Abstract
The gene encoding the alternative sigma factor σB in Listeria monocytogenes is induced upon exposure of cells to several stresses. In this study, we investigated the impact of a sigB null mutation on the survival of L. monocytogenes EGD-e at low pH, during high-hydrostatic-pressure treatment, and during freezing. The survival of ΔsigB mutant exponential-phase cells at pH 2.5 was 10,000-fold lower than the survival of EGD-e wild-type cells. Moreover, the ΔsigB mutant failed to show an acid tolerance response. Upon preexposure for 1 h to pH 4.5, the survival at pH 2.5 was 100,000-fold lower for the ΔsigB mutant than for the wild type. The glutamate decarboxylase (GAD) acid resistance system is important in survival and adaptation of L. monocytogenes in acidic conditions. The σB dependence of the gad genes (gadA, gadB, gadC, gadD, and gadE) was analyzed in silico. Putative σB-dependent promoter sites were found upstream of the gadCB operon (encoding a glutamate/γ-aminobutyrate antiporter and a glutamate decarboxylase, respectively) and the lmo2434 gene (gadD, encoding a putative glutamate decarboxylase). Reverse transcriptase PCR revealed that expression of the gadCB operon and expression of gadD are indeed σB dependent. In addition, a proteomics approach was used to analyze the protein expression profiles upon acid exposure. Although the GAD proteins were not recovered, nine proteins accumulated in the wild type but not in the ΔsigB strain. These proteins included Pfk, GalE, ClpP, and Lmo1580. Exposure to pH 4.5, in order to preload cells with active σB and consequently with σ B-dependent general stress proteins, also provided considerable protection against high-hydrostatic-pressure treatment and freezing. The combined data argue that the expression of σB-dependent genes provides L. monocytogenes with nonspecific multiple-stress resistance that may be relevant for survival in the natural environment as well as during food processing.
Listeria monocytogenes is a gram-positive, nonsporulating, facultatively anaerobic microorganism. It is the causative agent of listeriosis, a serious illness for which the young, elderly, and immunocompromised are especially at risk (31). L. monocytogenes is of particular concern for the food industry due to the severity of the illness, as well as the wide distribution of the pathogen in the environment and consequently its presence on raw and minimally processed foods. Moreover, L. monocytogenes is known for its ability to survive and proliferate in adverse environmental conditions, including acidic conditions, refrigeration temperatures, and high osmolarity (up to 10%) (4, 36). These characteristics of L. monocytogenes make it a food-borne pathogen that is problematic for certain kind of foods, especially minimally processed, ready-to-eat foods.
Adaptation to (sudden) adverse conditions in the environment of a bacterium requires the ability to respond rapidly. Such a response of a bacterium to environmental changes involves activation of existing enzymes and enhanced rates of transcription of genes, resulting in enhanced levels of (defensive) proteins. Initiation of transcription of mRNA from DNA is mediated by the holoenzyme RNA polymerase. The holoenzyme consists of a core enzyme and a sigma factor, and the sigma factor is primarily responsible for recognition and binding of the polymerase to the promoter sequence upstream of a gene. In the genome sequence of L. monocytogenes EGD-e there are genes encoding five sigma factors (15) (recovered by using the Ergo integrated genomics database). These genes include rpoD encoding σA, which regulates housekeeping genes; sigH encoding a sigma factor with an unknown specific function; rpoN encoding σ54 (SigL), which in L. monocytogenes has a role in resistance to the antibacterial peptide mesentericin Y105 (25); sigV (lmo0423) encoding an enzyme that regulates extracytoplasmic function genes; and the alternative sigma factor gene sigB encoding σB, which controls the transcription of genes involved in stress adaptation.
The role of σB in the stress responses of gram-positive bacteria, including Bacillus subtilis, Staphylococcus aureus, and L. monocytogenes, has received considerable attention in recent years. The best-studied system is that in B. subtilis. The activity of the σB protein is regulated by a posttranslational mechanism. The B. subtilis σB operon comprises rsbR, rsbS, rsbT, rsbU, rsbV, rsbW, sigB, and rsbX. The Rsb proteins are all involved in regulation of σB activity (32). RsbW binds under normal growth conditions with σB to form an inactive complex. However, the affinity of RsbW for its antagonists, σB and RsbV, can change via two independent processes that promote the binding of RsbW to RsbV, thus leaving free σB, which is then capable of forming holoenzyme complexes with core RNA polymerase (32). Carbon limitation and entry into the stationary phase correlate with a drop in the intracellular level of ATP, which may have a direct effect on the binding preference of RsbW, which shifts from σB to RsbV. Also, upon exposure to a number of environmental insults, such as ethanol treatment or salt or acid shock, RsbU dephosphorylates RsbV, which is then able to bind to RsbW, resulting in free σB (32). The other Rsb proteins have an indirect effect (positive or negative) on σB activity or gene expression. Activation of σB in L. monocytogenes might involve a similar process, as the structure and organization of the rsbU, rsbV, rsbW, and rsbX genes and the sigB gene exhibit a high level of similarity with the structure and organization of their B. subtilis counterparts; the encoded proteins exhibit predicted levels of identity of 53, 45, 47, 29, and 66%, respectively (2, 39).
In B. subtilis, the alternative sigma factor σB regulates expression of a large general stress operon that contributes to transcription of about 150 genes involved in heat, acid, ethanol, salt, and freezing stress resistance (23, 33, 34). These genes include the katE gene encoding a catalase (9); the opuE gene encoding a transporter dedicated to the transport of the osmoprotectant proline (35); the clpC gene, which encodes a protein similar to stress-induced ATPase subunits of ClpP-type proteases (18); and the gtaB gene encoding a UDP-glucose pyrophosphorylase believed to participate in trehalose biosynthesis (30).
In L. monocytogenes, a role for σB has been determined in response to several stresses; e.g., it plays a role in acid resistance of stationary-phase cells, in oxidative and osmotic stress resistance, in the response to carbon starvation, and in growth at low temperatures (3, 10, 39). L. monocytogenes displays an active acid tolerance response upon exposure to low, nonlethal pH values and subsequent exposure to a lethal pH. Recently, the contribution of σB to growth-phase-dependent acid resistance and to the adaptive acid tolerance response in L. monocytogenes 10403S was analyzed by Ferreira et al. (11). The survival of the ΔsigB strain upon exposure to brain heart infusion (BHI) at pH 2.5 (with and without prior acid adaptation) was consistently lower than the survival of the wild-type strain throughout all phases of growth. However, σB-mediated contributions to acquired acid tolerance appeared to be greatest in early logarithmic growth. The acid tolerance response has a great impact on food processing and the virulence of the bacterium, since exposure to acidic conditions not only enhances survival at a lethal pH but also provides protection against other challenges, such as heat, ethanol, oxidative, and osmotic stresses (6, 12, 19).
Here, we describe the role of σB in acid stress adaptation of exponentially growing L. monocytogenes EGD-e cells. Since the glutamate decarboxylase (GAD) acid resistance system is important in survival and adaptation of L. monocytogenes in acidic conditions (7), we tested whether this system plays a role in the acid adaptation of the wild-type strain and a strain with σB deleted. The putative σB dependence of the gad genes (gadA, gadB, gadC, gadD [lmo2434], and gadE [lmo0449]) was analyzed in silico, and subsequently reverse transcriptase PCR was performed to analyze the transcription of the gad genes. In addition, a proteomics approach was used to analyze the protein expression profiles after exposure of the wild-type and ΔsigB strains to acid. Additionally, the role of σB in two industrially important processes, high-hydrostatic-pressure (HHP) treatment and freezing, was assessed in combination with pretreatment at a low pH in order to assess the protective effect of adaptation to these food-processing methods.
MATERIALS AND METHODS
Bacterial strains and generation of mutant.
L. monocytogenes EGD-e and a ΔsigB mutant of this strain were used throughout this study. The ΔsigB mutant (U. Volk, S. S. Chatterjee, S. Otten, S. Wagner, S. Haas, B. Brors, T. Chakraborty, and T. Hain, submitted for publication) was constructed by using the temperature-sensitive suicide plasmid pAUL-A (5), which resulted in an in-frame chromosomal deletion.
Survival during acidic conditions and freeze-thaw cycles.
Cells were grown in BHI at 30°C to an optical density at 620 nm (OD620) of 0.4, centrifuged, and resuspended in fresh BHI at pH 2.5 (adjusted with HCl). Survival was measured during 3 h of incubation of this suspension at 30°C by determining the number of CFU. Survival in this acidic environment was also measured after adaptation for 1 h to BHI with a pH of 4.5 (adjusted with HCl). Survival of L. monocytogenes and survival of the ΔsigB mutant were also measured after freeze-thaw cycles. Exponential-phase cells, grown in BHI at 30°C, were centrifuged, resuspended in fresh BHI, and subsequently frozen for 24 h at −20°C. Then the preparation was thawed for 2 min at 30°C and the number of CFU was determined, and the preparation was subsequently frozen for another 24 h at −20°C. The numbers of CFU were determined after five freeze-thaw cycles. Freeze-thaw survival was also measured after exposure of the mid-exponential phase cells to BHI at pH 4.5 for 1 h and to BHI at 7°C for 4 h. The experiments were performed in duplicate, and standard errors are indicated below (see Fig. 1).
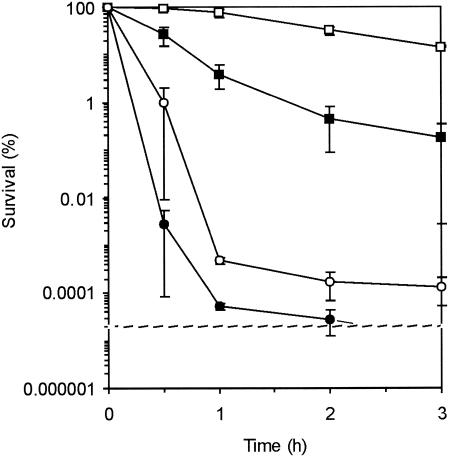
FIG. 1.
Survival of L. monocytogenes EGD-e (squares) and the ΔsigB mutant (circles) during incubation at pH 2.5 with (□ and ○) and without (▪ and •) preexposure to pH 4.5 for 1 h. The error bars indicate standard errors.
Analysis of protein extracts by 2D-E and matrix-assisted laser desorption ionization-time of flight analysis.
Samples (10 ml) of a bacterial suspension (OD620, 0.4) were removed, centrifuged, and suspended in water to an OD620 of 10. Total cellular proteins were extracted from the cells with a bead beater (B. Braun Biotech International, Melsungen, Germany) and zirconium beads (diameter, 0.1 mm; BioSpec Products, Bartlesville, Okla.) by using three 1-min treatments (with cooling on ice between treatments). After this the zirconium beads were allowed to sediment by gravity, and subsequently the supernatant, which contained the cellular proteins, was analyzed by two-dimensional gel electrophoresis (2D-E) by using a pI range of 4 to 7 and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (12 to 14% polyacrylamide) for the second dimension. 2D-E was performed essentially as described previously (38). Protein spots were visualized by silver staining, and the gels were analyzed with PDQuest software (Bio-Rad, Richmond, Calif.). Proteins that were differentially expressed (more than a threefold difference) were identified as proteins that increased expression upon acid exposure or as proteins that decreased expression upon acid exposure. For identification of protein spots gels were loaded with 200 μg of protein and stained with Coomassie blue. Differentially expressed proteins were cut out of the gel and analyzed at the Proteomics Center, Department of Human Biology, University of Maastricht, Maastricht, The Netherlands (J. Renes and F. Bouwman) by digestion with a MassPrep station (Micromass, Almere, The Netherlands) and subsequent analysis with a matrix-assisted laser desorption ionization-time of flight LR mass spectrometer (Micromass). The proteins were identified with PeptIdent.
Transcriptional analysis of the gad genes.
L. monocytogenes EGD-e and the ΔsigB mutant were grown to the mid-exponential phase at 30°C. A portion (10 ml) of each culture was centrifuged and resuspended in 1 ml of BHI (with and without the pH adjusted to 4.5). After 1 h of incubation, the cells were centrifuged, and the RNA was extracted with an RNeasy RNA extraction kit (QIAGEN, Hilden, Germany). cDNA was synthesized by adding 2 μl of total RNA to 1 μl of a reverse primer mixture (10 pmol/μl), 1 μl of 5× first strand buffer, and 1 μl of water. This mixture was incubated for 2 min at 75°C, after which 3 μl of first strand buffer, 2 μl of dithiothreitol (0.1 M), 4 μl of a preparation containing each deoxynucleoside triphosphate at a concentration of 2.5 mM, 0.4 μl of Superscript II reverse transcriptase enzyme, and 5.6 μl of water were added. The mixture was incubated for 1 h at 48°C, and then 3.55 μl of water, 0.2 μl of RNase H, and 1 μl of first strand buffer were added for cleavage of the RNA in RNA-DNA hybrids during incubation for 10 min at room temperature.
The PCR was carried out by using the primers listed in Table 1, and 20, 25, and 30 cycles were performed to allow optimal quantification of PCR products. Template cDNA was used in the reaction mixtures at levels that gave similar band intensities for 16S RNA reactions. The experiment was performed in duplicate, and representative results are described below.
TABLE 1.
Primers used in this study
| Gene | Primer | Sequence |
|---|---|---|
| gadA (GAD, lmo0447) | Forward | 5′-CGG TGT TTG GCT CTT TT GA-3′ |
| Reverse | 5′-CTC CGA TTC ATC CAC ATT CC-3′ | |
| gadB (GAD, lmo2363) | Forward | 5′-GGC ATG CAC CTA AGG ACC AAA AAT-3′ |
| Reverse | 5′-GAT ACC GAG GAT GCC GAC CAC AC-3′ | |
| gadC (antiporter, lmo2362) | Forward | 5′-AAA TGG CGA CGG TGG ATG GT-3′ |
| Reverse | 5′-TTT TGC GAT TTT AGC CGT GTT TT-3′ | |
| gadD (GAD, lmo2434) | Forward | 5′-ACT TGG CAA AAA CTG TAG AAA A-3′ |
| Reverse | 5′-TAG TGC GTA AAT CCG TAT GAA-3′ | |
| gadE (antiporter, lmo0448) | Forward | 5′-ATT CGG CGG CGG TGG TA-3′ |
| Reverse | 5′-AAA ACG GAA TTA AAA TAG TGA CGA-3′ | |
| 16S RNA | Forward | 5′-TTA GCT AGT TGG TAG GGT-3′ |
| Reverse | 5′-AAT CCG GAC AAC GCT TGC-3′ |
HHP treatment.
Cells were harvested by centrifugation, resuspended in 50 mM N-(2-acetamido)-2-aminoethanesulfonic acid (ACES buffer; Sigma-Aldrich, Steinheim, Germany) (pH 7.0), and subsequently packed in plastic bags. The survival after high-pressure treatment (150, 200, 250, 300, 350, and 400 MPa for 20 min) was determined for exponentially growing cells, cells exposed to pH 4.5 for 1 h, and cells exposed to 10°C for 4 h. Depressurization of the HHP apparatus (Resato, Roden, The Netherlands) was achieved within seconds. The pressurization was performed at 20°C, while the temperatures during pressurization did not exceed 32°C. In addition, L. monocytogenes wild-type and ΔsigB mutant cells that were preexposed to pH 4.5 were exposed to 350 MPa for 3, 8, 14, 18, 23, and 28 min at 20°C. The experiments were performed in duplicate, and the standard errors are indicated below.
RESULTS
Survival of L. monocytogenes EGD-e at low pH is σB dependent.
Survival of exponential-phase cells of L. monocytogenes EGD-e and the σB null mutant in BHI at pH 2.5 was determined in order to analyze the σB dependence of survival. About 5% of wild-type L. monocytogenes cells survived 1 h of exposure to the acidic conditions (Fig. 1). For the σB mutant cells the survival rate was about 0.00005%; i.e., there was a 5-log difference. This clearly shows that the survival of L. monocytogenes at pH 2.5 is σB dependent. In addition, we determined the σB dependence of the acid tolerance response in L. monocytogenes EGD-e. Cells were preexposed to a nonlethal pH (pH 4.5) prior to exposure to pH 2.5. After this preexposure to pH 4.5, almost 100% of the EGD-e wild-type cells survived exposure to pH 2.5 for 1 h. However, only about 0.0005% of the preexposed cells with a σB null mutation survived, indicating the importance of induction and/or activation of σB during adaptation to a low pH for subsequent survival at a lethal pH (Fig. 1). Notably, the mutant still displayed a small acid tolerance response, indicating that σB is not solely responsible for the acid tolerance response.
σB-Dependent expression of gad genes.
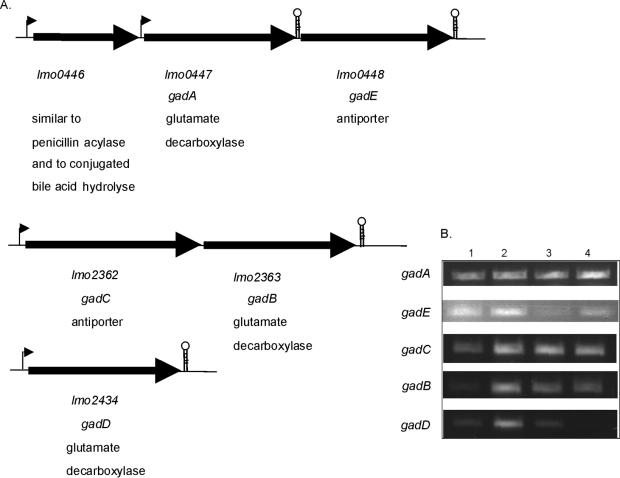
Adaptation of bacteria to acidic conditions involves maintenance of a relatively high intracellular pH (pHi) (21). A specific mechanism for acid adaptation in L. monocytogenes aimed at maintenance of the pHi is the GAD acid resistance system. This system involves an antiporter, which transports glutamate into the cell, and a glutamate decarboxylase (GAD), which converts glutamate into γ-aminobutyrate (GABA) upon consumption of a proton. The glutamate/GABA antiporter subsequently excludes GABA from the cell. Cotter et al. (7) identified the following three genes (in the pregenome era) involved in this system: gadA (lmo447), encoding a GAD; gadB (lmo2363), coding for another GAD; and gadC (lmo2362), encoding the associated glutamate/GABA antiporter. Using the genome data for L. monocytogenes EGD-e (http://genolist.pasteur.fr/ListiList/), Conte et al. (6) identified two other genes with high homology to the gad genes. lmo2434 (designated gadD in this study) codes for another GAD, and lmo448 (designated gadE in this study) encodes an additional antiporter. gadA and gadE are close to each other and are cotranscribed. However, a stem-loop-like structure within the intergenic spacer downstream of gadA stops readthrough transcription, resulting in single transcription of gadA (or in cotranscription with lmo0446) (6). Cotter et al. (7) described the cotranscription of gadC and gadB; this observation, together with the genome data, suggests that these two genes form an operon. gadD is oppositely oriented and is further downstream of the operon (Fig. 2A). Considering the low level of survival of a ΔsigB mutant at a low pH, the putative σB dependence of the gad genes was analyzed in silico. Comparison of the L. monocytogenes σB promoter sequence (GTTTTA-N14-GGGTAA, as described by Becker et al. [2]) revealed only three and two mismatches upstream (within 200 bp) of the gadCB operon and gadD, respectively (Table 2). Using reverse transcriptase PCR, we analyzed the transcription of the gad genes (gadA, gadB, gadC, gadD, and gadE) upon exposure to pH 4.5 for 1 h for both the wild-type and ΔsigB strains. For wild-type strain EGD-e all gad genes except gadA were induced after exposure to low pH (Fig. 2B). This is an indication that GadA plays a minor role in acid adaptation, which is supported by the results of the acid survival experiments of Cotter et al. (7) in which gad deletion mutant strains were used. Of the four induced transcripts in the wild type, only one, gadE, was induced in the ΔsigB mutant strain. This indicates that transcription of the other three genes, gadB, gadC, and gadD, is at least partially σB regulated under the conditions tested. The apparent constitutive expression of the gadCB operon in the ΔsigB strain suggests that additional regulators are involved. Slight induction of gadA transcripts could be detected upon acid exposure of the ΔsigB strain. This might be compensation for the reduced transcription of the other genes coding for GADs (e.g., gadD).
FIG. 2.
(A) Organization of gad genes in L. monocytogenes EGD-e. The hairpin structures indicate putative terminators. The numbering of the open reading frames is according to the L. monocytogenes EGD-e genome sequence (15). (B) Transcriptional analysis of gad genes in L. monocytogenes EGD-e (lanes 1 and 2) and the ΔsigB mutant (lanes 3 and 4) in exponential-phase cells (lanes 1 and 3) and cells exposed to pH 4.5 for 1 h (lanes 2 and 4).
TABLE 2.
Alignment of putative σB-dependent promoters in L. monocytogenes (based on in silico analysis)
| Gene | Promoter sequence
|
No. of mismatches | ||
|---|---|---|---|---|
| −35 Region | Spacing | −10 Region | ||
| sigB | GTTTTA | N14 | GGGTAA | |
| pfk | GTTTTG | N11 | GTTTAA | 3 |
| clpP | GTTTGA | N16 | GTGTAT | 3 |
| lmo1580 | GGTTCT | N13 | GGGTAA | 3 |
| galE | ATAGAT | N14 | GGGTCT | 4 |
| gadD | TTTTTA | N12 | CGGTAA | 2 |
| gadC | GTTTGT | N14 | GGGTAT | 3 |
| pykA | GTTTTA | N12 | TGGTAA | 1 |
| lmo1339 | GTTTAA | N16 | GGAGAA | 3 |
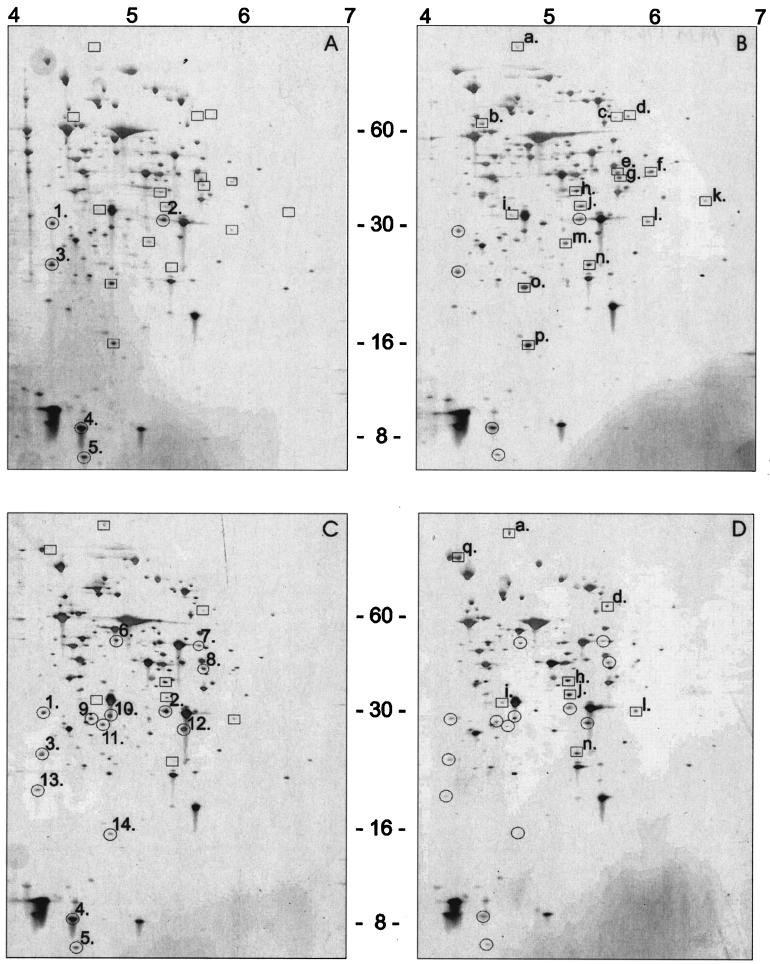
2D-E and identification of proteins involved in acid adaptation.
Previous research showed that the acid tolerance response of L. monocytogenes includes induction of several proteins (8, 22, 24). In this study the proteins present in the wild-type and ΔsigB strains after exposure to pH 4.5 were analyzed by 2D-E (Fig. 3) and were subsequently identified (Table 3). After exposure to pH 4.5 wild-type cells exhibited decreased accumulation of five proteins (spots 1 to 5), including Lmo1704; a protein similar to conserved hypothetical proteins, FbaA; a protein similar to fructose-1,6-biphosphate aldolase; and HPr, a phosphotransferase system (PTS) phosphocarrier protein (Table 3). However, the levels of 16 proteins were enhanced after exposure to low pH (spots a to p); these proteins included ValS, valyl-tRNA synthetase (Pfk), 6-phosphofructokinase (FlaA), flagellin protein (Lmo1709), a protein similar to methionine aminopeptidases (GalE), UDP-glucose-4-epimerase (ClpP), the proteolytic subunit of the ATP-dependent Clp protease, and Lmo1580, which is similar to an unknown protein. In the ΔsigB strain the levels of 14 proteins were reduced after exposure to low pH, whereas the levels of eight proteins were higher. Of the 16 proteins that accumulated in the wild type, 7 were also present at enhanced levels in the ΔsigB mutant, indicating that these proteins are not strictly σB regulated. Of these 16 proteins, 9 were present at enhanced levels in the wild type (Fig. 3), whereas they were not induced in the ΔsigB mutant strain. These putatively σB-regulated proteins, which may participate in acid adaptation, include Pfk, GalE, ClpP, and Lmo1580. Using the L. monocytogenes σB promoter sequence for putative σB-regulated promoter site recognition (2), we matched the putative promoter sequences of the genes encoding these proteins in silico. Three mismatches were found in the promoter regions of pfk, clpP, and lmo1580, and four mismatches were found for galE (Table 2).
FIG. 3.
Analysis of protein production in L. monocytogenes EGD-e exponential-phase cells (A), ΔsigB mutant exponential-phase cells (C), L. monocytogenes EGD-e cells exposed to pH 4.5 for 1 h (B), and ΔsigB mutant cells exposed to pH 4.5 for 1 h (D) by 2D-E. Differentially expressed proteins are indicated as follows: proteins whose levels were reduced are circled, and proteins that were induced upon exposure to low pH are enclosed in boxes.
TABLE 3.
Identification of differentially expressed proteins in acid-shocked L. monocytogenes EGD-e and the ΔsigB mutant
| Spot | Protein | Description | pI | Mol wt (103) |
|---|---|---|---|---|
| Reduced protein production upon acid shock of EGD-e wild-type strain | ||||
| 2 | FbaA | Similar to fructose-1,6-biphosphate aldolase | 5.1 | 30.0 |
| 4 | Hpr | PTS phosphocarrier protein Hpr | 4.5 | 9.4 |
| 5 | Lmo1704 | Similar to conserved hypothetical proteins | 5.8 | 14.4 |
| Increased protein production upon acid shock of EGD-e wild-type strain | ||||
| a | ValSa | Valyl-tRNA synthetase | 4.7 | 102 |
| f | Pfk | 6-Phosphofructokinase | 5.4 | 34.4 |
| i | FlaA | Flagellin protein | 4.7 | 30.4 |
| l | Lmo1709 | Similar to methionine aminopeptidases | 5.4 | 27.9 |
| m | GalE | UDP-glucose-4-epimerase | 4.9 | 36.2 |
| o | ClpP | ATP-dependent Clp protease proteolytic subunit | 4.7 | 21.6 |
| p | Lmo1580 | Similar to unknown protein | 4.7 | 16.9 |
| Reduced protein production upon acid shock of the EGD-e ΔsigB mutant | ||||
| 2 | FbaA | Similar to fructose-1,6-biphosphate aldolase | 5.1 | 30.0 |
| 4 | Hpr | PTS phosphocarrier protein Hpr | 4.5 | 9.4 |
| 5 | Lmo1704 | Similar to conserved hypothetical proteins | 5.8 | 14.4 |
| 10 | Lmo1011 | Hypothetical protein, similar to tetrahydrodipicolinate succinylase | 4.7 | 24.8 |
| 12 | ThiD | Phosphomethyl pyrimidine kinase | 5.0 | 28.8 |
| 14 | Lmo1580 | Similar to unknown protein | 4.7 | 16.9 |
| Increased protein production upon acid shock of the EGD-e ΔsigB mutant | ||||
| a | ValSa | Valyl-tRNA synthetase | 4.7 | 102 |
| i | FlaA | Flagellin protein | 4.7 | 30.4 |
| l | Lmo1709 | Similar to methionine aminopeptidases | 5.4 | 27.9 |
| Putative σB-regulated proteins involved in acid adaptation | ||||
| f | Pfk | 6-Phosphofructokinase | 5.4 | 34.4 |
| m | GalE | UDP-glucose-4-epimerase | 4.9 | 36.2 |
| o | ClpP | ATP-dependent Clp protease proteolytic subunit | 4.7 | 21.6 |
| p | Lmo1580 | Similar to unknown protein | 4.7 | 16.9 |
Only 6.2% of the sequence was covered.
Role of σB in survival after HHP treatment and freeze-thaw cycles.
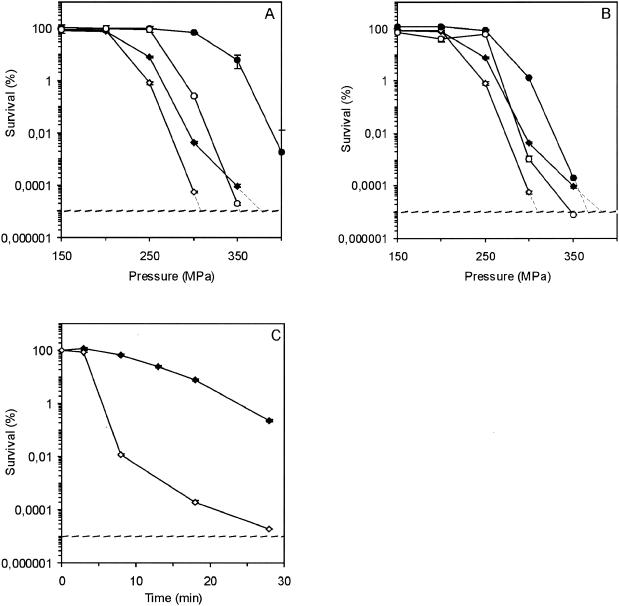
To determine the role of σB in other important food-manufacturing processes, we assessed the survival of the parent strain and the mutant after HHP treatment and freeze-thaw cycles. The parent strain was about 100-fold less susceptible to 20 min at 300 MPa than the ΔsigB mutant strain (Fig. 4A).
FIG. 4.
Survival of L. monocytogenes EGD-e (solid symbols) and the ΔsigB mutant (open symbols) after HHP treatment. (A) Survival of exponential-phase cells (♦ and ⋄) and cells exposed to pH 4.5 for 1 h (• and ○). (B) Survival of exponential-phase cells (♦ and ⋄) and cells exposed to 10°C for 4 h (• and ○). (C) Survival of preexposed (1 h, pH 4.5) L. monocytogenes EGD-e (♦) and the ΔsigB mutant (⋄) after exposure to 350 MPa. The error bars indicate standard errors.
Exposure to pH 4.5 for 1 h increased the survival of the parent strain after subsequent HHP exposure dramatically; i.e., treatment at 300 MPa did not significantly decrease the viable cell count. Notably, even treatment at 350 MPa only decreased the viable cell count to 10% of the initial count, whereas the cell counts for the ΔsigB mutant strain (preexposed to pH 4.5) were only just above the detection limit after treatment at 350 MPa; i.e., there was a >5-log difference. Additionally, we analyzed this difference in greater detail by varying the duration of exposure (Fig. 4C). There was a clear difference between the survival of EGD-e wild-type cells and the survival of the ΔsigB mutant; e.g., after 28 min of exposure to 350 MPa, there was a 4-log difference. These results indicate that acid-induced proteins encoded by σB-dependent genes provide considerable protection for cells exposed to HHP. Preexposure of the cells to a low temperature also resulted in increased survival after pressure treatment (more than 100-fold increase after treatment at 300 MPa) (Fig. 4B), as found previously for L. monocytogenes LO28 cells (38). Again, the ΔsigB mutant showed lower resistance to pressure treatment than the wild-type cells, indicating that the σB-dependent proteins formed during low-temperature adaptation are involved in tolerance to high pressure. However, the survival of both wild-type and ΔsigB mutant cells exposed to acid conditions was higher than the survival of refrigerated cells.
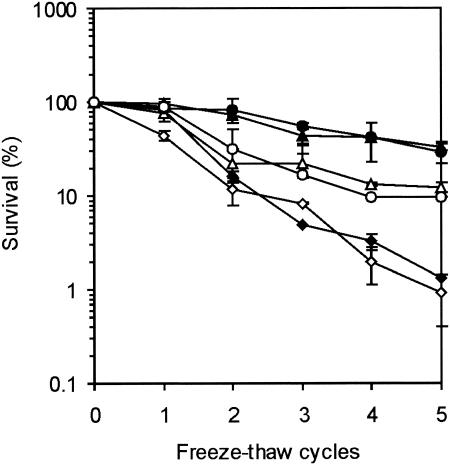
The survival of cells after exposure to freeze-thaw treatment increased after preexposure to another insult, and exposure for 4 h at 7°C was as effective as exposure to low pH (Fig. 5). The protection provided by preexposure before freezing was partially σB dependent, as the level of survival of wild-type strain EGD-e after five freeze-thaw cycles was threefold higher than the level of survival of the ΔsigB mutant.
FIG. 5.
Survival of L. monocytogenes EGD-e (solid symbols) and the ΔsigB mutant (open symbols) after freeze-thaw cycles. Freeze-thaw cycles were performed with exponential-phase cells (♦ and ⋄), cells preexposed to pH 4.5 for 1 h (▴ and ▵), and cells preexposed to 10°C for 4 h (• and ○). The error bars indicate standard errors.
DISCUSSION
σ B-Dependent expression of gad genes.
Here we show that survival upon exposure to lethal acidic conditions and the acid tolerance response of L. monocytogenes EGD-e exponential-phase cells are highly σB dependent. In L. monocytogenes, maintenance of a relatively high pHi is important for survival at a low pH. The GAD acid resistance system is a specific mechanism for acid adaptation in L. monocytogenes aimed at maintenance of the pHi. This system involves an antiporter, which transports glutamate into the cell, and a GAD, which converts glutamate into GABA upon consumption of a proton. The glutamate/GABA antiporter subsequently excludes GABA from the cell. As the ΔsigB mutant is significantly more acid sensitive than the wild type, the putative σB dependence of the gad genes was analyzed in silico. Putative σB-dependent promoter sites were found upstream of the gadCB operon and gadD and had three and two mismatches, respectively. Additionally, by performing transcription analysis with reverse transcriptase PCR, we confirmed the σB-dependent transcription of gadCB and gadD upon exposure to pH 4.5 for 1 h. This could partially explain the loss of acid tolerance by the ΔsigB mutant strain, as null mutations in the gad genes (gadA, gadB, and gadC) play an important role in acid resistance and adaptation (7). Ferreira et al. (11) did not find that σB contributes to the net proton movement across the cell membrane or to the GAD system. However, their experimental setup included incubation at pH 2.5 for 1 h of stationary phase cells that had not been preadapted to low pH; they showed that about 10% of the wild-type cells and only about 0.1% of the ΔsigB cells survived. Additionally, they analyzed the contribution of σB to growth-phase-dependent acid resistance and to the adaptive acid tolerance response in L. monocytogenes 10403S. Upon exposure to BHI at pH 2.5 (with and without prior acid adaptation) the survival of the ΔsigB strain was consistently lower than the survival of the wild-type strain throughout all phases of growth (11).
σB-Dependent proteins involved in acid adaptation.
The acid tolerance response induces de novo protein synthesis, as demonstrated previously by 2D-E (8, 22, 24). Moreover, treatment with chloramphenicol during acid adaptation eliminates the protective effect (22). 2D-E of proteins from cells of the wild-type strain and the ΔsigB mutant exposed to acidic pH conditions revealed several proteins whose levels were increased (16 and 8 proteins, respectively) or reduced (5 and 14 proteins, respectively) at least threefold. The proteins whose levels were reduced in both the wild-type strain and the ΔsigB mutant included Lmo1704 (a protein similar to conserved hypothetical proteins), FbaA (a protein which is similar to fructose-1,6-biphosphate aldolase), the PTS phosphocarrier protein HPr, and two unidentified proteins. Although not previously described for L. monocytogenes, the level of HPr is also reduced in Lactococcus lactis upon exposure to acid. HPr not only has a role in the phosphoenolpyruvate-dependent sugar PTS but also has a regulatory role in cell metabolism (20, 40).
For the ValS, FlaA, and Lmo1709 proteins, increased accumulation during exposure to low pH was observed for both the wild-type and mutant strains. FlaA, the structural protein of the flagellum, might be induced for increased motility of the bacterium and subsequent movement away from growth-inhibiting conditions. The σB-dependent proteins that accumulated in the wild type after acid adaptation include Pfk, GalE, ClpP, and Lmo1580. Pfk (6-phosphofructokinase) and GalE are enzymes that are involved in glycolysis and sugar metabolism, respectively. Pfk is one of the key enzymes involved in the control of glycolysis, together with hexokinase (glucose kinase) and pyruvate kinase. Strikingly, comparison of the promoter sites of pyruvate kinase (pykA) and glucose kinase (lmo1339) with the sequence recognized by σB revealed one and three mismatches (Table 2), respectively, suggesting that these enzymes might also be σB regulated. These data together may suggest that the rate of glycolysis is partially σB regulated. However, additional experiments (e.g., reverse transcriptase PCR or primer extension analysis) should be performed to test this hypothesis. ClpP is the proteolytic subunit of the ATP-dependent Clp protease. The ATPases ClpC and ClpE and the proteolytic subunit ClpP are all required for stress survival, growth at a high temperature, and virulence (13, 21, 26). Clp ATPases regulate ATP-dependent proteolysis, preventing accumulation of misfolded proteins, and they also play a role as molecular chaperones involved in protein folding and assembly (37). The clp genes in L. monocytogenes form part of the CtsR (class three stress gene repressor) stress response regulon, in which CtsR negatively regulates the clpP, clpC, and clpE genes. In B. subtilis, two transcriptional start sites upstream of the clpP gene were identified, and they were preceded by sequences resembling the consensus sequences of promoters recognized by σA and σB transcriptional factors of the B. subtilis RNA polymerase (14). Transcription initiation occurred predominantly at the putative σA-dependent promoter. However, after exposure to stress, initiation of transcription also increased at the σB-dependent promoter (14). Alignment of the promoter regions of clpP of L. monocytogenes EGD-e revealed a putative promoter with three mismatches to the σB consensus promoter sequence. Along with our proteomics findings, this indicates that clpP in L. monocytogenes might also be, in addition to CtsR regulated, σB regulated.
Survival after HHP treatment and freeze-thaw cycles.
Preexposure to a low pH not only provides protection against an otherwise lethal pH but also enhances survival during other challenges, such as heat, ethanol, oxidative, and osmotic stresses (12, 19). Here we determined the effect of preexposure of cells to acidic conditions on survival after HHP treatment and freezing. Cells exposed to a low pH prior to HHP treatment were significantly more piezotolerant. This effect was found to be conferred mainly by σB-dependent proteins, as ΔsigB mutant cells that were preexposed to a low pH were not as piezotolerant as wild-type cells. Piezotolerance was also observed for a mutant (AK01) that occurred naturally in an L. monocytogenes Scott A culture (16, 17). 2D-E showed that in this strain there were enhanced levels of ClpP that resulted from a single amino acid deletion in the highly conserved glycine-rich region of CtsR, a repressor of the clp genes. As we also found enhanced expression of ClpP in the wild type upon exposure to a low pH and not in the ΔsigB mutant, this protein may be an important factor in survival at high pressure. However, this could not be the only reason for increased pressure resistance, as the level of survival of AK01 was only about 2 logs higher than that of the wild type over a broad range of pressures (17), whereas the level of survival of acid-adapted wild-type EGD-e cells and the level of survival of ΔsigB mutant cells differed more than 5 logs at 350 MPa. Low-temperature adaptation of L. monocytogenes EGD-e cells also increased survival after HHP treatment. This is in agreement with previous results for HHP survival of low-temperature-adapted L. monocytogenes LO28 cells (38). Two cold shock proteins, Csp1 and Csp3, were found to be induced after a cold shock at 10°C. Strikingly, Csp1 was also induced after 10 min of exposure to 200 MPa (38). Analysis of the sequence of the putative promoter region of the cspL coding region revealed putative σB-regulated −10 and −35 promoter sequences. It can be speculated that the relative sensitivity of the ΔsigB mutant to high pressure after exposure to a low temperature may be due to the reduced expression of Csp1. Additionally, other (σB-regulated) proteins and other adaptation processes (e.g., membrane adaptations) might influence the survival during high-pressure treatment. In L. monocytogenes, the adaptation of membranes to low temperatures is accomplished by altering branching in the methyl end of the fatty acid from iso to anteiso and by shortening the fatty acid chains, resulting mainly in an increase in the level of anteiso-C15:0 fatty acids (1). In acid-adapted L. monocytogenes cells, the levels of the straight-chain C14:0 and C16:0 fatty acids were significantly increased, while the levels of C18:0 were decreased (29). In analogy, the fatty acids of barophilic microorganisms become more polyunsaturated with increases in the growth pressure (28). These observations together indicate that membrane adaptations that are induced in mildly acidic conditions may provide cross-protection against HHP. The influence of the membrane fluidity on piezotolerance may be exerted through the control of (ion) pumps in the membrane that are essential for maintaining pH homeostasis (27); e.g., the GAD system may have an impact on piezotolerance of acid-adapted cells, based on the importance of this system during adaptation to acidic conditions and its σB dependence.
The results of this study contribute to our understanding of survival under acid conditions and adaptation of L. monocytogenes. σB-Dependent protective mechanisms involved in survival and adaptation to low pH have been identified; these mechanisms include three genes of the GAD system and nine proteins induced at an acidic pH involved in stress protection and metabolism. Moreover, it is clear that acid adaptation provides, mainly in a σB-dependent manner, protection against HHP and freezing. The observed cross-adaptation phenomenon can have a significant impact on food processing (e.g., if acidification of food products is combined with a pressure treatment).
Acknowledgments
We thank Linda Kortenoeven and Marieke Pastink for expert technical assistance.
REFERENCES
- 1.Annous, B. A., L. A. Becker, D. O. Bayles, D. P. Labeda, and B. J. Wilkinson. 1997. Critical role of anteiso-C15:0 fatty acid in the growth of Listeria monocytogenes at low temperatures. Appl. Environ. Microbiol. 63:3887-3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becker, L. A., M. S. Çetin, R. W. Hutkins, and A. K. Benson. 1998. Identification of the gene encoding the alternative sigma factor σB from Listeria monocytogenes and its role in osmotolerance. J. Bacteriol. 180:4547-4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker, L. A., S. N. Evans, R. W. Hutkins, and A. K. Benson. 2000. Role of σB in adaptation of Listeria monocytogenes to growth at low temperature. J. Bacteriol. 182:7083-7087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buchanan, R. L., and L. A. Klawitter. 1990. Effects of temperature and oxygen on the growth of Listeria monocytogenes at pH 4.5. J. Food Sci. 55:1754-1756. [Google Scholar]
- 5.Chakraborty, T., M. Leimeister-Wachter, E. Domann, M. Hartl, W. Goebel, T. Nichterlein, and S. Notermans. 1992. Coordinate regulation of virulence genes in Listeria monocytogenes requires the product of the prfA gene. J. Bacteriol. 174:568-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conte, M. P., G. Petrone, A. M. Di Biase, C. Longhi, M. Penta, A. Tinari, F. Superti, G. Fabozzi, P. Visca, and L. Seganti. 2002. Effect of acid adaptation on the fate of Listeria monocytogenes in THP-1 human macrophages activated by gamma interferon. Infect. Immun. 70:4369-4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cotter, P. D., C. G. M. Gahan, and C. Hill. 2001. A glutamate decarboxylase system protects Listeria monocytogenes in gastric fluid. Mol. Microbiol. 40:465-475. [DOI] [PubMed] [Google Scholar]
- 8.Davis, M. J., P. J. Coote, and C. P. O'Byrne. 1996. Acid tolerance in Listeria monocytogenes: the adaptive acid tolerance response (ATR) and growth-phase-dependent acid resistance. Microbiology 142:2975-2982. [DOI] [PubMed] [Google Scholar]
- 9.Engelmann, S., and M. Hecker. 1996. Impaired oxidative stress resistance of Bacillus subtilis sigB mutants and the role of katA and katE. FEMS Microbiol. Lett. 145:63-69. [DOI] [PubMed] [Google Scholar]
- 10.Ferreira, A., C. P. O'Byrne, and K. J. Boor. 2001. Role of σB in heat, ethanol, acid, and oxidative stress resistance and during carbon starvation in Listeria monocytogenes. Appl. Environ. Microbiol. 67:4454-4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferreira, A., D. Sue, C. P. O'Byrne, and K. J. Boor. 2003. Role of Listeria monocytogenes σB in survival of lethal acidic conditions and in the acquired acid tolerance response. Appl. Environ. Microbiol. 69:2692-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gahan, C., B. O'Driscoll, and C. Hill. 1996. Acid adaptation of Listeria monocytogenes can enhance survival in acidic foods and during milk fermentation. Appl. Environ. Microbiol. 62:3128-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaillot, O., E. Pellegrini, S. Bregenholt, S. Nair, and P. Berche. 2000. The ClpP serine protease is essential for the intracellular parasitism and virulence of Listeria monocytogenes. Mol. Microbiol. 35:1286-1294. [DOI] [PubMed] [Google Scholar]
- 14.Gerth, U., E. kruger, I. Derre, T. Msadek, and M. Hecker. 1998. Stress induction of the Bacillus subtilis clpP gene encoding a homologue of the proteolytic component of the Clp protease and the involvement of ClpP and ClpX in stress tolerance. Mol. Microbiol. 28:787-802. [DOI] [PubMed] [Google Scholar]
- 15.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couvé, A. de Daruvar, P. Dehoux, E. Domann, G. Domínguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K.-D. Entian, H. Fsihi, F. Garcia-Del Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gómez-López, T. Hain, J. Hauf, D. Jackson, L.-M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueño, A. Maitournam, J. Mata Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J.-C. Pérez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vázquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 16.Karatzas, K. A., J. A. Wouters, C. G. M. Gahan, C. Hill, T. Abee, and M. H. J. Bennik. 2003. The CtsR regulator of Listeria monocytogenes contains a variant glycine repeat region that affects piezotolerance, stress resistance, motility, and virulence. Mol. Microbiol. 49:1227-1238. [DOI] [PubMed] [Google Scholar]
- 17.Karatzas, K. A. G., and M. H. J. Bennik. 2002. Characterization of a Listeria monocytogenes Scott A isolate with high tolerance towards high hydrostatic pressure. Appl. Environ. Microbiol. 68:3183-3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kruger, E., T. Msadek, and M. Hecker. 1996. Alternate promoters direct stress-induced transcription of the Bacillus subtilis clpC operon. Mol. Microbiol. 20:713-723. [DOI] [PubMed] [Google Scholar]
- 19.Lou, Y., and A. E. Yousef. 1997. Adaptation to sublethal environmental stresses protects Listeria monocytogenes against lethal preservation factors. Appl. Environ. Microbiol. 63:1252-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luesink, E. J., C. M. A. Beumer, O. P. Kuipers, and W. M. De Vos. 1999. Molecular characterization of the Lactococcus lactis ptsHI operon and analysis of the regulatory role of HPr. J. Bacteriol. 181:764-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nair, S., I. Derre, T. Msadek, O. Gaillot, and P. Berche. 2000. CtsR controls class III heat shock gene expression in the human pathogen Listeria monocytogenes. Mol. Microbiol. 35:800-811. [DOI] [PubMed] [Google Scholar]
- 22.O'Driscoll, B., C. G. M. Gahan, and C. Hill. 1997. Two-dimensional polyacrylamide gel electrophoresis analysis of the acid tolerance response in Listeria monocytogenes LO28. Appl. Environ. Microbiol. 63:2679-2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petersohn, A., M. Brigulla, S. Haas, J. D. Hoheisel, U. Völker, and M. Hecker. 2001. Global analysis of the general stress response of Bacillus subtilis. J. Bacteriol. 183:5617-5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Phan-Thanh, L., F. Mahouin, and S. Alige. 2000. Acid responses of Listeria monocytogenes. Int. J. Food Microbiol. 55:121-126. [DOI] [PubMed] [Google Scholar]
- 25.Robichon, D., E. Gouin, M. Debarbouille, P. Cossart, Y. Cenatiempo, and Y. Hechard. 1997. The rpoN (sigma54) gene from Listeria monocytogenes is involved in resistance to mesentericin Y105, an antibacterial peptide from Leuconostoc mesenteroides. J. Bacteriol. 179:7591-7594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rouquette, C., C. de Chastellier, S. Nair, and P. Berche. 1998. The ClpC ATPase of Listeria monocytogenes is a general stress protein required for virulence and promoting early bacterial escape from the phagosome of macrophages. Mol. Microbiol. 27:1235-1245. [DOI] [PubMed] [Google Scholar]
- 27.Russell, N. J. 2002. Bacterial membranes: the effects of chill storage and food processing. An overview. Int. J. Food Microbiol. 79:27-34. [DOI] [PubMed] [Google Scholar]
- 28.Smelt, J. P. P. M. 1998. Recent advances in the microbiology of high pressure processing. Trends Food Sci. Technol. 9:152-158. [Google Scholar]
- 29.van Schaik, W., C. G. M. Gahan, and C. Hill. 1999. Acid-adapted Listeria monocytogenes displays enhanced tolerance against the lantibiotics nisin and licticin 3147. J. Food Prot. 62:536-539. [DOI] [PubMed] [Google Scholar]
- 30.Varon, D., S. Boylan, K. Okamoto, and C. Price. 1993. Bacillus subtilis gtaB encodes UDP-glucose pyrophosphorylase and is controlled by stationary-phase transcription factor sigma B. J. Bacteriol. 175:3964-3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vazquez-Boland, J. A., M. Kuhn, P. Berche, T. Chakraborty, G. Dominguez-Bernal, W. Goebel, B. Gonzalez-Zorn, J. Wehland, and J. Kreft. 2001. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14:584-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Voelker, U., A. Voelker, B. Maul, M. Hecker, A. Dufour, and W. Haldenwang. 1995. Separate mechanisms activate sigma B of Bacillus subtilis in response to environmental and metabolic stresses. J. Bacteriol. 177:3771-3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Volker, U., S. Engelmann, B. Maul, S. Riethdorf, A. Volker, R. Schmid, H. Mach, and M. Hecker. 1994. Analysis of the induction of general stress proteins of Bacillus subtilis. Microbiology 140:741-752. [DOI] [PubMed] [Google Scholar]
- 34.Volker, U., B. Maul, and M. Hecker. 1999. Expression of the sigma B-dependent general stress regulon confers multiple stress resistance in Bacillus subtilis. J. Bacteriol. 181:3942-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Von Blohn, C., B. Kempf, R. M. Kappes, and E. Bremer. 1997. Osmostress response in Bacillus subtilis: characterization of a proline uptake system (OpuE) regulated by high osmolarity and the alternative transcription factor sigma B. Mol. Microbiol. 25:175-187. [DOI] [PubMed] [Google Scholar]
- 36.Walker, S. J., P. Archer, and J. G. Banks. 1990. Growth of Listeria monocytogenes at refrigeration temperatures. J. Appl. Bacteriol. 68:157-162. [DOI] [PubMed] [Google Scholar]
- 37.Wawrzynow, A., B. Banecki, and M. Zylicz. 1996. The Clp ATPases define a novel class of molecular chaperones. Mol. Microbiol. 21:895-899. [DOI] [PubMed] [Google Scholar]
- 38.Wemekamp-Kamphuis, H. H., A. K. Karatzas, J. A. Wouters, and T. Abee. 2002. Enhanced levels of cold shock proteins in Listeria monocytogenes LO28 upon exposure to low temperature and high hydrostatic pressure. Appl. Environ. Microbiol. 68:456-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wiedmann, M., T. J. Arvik, R. J. Hurley, and K. J. Boor. 1998. General stress transcription factor σB and its role in acid tolerance and virulence of Listeria monocytogenes. J. Bacteriol. 180:3650-3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wouters, J. A., H. H. Kamphuis, J. Hugenholtz, O. P. Kuipers, W. M. de Vos, and T. Abee. 2000. Changes in glycolytic activity of Lactococcus lactis induced by low temperature. Appl. Environ. Microbiol. 66:3686-3691. [DOI] [PMC free article] [PubMed] [Google Scholar]