:2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Houry WA (2001) Chaperone-assisted protein folding in the cell cytoplasm. Current Protein and Peptide Science 2:227–244. [DOI] [PubMed] [Google Scholar]
- 50. Brinker A, Scheufler C, von der Mülbe F, Fleckenstein B, Herrmann C, et al. (2002) Ligand discrimination by TPR domains relevance and selectivity of eevd-recognition in Hsp70· Hop· Hsp90 complexes. Journal of Biological Chemistry 277:19265–19275. [DOI] [PubMed] [Google Scholar]
- 51. Liu F-H, Wu S-J, Hu S-M, Hsiao C-D, Wang C (1999) Specific interaction of the 70-kDa heat shock cognate protein with the tetratricopeptide repeats. Journal of Biological Chemistry 274:34425–34432. [DOI] [PubMed] [Google Scholar]
- 52. Scheufler C, Brinker A, Bourenkov G, Pegoraro S, Moroder L, et al. (2000) Structure of TPR domain–peptide complexes: critical elements in the assembly of the Hsp70–Hsp90 multichaperone machine. Cell 101:199–210. [DOI] [PubMed] [Google Scholar]
- 53. Asea A, Rehli M, Kabingu E, Boch JA, Baré O, et al. (2002) Novel Signal Transduction Pathway Utilized by Extracellular HSP70: ROLE OF Toll-LIKE RECEPTOR (TLR) 2 AND TLR4. Journal of Biological Chemistry 277:15028–15034. [DOI] [PubMed] [Google Scholar]
- 54. Peatman E, Li C, Peterson BC, Straus DL, Farmer BD, et al. (2013) Basal polarization of the mucosal compartment in Flavobacterium columnare susceptible and resistant channel catfish (Ictalurus punctatus). Mol Immunol 56:317–327. [DOI] [PubMed] [Google Scholar]
- 55. Pridgeon JW, Russo R, Shoemaker CA, Klesius PH (2010) Expression profiles of toll-like receptors in anterior kidney of channel catfish, Ictalurus punctatus (Rafinesque), acutely infected by Edwardsiella ictaluri . Journal of Fish Diseases 33:497–505. [DOI] [PubMed] [Google Scholar]
- 56. Nair S, Kotrashetti VS, Nayak R, Bhat K, Somannavar P, et al. (2013) HSP70 induces TLR4 signaling in oral squamous cell carcinoma: An immunohistochemical study. J Cancer Res Ther 9:624–629. [DOI] [PubMed] [Google Scholar]
- 57. Asea A, Rehli M, Kabingu E, Boch JA, Baré O, et al. (2002) Novel Signal Transduction Pathway Utilized by Extracellular HSP70: role of toll-like receptor (TLR) 2 and TLR4. Journal of Biological Chemistry 277:15028–15034. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenetic tree of Hsp40s Type I. The phylogenetic tree was constructed by Mega5.2.2 using the Maximum Likelihood method based on the JTT matrix-based model of amino acid substitution as described in detail in Material and Method section. Numbers around the nodes correspond to bootstrap support values in percentages. A discrete Gamma distribution was used to model evolutionary rate differences among sites (5 categories (+G, parameter = 1.1981)). The rate variation model allowed for some sites to be evolutionarily invariable ([+I], 2.8567% sites). All positions with less than 95% site coverage were eliminated. That is, fewer than 5% alignment gaps, missing data, and ambiguous bases were allowed at any position. There were a total of 332 positions in the final dataset. Accession numbers for all protein sequences used in the analysis are provided in S1 Table. The black dots indicate catfish dnaja genes. Suffix “L” indicated “-like”, for instance, dnaja2L means dnaja2-like.
(TIF)
Phylogenetic tree of Hsp40s type II. The phylogenetic tree was constructed as in S1 Fig. Accession numbers for all sequences are provided in S1 Table. Suffix “L” indicated “-like”, for instance, dnajb5L means Dnajb5-like.
(TIF)
Phylogenetic tree of Hsp40s type III: Dnajc1 to Dnajc9. The phylogenetic tree was constructed as in S1 Fig. Accession numbers for all sequences are provided in S1 Table. The black dots indicate catfish Dnajc genes. Suffix “L” indicated “-like”, for instance, Dnajc7L means Dnajc7-like.
(TIF)
Phylogenetic tree of Hsp40s type III: Dnajc10 to Dnajc20. The phylogenetic tree was constructed as in S1 Fig. Accession numbers for all sequences are provided in S1 Table. The black dots indicate catfish Dnajc genes. Suffix “L” indicated “-like”.
(TIF)
Phylogenetic tree of Hsp40s type III: Dnajc21 to Dnajc30. The phylogenetic tree was constructed as in S1 Fig. Accession numbers for all sequences are provided in S1 Table. The black dots indicate catfish Dnajc genes. Suffix “L” indicated “-like”.
(TIF)
Gene names and accessions of reference Hsp40s used in this study.
(XLSX)
Fold changes of Hsp40s under each challenge. Bold font is the significant expression.
(XLSX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.