Abstract
Genome organization, plasmid content and localization of the pufLM genes of the photosynthesis reaction center were studied by pulsed-field gel electrophoresis (PFGE) in marine phototrophic Alphaproteobacteria. Both anaerobic phototrophs (Rhodobacter veldkampii and Rhodobacter sphaeroides) and strictly aerobic anoxygenic phototrophs from the Roseobacter-Sulfitobacter-Silicibacter clade (Roseivivax halodurans, Roseobacter litoralis, Staleya guttiformis, Roseovarius tolerans, and five new strains isolated from dinoflagellate cultures) were investigated. The complete genome size was estimated for R. litoralis DSM6996T to be 4,704 kb, including three linear plasmids. All strains contained extrachromosomal elements of various conformations (linear or circular) and lengths (between 4.35 and 368 kb). In strain DFL-12, a member of a putative new genus isolated from a culture of the toxic dinoflagellate Prorocentrum lima, seven linear plasmids were found, together comprising 860 kb of genetic information. Hybridization with probes against the pufLM genes of the photosynthesis gene cluster after Southern transfer of the genomic DNAs showed these genes to be located on a linear plasmid of 91 kb in R. litoralis and on a linear plasmid of 120 kb in S. guttiformis, theoretically allowing their horizontal transfer. In all other strains, the pufLM genes were detected on the bacterial chromosome. The large number and significant size of the linear plasmids found especially in isolates from dinoflagellates might account for the metabolic versatility and presumed symbiotic association with eukaryotic hosts in these bacteria.
Anoxygenic bacterial photosynthesis is an ancient process forming the evolutionary basis for all current types of photosynthesis (39, 54). It is thought to have been present in the ancestors of the present day Proteobacteria and was later lost in various lineages (53). In 1991, strictly aerobic marine bacteria containing bacteriochlorophyll a were described for the first time (43). They were termed aerobic anoxygenic phototrophs and later found in many marine, freshwater, and terrestrial habitats (55). In contrast to anaerobic phototrophs, they are unable to use light as sole source of energy, but they grow heterotrophically with light as an additional energy source. Such bacteria are much more diverse in the oceans than previously thought (4). It has been estimated that aerobic bacterial photosynthesis might be responsible for up to 5% of photosynthetic electron transport in the upper layers of the tropical oceans (20). Although the precise amount of energy generated by aerobic anoxygenic photosynthesis is still under discussion (9), it seems that in fact in the oligotrophic ocean phototrophic and heterotrophic modes of growth are being coupled in various ways (4, 19).
Analysis of whole-genome data from the five major photosynthetic prokaryotic phyla has shown that gene transfer of parts of the photosynthetic apparatus must have been a major driving force in their evolution (39). The same is assumed for the more recent evolution of aerobic anoxygenic phototrophs (31, 32). Phylogenetically, they belong into the alpha or beta subclass of the Proteobacteria (4, 55). Their photosynthesis genes are clustered into a superoperon comprising approximately 45 kb (15). Detailed analyses of the operon structure in Rubrivivax gelatinosus, a betaproteobacterium, suggested that it had been acquired by horizontal gene transfer from an alphaproteobacterium and later rearranged (15). Phylogenetic trees which were constructed from conserved photosynthesis genes, e.g., pufL or pufM genes, differed from 16S rRNA-based trees in that photosynthesis genes of betaproteobacteria were clustering among those of alphaproteobacteria (2, 31, 32), again providing indirect evidence for gene transfer. Nothing is known about actual mechanisms of gene transfer in phototrophic alphaproteobacteria. However, in 1981 a highly promiscuous plasmid related to RP1 was isolated in Rhodopseudomonas capsulata and shown to mobilize chromosomal markers, including the photosynthesis gene cluster, with high frequency (28).
At present, little is known about the genome organization of aerobic anoxygenic marine phototrophs, many of which have been isolated and described only recently (2, 22, 23). Here, we determined the genome size of Roseobacter litoralis DSM6996T as a representative of the ecologically important Roseobacter clade (42). In addition, plasmid content and pufLM gene localization in a relatively broad phylogenetic range of phototrophic Alphaproteobacteria were investigated to gain a better understanding of the structural flexibility of the genome and the potential for gene transfer in these organisms. The large number of both linear and circular plasmids which were discovered, which comprised in some instances a significant part of the total genome, indeed revealed a structural basis which would allow for the horizontal transfer of large pieces of genetic information. It might also be the reason for the observed physiological versatility of this group.
MATERIALS AND METHODS
Bacterial strains, media, and culture conditions.
Roseivivax halodurans DSM15395T (49), Rhodobacter veldkampii DSM11550T (10), Rhodobacter sphaeroides DSM159T (16), R. litoralis DSM6996T (43), Roseobacter denitrificans DSM7001T (43), Staleya guttiformis DSM11458T (22) and Roseovarius tolerans DSM11457T (23) were obtained from the German Collection of Microorganisms and Cell Cultures GmbH, Braunschweig, Germany. Strains designated DFL were isolated from dinoflagellate cultures of Alexandrium ostenfeldii, Alexandrium lusitanicum and Prorocentrum lima (2). Cells of R. halodurans DSM15395T and R. sphaeroides DSM159T were grown as described in reference 49 in liquid DSMZ medium no. 27 (www.dsmz.de). All other strains used in this study were cultured in marine broth 2216 (Difco).
Phylogenetic inferences.
The 16S rRNA gene sequences of strains DFL-13, DFL-43, DFL-42, DFL-44, DFL-33, and DFL-11 were determined as described in reference 2 and submitted to the EMBL database (see below). Sequences were analyzed with the ARB software package (27; www.arb-home.de). Almost complete 16S rRNA gene sequences were imported into the ARB database of ∼12,600 aligned reference sequences. Sequences were automatically aligned, and alignments were corrected manually. Sequences were integrated into the ARB phylogenetic tree by using the maximum-parsimony algorithm tool. This tool does not correct for evolutionary distances and does not change the tree topology. It served as a basis to identify the reference organisms. These were subsequently selected for tree calculation by means of the maximum-likelihood algorithm with the program fastDNAml.
Preparation and digestion of DNA.
Preparation of high-molecular-weight (HMW) total genomic DNA of the strains within agarose plugs followed the protocol of Bautsch (3). For each strain, various cell densities were embedded into agarose to yield optimal DNA concentrations for pulsed-field gel electrophoresis (PFGE) analysis. Plasmid DNA from the strains was extracted by a conventional plasmid isolation procedure. Cells were lysed with alkali, and plasmids were purified as indicated by the manufacturer of the Nucleobond PC 100 kit (Machery-Nagel, Düren, Germany). For PCR, the genomic DNA of R. litoralis DSM6996T, R. veldkampii DSM11550T, and strain DFL-12 was extracted from agarose plugs by using the Qiaex II gel extraction kit as described by the manufacturer (Qiagen, Hilden, Germany). To determine the chromosome size of R. litoralis DSM6996T, HMW genomic DNA of this strain inside agarose plugs was digested with 20 U of the restriction endonucleases PacI, PmeI (New England Biolabs, Beverly, Mass.), and SwaI (Roche Applied Science, Mannheim, Germany) and 5 U of the intron-encoded endonuclease I-CeuI (New England Biolabs) according to methods described by Pradella et al. (34).
Gel electrophoresis.
PFGE of the genomic DNA of the strains was performed in a contour-clamped homogeneous electric field (CHEF) system on a CHEF-DR III device (Bio-Rad Laboratories, Hercules, Calif.) with 0.6 or 1% agarose gels and modified 0.5× TBE buffer (45 mM Tris, 45 mM boric acid, 0.1 mM EDTA) at 14°C. To analyze the undigested HMW genomic DNA of the strains, PFGE parameters, namely pulse time ramps and run times, were varied both to resolve chromosomal and extrachromosomal DNA and to identify different plasmid conformations (12, 29, 45). At least two PFGE parameter sets were applied per strain to determine plasmid topology. PFGE parameter sets used with 1% agarose gels at 200 V (6 V/cm) were as follows: (i) set A, pulse times of 1 to 25 s for 22.5 h; (ii) set B, pulse times of 1 to 20 s for 22 h; (iii) set C, pulse times of 1 to 40 s for 21 h; (iv) set D, pulse times of 1 to 90 s for 24 h followed by pulse times of 1 to 10 s for 8 h. To calculate the chromosome size of R. litoralis DSM6996T, PFGE parameters in several gel runs were adapted to maximize the resolution of all generated macrorestriction fragments (8, 46). The PFGE conditions applied to documented experiments were as follows: (i) 0.6% (wt/vol) agarose gel with pulse times of 350 to 4,800 s for 120 h at 50 V (1.5 V/cm) (high-size range); (ii) 1% (wt/vol) agarose gel with pulse times of 1 to 90 s for 22.5 h at 200 V (6 V/cm) (medium-size range); (iii) 1% (wt/vol) agarose gel with pulse times of 1 to 22 s for 22 h at 200 V (6 V/cm) (low-size range). Conventional, unidirectional gel electrophoresis of DNA was done as described by Sambrook et al. (41) in 0.8% agarose gels and 1× TBE (89 mM Tris, 89 mM boric acid, 2 mM EDTA) at 10°C and 70 mA for 8.5 h. DNA banding patterns were stained in SYBR Green I solution as recommended by the manufacturer (Molecular Probes, Eugene, Oreg.) or with ethidium bromide (0.5 mg/ml), transilluminated with UV (λ = 254 nm), and photographed on Polaroid 667 films. Chromosomes of Schizosaccharomyces pombe 927 h, Hansenula wingei YB-4662-VIA (both Bio-Rad Laboratories), and Saccharomyces cerevisiae YPH80, concatemers of the bacteriophage lambda, and a low-molecular-weight PFGE marker (all New England Biolabs) served as standards for size calibration of linear DNA molecules. The supercoiled DNA ladder (from 2.067 to 16.210 kb) and the BAC Tracker supercoiled DNA ladder (from 38 to 120 kb; Epicentre, Madison, Wis.) were used to size plasmids with covalently closed circular (CCC) DNA topology.
Preparation and labeling of probes.
PCR-amplified pufLM genes from the bacterial photosynthesis reaction center (2) and 16S rRNA genes served as probes in this study. The PCR products were directly labeled with digoxigenin (DIG)-11-dUTP during PCR with the PCR DIG labeling mix (Roche Applied Science). The primer pair pufLF (5′-CTK TTC GAC TTC TGG GTS GG-3′) and pufMR (5′-CCA TSG TCC AGC GCC AGA A-3′) was used for the amplification of the pufLM genes (∼1.5 kb) from the genomic DNAs of R. litoralis DSM6996T, R. veldkampii DSM11550T, and strain DFL-12. The reaction mixture contained 1× Taq PCR buffer (10 mM Tris-HCl, 50 mM KCl, 1.5 mM MgCl2 [pH 8.3]), 200 μM (each) dATP, dCTP, and dGTP, 190 μM dTTP, 10 μM DIG-11-dUTP, 0.5 μM concentrations of each primer, 2.5 μl of template DNA, and 2 U of Taq DNA polymerase (Roche Applied Science) in a total volume of 50 μl. Hot-start PCR was performed in a PE 480 thermocycler (Perkin-Elmer, Boston, Mass.) with an initial denaturation step at 98°C (5 min), followed by 35 cycles of denaturation at 95°C (1 min), annealing at 56, 58, or 60°C (1 min) for genomic DNA of R. litoralis DSM6996T, R. veldkampii DSM11550T, and strain DFL-12, respectively, and extension at 72°C (2 min). A final extension at 72°C (10 min) and subsequent cooling at 4°C completed the reaction. Probes were designated pufRl, pufRv, and pufD12 for R. litoralis DSM6996T, R. veldkampii DSM11550T, and strain DFL-12, respectively. The primers 27f (5′-GAGTTTGATCCTGGCTCAG-3′) and 1525r (5′-AGAAAGGAGGTGATCCAGCC-3′) (24) were used to amplify nearly the complete 16S rRNA gene (∼1.5 kb) of R. litoralis DSM6996T. The reaction mixture was as described above, except that 1 μl of template DNA was used. Hot-start PCR was performed with an initial denaturation step at 94°C (3 min), followed by 32 cycles of denaturation at 93°C (1 min), annealing at 52°C (1 min), and extension at 72°C (2 min). Final extension and completion of the PCR program were as described above. The amplified 16S rDNA fragment was excised from a conventional 1% agarose gel under UV and extracted with the Nucleo Spin extract kit (Machery-Nagel). Bacteriophage lambda DNA was DIG labeled by using the DIG High-Prime DNA labeling kit (Roche Applied Science) to visualize the blotted low-range PFGE marker after Southern hybridization.
Southern blotting and hybridization.
For hybridization experiments, SYBR Green I-stained, UV-irradiated (2.5 min; λ = 254 nm), and alkaline-treated DNA was transferred from PFGE gels onto positively charged nylon membranes (Roche Applied Science) by using the VacuGene vacuum blotter (Amersham Biosciences, Piscataway, N.J.) as described previously (CHEF-DRr III PFGE system instruction manual and application guide, BioRad Laboratories). Nonradioactive hybridization and subsequent chemiluminescent signal detection with the DIG system were done according to the protocol provided in the DIG hybridization manual (DIG application manual for filter hybridization, Roche Applied Science). Prehybridization and hybridization were performed in roller tubes with 10 ml of DIG Easy Hyb buffer (Roche Applied Science) under stringent conditions at 42°C. Hybridization buffers contained 100 to 150 μl of the pufRl, pufRv, pufD12, or 16S rDNA probe, respectively, and 0.5 μl of DIG-labeled bacteriophage lambda DNA. Stringent washes were performed at 68°C with 0.5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) containing wash buffer. CDP-Star (Roche Applied Science) was used as a substrate for chemiluminescent signal detection.
Nucleotide sequence accession number.
The 16S rRNA gene sequences of strains DFL-13, DFL-43, DFL-42, DFL-44, DFL-33, and DFL-11 were submitted to the EMBL database and given accession numbers AJ582088 and AJ582083 to AJ582087.
RESULTS
Phylogenetic positions of investigated strains.
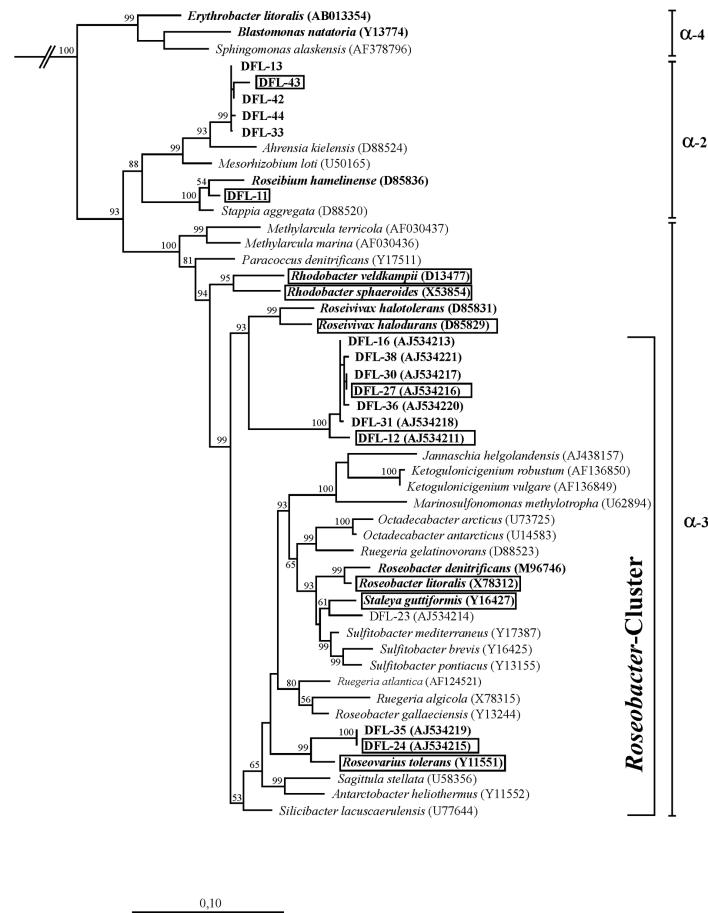
The 11 strains investigated here (Fig. 1) belonged into the α-2 and α-3 subgroups of the Alphaproteobacteria. They included two species of the facultative anaerobe Rhodobacter, namely R. veldkampii DSM11550T and R. sphaeroides DSM159T, of which the latter served as a reference for plasmid content. All other strains were obligate aerobic marine bacteria. We focused on the bacteriochlorophyll a containing type strains within the so-called Roseobacter clade. This is a heterogeneous group of organisms, representatives of which have been shown to have a high abundance in marine habitats (42, 52). The strains studied were R. halodurans DSM15395T, R. litoralis DSM6996T, S. guttiformis DSM11458T, and R. tolerans DSM11457T. In addition, five strains were included which have recently been isolated from dinoflagellate cultures and were shown to contain the pufLM genes of the photosynthesis reaction center and to produce bacteriochlorophyll a (2). DFL-43 and DFL-12 were isolated from cultures of P. lima, DFL-27 and DFL-24 were from A. ostenfeldii, and DFL-11 was from A. lusitanicum. These strains represent new genera or species. Their phylogenetic affiliation is shown in Fig. 1. DFL-43 is one representative from a cluster of strains related to Ahrensia kielensis. DFL-11 is affiliated to Roseibium hamelinense. DFL-27 and DFL-12 are representatives from a relatively deeply branching cluster of strains remotely related to Roseivivax. Based on 16S rRNA gene similarity, strain DFL-24 (and DFL-35, which was not studied here) is most closely related to R. tolerans.
FIG. 1.
Phylogenetic tree of Alphaproteobacteria showing the affiliation of facultative anaerobic and aerobic anoxygenic phototrophs (boldface type). Reference organisms are shown in italics; strains investigated here are boxed. EMBL accession numbers are indicated in parentheses. The tree is based on nearly complete 16S rRNA gene sequences and was calculated by using the maximum-likelihood algorithm. Bootstrap values above 50% are indicated. The bar corresponds to 10 base substitutions per 100 nucleotide positions. Flexibacter maritimus (M64629) was used as an out-group.
Genome size of R. litoralis DSM6996T.
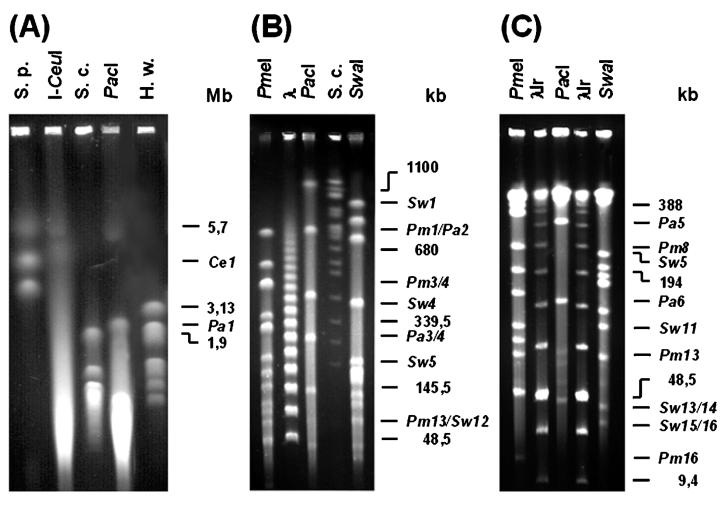
Macrorestriction analysis of the HMW genomic DNA of R. litoralis DSM6996T by PFGE was performed to estimate the chromosome size of this strain (Fig. 2). Of the restriction endonucleases tested (AseI, NotI, PacI, PmeI, SpeI, SwaI, and XbaI), PacI (5′-TTAAT/TAA-3′), PmeI (5′-GTTT/AAAC-3′), and SwaI (5′-ATTT/AAAT-3′) gave useful macrorestriction fragment patterns. In addition, the intron-encoded I-CeuI endonuclease, shown to recognize a sequence 26 bp present within the highly conserved 23S rRNA gene of bacteria and to cut several bacterial chromosomes at this site (26) was used to analyze the chromosome of R. litoralis DSM6996T. Various gel running parameters were performed to obtain optimal separation of the resulting DNA fragments (Fig. 2). The chromosome of R. litoralis DSM6996T harbors 7 PacI restriction sites and 16 PmeI and SwaI restriction sites. The sizes of the corresponding fragments ranged from 47 to 2,310 kb, 15 to 768 kb, and 31 to 1,013 kb, respectively (Table 1). The restriction enzymes used produced clearly visible doublets, e.g., the fragments Pm3 and Pm4 or Pa3 and Pa4. Digestion of the chromosome of R. litoralis DSM6996T with the intron-encoded I-CeuI endonuclease gave only one fragment with an estimated size of 4,462 kb, indicating that a single copy of the 23S rRNA gene might be located on the circular chromosome of this strain. Summing up the calculated I-CeuI, PacI, PmeI, and SwaI fragment lengths led to an estimated chromosome size of approximately 4,470 kb (Fig. 2D). Considering the presence of three extrachromosomal elements (91, 80, and 63 kb), which are not cleaved by the restriction enzymes used in this study, the genome size of R. litoralis DSM6996T amounts to 4,704 kb.
FIG. 2.
Macrorestriction analysis of the genomic DNA of R. litoralis DSM6996T hydrolyzed with the endonucleases I-CeuI, PacI, PmeI, and SwaI. Different size ranges of fragments were resolved on various gels, three of which are shown. (A) High size range; (B) medium size range; (C) low size range (see Materials and Methods for details). Restriction fragments are numbered and designated with respect to the endonucleases I-CeuI (Ce), PacI (Pa), PmeI (Pm), and SwaI (Sw). Chromosomes of S. pombe 927 h (S.p.), H. wingei YB-4662-VIA (H.w.), S. cerevisiae YPH80 (S.c.), concatemers of bacteriophage lambda (λ), and a low-range PFGE marker (λlr) were used as size standards.
TABLE 1.
Macrorestriction analysis of genomic DNA of R. litoralis DSM6996Ta
| Fragment no. | Fragment size (kb) forb:
|
|||
|---|---|---|---|---|
| I-Ceulc | Pacld | Pmeld | Swalf | |
| 1 | 4,462 | 2,310 | 768 | 1,013 |
| 2 | 770 | 610 | 844 | |
| 3 | 447* | 509* | 715 | |
| 4 | 447* | 509* | 415 | |
| 5 | 291 | 388 | 230 | |
| 6 | 149 | 340* | 200 | |
| 7 | 47 | 340* | 189 | |
| 8 | 235 | 180* | ||
| 9 | 194 | 180* | ||
| 10 | 163 | 141 | ||
| 11 | 121 | 123 | ||
| 12 | 102 | 91 | ||
| 13 | 89 | 45* | ||
| 14 | 51* | 45* | ||
| 15 | 51* | 31* | ||
| 16 | 15 | 31* | ||
The table represents a summation of the macrorestriction fragment sizes from Fig. 2.
Double fragments of identical size resulting from single bands of enhanced fluorescence intensity on the PFGE gels are marked with asterisks (for examples, see Fig. 2B, fragments Pm3 and Pm4).
Chromosome size, 4,462 kb.
Chromosome size, 4,461 kb.
Chromosome size, 4,485 kb.
Chromosome size, 4,473 kb.
Plasmid analyses.
Total undigested genomic DNA of each strain was analyzed by PFGE. PFGE parameters were adjusted to detect large (>10 kb) extrachromosomal elements and to determine their sizes and conformations. In addition, conventional plasmid preparation and separation on agarose gels were performed to detect small plasmids, to confirm the results of PFGE, and to determine the sizes of circular plasmids. The results of the plasmid analyses are shown in Fig. 3 and Table 2.
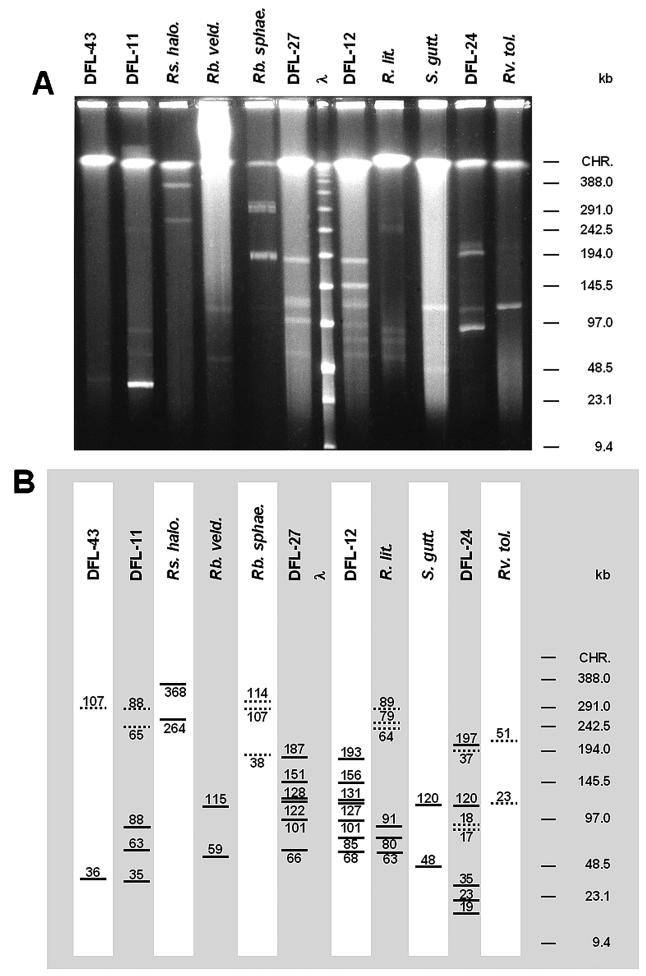
FIG. 3.
(A) PFGE of undigested HMW genomic DNA of strain DFL-43, strain DFL-11, R. halodurans DSM15395T (Rs. halo.), R. veldkampii DSM11550T (Rb. veld.), R. sphaeroides DSM159T(Rb. sphae), strain DFL-27, strain DFL-12, R. litoralis DSM6996T (R. lit.), S. guttiformis DSM11458T (S. gutt.), strain DFL-24, and R. tolerans DSM11457T (Rv. tol.) inside agarose plugs. λ, low-range PFGE marker; CHR, chromosomal DNA. PFGE parameter set A was a 1% (wt/vol) agarose gel, with pulse times of 0.1 to 25 s for 22.5 h at 6 V/cm and 14°C. (B) Schematic view of panel A. Linear plasmids are represented as bold lines; circular plasmids are represented as dotted lines. Sizes are given above or below the lines.
TABLE 2.
Compilation of extrachromosomal elements in phototrophic Alphaproteobacteria determined by PFGE and conventional plasmid analysis
| Band no. | DFL-43
|
DFL-11
|
R. halodurans DSM15395T
|
R. veldkampii DSM11550T
|
R. sphaeroides DSM159T
|
DFL-27
|
DFL-12
|
R. litoralis DSM6996T
|
S. guttformis DSM11458T
|
DFL-24
|
R. tolerans DSM11457T
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Size (kb) | Cona | Size (kb) | Con | Size (kb) | Con | Size (kb) | Con | Size (kb) | Con | Size (kb) | Con | Size (kb) | Con | Size (kb) | Con | Size (kb) | Con | Size (kb) | Con | Size (kb) | Con | |
| 1 | 36 | Lb | 35 | L | 264 | L | 59 | L | 38 | CCC | 66 | L | 68 | L | 63 | L | 4.35 | CCC | 19 | L | 23 | CCC |
| 2 | 107 | CCCc | 63 | L | 368 | L | 115 | L | 107 | CCC | 101 | L | 85 | L | 80 | L | 48 | L | 23 | L | 51 | CCC |
| 3 | 88 | L | 114 | CCC | 122 | L | 101 | L | 91 | L | 120 | L | 35 | L | ||||||||
| 4 | 65 | CCC | 128 | L | 127 | L | 64 | CCC | 17 | CCC | ||||||||||||
| 5 | 88 | CCC | 151 | L | 131 | L | 79 | CCC | 18 | CCC | ||||||||||||
| 6 | 187 | L | 156 | L | 89 | CCC | 120 | L | ||||||||||||||
| 7 | 193 | L | 37 | CCC | ||||||||||||||||||
| 8 | 197 | L | ||||||||||||||||||||
Con, conformation.
L, linear.
CCC, covalently closed circular.
Strain DFL-43.
Strain DFL-43 showed one linear extrachromosomal element of 36 kb in size which can only be weakly detected (Fig. 3A). In addition, a circular element of very low fluorescence intensity was found which had a size of 107 kb.
Strain DFL-11.
Five bands were present on PFGE gels of the genomic DNA of strain DFL-11. One band of high fluorescence intensity was identified as the 35-kb linear plasmid. Two weaker linear bands consisted of linear molecules with sizes of 63 and 88 kb, respectively. The remaining two bands are barely detectable (Fig. 3). Using PFGE parameter set A (Fig. 3A), they ran at 236 and 267 kb, respectively. Using PFGE parameter set B (data not shown), they ran at 208 and 233 kb, and with parameter set D (data not shown), they ran at 476 and 555 kb (sizes were calculated based on the position of the low-range PFGE marker). From this migration behavior, it can be concluded that these bands had a circular conformation. Their sizes were estimated, by using BAC Tracker, as 65 and 88 kb, respectively (data not shown). These sizes are the same as those of the two linear molecules, which could therefore be the same plasmids present in different conformations. The reason might be artifacts during DNA preparation. However, different conformations of the plasmid could also be present within the cell as a result of conformational changes during replication.
R. halodurans DSM15395T.
Two large linear plasmids were present in R. halodurans DSM15395T, which can clearly be seen in Fig. 3A. With sizes of 264 and 368 kb, respectively, they were the largest extrachromosomal elements found in this study. Their linear conformation was concluded from their behavior under different PFGE running parameters. Under parameter set A (Fig. 3A), they ran at 267 and 367 kb; under parameter set C (data not shown), they ran at 261 and 364 kb. Given the variability of size determination on gels, these sizes are identical, and thus, these elements were linear plasmids. No circular plasmids were found in R. halodurans, neither on PFGE nor by conventional plasmid preparations.
R. veldkampii DSM11550T.
The preparation of R. veldkampii DSM11550T DNA in agarose plugs resulted in a background smear of degraded DNA. Two linear plasmids (59 and 115 kb) were found. Additional plasmids or circular conformations of the two linear plasmids may have been undetectable because of the high background.
We repeated the isolation of HMW DNA of this strain several times, using both frozen cells and freshly grown cells. The DNA quality was the same in both cases. The background smear on the gel is usually attributed to the activity of nucleases. Why this effect was so strong for this particular strain is unknown. In addition to the bands of chromosomal DNA and the extrachromosomal elements, a diffuse HMW DNA band can be seen between gel poach and chromosomal DNA, which hybridized with the pufLM probes and thus should consist of chromosomal DNA. Directly below the gel poach, a band of chromosomal DNA can be seen, which is probably the result of the so-called trapping effect (8, 51). HMW DNA was trapped in the gel matrix because of a too-high voltage (more than 2 V per cm) and subsequently remained immobile.
R. sphaeroides DSM159T.
The facultative anaerobe R. sphaeroides has become a model organism for investigations of anoxygenic bacterial photosynthesis. It has a complex genome organization with two circular chromosomes, upon which the photosynthesis genes are located, and five circular endogenous plasmids (48). This strain was included as a reference because of its known plasmid composition (pRS141e, 42 kb; pRS141d, 99 kb; pRS141c, 100 kb; pRS141b, 104 kb; pRS141a, 114 kb) (5) (http://mmg.uth.tmc.edu/sphaeroides/). Here we detected three circular elements (Fig. 3A) with estimated sizes of 38, 107, and 114 kb. The 38-kb plasmid should be the same as the 42-kb plasmid pRS241e, and the 114-kb plasmid should be the same as the 114-kb plasmid pRS241a in reference 5. To find the missing plasmids, we extracted HMW DNA from cultures grown both aerobically and anaerobically in agarose plugs. We also extracted plasmids by a conventional plasmid preparation protocol. Both DNA preparations were analyzed and compared on pulsed-field gels as well as on standard agarose gels. We were unable to detect the 99- and 100-kb plasmids.
The DNA band at 107 kb found here, which is very broad with a strong band in the middle, could be composed of several plasmids: pRS241b (104 kb), pRS241c (100 kb), and pRS241d (99 kb). Because of the small differences in size, they might not have been separated under the conditions used. Alternatively, the band at 107 kb could be a single band, and the two missing plasmids may have been lost during the culturing and preservation of strain DSM159T.
Strain DFL-27.
In strain DFL-27, six linear plasmids (66, 101, 122, 128, 151, and 187 kb) were found, of which the 151-kb plasmid was difficult to detect. Together, these plasmids comprised 755 kb of extrachromosomal genetic information. No circular plasmids were detected.
Strain DFL-12.
Strain DFL-12 is very closely related to DFL-27 (Fig. 1), and the plasmid pattern showed high similarity to DFL-27. Seven linear plasmids were found, ranging from 68 to 193 kb (Table 2) and comprising a total of 861 kb of extrachromosomal genetic information. The 156-kb band showed enhanced fluorescence intensity, indicating that this plasmid might have been present in high copy numbers or that the band consisted of several bands.
R. litoralis DSM6996T.
Preparation of HMW genomic DNA inside agarose plugs did not work for R. denitrificans DSM7001T. On the PFGE gels, only a smear of degraded DNA could be seen (data not shown). Therefore, the closely related strain R. litoralis DSM6996T was analyzed here. Three linear and three circular bands were detected on PFGE gels, which had similar sizes (63 versus 64 kb, 80 versus 79 kb, and 91 versus 89 kb). Thus, the organism probably contains three plasmids in both conformations. Hybridization of the gel with pufLM gene probes after Southern transfer gave a signal with both the linear 91-kb band and the circular 89-kb band (see below), strongly supporting this interpretation.
S. guttiformis DSM11458T.
For S. guttiformis DSM11458T, two bands of 48 and 120 kb were clearly detected in spite of the relatively high background of degraded DNA. Conventional plasmid preparation did not reveal circular plasmids of that size, so these elements may be present as linear plasmids in the cell. In addition, a small circular plasmid was found (4.35 kb) (data not shown).
Strain DFL-24.
Strain DFL-24 had the most complex plasmid content of the investigated strains. Three bands of linear molecules are weakly visible (19, 23, and 35 kb) (Fig. 3). Three circular molecules were also found, which had similar estimated sizes (17, 18, and 37 kb) and thus might represent different conformations of the same plasmids. The circular 17-kb plasmid was hard to detect, since it is located directly below the strong 18-kb circular plasmid. In addition, two large linear plasmids were found (120 and 197 kb).
R. tolerans DSM11457T.
In R. tolerans DSM11457T, we found two circular plasmids. The small one (23 kb) was clearly visible, but the large one (51 kb) had a low intensity. No linear DNA bands were detected, but this could also have been caused by the small amount of DNA in the gel.
Southern blot hybridization.
The gel shown in Fig. 3 was blotted and hybridized with a probe against the 16S rRNA gene to clearly distinguish between the bacterial chromosomes, defined as replicons carrying ribosomal components like 16S rRNA genes (21) and extrachromosomal elements. For all strains, the blot (data not shown) showed the hybridization signal in the compressed HMW band, which could not be separated further under the PFGE conditions used here. Thus, these bands represented, as expected, the chromosomal DNA of the investigated strains.
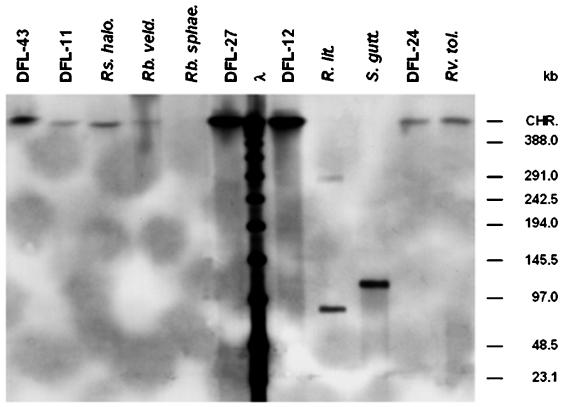
The blot was also hybridized with probes against the pufLM genes of the bacterial photosynthesis reaction center to localize them either on the chromosome or on extrachromosomal elements. The three probes used were homologous to R. litoralis DSM6996T (pufRl), R. veldkampii DSM11550T (pufRv), and strain DFL-12 (pufD12), respectively. The results were identical for all three probes and are shown in Fig. 4 for probe pufD12. The pufLM genes were localized on the chromosome of all strains, with the exception of R. litoralis and S. guttiformis. In R. litoralis, the puf operon was on a plasmid of 91 kb which was present both in linear and circular conformation. In S. guttiformis, the puf operon was located on a linear plasmid of 120 kb. No other hybridization signal was detected in this strain, confirming that this plasmid was not present in circular form. No hybridization signal was detected for R. sphaeroides, which had been shown previously to have the puf operon located on chromosome 1 of the organism (http://mmg.uth.tmc.edu/sphaeroides). Since a hybridization signal with the probe for R. veldkampii was not obtained, we tried to synthesize a homologous probe for R. sphaeroides by PCR amplification of the pufLM genes. An amplification product was not obtained. We conclude that the pufLM genes of R. sphaeroides are less homologous than expected.
FIG. 4.
Southern blot hybridization of the DIG-labeled probe pufD12 against PFGE-separated (Fig. 2) and immobilized HMW genomic DNA of strain DFL-43, strain DFL-11, R. halodurans DSM15395T (Rs. halo.), R. veldkampii DSM11550T (Rb. veld.), R. sphaeroides DSM159T(Rb. sphae), strain DFL-27, strain DFL-12, R. litoralis DSM6996T (R. lit.), S. guttiformis DSM11458T (S. gutt.), strain DFL-24, and R. tolerans DSM11457T (Rv. tol.). λ, low-range PFGE marker; CHR, chromosomal DNA.
DISCUSSION
Genome organization.
Prokaryotic genomes consist of one or several chromosomes, which can be circular or linear. In addition, they frequently contain extrachromosomal elements (plasmids) of various number and size. These plasmids can also be linear or circular and together may comprise a significant part of the organism's total genetic information. Extrachromosomal elements are defined as nonessential for the organism, since they do not contain housekeeping genes or rRNA genes. One of the megaplasmids of Sinorhizobium meliloti, which is ∼1.7 Mb in size, is an exception to this rule, since it contains the only copy of an essential tRNA gene of the strain and therefore could be viewed as a second chromosome (7). The total genome size of R. litoralis DSM6996T was determined here to be 4.70 Mb, which includes three extrachromosomal elements (91, 80, and 63 kb) present both in circular and linear conformation. This genome size is in the range of that of related bacteria, e.g., Rhodobacter capsulatus (3.70 Mb), R. sphaeroides (4.34 Mb), Rhizobium leguminosarum biovar viciae (3.5 Mb), Silicibacter pomeroyi (4.4 Mb), or Agrobacterium tumefaciens (4.9 Mb) (http://www.tigr.org), and does not belong to the size extremes of microbial genomes known today. The smallest genome of an organism known today is that of the newly discovered symbiont Nanoarchaeum equitans (0.49 Mb) in the domain Archaea (14). The smallest genome within the domain Bacteria is owned by Mycoplasma genitalium (0.59 Mb) (6). The largest prokaryotic genome is that of the myxobacterium Sorangium cellulosum (12.6 Mb) (35). Since digestion of the R. litoralis DSM6996T chromosome with the intron-encoded I-CeuI endonuclease resulted in a single large fragment, we conclude that the chromosome has a circular conformation and contains one copy of the 23S rRNA gene.
Linear plasmids in phototrophic Alphaproteobacteria.
Linear plasmids were discovered about two decades ago (reviewed in reference 30) and can be divided into two types which differ in their modes of replication. Hairpin plasmids with covalently closed ends go through a circular stage during replication. They are common in Borrelia strains that are pathogenic to humans. A total of 12 linear plasmids have been found in Borrelia burgdorferi, where they are associated with the infectivity and host specificity of the strains (17). The second type of linear plasmids have proteins attached to both ends at the 5′ DNA strand and start replication at the center of the plasmid (37). These are widespread in Streptomyces and Rhodococcus and code for ecologically important phenotypes, e.g., degradation of xenobiotics (44), metal resistance (38), production of antibiotics (33), or hydrogen autotrophy (18). In most of these bacteria, which are all gram positive, the chromosome also has a linear conformation. The size of the linear plasmids ranges from 5 to 450 kb.
Few reports exist about linear plasmids in gram-negative bacteria, with one example being Klebsiella oxytoca (47). The only linear plasmid in Escherichia coli is the linear plasmid prophage N15, with covalently closed hairpin termini (40). PY54 is a linear prophage in Yersinia enterocolitica which also has covalently closed ends (11). Within the Alphaproteobacteria, until now only circular plasmids have been found, e.g., in a nonphototrophic Ruegeria sp. (56) and in the anoxygenic phototroph R. sphaeroides (48). In the strain of Ruegeria sp., two large indigenous cryptic circular plasmids were recently sequenced which were 76 and 148 kb in size and whose replication systems were highly homologous to those of a plasmid from R. sphaeroides (56).
The phototrophic Alphaproteobacteria investigated here revealed a highly complex plasmid pattern. With the exception of R. tolerans and R. sphaeroides, each strain contained at least one and up to seven linear plasmids. The size of the linear plasmids ranged from 19 kb (DFL-24) to 369 kb (R. halodurans), justifying the term giant linear plasmid or megaplasmid for the latter. Giant linear plasmids (size, 120 to 430 kb) have also been reported in Streptomyces (for an example, see reference 33). None of the plasmids found here hybridized with the probe against the 16S rRNA gene, so they can all be categorized as true nonessential extrachromosomal elements.
Interestingly, the largest number of linear plasmids and the largest total amount of extrachromosomal linear plasmid DNA was found in the isolates DFL-12 (7 linear plasmids, 861 bp) and DFL-27 (6 linear plasmids, 755 bp). These bacteria were isolated from cultures of the toxigenic dinoflagellates P. lima and A. ostenfeldii, respectively. A close symbiotic relationship has been postulated between Roseobacter clade bacteria and dinoflagellates, and some of the dinoflagellate-associated Roseobacter clade bacteria have been shown to produce toxins and might be responsible for toxic algal blooms and shellfish poisoning (1, 13, 36, 50). The large number of linear plasmids found in these isolates may reflect the need to tune the specificity of symbiosis between the bacterium and the eukaryotic host in a flexible way, similar to observations made with the pathogen Borrelia.
Circular plasmids in phototrophic Alphaproteobacteria.
In addition to the linear plasmids, some strains contained circular plasmids. The size of the circular plasmids was in some cases identical to that of a linear plasmid present in the same strain. The pufLM genes detected on the 91-kb linear plasmid of R. litoralis were also found in the circular 89-kb band, suggesting that indeed the two plasmids represented different conformations of the same DNA molecule. Observations of Borrelia and Mycobacterium (25) suggest that these linear plasmids start replication from the ends and then go through a circular stage. The fact that circular plasmids corresponding in size to linear plasmids were only detected in some of the strains investigated here (DFL-11, R. litoralis, and DFL-24), whereas in the others the linear plasmids were present without the circular counterpart (R. halodurans, R. veldkampii, DFL-27, and DFL-12), may be due to differences in the growth stage of the investigated strains or indicate that these two groups of plasmids indeed have different modes of replication. Preparation artifacts may account for the conversion of circular plasmids to linear plasmids, but the conversion of linear plasmids into circular molecules during DNA preparation and pulsed-field analysis seems rather unlikely. Interestingly, only linear plasmids were found in R. veldkampii, whereas only circular plasmids have been found in the closely related R. sphaeroides by us and others. The highly complex and diverse composition of extrachromosomal elements discovered here in a closely related group of microbial genera suggests strong evolutionary pressures underlying the evolution of each organism's genome structure.
Localization of pufLM genes in phototrophic Alphaproteobacteria.
Southern hybridization of the genomic DNA of the strains clearly showed that the pufLM genes were present on the chromosome of all but 2 of the 11 investigated strains rather than on any of the extrachromosomal elements. For example, R. tolerans, which has been named for the variability of its pigment production, carries the pufLM genes on the chromosome. Thus, it cannot be the loss of a plasmid which is responsible for the occasional lack of pigment production in this organism, but rather cultivation-induced changes in gene expression are responsible (2, 23). However, there were two notable exceptions: R. litoralis and S. guttiformis, which are closely related, carried the pufLM genes on linear plasmids. The sizes of these plasmids (91 and 120 kb, respectively) would be sufficient to house the complete photosynthesis gene cluster, which comprises about 45 kb (15). All linear plasmids detected so far are conjugative (30). The localization of the pufLM genes on the linear plasmids of R. litoralis and S. guttiformis shows for the first time a possible mechanism of gene transfer for the photosynthesis gene cluster in proteobacteria, which has been heavily implicated from sequence comparisons of photosynthesis-related genes.
Acknowledgments
We thank the Biologische Anstalt Helgoland for providing M.A. with all of the necessary help and infrastructure during his stay as a guest researcher. Many thanks to Bettina Elxnat for growth of the bacteria and to Hanno Biebl for carefully commenting on the manuscript.
This work was supported by grants from the “Niedersächsischer Schwerpunkt Meeresbiotechnologie” to S.P. and M.A.
REFERENCES
- 1.Alavi, M., T. Miller, K. Erlandson, R. Schneider, and R. Belas. 2001. Bacterial community associated with Pfiesteria-like dinoflagellate cultures. Environ. Microbiol. 3:380-396. [DOI] [PubMed] [Google Scholar]
- 2.Allgaier, M., H. Uphoff, A. Felske, and I. Wagner-Döbler. 2003. Aerobic anoxygenic photosynthesis in Roseobacter clade bacteria from diverse marine habitats. Appl. Environ. Microbiol. 69:5051-5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bautsch, W. 1992. Bacterial genome mapping by two-dimensional pulsed-field gel electrophoresis (2D-PFGE), p. 185-201. In M. Burmeister and M. Ulanovsky (ed.), Pulsed-field gel electrophoresis: protocols, methods and theories. Humana Press, Totowa, N.J. [DOI] [PubMed]
- 4.Beja, O., M. T. Suzuki, J. F. Heidelberg, W. C. Nelson, C. M. Preston, T. Hamada, J. A. Eisen, C. M. Fraser, and E. F. DeLong. 2002. Unsuspected diversity among marine aerobic anoxygenic phototrophs. Nature 415:630-633. [DOI] [PubMed] [Google Scholar]
- 5.Fornari, C. S., M. Watkins, and S. Kaplan. 1984. Plasmid distribution and analyses in Rhodopseudomonas sphaeroides. Plasmid 11:39-47. [DOI] [PubMed] [Google Scholar]
- 6.Fraser, C. M., J. D. Gocayne, O. White, M. D. Adams, R. A. Clayton, R. D. Fleischmann, C. J. Bult, A. R. Kerlavage, G. Sutton, and J. M. Kelley. 1995. The minimal gene complement of Mycoplasma genitalium. Science 270:397-403. [DOI] [PubMed] [Google Scholar]
- 7.Galibert, F., T. M. Finan, S. R. Long, A. Puhler, P. Abola, F. Ampe, et al. 2001. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293:668-672. [DOI] [PubMed] [Google Scholar]
- 8.Gemmill, R. M. 1991. Pulsed-field gel electrophoresis, p. 1-48. In A. Chrambach, M. J. Dunn, and B. J. Radola (ed.), Advances in electrophoresis. VCH Verlagsgesellschaft mbH, Weinheim, Germany.
- 9.Goericke, R. 2002. Bacteriochlorophyll a in the ocean: is anoxygenic bacterial photosynthesis important? Limnol. Oceanogr. 47:290-295. [Google Scholar]
- 10.Hansen, T. A., and J. F. Imhoff. 1985. Rhodobacter veldkampii, a new species of phototrophic purple nonsulfur bacteria. Int. J. Syst. Bacteriol. 35:115-116. [Google Scholar]
- 11.Hertwig, S., I. Klein, R. Lurz, E. Lanka, and B. Appel. 2003. PY54, a linear plasmid prophage of Yersinia enterocolitica with covalently closed ends. Mol. Microbiol. 48:989-1003. [DOI] [PubMed] [Google Scholar]
- 12.Hightower, R. C., D. W. Metge, and D. V. Santi. 1987. Plasmid migration using orthogonal-field-alternating gel electrophoresis. Nucleic Acids Res. 15:8387-8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hold, G. L., E. A. Smith, T. H. Birkbeck, and S. Gallacher. 2001. Comparison of paralytic shellfish toxin (PST) production by the dinoflagellates Alexandrium lusitanicum NEPCC 253 and Alexandrium tamarense NEPCC 407 in the presence and absence of bacteria. FEMS Microbiol. Ecol. 36:223-234. [DOI] [PubMed] [Google Scholar]
- 14.Huber, H., M. J. Hohn, R. Rachel, T. Fuchs, V. C. Wimmer, and K. O. Stetter. 2002. A new phylum of Archaea represented by a nanosized hyperthermophilic symbiont. Nature 417:63-67. [DOI] [PubMed] [Google Scholar]
- 15.Igarashi, N., J. Harada, S. Nagashima, K. Matsuura, K. Shimada, and K. V. Nagashima. 2001. Horizontal transfer of the photosynthesis gene cluster and operon rearrangement in purple bacteria. J. Mol. Evol. 52:333-341. [DOI] [PubMed] [Google Scholar]
- 16.Imhoff, J. F., H. G. Trüper, and N. Pfennig. 1984. Rearrangements of the species and genera of the phototrophic purple nonsulfur bacteria. Int. J. Syst. Bacteriol. 34:340-343. [Google Scholar]
- 17.Iyer, R., O. Kalu, J. Purser, S. Norris, B. Stevenson, and I. Schwartz. 2003. Linear and circular plasmid content in Borrelia burgdorferi clinical isolates. Infect. Immun. 71:3699-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalkus, J., C. Dorrie, D. Fischer, M. Reh, and H. G. Schlegel. 1993. The giant linear plasmid pHG207 from Rhodococcus sp. encoding hydrogen autotrophy: characterization of the plasmid and its termini. J. Gen. Microbiol. 139:2055-2065. [DOI] [PubMed] [Google Scholar]
- 19.Karl, D. M. 2002. Hidden in a sea of microbes. Nature 415:590-591. [DOI] [PubMed] [Google Scholar]
- 20.Kolber, Z. S., C. L. Van Dover, R. A. Niederman, and P. G. Falkowski. 2000. Bacterial photosynthesis in surface waters of the open ocean. Nature 407:177-179. [DOI] [PubMed] [Google Scholar]
- 21.Kolstø, A.-B. 1997. Dynamic bacterial genome organization. Mol. Microbiol. 24:241-246. [DOI] [PubMed] [Google Scholar]
- 22.Labrenz, M., B. J. Tindall, P. A. Lawson, M. D. Collins, P. Schumann, and P. Hirsch. 2000. Staleya guttiformis gen. nov., sp. nov. and Sulfitobacter brevis sp. nov., alpha-3-Proteobacteria from hypersaline, heliothermal and meromictic antarctic Ekho Lake. Int. J. Syst. Evol. Microbiol. 50:303-313. [DOI] [PubMed] [Google Scholar]
- 23.Labrenz, M., M. D. Collins, P. A. Lawson, B. J. Tindall, P. Schumann, and P. Hirsch. 1999. Roseovarius tolerans gen. nov., sp. nov., a budding bacterium with variable bacteriochlorophyll a production from hypersaline Ekho Lake. Int. J. Syst. Evol. Microbiol. 49:137-147. [DOI] [PubMed] [Google Scholar]
- 24.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-148. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley and Sons Ltd., Chichester, England.
- 25.Le Dantec, C., N. Winter, B. Gicquel, V. Vincent, and M. Picardeau. 2001. Genomic sequence and transcriptional analysis of a 23-kilobase mycobacterial linear plasmid: evidence for horizontal transfer and identification of plasmid maintenance systems. J. Bacteriol. 183:2157-2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu, S.-L., A. Hessel, and K. E. Sanderson. 1993. Genomic mapping with I-CeuI, an intron-encoded endonuclease specific for genes for ribosomal RNA, in Salmonella spp., Escherichia coli, and other bacteria. Proc. Natl. Acad. Sci. USA 90:6874-6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ludwig, W., and O. Strunk. 1996. ARB: a software environment for sequence data. Technische Universität Muenchen, Munich, Germany.
- 28.Marrs, B. 1981. Mobilization of the genes for photosynthesis from Rhodopseudomonas capsulata by a promiscuous plasmid. J. Bacteriol. 146:1003-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mathew, M. K., C.-F. Hui, C. L. Smith, and C. R. Cantor. 1988. High-resolution separation and accurate size determination in pulsed-field gel electrophoresis of DNA. 4. Influence of DNA topology. Biochemistry 27:9222-9226. [DOI] [PubMed] [Google Scholar]
- 30.Meinhardt, F., R. Schaffrath, and M. Larsen. 1997. Microbial linear plasmids. Appl. Microbiol. Biotechnol. 47:329-336. [DOI] [PubMed] [Google Scholar]
- 31.Nagashima, K. V., A. Hiraishi, K. Shimada, and K. Matsuura. 1997. Horizontal transfer of genes coding for the photosynthetic reaction centers of purple bacteria. J. Mol. Evol. 45:131-136. [DOI] [PubMed] [Google Scholar]
- 32.Nagashima, K. V., K. Matsuura, N. Wakao, A. Hiraishi, and K. Shimada. 1997. Nucleotide sequences of genes coding for photosynthetic reaction centers and light-harvesting proteins of Acidiphilium rubrum and related aerobic acidophilic bacteria. Plant Cell Physiol. 38:1249-1258. [DOI] [PubMed] [Google Scholar]
- 33.Netolitzky, D. J., X. Wu, S. E. Jensen, and K. L. Roy. 1995. Giant linear plasmids of beta-lactam antibiotic producing Streptomyces. FEMS Microbiol. Lett. 131:27-34. [DOI] [PubMed] [Google Scholar]
- 34.Pradella, S., H. Hippe, E. Stackebrandt. 1998. Macrorestriction analysis of Desulfurella acetivorans and Desulfurella multipotens. FEMS Microbiol Lett. 159:137-144. [DOI] [PubMed] [Google Scholar]
- 35.Pradella, S., A. Hans, C. Sproer, H. Reichenbach, K. Gerth, and S. Beyer. 2002. Characterisation, genome size and genetic manipulation of the myxobacterium Sorangium cellulosum So ce56. Arch. Microbiol. 178:484-492. [DOI] [PubMed] [Google Scholar]
- 36.Prokic, I., F. Brummer, T. Brigge, H. D. Gortz, G. Gerdts, C. Schutt, M. Elbrachter, and W. E. G. Müller. 1998. Bacteria of the genus Roseobacter associated with the toxic dinoflagellate Prorocentrum lima. Protist 149:347-357. [DOI] [PubMed] [Google Scholar]
- 37.Qin, Z., and S. N. Cohen. 1998. Replication at the telomeres of the Streptomyces linear plasmid pSLA2. Mol. Microbiol. 28:893-903. [DOI] [PubMed] [Google Scholar]
- 38.Ravel, J., H. Schrempf, and R. T. Hill. 1998. Mercury resistance is encoded by transferable giant linear plasmids in two Chesapeake Bay Streptomyces strains. Appl. Environ. Microbiol. 64:3383-3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raymond, J., O. Zhaxybayeva, J. P. Gogarten, S. Y. Gerdes, and R. E. Blankenship. 2002. Whole-genome analysis of photosynthetic prokaryotes. Science 298:1616-1620. [DOI] [PubMed] [Google Scholar]
- 40.Rybchin, V. N., and A. N. Svarchevsky. 1999. The plasmid prophage N15: a linear DNA with covalently closed ends. Mol. Microbiol. 33:895-903. [DOI] [PubMed] [Google Scholar]
- 41.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 42.Selje, N., M. Simon, T. Brinkhoff. 2004. A newly discovered Roseobacter cluster in temperate and polar oceans. Nature 427:445-448. [DOI] [PubMed] [Google Scholar]
- 43.Shiba, T. 1991. Roseobacter litoralis gen. nov. spec. nov., and Roseobacter denitrificans sp. nov., aerobic pink-pigmented bacteria which contain bacteriochlorophyll a. Syst. Appl. Microbiol. 14:140-145. [Google Scholar]
- 44.Shimizu, S., H. Kobayashi, E. Masai, and M. Fukuda. 2001. Characterization of the 450-kb linear plasmid in a polychlorinated biphenyl degrader, Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 67:2021-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simske, J. S., and S. Scherer. 1989. Pulsed-field gel electrophoresis of circular DNA. Nucleic Acids Res. 17:4359-4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith, C. L., S. Klco, and C. R. Cantor. 1988. Pulsed field gel electrophoresis and the technology of large DNA molecules, p. 41-72. In K. Davies (ed.), Genome analysis: a practical approach. IRL Press, Oxford, England.
- 47.Stoppel, R. D., M. Meyer, and H. G. Schlegel. 1995. The nickel resistance determinant cloned from the enterobacterium Klebsiella oxytoca: conjugational transfer, expression, regulation and DNA homologies to various nickel-resistant bacteria. Biometals 8:70-79. [DOI] [PubMed] [Google Scholar]
- 48.Suwanto, A., and S. Kaplan. 1989. Physical and genetic mapping of the Rhodobacter sphaeroides 2.4.1 genome: presence of two unique circular chromosomes. J. Bacteriol. 171:5850-5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suzuki, T., Y. Muroga, M. Takahama, and Y. Nishimura. 1999. Roseivivax halodurans gen. nov., sp. nov. and Roseivivax halotolerans sp. nov., aerobic bacteriochlorophyll-containing bacteria isolated from a saline lake. Int. J. Syst. Bacteriol. 49:629-634. [DOI] [PubMed] [Google Scholar]
- 50.Vasquez, M., C. Gruttner, B. Moeller, and E. R. Moore. 2002. Limited selection of sodium channel blocking toxin-producing bacteria from paralytic shellfish toxin-contaminated mussels (Aulacomya ater). Res. Microbiol. 153:333-338. [DOI] [PubMed] [Google Scholar]
- 51.Viovy, J.-L., F. Miomandre, M.-C. Miquel, F. Caron, and F. Sor. 1992. Irreversible trapping of DNA during crossed-field gel electrophoresis. Electrophoresis 13:1-6. [DOI] [PubMed] [Google Scholar]
- 52.Wagner-Döbler, I., H. Rheims, A. Felske, R. Pukall, and B. Tindall. 2003. Jannaschia helgolandensis, gen. nov., sp. nov., a novel abundant member of the marine Roseobacter clade from the North Sea. Int. J. Syst. Evol. Microbiol. 53:731-738. [DOI] [PubMed] [Google Scholar]
- 53.Woese, C. R. 1987. Bacterial evolution. Microbiol. Rev. 51:221-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xiong, J., W. M. Fischer, K. Inoue, M. Nakahara, and C. E. Bauer. 2000. Molecular evidence for the early evolution of photosynthesis. Science 289:1724-1730. [DOI] [PubMed] [Google Scholar]
- 55.Yurkov, V. V., and J. T. Beatty. 1998. Aerobic anoxygenic phototrophic bacteria. Microbiol. Mol. Biol. Rev. 62:695-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhong, Z., R. Caspi, D. Helinski, V. Knauf, S. Sykes, C. O'Byrne, T. P. Shea, J. E. Wilkinson, C. DeLoughery, and A. Toukdarian. 2003. Nucleotide sequence based characterizations of two cryptic plasmids from the marine bacterium Ruegeria isolate PR1b. Plasmid 49:233-252. [DOI] [PubMed] [Google Scholar]