Abstract
In order to investigate the factors controlling the bacterial community composition (BCC) in reservoirs, we sampled three freshwater reservoirs with contrasted physical and chemical characteristics and trophic status. The BCC was analysed by 16S rRNA gene amplicon 454 pyrosequencing. In parallel, a complete dataset of environmental parameters and phytoplankton community composition was also collected. BCC in the analysed reservoirs resembled that of epilimnetic waters of natural freshwater lakes with presence of Actinobacteria, Alpha- and Betaproteobacteria, Cytophaga–Flavobacteria–Bacteroidetes (CFB) and Verrucomicrobia groups. Our results evidenced that the retrieved BCC in the analysed reservoirs was strongly influenced by pH, alkalinity and organic carbon content, whereas comparatively little change was observed among layers in stratified conditions.
Introduction
Microbes play a central role in global environmental processes and earth biogeochemistry [1], with bacteria being the most important component of microbial communities responsible, in aquatic ecosystems, for the organic matter mineralization and nutrient recycling processes [2], [3]. However, due to the intrinsic complexity of bacterial diversity and the small fraction of bacteria that can be cultivated, knowledge on the driving factors of bacterial community composition (BCC) has remained elusive until the last decades. In this sense, the introduction and wide use of molecular tools in microbial ecology has rapidly increased the knowledge on bacterial diversity, identity and involvement in key processes over many different environments [4]–[8]. As a consequence, microbial ecologists have been looking for drivers of the BCC fluctuations in relation with the functioning and the ecology of aquatic ecosystems.
Freshwater environments have been traditionally studied focusing on their physical and chemical characteristics and community composition, from algal assemblages to fish, and a considerable body of literature has been devoted to the ecology of these communities. In a context of environmental management, shifts in community composition have been used as indicative of shifts in the water bodies resulting from variation of both internal (i.e., physical and chemical properties of the water column or retention time) and external (i.e., climate conditions, organic matter and nutrient inputs) processes [9]–[13]. Concerning the microbial communities, it has been shown that abiotic (e.g., temperature, oxygen, pH, conductivity, water transparency, organic matter concentration and biodegradability, nutrients, and damming) as well as biotic (chlorophyll (Chl) a content, interactions with phyto- and zooplankton, grazing, competition) factors regulate temporal and spatial shifts of microbial communities in aquatic environments [9], [14]–[23].
Today, deep sequencing technologies such as 454 pyrosequencing provide an attractive avenue to explore complex microbial communities [4], [5], [24], [25] and evidenced the existence of a “rare biosphere” [26]–[28]. Moreover, ubiquitously distributed uncultured bacterial phylotypes have been also reported in freshwater lakes [29]–[33]. Newton and co-workers [34] reviewed the BCC present in epilimnetic waters of lakes worldwide, showing that over the most recovered phyla are: Proteobacteria, Actinobacteria, Cytophaga–Flavobacterium–Bacteroidetes (CFB), Cyanobacteria and Verrucomicrobia. In fact, Betaproteobacteria are by far the most studied and often the most abundant (up to 60 – 70% of total cells) bacteria in epilimnetic waters [34]. This bacterial group is mainly driven by high DOC and nitrate concentrations, and often appears associated with cyanobacteria or particles of different sizes [23], [35]–[38]. Nevertheless, most of the studies included in Newton and co-workers' review [34] were only based on surface or epilimnetic waters surveys from natural freshwater lakes or lagoons.
In spite of the knowledge recently gained on BCC and their ecological drivers in natural freshwater environments, there is a lack of knowledge on man-made reservoirs, with some exceptions [14], [20], [39]–[41]. Freshwater reservoirs present some specificities in comparison to natural lakes resulting in different biological, physical and chemical characteristics, such as shorter retention times, water level fluctuations, important shifts of nutrient and dissolved organic carbon (DOC) concentrations and scarcity of littoral macrophytes which all together affect the planktonic organisms present [20], [41]–[43].
In an attempt to identify the main drivers of the bacterial assemblage developing in freshwater reservoirs, we analysed three water bodies with contrasting physical and chemical characteristics, trophic status, DOC content and phytoplankton assemblage at three distinct periods of the year (spring, summer and fall). We hypothesised that those physical and chemical characteristics like pH, organic carbon content, or phytoplankton community composition may exert some influence on BCC in freshwater reservoirs.
Material and Methods
Sampling sites and sample collection
Three freshwater reservoirs located in Belgium (see Table 1 and S1 Fig. for exact locations) were sampled in April, July and October 2010. These reservoirs, named La Gileppe, Ry de Rome and Féronval, were selected in order to obtain contrasting physical, chemical, and trophic conditions. The characteristics of the studied reservoirs were described in [44] and classified according to the European classification of lakes [45]. La Gileppe reservoir is a deep (maximum depth 58 m), oligotrophic, cold-water body covering 130 ha, with a catchment area dominated by forests of conifers and deciduous trees. The pH of its waters is low, related to the geology of the watershed and the presence of peatbogs. It has a mean annual residence time of 315 days and belongs to the Central Baltic lake type (L–CB3; lowland, shallow lakes with an alkalinity between 0.2 and 1.0 meq L−1) albeit with lower pH and greater depth than the range of this lake type. These characteristics make it actually closer to Northern humic lakes of very low alkalinity. Ry de Rome reservoir is an oligo-/mesotrophic water body of 26 ha of surface area and a maximum depth of 25 m. It has been also classified as L–CB3 [44] and has a mean retention time of 200 days. The third reservoir (Féronval) belongs to the L–CB2 lake type (lowland, very shallow, calcareous, alkalinity> 0.5 meq L−1) with a surface area of 21 ha and a maximum depth of 12 m (mean depth 3.8 m). Féronval is a poly-eutrophic reservoir; with mean total phosphorus (TP) concentration of 55 µg L−1 and Chl a maxima higher than 100 µg L−1.
Table 1. General characteristics of the three analysed reservoirs and annual average values of some physical and chemical parameters.
| Parametersa | La Gileppe | Ry de Rome | Féronval |
| Latitude (N) | 50° 35′ 16.73′′ | 50° 01′ 21.67′′ | 50° 12′ 59.14′′ |
| Longitude (E) | 5° 58′ 34.75′′ | 4° 32′ 13.55′′ | 4° 23′ 25.34′′ |
| Altitude (m) | 310 | 280 | 220 |
| Surface of basin (km2) | 54 | 10 | 10 |
| Volume (hm3) | 26.4 | 2.2 | 0.8 |
| Surface (ha) | 130 | 26 | 21 |
| Retention time (years) | 0.86 | 0.55 | 0.31 |
| Max depth (m) | 58 | 25 | 12 |
| Mean depth (m) | 20.3 | 8.5 | 3.8 |
| Zeu (m) | 3.60 | 9.89 | 3.49 |
| Zm (m) | 5.67 | 4.33 | 5.33 |
| Zm/Zeu | 1.53 | 0.49 | 1.78 |
| pH | 5.71 | 6.81 | 7.86 |
| Conductivity (μS cm−1 at 25°C) | 57 | 59 | 392 |
| DO (mg O2 L−1) | 9.34 | 8.64 | 8.97 |
| Turbidity (NTU) | 3.57 | 1.90 | 6.79 |
| TAC (meq L−1) | 0.50 | 0.87 | 13.14 |
| DIN (mg L−1) | 0.63 | 0.72 | 2.21 |
| SRP (mg L−1) | 0.003 | 0.003 | 0.005 |
| TP (mg L−1) | 7.33 | 5.00 | 55.67 |
| Si (mg L−1) | 2.08 | 2.14 | 1.52 |
| BOD5 (mg O2 L−1) | 1.0 | 1.4 | 1.4 |
| COD (mg O2 L−1) | 19.0 | 5.0 | 11.9 |
| DOC (mg C L−1) | 6.74 | 2.66 | 4.00 |
| TOC (mg C L−1) | 8.39 | 3.12 | 5.50 |
| Alkalinity (meq L−1)b | 0.2 | 0.21 | 2.64 |
| Chlorophyll a (μg L−1) | 0.5 | 2.62 | 13.44 |
Values correspond to the average of six depths sampled in each reservoir, each month between March and October 2010.
, Zeu: Depth of euphotic zone, Zm: Depth of mixing zone, Cond: conductivity, TAC: total alkalinity, DIN: dissolved inorganic nitrogen, SRP: soluble reactive phosphate, TP: total phosphorus, Si: dissolved silica, BOD5: biological oxygen demand, COD: chemical oxygen demand, DOC: dissolved organic carbon, TOC: total organic carbon; b, [44].
During the three sampling campaigns, water samples representing the three main water compartments (i.e., epilimnion, metalimnion and hypolimnion, when the reservoirs were stratified) were collected in each reservoir using a 5.0 L Niskin bottle. Water samples for chemical and biological analyses were kept in a cooler until further analyses in the laboratory on the day of sampling. In all cases, the analysed reservoirs did not involve endangered or protected species thus no specific permissions were required for sample collection.
Physical and chemical analyses
The water column structure in the three reservoirs was determined using a multiparameter probe YSI 6000 V2 (Yellow Spring Instruments, USA) to determine vertical depth profiles of temperature (T), pH, conductivity (Cond), turbidity (Turb) and dissolved oxygen (DO). The depth of the mixed layer (Zm) was estimated from T and DO vertical profiles.
In addition, a large set of physical and chemical data was obtained from monthly sampling campaigns collecting water samples from six representative water depths, carried out by the ISSeP (Institut Scientifique de Service Public) from a water quality survey program in the context of a Water Framework Directive monitoring program. These data included Secchi disk depth measurements, TAC (total alkalinity), DIN (dissolved inorganic nitrogen; i.e., the sum of ammonium, nitrite, and nitrate concentrations), SRP (soluble reactive phosphate), TP (total phosphorous), Si (dissolved reactive silica), BOD5 (biological oxygen demand) and COD (chemical oxygen demand). All analyses were carried out following standard methods [46].
For dissolved organic carbon (DOC) analyses, water samples were filtered on GF/F glass fiber membranes (Whatman) previously combusted at 550°C. Glassware receiving water samples for DOC analyses were previously muffled at 550°C for 4 h after cleaning. Sample preservation was done by the addition of NaNO3 (0.05%, final concentration). DOC was measured after removal of inorganic carbon by bubbling in the presence of phosphoric acid using a total organic carbon analyser (Dohrman Apollo 2000, Dohrman) in which organic carbon was oxidised at high temperature and CO2 produced was detected by infrared spectrometry. The same procedure was carried out on unfiltered samples for total organic carbon (TOC) determination.
Phytoplankton pigment analyses
Samples for Chl a and secondary pigment analyses were treated following a procedure previously described [47] using High-Performance Liquid Chromatography (HPLC) with a Waters system comprising a NovaPak C18 HPLC column, a Waters 996 PDA detector and a Waters 470 fluorescence detector. Calibration was made using commercial external standards (DHI, Denmark).
Bacterial abundance and production
Water samples for bacterial abundance estimation were passed through 10.0 µm pore–size 47 mm diameter cellulose acetate filters (Whatman) and preserved by the addition of filtered (0.22 µm MILLEX) formaldehyde (2% final concentration) and stored at 4°C until analysis. Fixed water samples were subsequently passed through 0.22 µm pore-size black filters (Nucleopore) to retain free-living bacteria for enumeration. Bacterial abundance was determined using epifluorescence microscopy after DAPI (4,6 diamidino-2-phenylindole; 2 µg mL−1, final concentration) staining [48]. Briefly, after filtration of 1.0 mL of fixed water sample, 600 to 1200 cells were counted per sample.
Bacterial production was estimated from tritiated thymidine (3H-Thy) incorporation rates [49]. Briefly, 10 mL of water were incubated in triplicates with 3H-Thy (45 Ci mmol−1; ICN Pharmaceuticals) for one to two hours in the dark at in situ temperature and at saturation conditions (20 nM of 3H-Thy, [50]). After incubation, cold trichloroacetic acid (TCA) was added (5.0%, final concentration) and each sample was subsequently passed through 0.22 µm pore-size cellulose nitrate filters (Sartorius). Radioactivity associated with the filters was estimated by liquid scintillation in a Liquid Scintillator Analyser Tri-Carb 2100TR (Packard) apparatus. Cell production was calculated from 3H-Thy incorporation rates using the conversion factor of 0.5 × 1018 cells produced per mole of 3H-Thy incorporated into DNA experimentally determined for freshwater environments [50]. Bacterial production values (μg C L−1 h−1) based on thymidine incorporation were obtained by multiplying cell productions by the average carbon content (CC) per bacterial cell. In order to calculate the average CC per bacterial cell, image analysis was performed using the same microscopic preparations used for DAPI enumerations to carry out automatic measurement of bacterial cell size from digital images (Nikon DMX 1200). The LUCIA G program (Laboratory Imaging, [51]) was used for estimating cell dimension of around 1000 cells per slide and the biovolume of each cell was then calculated. Biomass was estimated from the abundance and biovolume distribution using the relationship CC = 92×V−0.598, relating carbon content per cell (CC; fg C cell−1) to biovolume (V; μm3) determined from the data of Simon and Azam [52].
Bacterial community composition: nucleic acid extraction and 16S rRNA gene amplicon 454 pyrosequencing
Samples for nucleic acids extraction were sequentially passed through 10.0 µm pore–size 47 mm diameter filters (Nuclepore), to remove particulate debris and large planktonic organisms, and through 0.22 µm pore–size 47 mm diameter DURAPORE® (Millipore, USA) filters to retain free–living prokaryotes. Filters were preserved in eppendorfs containing 2.0 mL Sucrose Buffer (Tris HCl 1 M pH 8.0, EDTA 0.5 M pH 8.0, Sucrose 1.5 M) at −20°C until further analyses. Total nucleic acids were extracted from 0.22 µm pore–size filters using enzymatic and mechanical cell lysis procedures [53], [54]. Nucleic acids concentration and purity were determined using a Nanodrop ND-2000 UV-Vis spectrophotometer (Nanodrop, DE). Finally, nucleic acid extracts were kept at −20°C until further analyses.
A total of 27 samples were individually processed for bacterial massive pyrosequencing. Bacterial tag-encoded 454 FLX amplicon pyrosequencing (b-TEFAP) was performed using a 454 FLX Titanium platform (Roche) with Titanium reagents at Research and Testing Laboratory (Lubbock, TX, USA) as previously described [55], [56]. The universal bacterial primers 28f (5′-gAg TTT gAT CNT ggC TCA g) and 519r (5′-gTN TTA CNg Cgg CKg CTg) were selected to span the V1–V3 variable region of the bacterial 16S rRNA gene [57].
Retrieved pyrosequencing data was subjected to further analyses by using the open-source software package mothur (v1.24.1, [58]). Data were decompressed and sequencing errors were reduced by trimming flows (i.e., denoising) and sequences by applying the following criteria: amplicons shorter than 200 bp in length, reads containing any unresolved nucleotides and more than 8 homopolimers were removed from the pyrosequencing-derived datasets. Subsequently, we processed improved sequences from individual files. Afterwards, putative chimeras (checked by using Uchime software [59]) and putative contaminants (understood as those unclassified or misclassified sequences after comparing candidate sequences against the latest Ribosomal Database Project training set database) were removed from our data set. Once files were considered free of chimeras or contaminants, alpha- and beta-diversity analyses by OTUs and phylogenetic relationships were conducted. For phylogenetic identification of b-TEFAP amplicons, latest available SILVA 16S rRNA gene database was uploaded into mothur and used to classify sequences and OTUs by using a confidence threshold of 80% (bootstrap). All sequences generated in the present study can be accessed through National Center for Biotechnology Information (NCBI) under the bioproject number: PRJNA241494.
Statistical analyses
Unless otherwise stated, all statistical analyses were performed using PRIMER 6 [60]. In order to analyse the influence of the discriminative environmental parameters on BCC, canonical correspondence analyses (CCA) were performed using the permutest option in the R package VEGAN [61]. The relative abundances of bacterial OTUs were square-transformed, and resemblances between samples were computed by using the Bray-Curtis similarity coefficient [60]. The effect of time, location and oxygen conditions on BCC was tested with one-way analysis of similarities (ANOSIM). In order to establish a relationship between the BCC and the environmental variables analysed, the BIOENV procedure [62] was used by computing Spearman rank correlations for each combination of environmental variables and community composition [63]. We also quantified the relative contribution of several environmental parameters (i.e., Zeu, Zm, temperature, pH, conductivity, DO, turbidity, total alkalinity, BOD5, COD, Chl a, DOC, TP, SRP, DIN concentrations; see Table 1) as well as bacterial production, phytoplankton species composition on the bacterial community shifts using variation partitioning analyses [64] by means of the varpart function in the VEGAN R package [61]. In addition, a Factorial Correspondence Analysis (FCA), based on the relative abundances of bacterial groups in the three reservoirs at each layer and sampling date, was performed using the ade-4 R package [65].
Results
Limnological characteristics of the reservoirs
The analysed reservoirs in the present study evidenced clear differences in their vertical structure of the water column. The three reservoirs presented a consistent thermal stratification from mid-April to mid-September (S2 Fig.). The whole water column remained oxic in La Gileppe reservoir all over the year, in accordance with its oligotrophic status, whereas in the poly-eutrophic reservoir Féronval the hypolimnion was oxygen depleted from beginning of summer to autumn. Ry de Rome reservoir presented hypolimnetic oxygen depletion only shortly in autumn. Conductivity and pH differed widely among reservoirs, in relation with their contrasting mineralisation and alkalinity (Table 1). The average concentrations of inorganic nutrients are also reported in Table 1. Dissolved phosphate concentrations were lower than 5.0 µg-P L−1 in the three reservoirs except in the hypolimnion of Féronval reservoir. Total P and DIN concentrations were also much higher in Féronval, in agreement with its reported poly-eutrophic status. DIN was dominated by nitrate in all reservoirs, with ammonium and nitrite often barely detected. The organic carbon content (DOC and TOC) was lowest in Ry de Rome reservoir and highest in La Gileppe reservoir, particularly in autumn samples. DOC and TOC values did not vary substantially with depth. Worth noticing is that BOD5 was lowest in La Gileppe reservoir despite its high DOC and TOC, indicating a high proportion of refractory organic carbon (e.g., humic acids), which is also reflected by the low pH and water transparency in La Gileppe.
Phytoplankton biomass and composition
The temporal variation in Chl a (average values of six depths sampled monthly in each reservoir from March to October 2010) highlighted the differences among the analysed reservoirs and periods (Fig. 1). Chl a in La Gileppe did not exceed 1.0 µg L−1 (Fig. 1), with a phytoplankton assemblage dominated by coccal green algae (Crucigenia sp.) and small chrysophytes and cryptomonads (Fig. 2). In Ry de Rome reservoir, Chl a ranged between 1.0 and 4.7µg L−1 (Fig. 1) and phytoplankton was co-dominated by diatoms (with a Tabellaria sp. peak in spring, followed by Asterionella formosa and Nitzschia acicularis in summer and autumn) and chrysophytes (Dinobryon sp.); peaks of Gonyostomum semen, a raphidophyte frequently reported in humic lakes, appeared in late summer and autumn (Fig. 2). In this small, stratified and clear reservoir, Chl a maxima occurred at 5 m depth (i.e., in the metalimnion). In contrast, in the poly-eutrophic Féronval reservoir, Chl a varied between 5.0 and 20.0 µg L−1 (Fig. 1), and the assemblage varied greatly over time, with peaks of diatoms (Nitzschia acicularis, Asterionella formosa as dominant species), coccal green algae (Coelastrum microporum, Pediastrum spp., Scenedesmus spp., among few others), cyanobacteria (mostly Aphanizomenon flos-aquae and Planktothrix agardhii), cryptomonads and chrysophytes (Fig. 2).
Figure 1. Chlorophyll a concentrations.
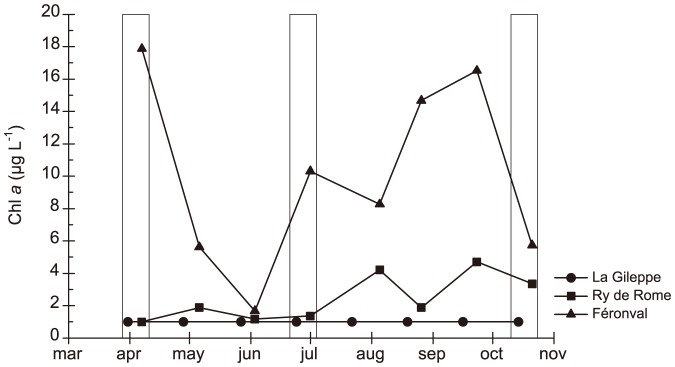
Chlorophyll a concentrations (average values from the three analysed water layers) from April to October 2010 measured in La Gileppe (dot), Ry de Rome (square) and Féronval (triangle). Boxes indicate sampling periods for both chemical and BCC analyses.
Figure 2. Phytoplantkon community composition.
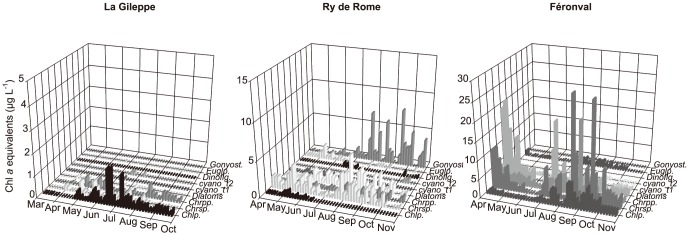
Phytoplantkon community composition over time (from April to October 2010) in the three analysed reservoirs by means of CHEMTAX pigment analyses and expressed as Chlorophyll a equivalents (μgr L−1).
Bacterial cell abundances and production
Total cells abundances ranged from 1.0×106 to 1.2×107 cells mL−1 with consistent differences among water layers and reservoirs (S3 Fig.). Féronval reservoir presented the highest cell abundances over the analysed periods whereas lower values were recovered for Ry de Rome and La Gileppe reservoirs. Interestingly, the oligotrophic La Gileppe reservoir evidenced higher mean values than the mesotrophic Ry de Rome reservoir during mid-April. In general, higher cell abundances were observed in epi- and metalimnetic samples independently of reservoirs and sampling date, with the exception of spring samples from La Gileppe and autumn ones from Féronval (S3 Fig.).
BP experiments were performed at three depths representing the water layers in stratified conditions. Higher BP values were observed in the poly-eutrophic Féronval reservoir (4.0 and ca. 40.0 times higher) in comparison to the lower ones obtained in the meso- and oligotrophic reservoirs (S3 Fig.). In Lake Féronval, BP clearly decreased from epilimnion to hypolimnion in April and July, whereas BP was fairly constant and lower in October. Interestingly, the lowest BP values were obtained in samples from the mesotrophic reservoir, especially in samples from epilimnetic waters (S3 Fig.).
Pyrosequencing data overview
Richness and diversity estimates
For all samples analysed by pyrosequencing, the quality criteria applied for reduction of sequencing noise, chimeric sequences and presence of contaminants omitted less than 10.0% of total sequences (data not shown). The curated (i.e., chimera-free and without contaminant sequences) 77815 bacterial 16S rRNA gene sequences (range: 995 – 4883 per sample) resulted in 4031 bacterial OTUs (0.03 cut-off; range: 118 – 654 OTUs per sample; see details in Table 2), which represented most of the richness in the analysed samples, thus supporting obtained coverage values (89.7% – 97.7% coverage estimates). Rarefaction curves (data not shown) reached saturation for all analysed samples. For all the samples analysed, values of richness (Chao1) and diversity (Shannon) estimators were higher in Ry de Rome and Féronval reservoirs than in La Gileppe (Table 2). Furthermore, rank abundance curves for each reservoir (S4 Fig.) showed dominance of one single OTU identified as a member of the Actinobacteria (hgcI_clade) representing between 9.9% and 38.6% of sequences retrieved from each reservoir when all samples of a given reservoir were pooled together (data not shown). Moreover, the second and third most abundant OTUs were affiliated to members of Actinobacteria (hcgI_clade), Alphaproteobacteria (SAR11), Betaproteobacteria (Polynucleobacter), and Verrucomicrobia (vadin HA64) clades with relative abundances ranging from 6.5% to 12.4% of retrieved sequences for each reservoir (data not shown).
Table 2. Observed bacterial richness and diversity estimates based on OTUs for La Gileppe, Ry de Rome, and Féronval reservoirs.
| Richnessd | Diversityd | |||||
| Reservoira | WCb | Seasonc | Filtered reads | OTUs | Chao1 (± sd) | Shannon (± sd) |
| G | E | Apr | 2606 | 203 | 445±97 | 2.9±0.1 |
| G | E | Jul | 2149 | 118 | 193±37 | 1.5±0.1 |
| G | E | Oct | 2938 | 131 | 210±37 | 2.1±0.1 |
| G | M | Apr | 2892 | 299 | 774±167 | 3.3±0.1 |
| G | M | Jul | 2819 | 234 | 444±82 | 2.4±0.1 |
| G | M | Oct | 2819 | 294 | 671±134 | 3.1±0.1 |
| G | H | Apr | 2058 | 255 | 581±117 | 3.3±0.1 |
| G | H | Jul | 1559 | 250 | 594±125 | 3.6±0.1 |
| G | H | Oct | 2033 | 303 | 507±72 | 4.1±019 |
| RR | E | Apr | 2952 | 340 | 908±201 | 3.9±0.1 |
| RR | E | Jul | 3211 | 235 | 590±140 | 2.9±0.1 |
| RR | E | Oct | 995 | 178 | 306±54 | 3.8±0.1 |
| RR | M | Apr | 3621 | 351 | 721±123 | 3.9±0.1 |
| RR | M | Jul | 3724 | 296 | 627±121 | 3.6±0.1 |
| RR | M | Oct | 1328 | 243 | 517±104 | 4.0±0.1 |
| RR | H | Apr | 3575 | 363 | 667±102 | 4.1±0.1 |
| RR | H | Jul | 4239 | 382 | 769±126 | 3.8±0.1 |
| RR | H | Oct | 1964 | 321 | 497±62 | 4.6±0.1 |
| F | E | Apr | 1782 | 190 | 289±43 | 3.7±0.1 |
| F | E | Jul | 3660 | 286 | 501±80 | 3.7±0.1 |
| F | E | Oct | 3117 | 381 | 678±98 | 4.2±0.1 |
| F | M | Apr | 3012 | 289 | 559±101 | 3.9±0.1 |
| F | M | Jul | 3053 | 451 | 793±101 | 4.5±0.1 |
| F | M | Oct | 4300 | 555 | 910±98 | 4.7±0.1 |
| F | H | Apr | 2833 | 408 | 830±134 | 4.6±0.1 |
| F | H | Jul | 4883 | 371 | 673±99 | 3.5±0.1 |
| F | H | Oct | 3693 | 654 | 1278±156 | 5.0±0.1 |
, G, Gileppe; RR, Ry de Rome; F, Féronval.
, Water layer: E, epilimnion; M, metalimnion; H, hypolimnion.
, Apr, April 2010; Jul, July 2010; Oct, October 2010.
, data for OTU level (0.03 cut-off).
Bacterial Community Composition
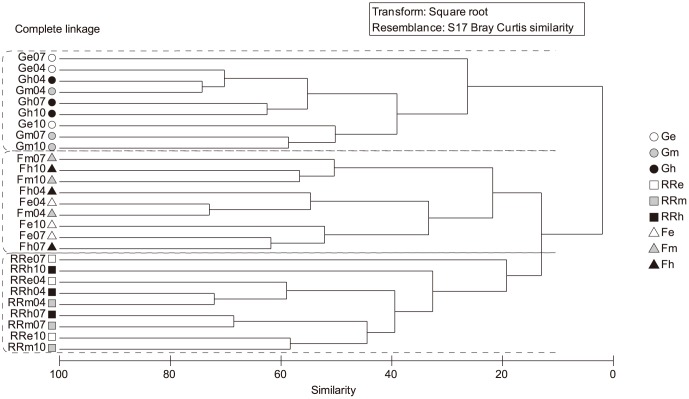
Hierarchical clustering based on the relative abundances of bacterial OTUs showed a clear structuring effect of water sample origin, resulting in three major clusters related to the analysed reservoirs (Fig. 3). Accordingly, samples from La Gileppe reservoir were significantly different and distinct from those of Ry de Rome and Féronval reservoirs (Fig. 3; p<0.01) as was also evidenced by ANOSIM analyses (Table 3). When comparing the BCC retrieved in the three reservoirs at different seasons, ANOSIM analyses showed differences in the BCC between April and July and between April and October, but not between July and October. However, clustering results presented in Fig. 3 clearly show that the origin of the samples (i.e., reservoir in which the sample was collected) clearly dominated the seasonal effect.
Figure 3. Clustering analyses.
Hierarchical clustering analyses of Bray-Curtis similarities of the community composition at OTU level (0.03 cut-off) in the three reservoirs. Legend: G, La Gileppe (dots); RR, Ry de Rome (square); F, Féronval (triangles); e, epilimnion (white); m, metalimnion (grey); h, hypolimnion (black); 04, April 2010; 07, July 2010; 10, October 2010.
Table 3. ANOSIM significance values based on OTUs (0.03 cut-off value).
| Test for differences between sampling locations Across all layer groups | R | p |
| Global test | 0.989 | 0.001 |
| Pairwise test | ||
| La Gileppe – Ry de Rome | 1.000 | 0.002 |
| Féronval – Ry de Rome | 0.975 | 0.002 |
| Féronval – La Gileppe | 1.000 | 0.001 |
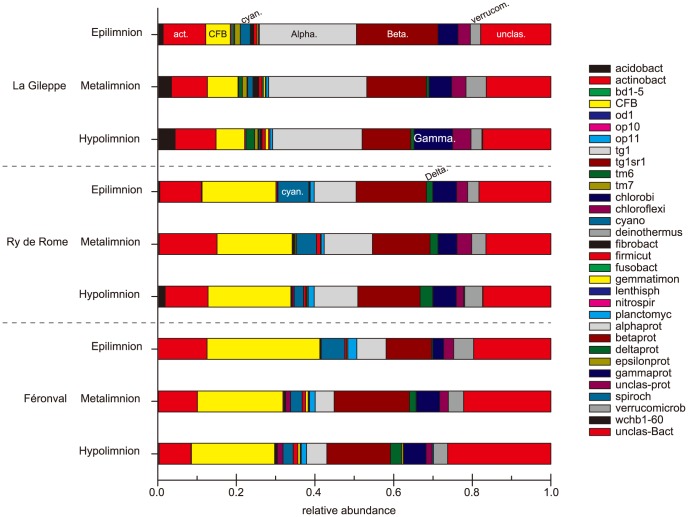
At phylum level, the three analysed reservoirs presented similar BCC with the dominance of few but typical freshwater bacterial groups (Fig. 4 and S1 Table) even though conspicuous variations in their relative abundance were also observed. Taxonomic affiliations evidenced Betaproteobacteria (range: 10.1% – 26.6%), CFB (range: 5.4% – 32.9%), Alphaproteobacteria (range: 3.5% – 32.0%), and Actinobacteria (range: 5.6% – 22.6%) as the four most abundant bacterial groups recovered in each reservoir (from first to fourth). The relative abundance of CFB-related OTUs was higher in Ry de Rome (deep waters) and Féronval (epilimnetic waters) reservoirs, whereas relative abundance of Alphaproteobacteria showed an opposite pattern (higher recovery in La Gileppe reservoir). Betaproteobacteira presented higher relative abundance in the epilimnetic waters of the less productive reservoirs (i.e., oligotrophic and mesotrophic) mainly due to Burkholderiales and unclassified betaproteobacterial OTUs contribution (S1 Table), which contrasted with their lower recovery in the deep waters of the poly-eutrophic reservoir (S1 Table). In turn, Actinobacteria-related OTUs showed similar values all over the three water layers, except for markedly higher values in metalimnetic waters of Ry de Rome reservoir and in the epilimnion of Féronval reservoir. Furthermore, unclassified bacterial OTUs (range: 8.4% – 30.1% of retrieved amplicons) also represented a large fraction in the analysed reservoirs, especially in meta- and hypolimnetic waters of Féronval reservoir. Some less abundant groups also exhibited a contrasted pattern. For instance, Acidobacteria were slightly more abundant in La Gileppe reservoir (average for all depths and sampling dates ca. 2.6% of the BCC) than in Ry de Rome (ca. 0.7%) and Féronval (ca. 0.2%) reservoirs. In contrast, an inverse pattern for Deltaproteobacteria was observed (average values of ca. 0.3%, 1.9%, and 1.0% for La Gileppe, Ry de Rome and Féronval; respectively).
Figure 4. Bacterial community composition.
Relative proportion of bacterial retrieved groups (by b-TEFAP) at OTU level (0.03 cut-off) in the water layers of the three analysed reservoirs.
In La Gileppe reservoir, the three water layers showed similar BCC (Fig. 4 and S1 Table) all over the three seasons, with dominance of Alpha-, Betaproteobacteria, Actinobacteria and unclassified OTUs. Alphaproteobacteria represented the main bacterial group in the oligotrophic reservoir although a lower relative abundance was observed in epilimnetic waters during mid-October. Moreover, slight changes in the hypolimnion were detected, including a higher relative contribution of Gammaproteobacteria (range: 7.6% – 10.2%) in comparison with the other retrieved groups. Furthermore, inverse patterns between Betaproteobacteria (decreasing values from epi- to hypolimentic waters ranging from 25.4% to 11.5%) and Gammaproteobacteria (increasing values from epi- to hypolimnetic waters; range: 4.2% – 10.2%) were also observed. No clear trend was observed for other typical freshwater groups like Verrucomicrobia and Acidobacteria (only detected in La Gileppe), even though slightly higher values were observed in all samples from meta- and hypolimnetic waters (range: 2.0% – 6.4% and 2.1% – 4.9%, respectively).
In the mesotrophic reservoir, the three water layers showed a BCC dominated by Betaproteobacteria, CFB, Actinobacteria and Alphaproteobacteria. No clear seasonal trend could be observed. Higher values of CFB (range: 13.5% – 25.2%) were obtained in hypolimnetic waters, whereas a higher relative abundance of Actinobacteria was observed in metalimnetic (range: 17.1% – 22.6%) samples. Other minor groups like Cyanobacteria (range: 2.0% – 11.5%) were also recovered from epi- and metalimnetic waters in Ry de Rome reservoir with higher relative abundance in mid-July samples (11.5% and 7.7% for epi- and metalimentic water samples, respectively). Slightly increasing relative abundance values with depth were observed for Verrucomicrobia in mid-July samples, but never exceeding 9.0% of the total BCC. Deltaproteobacteria evidenced higher relative abundance values in the hypolimnion, but never exceeded ca. 6.0% of total BCC.
CFB, Actinobacteria, Betaproteobacteria, Alphaproteobacteria and unclassified bacterial OTUs were also the main members of the BCC of the poly-eutrophic reservoir, as for the other studied reservoirs, but Lake Féronval harboured the highest contribution of CFB and the lowest one of Alphaproteobacteria. A slight decreasing trend of CFB relative abundance with depth and sampling period (from April to October) was observed. This trend was opposite for unclassified bacterial OTUs. As for Actinobacteria, a marked decreasing pattern with respect to depth (from epilimnion to hypolimnion) and period (from April to October) was observed. In the case of Betaproteobacteria, higher relative abundances were observed in meta- and hypolimnetic waters at the three seasons. Alphaproteobacteria represented a minor fraction of Féronval reservoir BCC with relative abundance values ranging from 3.5% to 7.3%. Delta- and Gammaproteobacteria (Methylococcales in poly-eutrophic reservoir Féronval and Legionellaceae in La Gileppe and Ry de Rome) showed increasing relative abundances with depth.
Overlapping microbial communities
Analyses of shared OTUs among the three reservoirs and water compartments were conducted by considering each environment as a whole (data not shown). In this sense, only 1.5% (59) of the OTUs were shared among the three freshwater reservoirs thus representing the core bacterial community of the analysed reservoirs microbiome (data not shown). Of those 59 OTUs, nine were highly abundant (1.4% – 11.6% of the reads) and were assigned to hgcl-clade (Actinobacteria), Methylophilaceae and Burkholderiales (Betaproteobacteria), and Chitinophagaceae (genera Sediminibacterium of CFB) families and to the uncultured vadinHA64 (Opitutae class of Verrucomicrobia) group. Besides, 8.75% of retrieved OTUs were present at least in two microbiomes (i.e., two reservoirs). A higher number of OTUs (233; 5.78%) was shared between the mesotrophic and the poly-eutrophic reservoirs whereas only 104 OTUs (2.58%) were shared between those reservoirs within the lower nutrient content range (data not shown). Interestingly, those reservoirs with the most different features only shared 16 OTUs (0.40%, data not shown). Besides, the highest number of unique OTUs (1613, 40.0%) was found in the poly-eutrophic reservoir whereas the oligotrophic, acid reservoir contained the lowest number (971 unique OTUs, 24.1%). Highly abundant and unique OTUs in La Gileppe reservoir belonged to Acetobacteraceae, Betaproteobacteria, Burkholderiales, Alphaproteobacteria, hgcl-clade and Acidobacteria, whereas in Ry de Rome reservoir belonged to Flavobacterium, hgcl-clade and Comamonadaceae. In turn, in the poly-eutrophic Féronval reservoir, abundant OTUs were more equally distributed between the Actinomycetales, Flavobacterium, Polynucleobacter, Methylococcales, hgcl-clade, and Planctomycetales. Venn diagrams were constructed in order to analyse the shared microbiome in each water compartment of the three reservoirs (Fig. 5A – C) clearly showing an increase in the number of OTUs with depth. Higher numbers of unique and shared OTUs were observed in relation to increasing nutrient loading. In this sense, La Gileppe contained less unique OTUs than Féronval reservoir and at the same time La Gileppe and Féronval shared less OTUs than Ry de Rome and Féronval, when comparing the three epilimnia and hypolimnia between them (Fig. 5A and C). However and due to the transition status that could characterize metalimnetic waters, Ry de Rome presented the lower number of exclusive OTUs (22.6%).
Figure 5. Shared bacterial community.
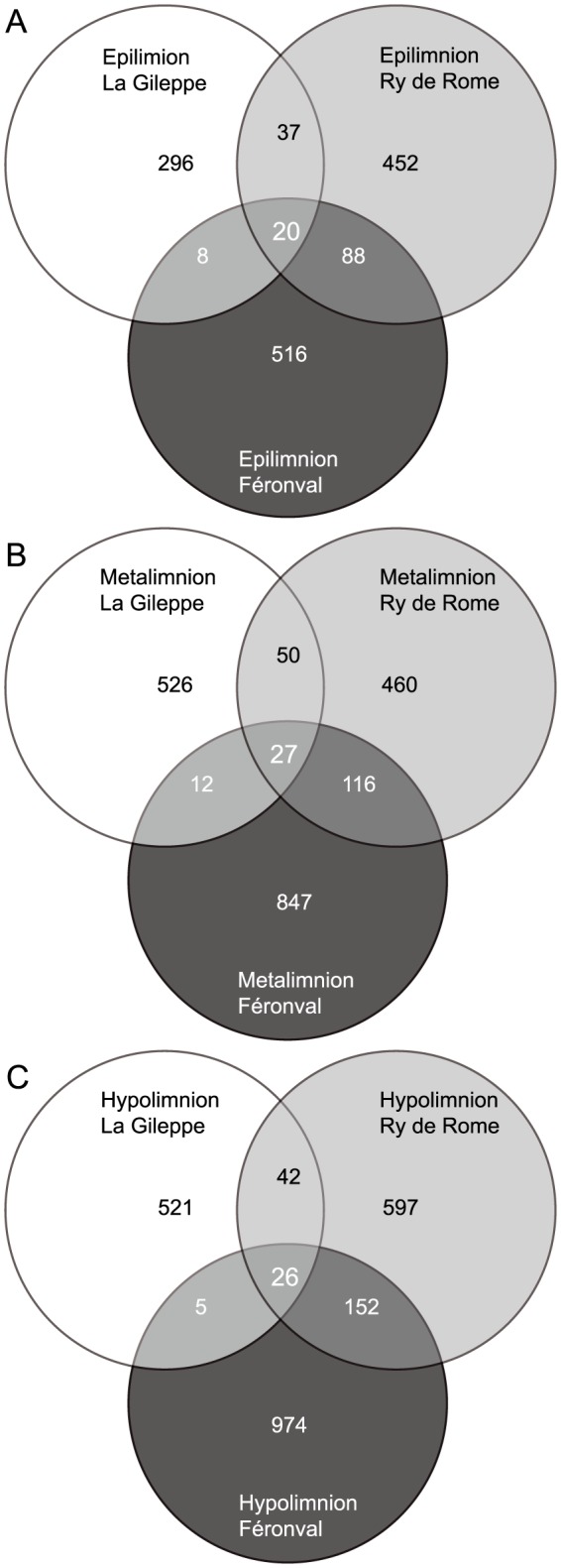
Venn diagrams highlighting shared OTUs for each analysed water compartment (A, epilimnion; B, metalimion; and C, hypolimnion) and reservoir (i.e., La Gileppe, Ry de Rome and Féronval).
Environmental factors driving BCC
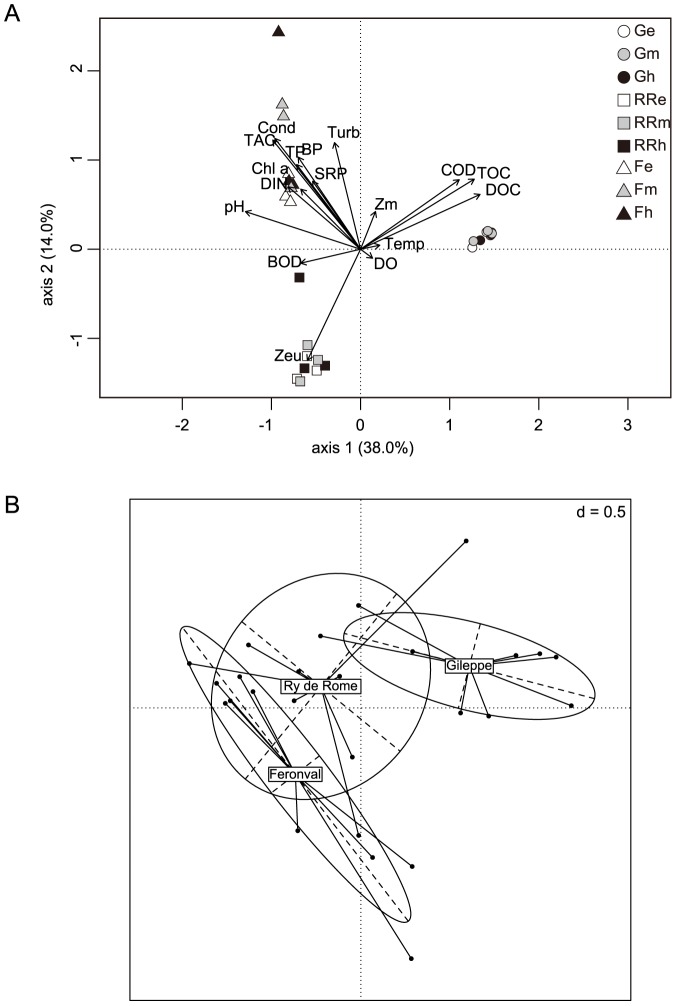
Statistical analyses of the analysed environmental parameters by means of CCA showed a clear segregation among lakes (permutest, F = 2.19, p = 0.001). In particular, samples from La Gileppe reservoir formed a cluster well separated from those of the two other reservoirs in the first axis; as Ry de Rome and Féronval reservoirs grouped separately mainly along the second axis with a smaller covariance (Fig. 6A and B). The DOC, TOC and COD were shown to be associated with BCC in La Gileppe, whereas BCC in Féronval was correlated with multiple parameters such as pH, Chl a, TP, BP and total alkalinity. The effect of environmental parameters on BCC was confirmed by Mantel tests (BIO-ENV, Table 4), where DOC and TOC together with pH and TA were evidenced as the strongest factors structuring BCC (Fig. 6B).
Figure 6. Canonical and Factorial correlation analyses.
Canonical correlation analyses (A) and factorial correspondence analysis (B) of bacterial communities based on relative abundance at OTU level (0.03 cut-off) and environmental parameters in the analysed reservoirs and all sampling dates. Legend: G, La Gileppe (dots); RR, Ry de Rome (square); F, Féronval (triangles); e, epilimnion (white); m, metalimnion (grey); h, hypolimnion (black); 04, April 2010; 07, July 2010; 10, October 2010.
Table 4. Results from BIO-ENV analyses of bacterial community composition and environmental parameters.
| Variables | ρ |
| DOC | 0.669 |
| pH | 0.582 |
| TOC | 0.572 |
| TAC | 0.526 |
| pH, TAC, DOC, TOC | 0.722 |
| pH, TAC, DOC | 0.764 |
| pH, Cond, DOC, TOC | 0.763 |
The Spearman rank ρ values indicate rank correlation between the matrix of bacterial community composition similarity and the similarity matrices from environmental variables. Only significant p values are shown.
Furthermore, the abundance profile of each OTU was correlated with each measured environmental parameter and those OTUs that were significantly linked to any environmental parameter were identified (Pearson correlation ρ>0.85, p<0.05, S2 Table). In this sense, a strong correlation between Chl a and OTUs related to Flavobacteriales, Comamonadaceae, Cyanobacteria and hgcl-clade (Actinobacteria) were observed; whereas, positive correlations between DIN and Cyanobacteria were also observed, especially in Féronval reservoir.
Discussion
In the present study, three distinct freshwater reservoirs were analysed covering a wide range of physical and chemical contrasting conditions (Table 1) resulting in distinct phytoplankton (Fig. 2) and bacterioplankton (Fig. 5 and S1 Table) community compositions. The bacterial diversity identified in the three freshwater reservoirs further extends the presence and dominance of few bacterial groups previously detected in epilimnetic waters of natural freshwater lakes [34]. In our study, we examined the recovery of these cosmopolitan bacterial groups according to water physical and chemical properties, water layers and seasons in man-constructed lakes.
In concordance with previous works on freshwater lakes [21], [34], the three reservoirs presented a typical freshwater bacterial community with a core microbiome mainly composed by Actinobacteria (hgcI_clade), Betaproteobacteria (Rhodoferax and Polynucleobacter) and Verrucomicrobia (vadinHA64) groups. In this sense and in agreement with their stable [66] and cosmopolitan [34] behaviour, members of Actinobacteria have been detected through time and space in all three analysed water bodies (relative abundance never exceeding 20.0%, Fig. 5 and S1 Table). A possible explanation for the widespread detection of Actinobacteria could be the reduction of grazing losses due to their reduced cell size and cell wall type [34], [67]–[69]. Accordingly, no clear trend for Actinobacteria could be detected depending on reservoir trophic status. However, lower relative abundance of Actinobacteria was observed in the meta- and hypolimnion than in the epilimnion of the studied poly-eutrophic reservoir as observed in several lakes of contrasting limnological characteristics [70], [71].
Data from the present study showed high relative abundance of CFB in the meso- and poly-eutrophic reservoirs, agreeing with current knowledge on CFB showing that this bacterial group was mainly found in environments containing high autochthonous organic carbon [72], [73]. These two reservoirs are prone to phytoplankton blooms, with blooms of Gonyostomum semen in the mesotrophic Ry de Rome reservoir and a rich community of diatoms, cryptophytes and chrysophytes in the poly-eutrophic Féronval reservoir. Accordingly, a low contribution of CFB to the BCC was observed in the oligotrophic La Gileppe reservoir, where DOC is mostly allochthonous, supporting previous evidences of low relative contribution of CFB in lakes characterized by low primary productivity [72]–[74].
Alphaproteobacteria have been described to be dominant in oligotrophic and in acid humic rich environments [75]. In the oligotrophic reservoir studied, a higher relative abundance of Rhizobiales and Rhodospirillales was obtained, whereas lower relative abundances were observed in the meso- and poly-eutrophic reservoirs, with much lower content of humic acids. However, our data could not establish a clear link between Alphaproteobacteria and Cyanobacteria as previously evidenced for marine and freshwater environments [76]. Betaproteobacteria usually represent the most abundant bacteria (up to 60 – 70% of total cells numbers) inhabiting lacustrine epilimnetic waters [34], with BetI and BetII groups being responsible for the overall Betaproteobacteria dominance in freshwater habitats [77]. BetI tribe (e.g., BAL47, Rhodoferax, GSK and Limnohabitants clades) are known to prefer algal-derived DOC [20] while members of the BetII tribe (e.g., Polynucleobacter) mostly consume photo-oxidised products from humic acids [78], [79]. Data retrieved in the present study evidence similar relative abundance of Betaproteobacteria in the three studied reservoirs. However, a dominance of Burkholderia-related OTUs was observed in the epilimia of La Gileppe and Ry de Rome while a dominance of Rhodocyclales-related OTUs was observed in meta- and hypolimnetic waters of Féronval reservoir in relation to its high nitrogen content (Table 1 and Fig. 6A). Furthermore, Gammaproteobacteria were not particularly abundant in the three studied reservoirs as was also reported for freshwater lakes [31] even if culture studies [39], [80] suggest better growth of this group under N- and P-enriched media. Our results, on the contrary, evidenced a slightly higher relative abundance of Gammaproteobacteria in the hypolimnion of La Gileppe reservoir (Fig. 4), which has lower TP content than the poly-eutrophic Féronval reservoir (Table 1). Furthermore, the most recovered Gammaproteobacteria (Methylococcales, representing up to 50% of all Gammaproteobacteria) were more abundant in the poly-eutrophic Féronval reservoir, which could indicate putative methane oxidation processes in this water body.
Another cosmopolitan bacterial inhabitant of freshwater lakes are Verrucomicrobia, which showed a wide geographical distribution [34]. This bacterial group was recovered from the three reservoirs studied supporting the idea of Verrucomicrobia as a bacterial group possessing a wide variety of metabolic strategies [81].
The present study reveals significant differences at BCC level per reservoir. In this sense, the BCC of the mesotrophic Ry de Rome reservoir was more similar to the BCC of the poly-eutrophic reservoir Féronval than to the oligotrophic one (i.e., La Gileppe; Fig. 3). By contrast, a fingerprinting analysis conducted in several Portuguese water bodies, including reservoirs, lakes and rivers, evidenced more similarities among oligo- and mesotrophic water bodies than with respect to eutrophic ones [82]. The observed BCC similarities in the present study between Féronval and Ry de Rome reservoirs might be related to the origin of their organic carbon (i.e., autochthonous) and to phytoplankton community composition. Several studies evidenced that freshwater lakes from the same catchment area (as the case of Féronval and Ry de Rome) were more similar in terms of BCC than isolated and not-connected lakes, in which these similarities could not be related to water transport of bacterial cells [83], [84]. Interestingly, the closest reservoirs in terms of distance also showed strong differences with respect to BP, cell abundances and nutrient content (S3 Fig. and Table 1). The differences mentioned above might also contribute to explain the differences observed in the BCC in relation to those heterotrophic microbes like CFB and Alpha- and Betaproteobacteria (Fig. 4 and S1 Table). BP values obtained in the present study were in agreement with previously reported values for temperate lakes [85].
In contrast with other studies carried out in freshwater lakes that evidenced a strong effect of depth in BCC structure [15], [86], no clear vertical stratification of the BCC within the water column was observed. The similarity among water layers might result from the fact that present microbial groups have broad metabolic capabilities, and were able to develop in both oxic and illuminated epilimnetic waters as well as in anoxic hypolimna. Besides, reservoirs strongly differ from natural lakes in their shorter water residence time and in the fact that they present a longitudinal gradient from their inlet, where river-like conditions prevail, to the dam, where lake-like conditions develop. Accordingly, plankton communities differ along this gradient [87], [88] and it is likely that BCC in reservoirs depends on inputs from the inflowing rivers thus reacting more to the variations along the longitudinal gradients than to the variations among water layers during the stratification period.
The statistical analyses revealed that organic carbon (either total or dissolved) and its origin together with pH and alkalinity exert the strongest segregation effect on the BCC in the studied reservoirs (Fig. 6). The seasonal succession of the BCC observed in the present study was associated with changes in key physical and chemical parameters and also to changes in phytoplankton biomass and community composition, which might reflect interactions between phyto- and bacterioplankton communities. Such correlations between BCC and phytoplankton community were also previously observed across lakes and seasons [89], [90]. Nutrient content in freshwater environments has long been seen as an important environmental factor controlling planktonic community composition [13], [19] as revealed in the present study where differences in both phyto- and bacterioplankton community composition level according to nutrient status (i.e., organic carbon content) were observed but also influenced by pH (understood as a proxy for humic acid) and alkalinity (Fig. 6). Furthermore, other parameters like presence of aquatic vegetation, zooplankton community composition, and viruses might also exert some influence in BCC in the analysed reservoirs [3], [91], [92].
Conclusions
The present study evidenced differences in terms of physical and chemical data and phytoplankton community composition in the three freshwater reservoirs, whereas they harboured a core microbiome composed of cosmopolitan bacterial groups (Actinobacteria (hgcI_clade), Betaproteobacteria (Rhodoferax and Polynucleobacter) and Verrucomicrobia (vadinHA64)) as recently stated for the epilimnion of freshwater lakes [34]. The dissimilarities observed in the BCC of the analysed reservoirs mainly resulted from differences in the typology of organic carbon content (i.e., La Gileppe reservoir is rich in allochthonous organic carbon). As for freshwater lakes, data presented here for reservoirs also evidenced strong relationships between trophic status, phytoplankton community and productivity. Future studies might focus on analysing the DOC fluxes, the archaeal counterparts, as well as the bacterial grazing and lysis due to viruses in the reservoirs in order to decipher the potential role in nutrients cycling of the microbial communities present.
Supporting Information
Sample locations. Map of Belgium showing the location of analysed reservoirs (G, for La Gileppe, RR, for Ry de Rome and F, for Féronval) described in this study.
(PDF)
Temperature depth profiles. Vertical depth profiles of temperature over time in the three analysed reservoirs (A, La Gileppe; B, Ry de Rome; and C, Féronval) during 2010.
(PDF)
Bacterial production and abundances. Bacterial production (BP; bars) and bacterial abundances (DAPI stained cells; dots) in the epilimnion (E), metalimnion (M) and hypolimnion (H) of La Gileppe, Ry de Rome and Féronval reservoirs. Legend: Black symbols correspond to April 2010 samples, white symbols correspond to July 2010 samples and grey symbols correspond to October 2010 samples.
(PDF)
Rank abundance. Rank abundance plots of retrieved OTUs (0.03 cut-off) from La Gileppe (black dots), Ry de Rome (grey dots) and Féronval (red dots) reservoirs.
(PDF)
Bacterial diversity. Bacterial groups retrieved after bTEFAP from the three analysed reservoirs (full depths).
(XLSX)
Pearson correlations. Pearson correlation between OTUs (0.03 cut-off level) and environmental factors (ρ>0.85, p<0.05).
(XLSX)
Acknowledgments
The authors thank S Crevecoeur for assistance and collaboration in laboratory and fieldwork.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files. Furthermore, all sequences generated in the present study can be accessed through National Center for Biotechnology Information (NCBI) under the bioproject number: PRJNA241494.
Funding Statement
The Fonds National de la Recherche Scientifique (FNRS, Belgium; project 2.4.547.09) funded this work. MLl was recipient of a postdoctoral CERUNA grant from the Université de Namur (Belgium). ÖI benefits from a FNRS postdoctoral grant. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Falkowski PG, Fenchel T, DeLong EF (2008) The microbial engines that drive earth's biogeochemical cycles. Science 320:1034–1039 10.1126/science.1153213 [DOI] [PubMed] [Google Scholar]
- 2. Cole JJ, Findlay S, Pace ML (1988) Bacterial production in fresh and saltwater ecosystems: a cross-system overview. Mar Ecol Prog Ser 43:1–10. [Google Scholar]
- 3. Berdjeb L, Ghiglione JF, Domaizon I, Jacquet S (2011) A 2-year assessment of the main environmental factors driving the free-living bacterial community structure in Lake Bourget (France). Microb Ecol 61:941–954 10.1007/s00248-010-9767-6 [DOI] [PubMed] [Google Scholar]
- 4. Edwards RA, Rodriguez-Brito B, Wegley L, Haynes M, Breitbart M, et al. (2006) Using pyrosequencing to shed light on deep mine microbial ecology. BMC Genomics 7:57 10.1186/1471-2164-7-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Roesch LF, Fulthorpe RR, Riva A, Casella G, Hadwin AKM, et al. (2007) Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J 1:283–290 10.1038/ismej.2007.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Frias-Lopez J, Shi Y, Tyson GW, Coleman ML, Schuster SC, et al. (2008) Microbial community gene expression in ocean surface waters. Proc Natl Acad Sci USA 105:3805–3810 www.pnas.org/cgi/doi/.10.1073/pnas.0708897105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Huse SM, Dethlefsen L, Huber JA, Welch DM, Relman DA, et al. (2008) Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet 4:e1000255 10.1371/journal.pgen.1000255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Huse SM, Welch DM, Morrison HG, Sogin ML (2010) Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ Microbiol 12:1889–1898 10.1111/j.1462-2920.2010.02193.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lindström ES (2000) Bacterioplankton community composition in five lakes differing in trophic status and humic content. Microb Ecol 40:104–113 10.1007/s002480000036 [DOI] [PubMed] [Google Scholar]
- 10. Lindström ES (2001) Investigating influential factors on bacterioplankton community composition: results from a field study of five mesotrophic lakes. Microb Ecol 42:598–605. [DOI] [PubMed] [Google Scholar]
- 11. Ptacnik R, Solimini AG, Andersen T, Tamminen T, Brettum P, et al. (2008) Diversity predicts stability and resource use efficiency in natural phytoplankton communities. Proc Natl Acad Sci USA 105:5134–5138 10.1073/pnas.0708328105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Reiss J, Bridle JR, Montoya JM, Woodward G (2009) Emerging horizons in biodiversity and ecosystem functioning research. TRENDS in Ecology and Evolution 24:505–514 10.1016/j.tree.2009.03.018 [DOI] [PubMed] [Google Scholar]
- 13. Weyhenmeyer GA, Peter H, Willén E (2012) Shifts in phytoplankton species richness and biomass along a latitudinal gradient – consequences for relationships between biodiversity and ecosystem functioning. Freshwater Biol 58:612–623. [Google Scholar]
- 14. Dumestre JF, Casamayor EO, Massana R, Pedrós-Alió C (2001) Changes in bacterial and archaeal assemblages in an equatorial river induced by the water eutrophication of Petit Saut dam reservoir (French Guiana). Aquat Microb Ecol 26:209–221. [Google Scholar]
- 15. Yannarell AC, Kent AD, Lauster GH, Kratz TK, Triplett EW (2003) Temporal patterns in bacterial communities in three temperate lakes of different trophic status. Microb Ecol 46:391–405 10.1007/s00248-003-1008-9 [DOI] [PubMed] [Google Scholar]
- 16. Yannarell AC, Triplett EW (2004) Geographic and environmental sources of variation in lake bacterial community composition. Appl Environ Microbiol 71:227–239 10.1128/AEM.71.1.227-239.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yannarell AC, Triplett EW (2004) Within- and between-lake variability in the composition of bacterioplankton communities: investigations using multiple spatial scales. Appl Environ Microbiol 70:214–223 10.1128/AEM.70.1.214-223.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lindström ES, Kamst-Van Agterveld MP, Zwart G (2005) Distribution of typical freshwater bacterial groups is associated with pH, temperature, and lake water Retention Time. Appl Environ Microbiol 71:8201–8206 10.1128/AEM.71.12.82018206.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kritzberg ES, Langenheder S, Lindström ES (2006) Influence of dissolved organic matter source on lake bacterioplankton structure and function - implications for seasonal dynamics of community composition. FEMS Microb Ecol 56:406–417 10.1111/j.1574-6941.2006.00084.x [DOI] [PubMed] [Google Scholar]
- 20. Simek K, Hornák K, Jezbera J, Nedoma J, Znachor P, et al. (2008) Spatio-temporal patterns of bacterioplankton production and community composition related to phytoplankton composition and protistan bacterivory in a dam reservoir. Aquat Microb Ecol 51:249–262. [Google Scholar]
- 21. Jones SE, Newton RJ, Mcmahon KD (2009) Evidence for structuring of bacterial community composition by organic carbon source in temperate lakes. 11:2463–2472 10.1111/j.1462-2920.2009.01977.x [DOI] [PubMed] [Google Scholar]
- 22. Lindström ES, Feng XM, Granéli W, Kritzberg ES (2010) The interplay between bacterial community composition and the environment determining function of inland water bacteria. Limnol Oceanogr 55:2052–2060 10.4319/lo.2010.55.5.2052 [DOI] [Google Scholar]
- 23. Ruiz-González C, Proia L, Ferrera I, Gasol JM, Sabater S (2013) Effects of large river dam-regulation on bacterioplankton community structure. FEMS Microb Ecol 84:316–331 10.1111/1574-6941.12063 [DOI] [PubMed] [Google Scholar]
- 24. Andersson AF, Riemann L, Bertilsson S (2009) Pyrosequencing reveals contrasting seasonal dynamics of taxa within Baltic Sea bacterioplankton communities. ISME J 4:171–181 10.1038/ismej.2009.108 [DOI] [PubMed] [Google Scholar]
- 25. Logue JB, Langenheder S, Andersson AF, Bertilsson S, Drakare S, et al. (2012) Freshwater bacterioplankton richness in oligotrophic lakes depends on nutrient availability rather than on species–area relationships. ISME J 6:1127–1136 10.1038/ismej.2011.184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pedrós-Alió C (2006) Marine microbial diversity: can it be determined? Trends Microbiol 14:257–263 10.1016/j.tim.2006.04.007 [DOI] [PubMed] [Google Scholar]
- 27. Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, et al. (2006) Microbial diversity in the deep sea and the underexplored “rare biosphere.”. Proc Natl Acad Sci USA 103:12115–12120 10.1073/pnas.0605127103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pedrós-Alió C (2007) Dipping into the Rare Biosphere. Science 315:192–193 10.1126/science.1135933 [DOI] [PubMed] [Google Scholar]
- 29. Glöckner FO, Zaichikov E, Belkova N, Denissova L, Pernthaler J, et al. (2000) Comparative 16S rRNA analysis of lake bacterioplankton reveals globally distributed phylogenetic clusters including an abundant group of actinobacteria. Appl Environ Microbiol 66:5053–5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Simek K, Pernthaler J, Weinbauer MG, Hornák K, Dolan JR, et al. (2001) Changes in bacterial community composition and dynamics and viral mortality rates associated with enhanced flagellate grazing in a mesoeutrophic reservoir. Appl Environ Microbiol 67:2723–2733 10.1128/AEM.67.6.2723-2733.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zwart G, Crump BC, Kamst-Van Agterveld MP, Hagen F, Han S-K (2002) Typical freshwater bacteria: an analysis of available 16S rRNA gene sequences from plankton of lakes and rivers. Aquat Microb Ecol 28:141–155. [Google Scholar]
- 32. Newton RJ, Jones SE, Helmus MR, Mcmahon KD (2007) Phylogenetic ecology of the freshwater Actinobacteria acI lineage. Appl Environ Microbiol 73:7169–7176 10.1128/AEM.00794-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fortunato CS, Crump BC (2011) Bacterioplankton community variation across river to ocean environmental gradients. Microb Ecol 62:374–382 10.1007/s00248-011-9805-z [DOI] [PubMed] [Google Scholar]
- 34. Newton RJ, Jones SE, Eiler A, Mcmahon KD, Bertilsson S (2011) A guide to the natural history of freshwater lake bacteria. Microbiol Mol Biol Rev 75:14–49 10.1128/MMBR.00028-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Weiss P, Schweitzer B, Amann R, Simon M (1996) Identification in situ and dynamics of bacteria on limnetic organic aggregates (lake snow). Appl Environ Microbiol 62:1998–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Simek K, Hornák K, Jezbera J, Masin M, Nedoma J, et al. (2005) Influence of top-down and bottom-up manipulations on the R-BT065 subcluster of beta-proteobacteria, an abundant group in bacterioplankton of a freshwater reservoir. Appl Environ Microbiol 71:2381–2390 10.1128/AEM.71.5.2381-2390.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lemarchand C, Jardillier L, Carrias J-F, Richardot M, Debroas D, et al. (2006) Community composition and activity of prokaryotes associated to detrital particles in two contrasting lake ecosystems. FEMS Microb Ecol 57:442–451 10.1111/j.1574-6941.2006.00131.x [DOI] [PubMed] [Google Scholar]
- 38. Salcher MM, Pernthaler J, Zeder M, Psenner R, Posch T (2008) Spatio-temporal niche separation of planktonic Betaproteobacteria in an oligo-mesotrophic lake. Environ Microbiol 10:2074–2086 10.1111/j.1462-2920.2008.01628.x [DOI] [PubMed] [Google Scholar]
- 39. Gasol JM, Comerma M, García JC, Armengol J (2002) A transplant experiment to identify the factors controlling bacterial abundance, activity, production, and community composition in a eutrophic canyon-shaped reservoir. Limnol Oceanogr 47:62–77. [Google Scholar]
- 40. Trusova MY, Gladyshev MI (2002) Phylogenetic diversity of winter bacterioplankton of eutrophic siberian reservoirs as revealed by 16S rRNA gene sequence. Microb Ecol 44:252–259 10.1007/s00248-002-2020-1 [DOI] [PubMed] [Google Scholar]
- 41. Lymperopoulou DS, Kormas KA, Karagouni AD (2011) Variability of prokaryotic community structure in a drinking water reservoir (Marathonas, Greece). Microbes Environ 27:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Naselli-Flores L, Barone R (2005) Water-level fluctuations in mediterranean reservoirs: setting a dewatering threshold as a management tool to improve water quality. Hydrobiologia 548:85–99 10.1007/s10750-005-1149-6 [DOI] [Google Scholar]
- 43. Katsiapi M, Moustaka-Gouni M, Michaloudi E, Kormas KA (2011) Phytoplankton and water quality in a Mediterranean drinking-water reservoir (Marathonas Reservoir, Greece). Environ Monit Assess 181:563–575 10.1007/s10661-010-1851-3 [DOI] [PubMed] [Google Scholar]
- 44. Sarmento H, Descy J-P (2008) Use of marker pigments and functional groups for assessing the status of phytoplankton assemblages in lakes. J Appl Phycol 20:1001–1011 10.1007/s10811-007-9294-0 [DOI] [Google Scholar]
- 45.Poikane S (2009) Water Framework Directive intercalibration technical report. JRC Scientific and Technical Reports: 1–178.
- 46.American Public Health Association (2005) Standard methods for the examination of water and wastewater. Washingotn DC: Water Environmental Association. 1 pp.
- 47. Descy J-P, Higgins HW, Mackey DJ, Hurley JP, Frost TM (2000) Pigment ratios and phytoplankton assessment in northern Wisconsin lakes. J Phycol 36:274–286 10.1046/j.1529-8817.2000.99063.x [DOI] [Google Scholar]
- 48. Porter KG, Feig YS (1980) The use of DAPI for identifying and counting aquatic microflora. Limnol Oceanogr 25:943–948. [Google Scholar]
- 49. Fuhrman JA, Azam F (1982) Thymidine incorporation as a measure of heterotrophic bacterioplankton production in marine surface waters: Evaluation and field results. Mar Biol 66:109–120. [Google Scholar]
- 50. Servais P (1989) Bacterioplanktonic biomass and production in the river Meuse (Belgium). Hydrobiologia 174:99–110 10.1007/BF00014058 [DOI] [Google Scholar]
- 51. Posch T, Franzoi J, Prader M, Salcher MM (2009) New image analysis tool to study biomass and morphotypes of three major bacterio-plankton groups in an alpine lake. Aquat Microb Ecol 54:113–126 10.3354/ame01269 [DOI] [Google Scholar]
- 52. Simon M, Azam F (1989) Protein content and protein synthesis rates of planktonic marine bacteria. Mar Ecol Prog Ser 51:201–213. [Google Scholar]
- 53. García-Armisen T, Papalexandratou Z, Hendryckx H, Camu N, Vrancken G, et al. (2010) Diversity of the total bacterial community associated with Ghanaian and Brazilian cocoa bean fermentation samples as revealed by a 16 S rRNA gene clone library. Appl Microbiol Biotechnol 87:2281–2292 10.1007/s00253-010-2698-9 [DOI] [PubMed] [Google Scholar]
- 54. Yuan S, Cohen DB, Ravel J, Abdo Z, Forney LJ (2012) Evaluation of methods for the extraction and purification of DNA from the human microbiome. PLoS ONE 7:e33865 10.1371/journal.pone.0033865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dowd SE, Callaway TR, Wolcott RD, Sun Y, McKeehan T, et al. (2008) Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP). BMC Microbiol 8:125 10.1186/1471-2180-8-125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lenchi N, Inceoğlu O, Kebbouche-Gana S, Gana ML, Llirós M, et al. (2013) Diversity of microbial communities in production and injection waters of Algerian oilfields revealed by 16S rRNA gene amplicon 454 pyrosequencing. PLoS ONE 8:e66588 10.1371/journal.pone.0066588.s005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Shah V, Shah S, Kambhampati MS, Ambrose J, Smith N, et al. (2011) Bacterial and archaea community present in the Pine Barrens Forest of Long Island, NY: unusually high percentage of ammonia oxidizing bacteria. PLoS ONE 6:e26263 10.1371/journal.pone.0026263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, et al. (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200 10.1093/bioinformatics/btr381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Clarke KR, Gorley RN (2006) PRIMER V6: User Manual/Tutorial. Plymouth.
- 61.Oksanen J, Kindt R, Legendre P, O'hara B, Simpson GL, et al. (2008) Vegan: community ecology package. veganr-forger-projectrog. Available: http://vegan.r-forge.r-project.rog. Accessed 15 July 2013.
- 62.Clarke KR, Warwick RM (2001) Changes in marine communities: an approach to statistical analysis and interpretation.
- 63. Clarke KR, Ainsworth M (1993) A method of linking multivariate community structure to environmental variables. Mar Ecol Prog Ser 92:205–219. [Google Scholar]
- 64. Peres-Neto PR, Legendre P, Dray S, Borcard D (2006) Variation partitioning of species data matrices: estimation and comparison of fractions. Ecology 87:2614–2625. [DOI] [PubMed] [Google Scholar]
- 65. Thioulouse J, Chessel D, dec SD, Olivier J-M (1997) ADE-4: a multivariate analysis and graphical display software. Stat Comput 7:75–83 10.1023/A:1018513530268 [DOI] [Google Scholar]
- 66. Youngblut ND, Shade A, Read JS, Mcmahon KD, Whitaker RJ (2013) Lineage-specific responses of microbial communities to environmental change. Appl Environ Microbiol 79:39–47 10.1128/AEM.02226-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Pernthaler J, Posch T, Simek K, Vrba J, Pernthaler A, et al. (2001) Predator-specific enrichment of actinobacteria from a cosmopolitan freshwater clade in mixed continuous culture. Appl Environ Microbiol 67:2145–2155 10.1128/AEM.67.5.2145-2155.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hahn MW, Lünsdorf H, Wu Q, Schauer M, Höfle MG, et al. (2003) Isolation of novel ultramicrobacteria classified as actinobacteria from five freshwater habitats in Europe and Asia. Appl Environ Microbiol 69:1442–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Tarao M, Jezbera J, Hahn MW (2009) Involvement of cell surface structures in size-independent grazing resistance of freshwater Actinobacteria . Appl Environ Microbiol 75:4720–4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Allgaier M, Grossart H-P (2006) Diversity and seasonal dynamics of Actinobacteria populations in four lakes in northeastern Germany. Appl Environ Microbiol 72:3489–3497 10.1128/AEM.72.5.3489-3497.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Taipale S, Jones RI, Tiirola M (2009) Vertical diversity of bacteria in an oxygen-stratified humic lake, evaluated using DNA and phospholipid analyses. Aquat Microb Ecol 55:1–16. [Google Scholar]
- 72. Kirchman DL (2002) The ecology of Cytophaga-Flavobacteria in aquatic environments. FEMS Microb Ecol 39:91–100. [DOI] [PubMed] [Google Scholar]
- 73. Eiler A, Bertilsson S (2007) Flavobacteria blooms in four eutrophic lakes: linking population dynamics of freshwater bacterioplankton to resource availability. Appl Environ Microbiol 73:3511–3518 10.1128/AEM.02534-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. van der Gucht K, Vandekerckhove T, Vloemans N, Cousin S, Muylaert K, et al. (2005) Characterization of bacterial communities in four freshwater lakes differing in nutrient load and food web structure. FEMS Microb Ecol 53:205–220 10.1016/j.femsec.2004.12.006 [DOI] [PubMed] [Google Scholar]
- 75. Hutalle-Schmelzer KML, Zwirnmann E, Krüger A, Grossart H-P (2010) Enrichment and cultivation of pelagic bacteria from a humic lake using phenol and humic matter additions. FEMS Microb Ecol 72:58–73 10.1111/j.1574-6941.2009.00831.x [DOI] [PubMed] [Google Scholar]
- 76. Berg KA, Lyra C, Sivonen K, Paulin L, Suomalainen S, et al. (2009) High diversity of cultivable heterotrophic bacteria in association with cyanobacterial water blooms. ISME J 3:314–325 10.1038/ismej.2008.110 [DOI] [PubMed] [Google Scholar]
- 77. Jezbera J, Jezberová J, Koll U, Hornák K, Simek K, et al. (2012) Contrasting trends in distribution of four major planktonic betaproteobacterial groups along a pH gradient of epilimnia of 72 freshwater habitats. FEMS Microb Ecol 81:467–479 10.1111/j.1574-6941.2012.01372.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Watanabe K, Komatsu N, Ishii Y, Negishi M (2009) Effective isolation of bacterioplankton genus Polynucleobacter from freshwater environments grown on photochemically degraded dissolved organic matter. FEMS Microb Ecol 67:57–68 10.1111/j.1574-6941.2008.00606.x [DOI] [PubMed] [Google Scholar]
- 79. Hahn MW, Scheuerl T, Jezberová J, Koll U, Jezbera J, et al. (2012) The passive yet successful way of planktonic life: genomic and experimental analysis of the ecology of a free-living polynucleobacter population. PLoS ONE 7:e32772 10.1371/journal.pone.0032772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Simek K, Hornák K, Jezbera J, Nedoma J, Vrba J, et al. (2006) Maximum growth rates and possible life strategies of different bacterioplankton groups in relation to phosphorus availability in a freshwater reservoir. 8:1613–1624 10.1111/j.1462-2920.2006.01053.x [DOI] [PubMed] [Google Scholar]
- 81. Lindström ES, Vrede K, Leskinen E (2004) Response of a member of the Verrucomicrobia, among the dominating bacteria in a hypolimnion, to increased phosphorus availability. J Plankton Res 26:241–246. [Google Scholar]
- 82. de Figueiredo DR, Pereira MJ, Moura A, Silva L, Bárrios S, et al. (2007) Bacterial community composition over a dry winter in meso- and eutrophic Portuguese water bodies. FEMS Microb Ecol 59:638–650 10.1111/j.1574-6941.2006.00241.x [DOI] [PubMed] [Google Scholar]
- 83. Lindström ES, Bergström A-K (2005) Community composition of bacterioplankton and cell transport in lakes in two different drainage areas. Aquat Sci 67:210–219 10.1007/s00027-005-0769-2 [DOI] [Google Scholar]
- 84. Crump BC, Adams HE, Hobbie JE, Kling G (2007) Biogeography of bacterioplankton in lakes and streams of an Arctic tundra catchment. Ecology 88:1365–1378 Available: 10.1890/06-0387 [DOI] [PubMed] [Google Scholar]
- 85. Amado AM, Meirelles-Pereira F, Vidal LO, Sarmento H, Suhett AL, et al. (2013) Tropical freshwater ecosystems have lower bacterial growth efficiency than temperate ones. Front Microbiol 4:1–8 10.3389/fmicb.2013.00167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Peura S, Eiler A, Bertilsson S, Nykänen H, Tiirola M, et al. (2012) Distinct and diverse anaerobic bacterial communities in boreal lakes dominated by candidate division OD1. ISME J 6:1640–1652 10.1038/ismej.2012.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Masin M, Jezbera J, Nedoma J, Straškrabova V, Hejzlar J, et al. (2003) Changes in bacterial community composition and microbial activities along the longitudinal axis of two canyon-shaped reservoirs with different inflow loading. Hydrobiologia 504:99–113 10.1023/B:HYDR.0000008512.04563.0b [DOI] [Google Scholar]
- 88. Simek K, Armengol J, Comerma M, García JC, Kojecká P, et al. (2001) Changes in the epilimnetic bacterial community composition, production, and protist-induced mortality along the longitudinal axis of a highly eutrophic reservoir. Microb Ecol 42:359–371 10.1007/s00248-001-0014-z [DOI] [PubMed] [Google Scholar]
- 89. Kent AD, Jones SE, Lauster GH, Graham JM, Newton RJ, et al. (2006) Experimental manipulations of microbial food web interactions in a humic lake: shifting biological drivers of bacterial community structure. 8:1448–1459 10.1111/j.1462-2920.2006.01039.x [DOI] [PubMed] [Google Scholar]
- 90. Kent AD, Yannarell AC, Rusak JA, Triplett EW, Mcmahon KD (2007) Synchrony in aquatic microbial community dynamics. ISME J 1:38–47 10.1038/ismej.2007.6 [DOI] [PubMed] [Google Scholar]
- 91. Pernthaler J (2005) Predation on prokaryotes in the water column and its ecological implications. Nature Rev Microbiol 3:537–546 10.1038/nrmicro1180 [DOI] [PubMed] [Google Scholar]
- 92. Berdjeb L, Ghiglione JF, Jacquet S (2011) Bottom-up versus top-down control of hypo- and epilimnion free-living bacterial community structures in two neighboring freshwater lakes. Appl Environ Microbiol 77:3591–3599 10.1128/AEM.02739-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sample locations. Map of Belgium showing the location of analysed reservoirs (G, for La Gileppe, RR, for Ry de Rome and F, for Féronval) described in this study.
(PDF)
Temperature depth profiles. Vertical depth profiles of temperature over time in the three analysed reservoirs (A, La Gileppe; B, Ry de Rome; and C, Féronval) during 2010.
(PDF)
Bacterial production and abundances. Bacterial production (BP; bars) and bacterial abundances (DAPI stained cells; dots) in the epilimnion (E), metalimnion (M) and hypolimnion (H) of La Gileppe, Ry de Rome and Féronval reservoirs. Legend: Black symbols correspond to April 2010 samples, white symbols correspond to July 2010 samples and grey symbols correspond to October 2010 samples.
(PDF)
Rank abundance. Rank abundance plots of retrieved OTUs (0.03 cut-off) from La Gileppe (black dots), Ry de Rome (grey dots) and Féronval (red dots) reservoirs.
(PDF)
Bacterial diversity. Bacterial groups retrieved after bTEFAP from the three analysed reservoirs (full depths).
(XLSX)
Pearson correlations. Pearson correlation between OTUs (0.03 cut-off level) and environmental factors (ρ>0.85, p<0.05).
(XLSX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files. Furthermore, all sequences generated in the present study can be accessed through National Center for Biotechnology Information (NCBI) under the bioproject number: PRJNA241494.