Abstract
The aim of the study was to evaluate the presence of pathogenic viruses in the Moselle River and to compare the usefulness of thermotolerant coliforms and somatic coliphages as tools for river water quality assessment in terms of viral contamination. Thermotolerant coliforms and somatic coliphages were enumerated by standardized methods in 170 samples of river water drawn from five sampling sites along the Moselle River (eastern France). BGM cell culture and integrated cell culture-reverse transcription-PCR DNA enzyme immunoassay were used to determine the presence of pathogenic viral genome (Enterovirus and Norovirus genogroup II [GGII]) and infectious Enterovirus spp. in 90 1-liter samples. No infectious Enterovirus spp. were isolated, but Enterovirus and Norovirus GGII genomes were detected in 38% of the samples. Norovirus GGII genome was mostly detected in winter, whereas Enterovirus genome was mostly detected in summer and fall. Somatic coliphages appeared to be less sensitive to higher river water temperature than thermotolerant coliforms. Furthermore, the number of river water samples positive for pathogenic viral genome increased with increasing concentration of somatic coliphages, whereas coliform concentration was unrelated to viral genome contamination. Consequently somatic coliphages, which are less sensitive to environmental factors than thermotolerant coliforms in river water, would provide a promising tool for assessment of river water quality in terms of fecal and viral pollution.
Many factors influence viral pollution of surface water. Factors involved include the distance from wastewater discharge traveled by the virus (30), the epidemic period and thus the initial viral concentration (16), the survival (6) and transport (42) properties of the virus, the fact that the viruses are free or attached to suspended solids (31), and the environmental conditions (temperature, flow rate, salinity, and pH, etc.) (35). As a consequence, the crucial task of assessing viral contamination of river water, a major source of drinking water, is most difficult. Currently, viral contamination can be estimated either by specifically detecting pathogenic viruses or by evaluating the level of fecal contamination using indicators. Specific detection of pathogenic viruses is not adapted to routine analysis. Cell culture, which is the reference technique for the detection of environmental viruses, is time-consuming and does not allow the detection of all viral serotypes (e.g., Norovirus). Molecular biology techniques overcome some of the disadvantages of cell culture, but protocols are not standardized and the high genetic variability of some viruses (e.g., Norovirus) can be a source of underestimation. Inversely, molecular biology techniques can yield an overestimation since no information is obtained concerning viral infectivity (7). Furthermore, no matter which technique is used, it is not realistic to search for all virus serotypes that might be present in water. For this reason, some authors have suggested Enterovirus spp. could be used as indicators of the global viral pollution (19, 26). Using this approach however, other viruses such as hepatitis viruses (hepatitis E or A virus) or gastroenteritis viruses (Norovirus and Rotavirus, etc.) are not taken into consideration despite their high prevalence (18, 23). Epidemic peaks also influence the density of pathogenic viruses in surface waters. For example, enteroviruses are mainly present in summer or fall (16, 20, 36) whereas noroviruses are principally detected in winter (27). This difference in seasonal distribution of viruses may seriously discredit the use of a single pathogenic virus as an indicator of global contamination. Several authors have shown that Enterovirus spp. are not representative of all viruses (4, 6, 28, 29, 38).
An alternative to specific search for pathogenic viruses is to use indicators of fecal contamination which are supposed to be representative of all pathogenic microorganisms of enteric origin and, among them, viruses. Bacterial indicators, and more particularly coliforms and streptococci, have been used for this purpose for a hundred years, but in some cases they are less resistant than viruses and consequently underestimate viral pollution (10, 34). The search has focused on other indicators better correlated with viral contamination. Some studies have shown that bacteriophages, viral particles similar in size and structure to pathogenic viruses, could be good indicators of viral contamination. The bacteriophages most frequently studied in this context are somatic coliphages (3, 11, 13), F-specific phages (9, 13, 39), and Bacteroides fragilis phages (13, 17, 24, 37). It is noteworthy that unlike methods specifically designed to detect pathogenic viruses, methods used to detect phages and bacteria are inexpensive, rapid, and easy to perform and thus applicable in both industrialized and developing countries. The diversity of indicators already in use or still in the research phase highlights the fact that no universal indicator has been identified. Consequently, it may be useful to determine which indicator(s) is best suited to practical conditions of application (e.g., type of water to analyze). In earlier work on a 30-km stretch of the Moselle River, we compared the behavior of three bacterial indicators of fecal pollution (thermotolerant coliforms, enterococci, and spores of sulfite-reducing anaerobes) with that of three bacteriophages (somatic coliphages, F-specific phages and B. fragilis phages) (35). The results showed that spores of sulfite-reducing anaerobes, F-specific phages and B. fragilis phages can underestimate fecal pollution in comparison with thermotolerant coliforms. Enterococus concentrations are highly correlated to thermotolerant coliform concentrations, while somatic coliphages give original information tracking fecal pollution longer and farther. This difference in the behavior of coliforms and coliphages has been confirmed in a recent study of 392 water samples coming from 10 different rivers located in four countries (Argentina, Colombia, Spain, and France) (25). Thus, in river water, somatic coliphages could be valuable indicators of fecal pollution, taking into account viral contamination. In this context, the aim of this study was to define which indicator of fecal pollution, thermotolerant coliforms or somatic coliphages, is the most appropriate for assessing viral contamination in river water. To achieve this objective, we worked on the same sample sites along the Moselle River as in our previous study (35) looking for fecal indicators (thermotolerant coliforms and somatic coliphages) and pathogenic viruses (Enterovirus and Norovirus genogroup II [GGII]). Standardized methods were used to enumerate fecal indicators. Enterovirus spp. were identified by cell culture, integrated cell culture (ICC)-reverse transcription (RT)-PCR, and RT-PCR, while Norovirus GGII spp., which are not cultivable, were identified by RT-PCR alone.
MATERIALS AND METHODS
River water samples.
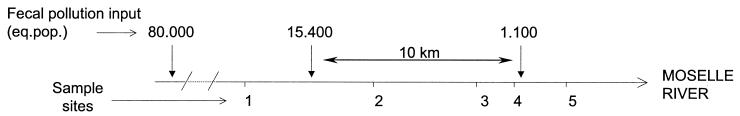
All of the river water samples were taken from the Moselle River in eastern France. This river is about 70 m wide and is open to navigation. Its flow rate can exceed 130 m3/s in winter, decreasing to less than 20 m3/s in summer. Sampling sites (Fig. 1) were selected on the basis of levels of fecal contamination. Sites 1 to 5 correspond to the following towns, respectively: Méréville, Maron, Villey le Sec, Pierre la Treiche, and Chaudenay (see also reference 35). It can be noticed that there is a stretch of water between sites 2 and 4 where there is no source of human or animal fecal contamination over a distance of more than 10 km. Thirty-four river water sampling campaigns were undertaken between February 2000 and May 2002 at each of the five sampling sites (n = 170). The physicochemical characteristics of the water are given in Table 1. Samples were taken from the middle of the river, at a depth of 1 m. The samples were maintained at 4°C. They were transported to the laboratory and analyzed within 2 h.
FIG. 1.
The five sampling sites along the Moselle River.
TABLE 1.
Physicochemical characteristics of Moselle River water samples (n = 170)
| Parameter | Flow rate (m3/s) | Temp (°C) | pH | Turbidity (NTU) | Conductivity (μS/cm) | Coliforms log CFU/100 ml | Coliphages log PFU/100 ml |
|---|---|---|---|---|---|---|---|
| Mean | 51 | 13.6 | 7.9 | 6.1 | 280 | 2.53 | 3.06 |
| SD | 35 | 6.0 | 0.3 | 4.3 | 95 | 0.78 | 0.51 |
| Minimum | 8 | 1.5 | 7.1 | 1.1 | 40 | 0.88 | 1.30 |
| Maximum | 133 | 24.0 | 9.2 | 30.1 | 464 | 3.75 | 4.14 |
Pathogenic viruses.
Ninety samples out of 170 were analyzed for pathogenic viruses. The concentration of pathogenic viruses was measured in 1 liter of river water by filtration using a protocol adapted from Gilgen et al. (8). Briefly, water was filtered through a positively charged membrane with 0.45-μm pores (CUNO NM04701 045SP). An 8-μm-porosity cellulose ester prefilter (Millipore SCWP04700) was used to limit clogging. The filter and prefilter were then eluted by immersion in 4.6 ml of sterile glycine buffer (0.4%) meat extract (1%) at pH 9.5. After stirring for 10 min at 100 rpm, the eluate was centrifuged at 1,500 × g for 10 min and the volume of the supernatant was measured with precision. The pH was adjusted to 7.2. The eluate was then divided into two equal volumes in cryotubes (2 ml/tube) and held at −80°C.
Detection of infectious Enterovirus by cell culture.
Each 2 ml of eluate was treated with 10% antibiotic and antimycotic solution (catalog no. A-5955; Sigma). Qualitative detection of infectious Enterovirus was performed by adding treated samples on monolayer BGM cells. After 2 h of contact, cells were covered with MEM solution (catalog no. M-5650; Sigma) containing 2% fetal calf serum. After 6 days of incubation at 37°C under 5% CO2 atmosphere, the flasks were examined under microscope for cytopathogenic effect, and 140 μl of triple freeze-thaw cell culture were analyzed for Enterovirus genome by ICC-RT-PCR.
RT-PCR detection of pathogenic viral genome. (i) Ultrafiltration.
Each 2 ml of eluate was introduced into an ultrafiltration tube (catalog no. UFV4BHK00; Millipore) and centrifuged 5 min at 3,100 × g. One hundred forty microliters of sample was collected for viral RNA extraction.
(ii) Viral RNA extraction.
Viral genome was extracted from 140 μl of ultrafiltrated sample with the Qiamp viral RNA kit (Qiagen, 52904) according to the manufacturer's instructions. Hence, 60 μl of final extracted RNA solution were obtained and conserved at −80°C.
(iii) Primers, probes, and standard RNA.
Primers and probes are presented in Table 2. To avoid false-negative results, 20 μl of a standard RNA solution was added during the viral RNA extraction step to each sample. This standard was a 281-nucleotide RNA fragment which was amplified using the same primers as for Enterovirus and detected using a specific probe (Std).
TABLE 2.
RT and PCR primers used in this study
| Virus | Primer | Sequencea | Position | Reference |
|---|---|---|---|---|
| Enterovirus | Ent2 | 5′-ATTGTCACCATAAGCAGCCA-3′ | 578-597 | 29 |
| Ent1 | 5′-CGGTACCTTTGTACGCCTGT-3′ | 64-83 | 29 | |
| Norovirus GGII | NVP110 | 5′-ACDATYTCATCATCACCATA-3′ | 4865-4884 | 22 |
| SR46 | 5′-TGGAATTCCATCGCCCACTGG-3′ | 4754-4773 | 1 |
Single letter code: D = G, A, or T; Y = C or T.
Standard RNA solution was added to each sample at the limit of detection concentration.
(iv) cDNA synthesis.
Five microliters of extracted sample was mixed with either 1 μl of 10 μM Enterovirus primer (Ent2) (final concentration of 0.5 μM in 20 μl of mixture) or 1 μl of 66 μM Norovirus GGII primer (NVP110) (final concentration of 3.3 μM in 20 μl of mixture) and heated at 95°C for 3 min. To this 6-μl mixture was added 14 μl of a mixture containing 4 μl of 5× RT buffer (250 mM Tris-HCl [pH = 8.4]), 50 mM MgCl2, 350 mM KCl, 15 mM dithiothreitol, 2.5 mM spermidine (catalog no. M515A; Promega), 1 μl of RNase inhibitor (40 U · μl−1; catalog no. N211A; Promega), 2 μl of 10 mM each deoxynucleoside triphosphate (catalog no. N8080260; Perkin-Elmer), 1 μl of reverse transcriptase (8 U · μl−1; catalog no. M510A; Promega), 4.6 μl of DNase- and RNase-free water (catalog no. W4502; Sigma), and 1.4 μl of T4gene32 (1 μg · μl−1; catalog no. 972991; Roche). RT was performed at 42°C for 60 min. Finally, RNA-DNA hybrids were denatured, and reverse transcriptase was inactivated by heating to 95°C for 5 min. The resulting cDNA was then amplified by PCR.
(v) PCR amplification.
PCR assay was performed with 5 μl of cDNA and 45 μl of the mixture containing 0.5 μl of each 10 μM Enterovirus primer (Ent1 and Ent2) or 0.5 μl of each Norovirus GGII primer (SR46 and NVP110, each at a concentration of 66 μM), 0.3 μl of Hotstart DNA polymerase (5 U · μl−1; catalog no. 1007837; Qiagen), 23 μl of nuclease-free water (Sigma W4502), 11 μl of a 20% glycerol solution (catalog no. 101186 M; BDH), 5 μl of 10× PCR Mix (catalog no. 1005479; Qiagen), and 5 μl of each deoxynucleoside triphosphate (each at a concentration of 10 mM; catalog no. N808-0260; Perkin-Elmer). Amplification was performed using GeneAmp 2700 (Applied Biosystems). The amplification included (i) 15 min at 95°C to release the activity of the Hotstart DNA polymerase; (ii) 45 cycles of 30 s at 94°C, 30 s at 50°C, and 30 s at 72°C; and (iii) 10 min at 72°C. Amplified DNA was kept at 4°C until the DNA enzyme immunoassay (DEIA) detection which was performed the same day. RT and PCR mixture with 5 μl of phosphate-buffered saline buffer instead of sample were used as negative control.
(vi) DEIA detection.
Amplified DNA samples were analyzed for the presence of viral genome (Enterovirus and Norovirus GGII) using the DEIA kit (DiaSorin PS0001) according to the manufacturer's instructions. The different probes are described in Table 3 and were used at a concentration of 100 pg/μl.
TABLE 3.
Probes for viral genome detection (DEIA)
| Virus | Probe | Sequence | Position | Reference |
|---|---|---|---|---|
| Enterovirus | Ent | 5′-TCCTCCGGCCCCTGAATGCG(biotin)-3′ | 445-464 | 43 |
| Norovirus GGII | SR47 | 5′-ATGTCAGGGGACAGGTTTGT(biotin)-3′ | 1804-4823 | 1 |
| SR61 | 5′-ATGTCGGGGCCTAGTCCTGT(biotin)-3′ | 1804-4823 | 1 | |
| SR67 | 5′-ACATCTGGTGAGAGACCTGA(biotin)-3′ | 1804-4823 | 1 | |
| Standard | Std | 5′-GACGGCCAGGTCCGGATG(biotin)-3′ |
Indicators.
Thermotolerant coliforms were quantified on m-FC medium after membrane filtration according to standard methods (15). Somatic coliphages were quantified using the bacterial host strain Escherichia coli (WG5) according to standard methods (14).
RESULTS
Influence of temperature on coliform and coliphage concentrations in river water.
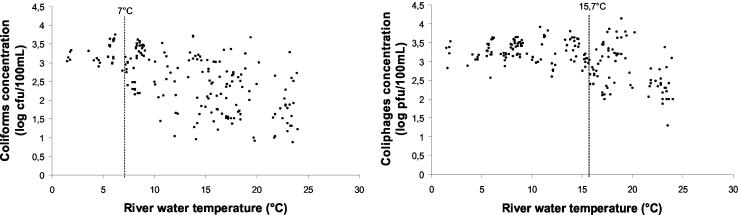
Among the 170 river water samples analyzed between February 2000 and May 2002, all were positive for both thermotolerant coliforms and somatic coliphages. The concentration-water temperature relationships are shown in Fig. 2 (river water temperatures varied from 1.5 to 24.0°C). Concentrations of thermotolerant coliforms were relatively homogeneous below 7°C (mean value: 3.29 ± 0.26 log CFU/100 ml; n = 25) but decreased and became highly heterogeneous at higher temperature (2.40 ± 0.77 log CFU/100 ml; n = 145). For somatic coliphages, the same phenomenon was observed, but at a higher temperature: below 15.7°C, concentrations were relatively homogeneous (3.29 ± 0.28 log PFU/100 ml; n = 101), whereas above 15.7°C, concentrations decreased and became heterogeneous (2.73 ± 0.59 log PFU/100 ml; n = 69). Those variations above 7°C for thermotolerant coliforms and above 15.7°C for somatic coliphages were mainly due to a significant decrease between sites 2 and 5 (Student's test, P < 0.05) (data not shown). Indeed, for each sampling date, the highest value of concentrations for both coliforms and coliphages always corresponded to the most polluted sample site (site 2) located 3 km upstream from an urban wastewater discharge. Thermotolerant coliform concentrations were stable for temperatures under 7°C and started decreasing between samples sites 2 and 5 for higher temperatures with a 0.66-log reduction for temperatures between 7 and 15.7°C and a >1.31-log reduction for temperatures over 15.7°C. Regarding somatic coliphages, concentrations were stable for temperature less than 15.7°C and started decreasing when temperature rose above that value with a 0.62-log reduction between samples sites 2 and 5. Results show clearly that thermotolerant coliforms were more sensitive than coliphages to the temperature effect. The two indicators thus described different patterns of fecal pollution.
FIG. 2.
Coliform and coliphage concentrations in river water as a function of water temperature (n = 170).
Presence of pathogenic viruses in river water.
Ninety samples out of 170 were analyzed for pathogenic viruses between December 2000 and May 2002. We looked for Enterovirus and Norovirus (GGII) contamination. Enterovirus spp. were identified using cell culture, ICC-RT-PCR, and RT-PCR while Norovirus GGII spp., which are not cultivable, were identified with only RT-PCR. Cell culture and ICC-RT-PCR failed to isolate infectious Enterovirus spp. from any of the 90 river water samples. Nevertheless, using RT-PCR-DEIA which is a more sensitive method, 34 samples out of 90 (38%) were positive for Enterovirus (13%) and/or Norovirus GGII (27%) genome. These results are detailed as a function of water temperature range in Table 4. Norovirus GGII genome was largely detected when temperature was below 7°C with 93% of the samples being positive while the proportion of positive samples fell to 7% for temperature over 15.7°C. This observation can be explained by the winter prevalence of Norovirus. Conversely, Enterovirus genome was detected less often for temperature under 7°C whereas 16% of the above 15.7°C samples were positive. This observation can also be explained by the summer-autumn prevalence of Enterovirus. Our results show that the viral contamination of river water was mainly represented by Norovirus GGII in winter and mainly by Enterovirus in summer. When considering the sampling sites as well as the temperature (Table 5), viral contamination expressed as percentage of samples positive for viral genome did not vary with sampling site irrespective of the temperature range.
TABLE 4.
Enterovirus and Norovirus GGII genome-positive samples as a function of river water temperature
| Temp (°C) | n | No. (%) of samples positive for:
|
|
|---|---|---|---|
| Enterovirus | Norovirus GGII | ||
| <7 | 15 | 1 (7) | 14 (93) |
| ≥7-≤15.7 | 30 | 4 (13) | 7 (23) |
| >15.7 | 45 | 7 (16) | 3 (7) |
TABLE 5.
Number of samples positive for pathogenic viral genome in river water as a function of sampling site and river water temperature range
| Site no. | n | No. of samples positive in water temp range
|
Total no. of positive samples (n = 90) | ||
|---|---|---|---|---|---|
| <7°C (n = 15) | ≥7°C-≤15.7°C (n = 30) | >15.7°C (n = 45) | |||
| 1 | 18 | 3 | 3 | 2 | 8 |
| 2 | 18 | 3 | 2 | 2 | 7 |
| 3 | 18 | 3 | 2 | 2 | 7 |
| 4 | 18 | 3 | 2 | 2 | 7 |
| 5 | 18 | 2 | 1 | 2 | 5 |
| Total | 90 | 14 | 10 | 10 | 34 |
Indicators and pathogenic viruses.
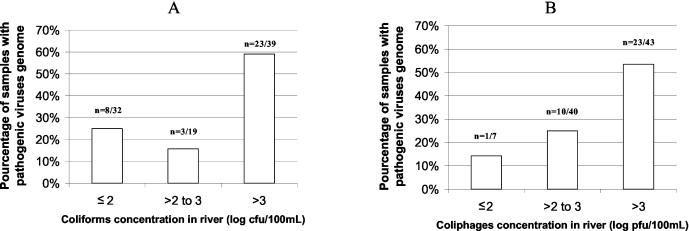
Among the 90 river water samples analyzed for pathogenic viruses, no infectious enteroviruses were detected by either cell culture or ICC-RT-PCR. While the absence of genome can be interpreted as an absence of virus, the presence of genome cannot be construed as a guarantee of the presence of infectious particles. Nevertheless, as we did not detect any infectious enterovirus, we compared the results for thermotolerant coliforms and somatic coliphages to those of Enterovirus and Norovirus GGII genome and thus classed the concentrations of both indicators into three categories (100 or less, over 100 to 1,000, and more than 1,000 PFU or CFU/100 ml). Results given in Fig. 3 clearly show that coliforms were not linked to the viral contamination represented by the presence of virus genome. Indeed, when the level of coliforms fell below 102 CFU/100 ml, 8 samples out of 32 (25%) contained viral genome (6 samples were positive for Enterovirus and 2 were positive for Norovirus GGII). Conversely, the number of samples positive for viral genome decreased with decreasing concentration of coliphages. Nevertheless, it must be underlined that only 7 samples out of 90 contained 102 PFU/100 ml or less, and one of them was positive for Enterovirus genome.
FIG. 3.
Number of samples containing pathogenic viruses genome as a function of coliform (A) or coliphage (B) concentration.
DISCUSSION
To determine which indicator of fecal pollution, thermotolerant coliforms or somatic coliphages, is the most appropriate for assessment of viral contamination in river water, we propose to analyze our results in three distinct parts.
First, following up on earlier work (35), we compared indicators of fecal contamination. Our earlier results showed that thermotolerant coliforms and somatic coliphages were the most representative indicators of the fecal pollution along the river. Here, we examined their respective behavior over a longer period of time and a wider range of temperature. Our present results confirm that between sites 2 and 5 concentrations of thermotolerant coliforms decline faster than those of somatic coliphages. Tracking fecal pollution of the river over a longer distance with somatic coliphages thus corroborated the difference in fecal contamination estimated from bacteria and bacteriophages (25). This self-clearance capacity of the river could be related to water temperature, in agreement with our previous results.
Secondly, cell culture and ICC-RT-PCR detection of pathogenic viruses did not reveal any infectious Enterovirus spp. This could be explained either by the distance (in time and space) between the sample sites and the potential sources of viable viruses and/or by the small volume analyzed (equivalent to 500 ml). Nevertheless, considering the molecular biology results, 34 samples out of 90 (38%) were positive for Norovirus GGII (27%) and/or Enterovirus (13%) genome. These positive results could be explained by the fact that RT-PCR is known to be more sensitive than cell culture (19, 29, 41), possibly because RT-PCR allows detecting genome from both infectious and noninfectious particles, which increases the number of targets for the detection technique. Moreover, our observation is consistent with previous results showing the presence in surface water of Enterovirus genome (2, 5, 8, 33) or Norovirus genome (12, 32, 40). Looking at the qualitative results, overall viral pollution of the river appeared to remain constant at the different sampling sites, while individual viral distributions exhibited seasonal patterns. These results are in agreement with previous studies showing that Enterovirus spp. are mostly detected in summer (16, 20, 36), whereas noroviruses are mainly isolated in winter (27). As previously noted (16, 21), this observation argues against proposing the use of a single pathogenic virus as an indicator of global viral contamination in river water.
Finally, we compared thermotolerant coliform and somatic coliphage concentrations for each sample positive for Enterovirus or Norovirus GGII genome. Our results show that the number of virus genome-positive samples decreased with decreasing concentration of coliphages, while no such relation was observed for thermotolerant coliforms. In fact, concentrations of thermotolerant coliforms fell off between sites 2 and 5 when temperature increased (temperature > 7°C), whereas the number of samples positive for pathogenic viral genome did not vary in the same way. Inversely, somatic coliphages appear to be interesting because of the following observations.
(i) A relation seems to exist between the concentrations of these indicators and viral contamination of river water in terms of pathogenic viral genome. This apparent relation is based on qualitative results and would have to be confirmed by quantitative molecular biology. Nevertheless, this is in agreement with another study (2) which concludes that bacteriophages (somatic coliphages and F-specific phages) but not fecal indicators are correlated with different enteric viral genomes in river water downstream from a wastewater discharge.
(ii) The number of samples positive for viral genome, and as a result for infectious particles, was low when the concentrations of somatic coliphages remained under the value of 100 PFU/100 ml. This threshold might be very useful for forecasting virological contamination in river water, but further confirmation would be necessary since this condition was only found for seven samples in our study.
In conclusion, somatic coliphages appear to be a promising tool for river water quality assessment in terms of fecal pollution that takes into account the virological parameter.
Acknowledgments
This work was funded by an E.U. International Cooperation with Developing Countries (INCO-DC) contract (number ERB 3514 PL 972471) and the Conseil Régional de Lorraine (ZAbM [Zone Atelier du bassin de la Moselle]).
REFERENCES
- 1.Ando, T., Q. Jin, J. R. Gentsch, S. S. Monroe, J. S. Noel, S. F. Dowell, H. G. Cicirello, M. A. Kohn, and R. I. Glass. 1995. Epidemiologic applications of novel molecular methods to detect and differentiate small round structured viruses (Norwalk-like viruses). J. Med. Virol. 47:145-152. [DOI] [PubMed] [Google Scholar]
- 2.Baggi, F., A. Demarta, and R. Peduzzi. 2001. Persistence of viral pathogens and bacteriophages during sewage treatment: lack of correlation with indicator bacteria. Res. Microbiol. 152:743-751. [DOI] [PubMed] [Google Scholar]
- 3.Borrego, J. J., M. A. Morinigo, A. De Vicente, R. Cornax, and P. Romero. 1987. Coliphages as an indicator of faecal pollution in water. Its relationship with indicator and pathogenic microorganisms. Water Res. 21:1473-1480. [Google Scholar]
- 4.Castignolles, N., F. Petit, I. Mendel, L. Simon, L. Cattolico, and C. Buffet-Janvresse. 1998. Detection of Adenovirus in the waters of the Seine River estuary by nested-PCR. Mol. Cell. Probes 12:175-180. [DOI] [PubMed] [Google Scholar]
- 5.Cho, H. B., S. H. Lee, J. C. Cho, and S. J. Kim. 2000. Detection of adenoviruses and Enteroviruses in tap water and river water by reverse transcription multiplex PCR. Can. J. Microbiol. 46:417-424. [PubMed] [Google Scholar]
- 6.Enriquez, C. E., C. J. Husrt, and C. P. Gerba. 1995. Survival of the enteric Adenoviruses 40 and 41 in tap, sea, and wastewater. Water Res. 29:2548-2553. [Google Scholar]
- 7.Gassilloud, B., L. Schwartzbrod, and C. Gantzer. 2003. Presence of viral genomes in mineral water: a sufficient condition to assume infectious risk? Appl. Environ. Microbiol. 69:3965-3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gilgen, M., D. Germann, J. Lüthy, and P. Hübner. 1997. Three-step isolation method for sensitive detection of Enterovirus, rotavirus, hepatitis A virus, and small round structured viruses in water samples. Int. J. Food Microbiol. 37:189-199. [DOI] [PubMed] [Google Scholar]
- 9.Havelaar, A. H., and W. H. Hogeboom. 1984. A method for the enumeration of male specific bacteriophages in sewage polluted waters. J. Appl. Bacteriol. 56:439-447. [DOI] [PubMed] [Google Scholar]
- 10.Havelaar, A. H., M. Van Olphen, and Y. C. Drost. 1993. F-specific RNA bacteriophages are adequate model organism for enteric viruses in freshwater. Appl. Environ. Microbiol. 59:2956-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hilton, M. C., and C. Stotzky. 1973. Use of coliphages as indicators of water pollution. Can. J. Microbiol. 19:747-751. [DOI] [PubMed] [Google Scholar]
- 12.Huang, P. W., D. Laborde, V. R. Land, D. O. Matson, A. W. Smith, and X. Jiang. 2000. Concentration and detection of caliciviruses in water samples by reverse transcription-PCR. Appl. Environ. Microbiol. 66:4383-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.IAWPRC Study Group on Health Related Water Microbiology. 1991. Bacteriophages as model of viruses in water quality control. Water Res. 25:529-545. [Google Scholar]
- 14.International Organization for Standardization. 1999. Water quality—detection and enumeration of bacteriophages. Part 2: enumeration of somatic coliphages. Document ISO/FDIS 10705-2. International Organization for Standardization, Geneva, Switzerland.
- 15.International Organization for Standardization. 1990. Document ISO 9308/1. Water quality—detection and enumeration of coliform organisms, thermotolerant coliform organisms and presumptive Escherichia coli. International Organization for Standardization, Geneva, Switzerland.
- 16.Irving, L. G., and F. A. Smith. 1981. One-year survey of enteroviruses, adenoviruses and reoviruses isolated from effluent at an activated-sludge purification plant. Appl. Environ. Microbiol. 41:51-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jofre, J., A. Bosch, F. Lucena, R. Girones, and C. Tartera. 1986. Evaluation of Bacteroides fragilis bacteriophages as indicators of the virological quality of water. Water Sci. Tech. 18:167-173. [Google Scholar]
- 18.Koopmans, M., C. H. von Bonsdorff, J. Vinjé, D. de Medici, and S. Monroe. 2002. Foodborne viruses. FEMS Microbiol. Rev. 26:187-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kopecka, H., S. Dubrou, J. Prevot, J. Marechal, and J. M. Lopez-Pila. 1993. Detection of naturally occurring enteroviruses in waters by reverse transcription, polymerase chain reaction, and hybridization. Appl. Environ. Microbiol. 59:1213-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krikelis, V., N. Spyrou, P. Markoulatos, and C. Serie. 1984. Seasonal distribution of Enteroviruses and adenoviruses in domestic sewage. Can. J. Microbiol. 31:24-25. [DOI] [PubMed] [Google Scholar]
- 21.Lee, S. H., and S. J. Kim. 2002. Detection of infectious Enteroviruses and Adenoviruses in tap water in urban areas in Korea. Water Res. 36:248-256. [DOI] [PubMed] [Google Scholar]
- 22.Le Guyader, F., F. H. Neill, M. K. Estes, S. S. Monroe, T. Ando, and R. L. Atmar. 1996. Detection and analysis of a small round-structured virus strain in oysters implicated in an outbreak of acute gastroenteritis. Appl. Environ. Microbiol. 62:4268-4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lopman, B. A., D. W. Brown, and M. Koopmans. 2002. Human caliciviruses in Europe. J. Clin. Virol. 24:137-160. [DOI] [PubMed] [Google Scholar]
- 24.Lucena, F., R. Araujo, and J. Jofre. 1996. Usefulness of bacteriophages infecting Bacteroides fragilis as index microorganisms of remote faecal pollution. Water Res. 30:2812-2816. [Google Scholar]
- 25.Lucena, F., X. Méndez, A. Morón, E. Calderón, C. Campos, A. Guerrero, M. Cárdenas, C. Gantzer, L. Schwartzbrod, S. Skraber, and J. Jofre. 2003. Occurrence and densities of bacteriophages proposed as indicators and bacterial indicators in river waters from Europe and South America. J. Appl. Microbiol. 94:808-815. [DOI] [PubMed] [Google Scholar]
- 26.Metcalf, T. G., J. L. Melnick, and M. K. Estes. 1995. Environmental virology: from detection of virus in sewage and water by isolation to identification by molecular biology—a trip of over 50 years. Annu. Rev. Microbiol. 49:461-487. [DOI] [PubMed] [Google Scholar]
- 27.Mounts, A. W., T. Ando, M. Koopmans, J. S. Bresee, J. Noel, and R. I. Glass. 2000. Cold weather seasonality of gastroenteritis associated with Norwalk-like viruses. J. Infect. Dis. 181:284-287. [DOI] [PubMed] [Google Scholar]
- 28.Pina, S., M. Puig, F. Lucena, J. Jofre, and R. Girones. 1998. Viral pollution in the environment and in shellfish: human Adenovirus detection by PCR as an index of human viruses. Appl. Environ. Microbiol. 64:3376-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Puig, M., J. Jofre, F. Lucena, A. Allard, G. Wadell, and R. Girones. 1994. Detection of Adenoviruses and Enteroviruses in polluted waters by nested PCR amplification. Appl. Environ. Microbiol. 60:2963-2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rajala, R. L., and H. Tanski. 1998. Survival and transfer of faecal indicator organisms of wastewater effluents in receiving lake waters. Water Sci. Technol. 38:191-194. [Google Scholar]
- 31.Schernewski, G., and W. D. Jülich. 2001. Risk assessment of virus infections in the Oder estuary (southern Baltic) on the basis of spatial transport and virus decay simulations. Int. J. Hyg. Environ. Health 203:317-325. [DOI] [PubMed] [Google Scholar]
- 32.Schvoerer, E., F. Bonnet, V. Dubois, G. Cazaux, R. Serceau, H. J. Fleury, and M. E. Lafon. 2000. PCR detection of human enteric viruses in bathing areas, waste waters and human stools in Southwestern France. Res. Microbiol. 151:693-701. [DOI] [PubMed] [Google Scholar]
- 33.Schvoerer, E., M. Ventura, O. Dubos, G. Cazaux, R. Serceau, N. Gournier, V. Dubois, P. Caminade, H. J. A. Fleury, and M. E. Lafon. 2001. Qualitative and quantitative molecular detection of enteroviruses in water from bathing areas and from a sewage treatment plant. Res. Microbiol. 152:179-186. [DOI] [PubMed] [Google Scholar]
- 34.Schwartzbrod, L., C. Finance, M. Aymard, M. Brigand, and F. Lucena. 1985. Recovery of reovirus from tap water. Zentbl. Bakteriol. Mikrobiol. Hyg. 181:383-389. [PubMed] [Google Scholar]
- 35.Skraber, S., C. Gantzer, A. Maul, and L. Schwartzbrod. 2002. Fates of bacteriophages and bacterial indicators in the Moselle river (France). Water Res. 36:3629-3637. [DOI] [PubMed] [Google Scholar]
- 36.Tani, N., Y. Dohi, N. Kurumatani, and K. Yonemasu. 1995. Seasonal distribution of adenoviruses, enteroviruses and reoviruses in urban river water. Microbiol. Immunol. 39:577-580. [DOI] [PubMed] [Google Scholar]
- 37.Tartera, C., and J. Jofre. 1987. Bacteriophages active against Bacteroides fragilis in sewage-polluted waters. Appl. Environ. Microbiol. 53:1632-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vantarakis, A. C., and M. Papapetropoulou. 1998. Detection of enteroviruses and adenoviruses in coastal waters of SW Greece by nested polymerase chain reaction. Water Res. 32:2365-2372. [Google Scholar]
- 39.Woody, M. A., and D. O. Cliver. 1995. Effects of temperature and host cell growth phase on replication of F-specific RNA coliphage QBeta. Appl. Environ. Microbiol. 61:1520-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wyn-Jones, A. P., R. Pallin, C. Dedoussis, J. Shore, and J. Sellwood. 2000. The detection of small round-structured viruses in water and environmental materials. J. Virol. Methods 87:99-107. [DOI] [PubMed] [Google Scholar]
- 41.Young, P. P., R. S. Buller, and G. A. Storch. 2000. Evaluation of a commercial DNA enzyme immunoassay for detection of enterovirus reverse transcription-PCR products amplified from cerebrospinal fluid specimens. J. Clin. Microbiol. 38:4260-4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhuang, J., and J. Yan. 2003. Virus retention and transport as influenced by different forms of soil organic matter. J. Environ. Qual. 32:816-823. [DOI] [PubMed] [Google Scholar]
- 43.Zoll, G. J., W. J. Melchers, H. Kopecka, G. Jambroes, H. J. van der Poel, and J. M. Galama. 1992. General primer-mediated polymerase chain reaction for detection of enteroviruses: application for diagnostic routine and persistent infections. J. Clin. Microbiol. 30:160-165. [DOI] [PMC free article] [PubMed] [Google Scholar]