SUMMARY
Post-transcriptional gene regulation by miRNAs and RNA binding proteins (RBP) is important in development, physiology and disease. To examine the interplay between miRNAs and the RBP ELAVL1 (a.k.a. HuR), we mapped miRNA binding sites at the transcriptome-wide scale in WT and Elavl1 knockout murine bone marrow-derived macrophages. Proximity of ELAVL1 binding sites attenuated miRNA binding to transcripts and promoted gene expression. Transcripts that regulate angiogenesis and macrophage/endothelial cross talk were preferentially targeted by miRNAs, suggesting that ELAVL1 promotes angiogenesis, at least in part, by antagonism of miRNA function. We found that ELAVL1 antagonized binding of miR-27 to the 3′UTR of Zfp36 mRNA and alleviated miR-27-mediated suppression of the RBP ZFP36 (a.k.a. Tristetraprolin). Thus the miR-27-regulated mechanism synchronizes the expression of ELAVL1 and ZFP36. This study provides a resource for systems-level interrogation of post-transcriptional gene regulation in macrophages, a key cell type in inflammation, angiogenesis and tissue homeostasis.
INTRODUCTION
In multicellular eukaryotes, post-transcriptional gene regulatory mechanisms are critical for coordinating complex cellular behavior during development, homeostasis and disease (Chang and Hla, 2011; Chekulaeva and Filipowicz, 2009; Farazi et al., 2011; Keene, 2007). Recent work has highlighted the essential roles played by microRNAs (miRNAs) and RNA binding proteins (RBPs) that interact with the cis-regulatory elements of transcripts to regulate gene expression (Ciafre and Galardi, 2013). Among the RBPs, ELAVL1 (also known as HuR) is ubiquitously expressed and binds to AU rich element (ARE)- and U-rich element (URE)-containing mRNAs (Abdelmohsen et al., 2007). ELAVL1 has been implicated in the regulation of cell cycle, cell migration, tumorigenesis, apoptosis, immunity, inflammation and angiogenesis (Abdelmohsen and Gorospe, 2010; Gorospe et al., 2011; Katsanou et al., 2009). Indeed, ELAVL1 expression is elevated in cancers of the breast, ovary, colon and brain (Denkert et al., 2006; Vo et al., 2012) and associated with poor prognosis (Abdelmohsen and Gorospe, 2010; Wang et al., 2013).
ELAVL1, located primarily in the nucleus, is translocated into the cytoplasm upon cellular perturbation and returns back into the nucleus (Abdelmohsen et al., 2007). ELAVL1 function is implicated in gametogenesis (Levadoux-Martin et al., 2003), placental development (Katsanou et al., 2009) and stem/progenitor cell survival (Ghosh et al., 2009). In macrophages, ELAVL1 regulates inflammatory and angiogenic processes (Zhang et al., 2012).
miRNAs are small RNAs of 21–24 nt that associate with Argonaute (Ago) proteins in the RNA induced silencing complex (RISC), and bind to partially complementary sites predominantly located in the 3′ untranslated region (UTR) of target mRNAs. RISC binding leads to repression of target gene expression by deadenylation and subsequent mRNA destabilization and/or translational repression (Chekulaeva and Filipowicz, 2009; Fabian et al., 2010). Recent work suggests dynamic regulation of miRNA-mediated gene silencing, including cooperative/antagonistic interactions with RBPs, such as ELAVL1, PUM1, and DND1. For example, let-7b/c binding to the 3′-UTR of c-Myc was strongly induced when ELAVL1 binds to a nearby ARE site (Kim et al., 2009). In contrast, ELAVL1 binding to the CAT-1 (cationic amino acid transporter 1) 3′-UTR blocks the miR-122 binding on the CAT-1 mRNA, allowing the exit of the CAT-1 mRNA from cytoplasmic processing bodies (P bodies) to induce expression of the CAT-1 protein (Bhattacharyya et al., 2006). Further, ELAVL1 and miR-200b antagonistically regulate Vegfa and angiogenesis in murine macrophages (Chang et al., 2013). PUM1-binding to a canonical site in the 3′UTR of the p27 (Cdkn1b) mRNA exposes a miRNA binding site and allowed miR-221/-222-mediated suppression of this critical cell cycle regulator (Kedde et al., 2010). In contrast, DND1 was found to antagonize the interaction of miR-221/222 with p27 mRNA, miR-430 with Nos1 mRNA (Kedde et al., 2007) and miR-21 with Msh2 (Bhandari et al., 2013). These studies indicate that miRNA suppression of gene expression is regulated by RBPs. Transcriptome-wide characterization of miRNA/RBP interaction has not been reported.
A high-throughput method called photoactivatable ribonucleoside-enhanced cross-linking and immunoprecipitation (PAR-CLIP) was developed recently which provided a means to precisely define binding sites of miRNAs and RBPs with their target transcripts at the global scale (Hafner et al., 2010a, b; Lebedeva et al., 2011). To gain a comprehensive understanding of ELAVL1-mediated modulation of miRNA/mRNA interaction sites in macrophages, we performed Ago2 PAR-CLIP experiments in WT and Elavl1 KO BMDM. We combined these data with miRNA and mRNA expression profiling to provide a global view of miRNA/ELAVL1 regulatory networks in vivo. From this, we found that in macrophages, genes that regulate angiogenesis and blood vessel development were highly targeted by miRNAs and ELAVL1. In addition, we identified a novel mechanism by which ELAVL1 controls the expression of ZFP36, a major RBP that destabilizes ARE-containing transcripts, thus achieving homeostasis of the ARE-containing transcriptome.
RESULTS
PAR-CLIP reveals the global landscape of ELAVL1-dependent miRNA targets in bone marrow derived macrophages (BMDM)
In order to define miRNA/mRNA interaction sites at the global scale, and to examine regulation of these sites by ELAVL1, we conducted Ago2 PAR-CLIP analysis in pooled BMDM (~ 108 cells) isolated from WT (ELAVL1f/f) and macrophage-specific Elavl1 KO (ELAVL1f/f LysM-Cre) mice. The outline of Ago2 PAR-CLIP experiments is described in Figure 1A.
Figure 1. Ago2 PAR-CLIP experiments in BMDM identify ELAVL1-regulated miRNA binding sites.
(A) Experimental outline for Ago2 PAR-CLIP in BMDM. WT and Elavl1 KO BMDM were labeled with 4-thiouridine (4-SU) and the PAR-CLIP experiments and bioinformatic pipeline are schematically represented. See also Figure S1. (B, C) Distribution of PAR-CLIP binding sites (Total sites (B) and high-read PAR-CLIP sites (C) were determined by aligning the PARalyzer results with the mouse MM9 genome.
PARalyzer was used to map reads to the mouse genome (mm9), and identify groups of overlapping reads, which represent Ago2-binding sites on the transcriptome (Corcoran et al., 2011). Approximately 3.5×106 reads from WT and ELAVL1 KO of the sequence datasets met our criteria and were further analyzed. In total we obtained ~25,500 clusters representing predicted Ago2 binding sites from both WT and Elavl1 KO. PAR-CLIP sites are most frequently associated with 3′UTRs (52%), followed by coding regions (20%), intergenic (17%) and intronic (7%) regions (Figure 1B). Further, more than 58% of high depth PAR-CLIP sites (>100 reads; 5,362 sites) are in the 3′ UTR (Figure 1C). We also searched the PAR-CLIP 3′UTR sites for the presence of G-rich element, which was purported to represent Ago2 binding sites on transcripts independently of miRNAs (Leung et al., 2011). However, the occurrence of G rich element was lower then random chance, suggesting that 3′UTR PAR-CLIP sites are likely to be primarily targeted by the RISC complex and were further analyzed as described below.
Characterization of the miRNome, transcriptome and mapping transcriptome-wide miRNA binding sites
Next, we quantified miRNA expression levels by RNA sequencing in BMDM of WT and Elavl1 KO mice and identified 211 expressed miRNA species (Table S1). Only 5 miRNAs showed more than 2 fold alteration in expression upon Elavl1 deletion (Figure S2).
We performed a similar analysis on the PAR-CLIP dataset to identify Ago2-associated miRNA species and quantified their relative abundance (Figure 2A), which revealed significant differences of miRNAs in this population with that of total miRNAs. However, differential cross-linking efficiency could result in erroneous abundance assignment in this population of miRNAs. Therefore, we used the total miRNA abundance data in the assignment of particular species of miRNAs to PAR-CLIP sites.
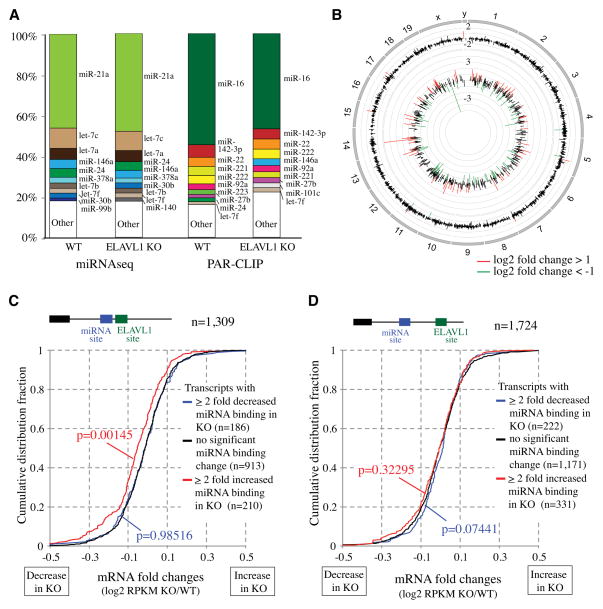
Figure 2. Transcriptome-wide view of ELAVL1 regulation of miRNA binding sites in BMDM.
(A) miRNA sequencing profile of total miRNAs and PAR-CLIP miRNAs in WT and Elavl1 KO BMDM. Colored sections indicate top 10 highly expressed miRNA. (B) The Circos plot shows transcriptome-wide view of mRNA levels and miRNA binding activity on transcripts. Results are presented as the ratio between Elavl1 KO/WT. The outer circle indicated the position of chromosomes 1–19 and sex chromosomes. The middle circle represents mRNA expression determined by RNAseq experiments. Log2 relative change for transcripts that have more than 0.1 RPKM in mRNA-seq (11,033 genes) are shown (n=4). The innermost track shows the ratio (KO/WT) of log2 relative reads of top 1000 PAR-CLIP sites at 3′ UTRs. Color code is as above. (C)(D) ELAVL1 depletion affects gene expression via interactions with miRNA binding sites on transcripts. (C) Among the transcripts with a predicted ELAVL1 site near miRNA binding sites (≥ 100 PAR-CLIP reads/ site), those in which miRNA binding is increased in the KO (≥ 2 fold) showed decreased expression (Red line). In contrast, genes with decreased miRNA binding (KO/WT ≤ 0.5, blue line) showed no change in expression level. (D) Among the transcripts with no predicted ELAVL1 site near miRNA binding sites (≥ 100 PAR-CLIP reads/ site), even when miRNA binding is increased (KO/WT ≥2, red line) or decreased (KO/WT < 0.5, blue line), there was no significant change in expression.
We also characterized the BMDM transcriptome by mRNAseq analysis. Sequencing analysis from WT and Elavl1 KO samples detected 11,033 mRNA species expressed in BMDM (RPKM ≥ 0.1) (Table S2). Only 14 of the mRNAs exhibited ≥ 2 fold change (p < 0.05). This dataset contains 8,043 transcripts that were also present in the PAR-CLIP data set. Among those, 12,323 miRNA binding sites in the 3′UTR region of 4,145 transcripts were mapped.
In order to identify high confidence miRNA binding sites, we selected PAR-CLIP sites with high reads (≥ 100) in 3′UTR sequences. As shown (Figure S3), 3,033 sites out of ~ 25,000 sites meet our stringent criteria; which is PAR-CLIP sites with ≥100 combined reads and > 98% sequence overlap at the binding site between WT and Elavl1 KO. Using this data set, the transcriptome-wide representation of miRNA binding sites and the associated mRNA expression profiles are represented in the Circos plot (Figure 2B). Significant number of miRNA binding sites showed altered binding strength in Elavl1 KO BMDM; 466 (15.36%) showed reduction and 432 (14.24%) showed increase (± two-fold or more). However, corresponding mRNAs did not show similar degree of changes in expression. Indeed, very few (0.03%) of the high miRNA-bound transcripts showed greater than 2 fold change in expression in the absence ELAVL1.
We categorized 3,033 sites into two groups based on the distance between Ago2 binding sites and ELAVL1 consensus sites. In the first group of transcripts in which Ago2 binding site and ELAVL1 consensus site are ≤ 30nt (total number = 1,309), increased miRNA binding significantly correlated with reduced mRNA expression (Figure 2C, red line). However, transcripts with decreased miRNA binding did not change mRNA expression levels (Figure 2E, blue line). In contrast, transcripts containing Ago2 binding site without or > 30 nt (total number = 1,724) away from ELAVL1 binding site did not show any changes in mRNA expression levels regardless of miRNA binding (Figure 2D). These data suggest that ELAVL1 competes with proximal miRNA binding on target mRNAs.
Prediction of miRNA/mRNA interactions from the PAR-CLIP dataset
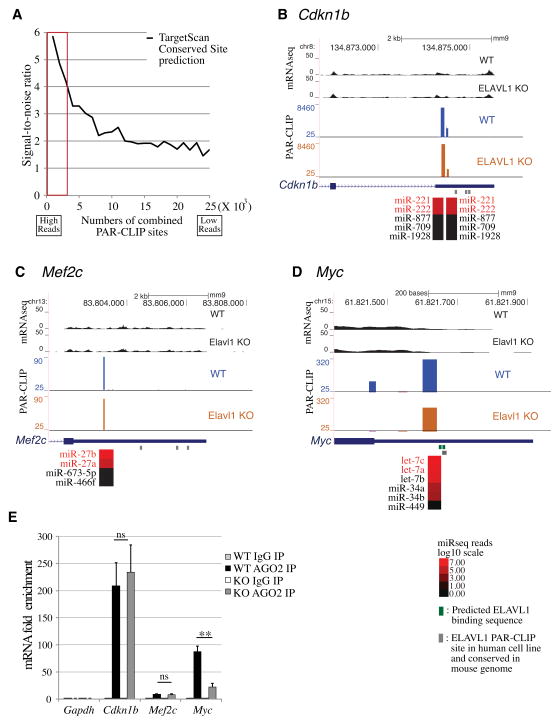
We annotated PAR-CLIP sites with predicted miRNA target sites using the TargetScan (TargetScan 6.2) (Lewis et al., 2005). Approximately 34% of high confidence PAR-CLIP sites at 3′ UTR region were assigned to at least one miRNA by TargetScan. We noticed that the high-read miRNA binding sites were more likely to be conserved among species. In addition, conserved miRNA binding sites with high reads have a high signal (bona fide PAR-CLIP signal) to noise (occurrence of a random 28 nt fragment from any 3′UTR that contain a TargetScan-predicted seed region) ratio (Figure 3A, red box). This indicates that our PAR-CLIP dataset is highly correlated with miRNA binding sites identified by computational methods. We also used seed sequences from the miRNA list that we identified in BMDM (Table S1) to predict miRNA/mRNA binding. Since several confirmed miRNA target genes contain G:U base-pairs or single nucleotide bulges, we used 7mer seed sequence and allowed one G:U wobble in our prediction. Using this work flow, we were able to assign at least one miRNA to 87.9% of PAR-CLIP sites in the 3′UTR region. These predicted miRNA/mRNA pairs for PAR-CLIP sites are shown in table S3. If more than 1 miRNAs were identified by miRNA target prediction, preference was given to the more abundant miRNA species.
Figure 3. Detection of constitutive and ELAVL1-modulated miRNA binding on 3′ UTR.
(A) BMDM Ago2 PAR-CLIP sites with high reads contain miRNA binding sites predicted by TargetScan. PAR-CLIP sites were sorted by the number of combined reads (WT and Elavl1 KO) from high to low and plotted in the X axis. The signal to noise ratio on Y axis represents the probability of finding miRNA seed sequences in the PAR-CLIP sites normalized to the probability of TargetScan prediction of a random 3′UTR sequence of similar length. (B) miRNA binding sites (PAR-CLIP signals) on Cdkn1b (p27) mRNA 3′ UTR. Two conserved miR-221/222 binding sites across human and mouse were reported on Cdkn1b 3′ UTR regions and detected in our PAR-CLIP data with very high reads. (C) miR-27 binding on Mef2c 3′ UTR was reported in mouse embryonic cardiomyocytes and detected in our dataset. (D) miRNA binding sites on the c-Myc mRNA. UCSC genome browser screen shot was annotated with predicted miRNA species (red). Potential ELAVL1 binding sites are indicated with green bars. Actual ELAVL1 binding sites from previous PAR-CLIP in human HEK293 cell are indicated with gray bars. MiRNA expression levels in BMDM were shown as a heatmap in log10 scale. (E) Validation of miRNA binding to Cdkn1b, Mef2c and Myc transcripts. WT or Elavl1 KO cell lysates were immunoprecipitated with control IgG or anti-Ago2 antibody and subjected to qRT-PCR to quantify Cdkn1b, Mef2c and Myc transcripts. Hprt was used for normalization and Gapdh was used as negative control. t-Test for significance between Ago2 IP and control (IgG): **P < 0.01). Error bars indicate s.d.
This analysis identified previously characterized miRNA/mRNA interactions correctly. For example, the Cdkn1b transcript, which encodes the cyclin-dependent kinase inhibitor p27, showed two tandem miR-221/222 binding sites (Figure 3B), which is consistent with previous work (Kedde et al., 2010). We did not see alterations in miR-221 binding to Cdkn1b transcript in Elavl1 KO BMDM. Similarly, miR-27 interaction with the Mef2c transcript was identified (Figure 3C) (Chinchilla et al., 2011). For the c-Myc transcript, a major let-7 binding site which is 11 nt downstream of an ELAVL1 consensus motif was identified. Let-7 binding was decreased 1.4 fold in Elavl1 KO cells, suggesting that ELAVL1 binding to the c-Myc mRNA facilitates let-7/RISC complex binding to the transcript (Figure 3D). This is analogous to human ELAVL1 which cooperated with the let-7 miRNA to suppress C-MYC expression (Kim et al., 2009). To validate miRNA interaction with these transcripts, we immunoprecipitated WT and Elavl1 KO BMDM lysates with anti-Ago2 antibody and examined the amount of mRNAs (Figure 3E). Cdkn1b, Myc and Mef2c transcripts were associated with Ago2 complex and immunoprecipated mRNA level correlated with the strength of PAR-CLIP signal. Moreover, Ago2-associated Myc mRNA was significantly reduced in Elavl1 KO BMDM whereas Cdkn1b and Mef2c mRNAs were unchanged. These data validate our identification of specific miRNA binding sites and modulation of such interactions by ELAVL1 in BMDM.
miRNA-regulated angiogenic genes expression in BMDM and modulation by ELAVL1
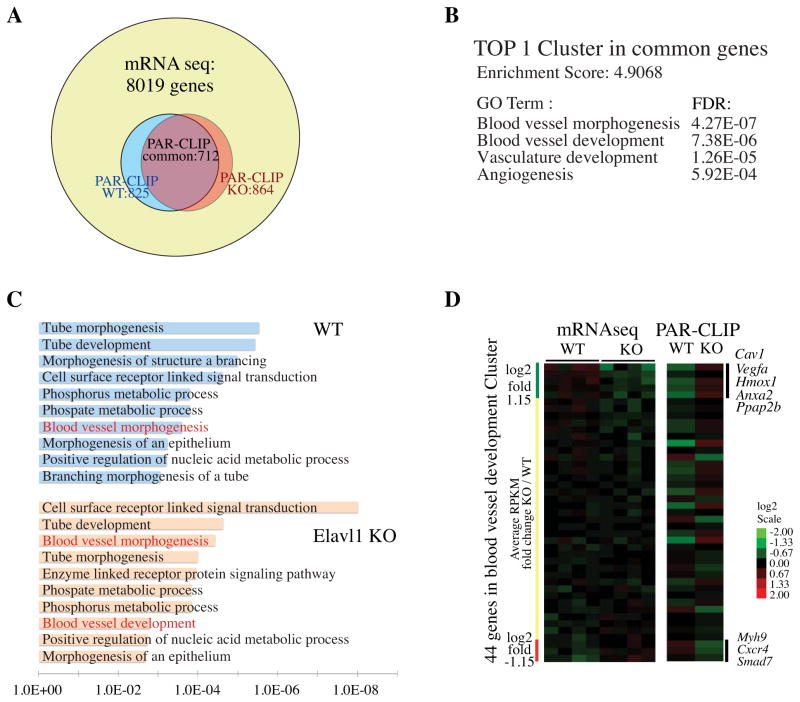
We analyzed transcripts that show strong miRNA binding (≥ 200 Ago2 PAR-CLIP reads) by Gene Ontology (GO) analysis to identify cellular processes that are regulated by miRNA-mediated gene regulation. Using these criteria, we identified 825 and 864 genes from WT and Elavl1 KO, respectively, among which 712 genes are common (Figure 4A). We performed GO analysis with 712 genes in the background of 8,019 expressed genes in BMDM. The analysis identified blood vessel morphogenesis/development, vasculature development and tube development/branching (collectively referred to as the angiogenesis cluster) as highly significant biological processes (Figure 4B, C). In the Elavl1 KO BMDM, cell surface receptor-linked signal transduction process was identified as most significant.
Figure 4. Gene ontology (GO) analysis for biological processes targeted by miRNAs in WT and Elavl1 KO BMDM.
(A) The Venn diagram shows the number of transcripts with high miRNA binding (≥ 200 PAR-CLIP / transcript) on 3′ UTR from WT and Elavl1 KO BMDM that were used for GO analysis using the DAVID bioinformatics program. An expressed gene pool (RPKM ≥ 0.1) from mRNAseq data was used as the background for GO analysis. (B) GO analysis identified 4 categories of GO terms that were related to blood vessel development from the top first GO cluster in common transcripts (n=712) that are highly targeted by miRNAs. (FDR: False discovery rate). (C) Bar plot of GO analysis results for the top 10 GO terms from WT and Elavl1 KO BMDM. GO terms that are common with those in (B) shown above were labeled in red. (D) Heat map of mRNA expression level (left panel) of 44 genes from blood vessel development/ angiogenesis cluster in (B) were compared with respective total PAR-CLIP signals on the 3′ UTR (right panel).
Even though miRNA binding of transcripts involved in the blood vessel development/angiogenesis cluster is modulated by ELAVL1, steady state mRNA levels do not show similar degree of fluctuations (Figure 4D). However, in general, there appears to be negative correlation between miRNA binding intensity and transcript expression levels. These data suggest that ELAVL1 competes with miRNA binding of a subset of mRNAs and that such interactions influence steady state mRNA levels in a reciprocal manner.
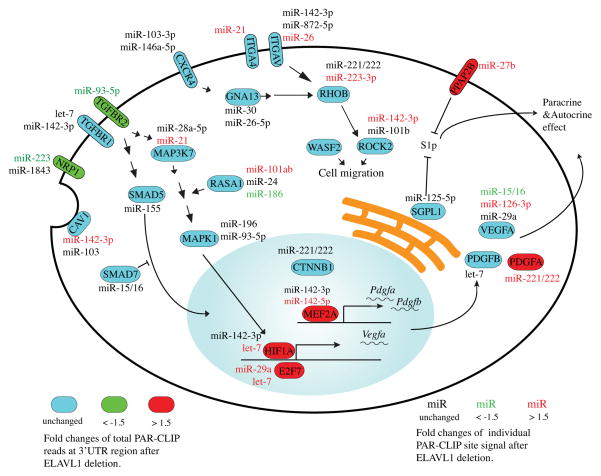
The miRNA-targeted transcripts in BMDM that are involved in angiogenesis/blood vessel development processes are shown in table S4. Transcripts that encode cell surface receptors involved in signal transduction, namely, CXC chemokine receptor-4 (Cxcr4), TGF-β receptor-1 and -2 (Tgfbr1, Tgfbr2), integrin α4 (Itga4), integrin αv (Itgav), VEGF-A co-receptor neuropilin-A (Nrp1), raft-specific protein caveolin (Cav1), cell-surface phosphatase specific for lipid lysophosphates (Ppap2b) were targeted by miRNAs. In addition, transcripts that encode the cell surface protease Adam8 (Adamts8) and phospholipid binding protein Annexin A2 (Anxa2), which acts as a cell-surface receptor for tissue-type plasminogen activator, were also identified. Transcripts encoding cytosolic signaling proteins, namely, the heterotrimeric Gα13 protein (Gna13), small G protein RhoB (Rhob), small GTPase activating protein Rasa (Rasa), cytoskeletal regulators – myosin H9 (Myh9), myosin 1e (Myo1e), Wasf2, signaling kinases – Mapk1, TGFβ-activated kinase-1 (Map3k7 aka Tak1), TGFβ–regulated signaling proteins (Smad5 and Smad7), RNA binding proteins – Quaking (Qk), Zinc finger protein 36-like 1 (Zfp36l1) and Rbm15, ubiquitin E3 ligase involved in Notch signaling Mindbomb (Mib1) and Bcl-2 family member Bak1 were targeted. ER/Golgi-resident proteins, inducible heme oxygenase (Hmox1), Nogo4 (Rtn4) and sphingosine 1-phosphate lyase (Sgpl1), ER-specific protein (Erap1) and β-galactosidase (B4galt1) involved in cell-surface glycan composition were also targeted by miRNAs. Nuclear proteins such as transcription factors – E2F7 (E2f7), hypoxia-inducible factor (HIF)-1α (Hif1a), myocyte enhancer factor (MEF)2a (Mef2a), myocardin-like (Mkl)-2 (Mkl2), upstream binding protein (UBP)-1 (Ubp1), zinc finger MIZ type containing (Zmiz-1) (Zmiz1) and β-catenin (Ctnnb1) were targeted by miRNAs. In addition, TCDD-inducible poly-A polymerase (TipArp), an enzyme involved in RNA metabolism was also targeted. Five of these mRNAs (E2f7, Hif1a, Mef2a, Pdgfa and Ppap2b) showed higher miRNA binding in the absence of ELAVL1 whereas Erap1and Nrp1 showed reduced targeting of miRNAs in the absence of ELAVL1. The schematic of biological roles and subcellular localization of proteins encoded by miRNA-targeted transcripts identified in BMDM is shown in Figure 5.
Figure 5. Schematic of miRNA-targeted transcripts in BMDM that regulate vessel development/ angiogenesis.
25 out of 44 transcripts in the vessel development/ angiogenesis cluster from GO analysis are depicted schematically to illustrate the signaling network targeted by specific miRNAs that are modulated by ELAVL1. Transcripts labeled in red indicate those in which miRNA binding in the 3′ UTR is antagonized by ELAVL1 (≥ 1.5 fold increase in PAR-CLIP reads in KO). Those labeled in green indicate that miRNA binding in the 3′ UTR is promoted by ELAVL1 (≤ 1.5 fold decreased in PAR-CLIP reads in KO). Individual miRNA ID with red and green colors indicate that miRNA binding sites on 3′ UTR was predicted to this miRNA and shows more than 1.5 fold increase or decrease. Those miRNAs indicated in black did not show alterations in WT and KO BMDM.
Macrophage transcripts involved in chemotaxis, paracrine interactions with vascular cells and TGFβ signaling are targeted by miRNAs and ELAVL1
Interestingly, miRNA targeting is prominent in macrophage genes important for migration and chemotaxis. For example, mRNA encoding integrins α4 and αv, chemokine receptor CXCR4, cell-surface lysophospholipid phosphatase LPP3, heterotrimeric Gα13 protein, small GTPase RhoB, small GTPase regulator Rasa1, Rho kinase Rock2, cytoskeletal regulators Myosin H9 and myosin 1e, S1P lyase (Sgpl1) and raft-resident Caveolin-1 proteins were targeted. These genes are involved in migration of macrophages in response to chemokines and lipid mediators as well as extracellular matrix molecules (Giannice et al., 2013; Grande-Garcia and del Pozo, 2008; Humtsoe et al., 2010; Lue et al., 2011; Mai et al., 2011; Mao et al., 2013; Rasheed et al., 2013; Wheeler and Ridley, 2007; Yester et al., 2011).
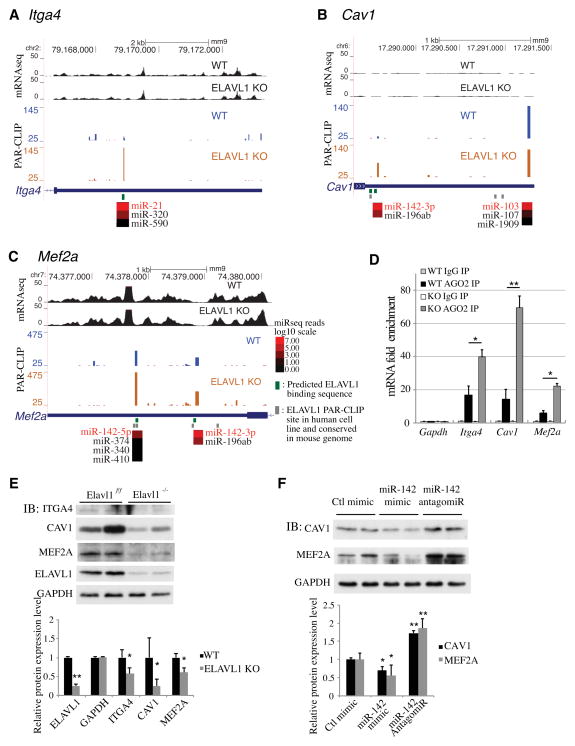
These mRNAs were further examined in detail for miRNA interaction sites. In the integrin alpha 4 gene (Itga4) 3′ UTR region, miR-21 binding site at position 2,100nt is induced ~ 3.6 fold in the Elavl1 KO BMDM. Indeed, an overlapping ELAVL1 binding motif was identified suggesting that miR-21 and ELAVL1 compete for the expression of ITGA4 polypeptide (Figure 6A). Similarly, miR-21 binding sites were predicted in the Map3k7 3′UTR (Figure S4A). This site is also located close to the ELAVL1 consensus site and Ago2 PAR-CLIP signal was increased in the Elavl1 KO BMDM. These data suggest that multiple transcripts in the macrophage migration pathway are targeted by miR-21, which are antagonized by ELAVL1 binding to mRNAs. In the Cav1 3′UTR region, two major miRNA binding sites for miR-103 and miR-142-3p were predicted (Figure 6B). miR-103 binding site showed strong Ago-2 PAR-CLIP signal both in WT and Elavl1 KO. Interestingly, miR-142-3p binding site where two potential ELAVL1 binding sites are closely located showed increased Ago2 PAR-CPIP signal (2.32 fold) in Elavl1 KO BMDM. This result suggests that competition of miR-142-3p and ELAVL1 binding regulates CAV1 expression. In a similar manner, miR-27b targeting of Tgfbr1 and miR-26a targeting of Itgav were also antagonized by ELAVL1 in BMDM (Figure S4B, C). These data suggest that miRNAs regulate the transcripts that encode proteins required for macrophage chemotactic and migratory responses and that these interactions are regulated by ELAVL1.
Figure 6. Modulation of miRNA binding sites on angiogenic regulatory transcripts by ELAVL1.
UCSC genome browser views of PAR-CLIP data and mRNA-seq data on Itga4 (A), Cav1 (B), and Mef2a (C) transcripts. Labeling is similar to figure 2 (above). The color code represents log10 scale of average miRNA reads from miRNA-seq data. Green bars represents potential ELAVL1 binding sequences that contain UUUNUUU or less than 2 A base in 7 poly U region (ex. UUAUUAU or UAUUUAU). Red-labeled miRNA ID represent the most likely miRNAs for a given PAR-CLIP site. (D) Itga4, Cav1 and Mef2a have higher miRNA binding in Elavl1 KO BMDM. BMDM lysates were immunoprecipitated with control IgG or Ago2 antibody and gene enrichment fold changes were measured by qRT-PCR. Hprt was used for normalization and Gapdh was used as a negative control. (E) The protein expression of ITGA4, CAV1 and MEF2A in WT and Elavl1 KO BMDM. The representative immunoblot image (top) and densitometry analysis (bottom) were shown. The value was expressed as the mean ratio to GAPDH expression of five independent experiments. (F) miR-142-3p regulates CAV1 and MEF2A protein expression in BMDM. Cells were transfected with 50 nM of control mimic, miR-142-3p mimic, or miR-142 antagomiR and protein lysates were immunoblotted with CAV1, MEF2A and GAPDH antibody. The representative immunoblot image (top) and densitometry analysis (bottom) were shown. The value was expressed as the mean ratio to GAPDH expression of five independent experiments. All error bars show s.d. P values were determined by Student’s t-test. *P < 0.05, **P < 0.01.
Transcripts that regulate paracrine signaling factors between macrophages and endothelial cells were also targeted by miRNAs. For example, secreted growth factors, VEGF-A, PDGF-A and PDGF-B and transcription factors that control their expression, HIF1α, E2F7 and MEF2A were subject to miRNA-mediated gene regulation. Interestingly, let-7 was predicted to target both E2f7 and Hif1a and let-7 binding to the E2f7 and Hif1a mRNAs was increased in ELAVL1 KO (Figure S4D, E). Since E2f7 can form a transcriptional complex with Hif1α to stimulate Vegfa transcription (Weijts et al., 2012), these data suggest that ELAVL1 allows VEGF-A expression by antagonizing miRNA-mediated repression of transcription factors for this potent angiogenic factor in macrophages. In addition, transcript for Mef2a, a transcription factor that regulates PDGF-B expression (Yablonka-Reuveni and Rivera, 1997) was found to be targeted by miR-142 family and the binding sites of miR-142-5p and miR-142-3p was antagonized by ELAVL1 (figure 6C). Furthermore, mRNAs for paracrine growth factors themselves, namely, Pdgfa, Pdgfb and Vegfa were targeted by miRNAs, and in particular, mir221/222 that targets Pdgfa and miR-126-3p that targets Vegfa were affected by Elavl1 deletion (Figure S4F, G). These data provide novel information on impact of miRNA and ELAVL1 regulation of macrophage paracrine angiogenic factor expression and transcription factors that regulate the expression of such factors.
Our PAR-CLIP data also revealed that components of the TGFβ signal transduction pathway are targeted by miRNAs in BMDM. Several studies have indicated that TGFβ receptors and their signal transduction through Smad family and MAPK cascade can regulate macrophage alternative activation phenotypes associated with anti-inflammation, wound healing and angiogenesis (Koide et al., 2013; Valluru et al., 2011). In our PAR-CLIP dataset, Smad5 and Smad7 transcripts were targeted by miR-155 and miR-15/16 in respectively (Figure S5H, I) while Map3k7 (TAK1) and Mapk1 (ERK) mRNAs were regulated by miR-21 and several other miRNAs (miR-28a-5p, miR-196, miR-93-5p) (Figure S4A). Transcripts encoding TGFβ receptors, Tgfbr1 and Tgfbr2, were targeted by Let-7, miR-142-3p and miR-93-5p (Figure S4B, J). Interestingly, miR-21 binding to Map3k7 mRNA was antagonized by ELAVL1 whereas miR-93-5p binding to Tgfbr2 mRNA was cooperative with ELAVL1.
Consistent with the PAR-CLIP data, Itga4, Cav1 and Mef2a transcripts were associated with Ago2 complex and the level of Ago2- enriched Itga4, Cav1 and Mef2a transcripts were significantly induced in Elavl1 KO BMDM (Figure 6D). Accordingly, expression of ITGA4, CAV1 and MEF2A polypetides were reduced in Elavl1 KO cells (Figure 6E). Furthermore, transfection of miR-142-3p mimic decreased expression whereas inhibition of miR-142-3p by siRNA increased expression of both CAV1 and MEF2A (Figure 6F). These experiments confirmed of our prediction of miRNA/mRNA interactions.
MiR-142-3p and miR-27 were the most represented in targeting the 42 transcripts involved in the angiogenesis cluster. For example, 9 transcripts (Hif1a, Rhob, Tgfbr1, Tiparp, Mef2a, Itgav, Cav1, Adam8 and Qk) were predicted as the target of miR-142-3p. Several studies indicate that miR-142-3p is critical for myeloid development and differentiation and tumor progression (Saito et al., 2012; Sonda et al., 2013).
ELAVL1 regulates ZFP36 expression via a miRNA-mediated regulatory loop
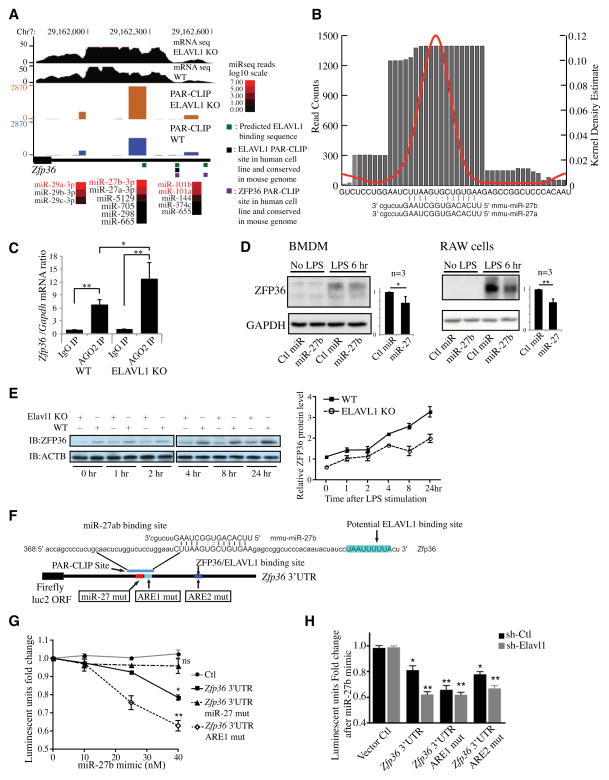
ZFP36 or tristetraprolin (TTP), a ubiquitously-expressed ARE-binding protein, promotes the decay of mRNAs (Blackshear et al., 2003). Both ELAVL1 and ZFP36 target the ARE on Zfp36 3′UTR and ZFP36 autoregulates its own expression through a negative feedback loop (Al-Ahmadi et al., 2009). In our Ago2 PAR-CLIP analysis of BMDM, one of the top 100 miRNA-binding sites with an adjacent/ overlapping ELAVL1-binding sequence was identified in the 3′UTR of Zfp36 mRNA (Figure 7A). This interaction was increased ~1.9 fold in Elavl1 KO BMDM (figure 7B). Two other less prominent PAR-CLIP signals were not altered by Elavl1 deletion. Expression of the Zfp36 mRNA was increased ~1.26 fold after Elavl1 deletion (Table S2). After normalization of the mRNA levels, miR-27b binding to the Zfp36 transcript was increased ~1.5 fold in Elavl1 KO BMDM. Ago2-associated Zfp36 mRNA was increased in Elavl1 KO BMDM (Figure 7C). To test if increased miR-27b binding regulates the ZFP36 protein expression, we transfected miRNA-27b mimic in BMDM and measured ZFP36 expression by Western blot analysis. ZFP36 expression in nonstimulated cells was barely detectable and strongly induced with LPS treatment. As shown in figure 7D, miR-27b mimic suppressed about ~30% LPS-induced ZFP36 expression. This suppression was also observed in RAW cells, a mouse macrophage cell line. These data suggest that Elavl1 antagonizes miR-27b binding to the 3′UTR of Zfp36 mRNA and allows robust expression of this mRNA instability factor.
Figure 7. Loss of ELAVL1 suppresses the synthesis of ZFP36 polypeptide.
(A) miRNA binding sites from PAR-CLIP dataset and mRNA expression levels for the Zfp36 gene is indicated on the USCS genome browser screen shot. The labeling is the same as figures 3 and 6 (above), Actual ZFP36 binding sites from previous PAR-CLIP in human HEK293 cell are indicated with purple bars. (B) Visualization of individual miRNA binding sites with the T-to-C conversion density estimate (red line) and read counts (grey bar) of the strongest PAR-CLIP signal area within the Zfp36 3′UTRs. Predicted miRNA:mRNA alignment for miR-27a and miR-27b is listed below. (C) Effect of Elavl1 deletion on miRNA binding with Zfp36 mRNA. WT or Elavl1 KO BMDM were lysed and immunoprecipitated with anti-Ago2 or IgG control antibody. Zfp36 mRNA physically associated with the Ago2 complex was quantified by qPCR. (D) Western blot analysis of ZFP36 polypeptide levels in control or miR-27b mimic transfected BMDM or Raw 246.7 cell with or without LPS stimulation. Results show a representative experiment that was repeated 3 times. Densitometry analysis of ZFP36 expression levels normalized to GAPDH (N=3, right panel). (E) Western blot analysis of ZFP36 polypeptide levels in WT and Elavl1 KO BMDM after LPS stimulation. Results show a representative experiment that was repeated at least 3 times. Densitometry analysis of ZFP36 expression levels normalized to beta-actin (N=3, bottom panel). All error bars indicate the standard deviation. P value were determined by Student’s t-test. *P < 0.05, **P < 0.01. (F) Schematic of different mutation sites on the Zfp36 3′UTR construct in a dual luciferase assay. (G) Luciferase reporter containing WT or mutated 3′UTR of Zfp36 was transfected into HEK293T cells, and the expression of luciferase reporter in response to miR-27b mimic in dose dependent manner was quantified. For each 3′UTR, renilla luciferase values were first normalized to firefly luciferase activity, and the calculated relative luciferase activity of each 3′UTR reporter was further normalized to that of the control reporter without any miR-27b co-transfection (N=3). (H) HEK293T cells expressing control or Elavl1 shRNA were transfected with luciferase reporter containing Zfp36 3′ UTR or mutants as above. Knockdown efficiency of Elavl1 with shRNA (~90% ) was confirmed by an immunoblot analysis (Figure S6). All 3′UTR luciferase reporters were co-transfected with 40 nM of miR-27b mimic in both cell lines and reporter activity was determined (N=3). All error bars indicate the standard deviation. P value were determined by Student’s t-test. *P < 0.05, **P < 0.01.
To further confirm this model, we measured ZFP36 protein expression in WT and Elavl1 KO BMDM. As shown in Figure 7E, ZFP36 expression level is reduced ~50% in Elavl1 KO BMDM treated or not with LPS (Figure 7E). The modulation of Zfp36 mRNA levels by LPS treatment was not altered by Elavl1 deletion (Figure S5A). On the other hand, the expression of miR-27b was strongly suppressed by LPS between 8–24 h, an effect that occurred in both WT and Elavl1 KO BMDM (Figure S5B).
Zfp36 3′UTR-driven luciferase reporter assays showed that the miR-27b mimic suppressed reporter activity (Figure 7G). When the miR-27b binding seed region on Zfp36 3′UTR was mutated, the suppressive effect of miR-27b was lost. Further, if the ELAVL1 binding sequence near the miR-27b site was mutated, the suppressive effect of the miR-27b mimic was more efficient (Figure 7G). These data confirm that miR-27 suppression of ZFP36 expression is antagonized by ELAVL1. Knockdown of Elavl1 in HEK293T cell by shRNA was used to further confirm the interactions between ELAVL1 and miR-27b (Figure S6). The suppressive effect of miR-27b was more efficient in Elavl1 knockdown cells (Figure 7H). Mutation in the ARE1 of the Zfp36 3′UTR resulted in efficient suppression of luciferase activity in both control and Elavl1 knockdown cells in response to miR-27b mimic, further confirming competitive interactions of miR-27b and ELAVL1 at this site. In contrast, mutation in the ARE2 did not alter reporter activity of Zfp36 3′UTR in response to miR-27b mimic. Taken together, we identified a key regulatory element in the Zfp36 3′UTR and the competitive interaction of miR-27 and ELAVL1.
DISCUSSION
PAR-CLIP analysis allows systems level mapping of miRNA binding sites
In this report, we describe a comprehensive mapping of miRNA binding sites in murine BMDM, which have been used extensively in the studies of inflammation, innate immunity, tumor biology and angiogenesis. Our primary purpose was to examine the biology of the RBP ELAVL1, which regulates gene expression at the post-transcriptional level by binding to ARE and URE sequences. ELAVL1 is primarily localized in the nucleus even though it can be exported to the cytoplasm and shuttles between the nucleus and the cytoplasm. Since miRNA-mediated gene suppression is thought to occur primarily in the cytoplasm, our aim was to comprehensively define modulation of miRNA binding sites by ELAVL1 using BMDM that are from WT and Elavl1 KO mice. Our rationale was to define miRNA binding sites induced or suppressed by ELAVL1 as well as those that are not affected (Chang et al., 2013). We also developed conditions to specifically immuoprecipitate endogenous murine Ago2 after cross-linking of the RISC complex. For this approach, we employed the PAR-CLIP technique which uses 4-SU incorporation followed by UV-crosslinking and immunoprecipitation (Hafner et al., 2010b).
Our dataset was comprehensive and defined miRNA binding sites in the BMDM transcriptome with high accuracy. Since we considered only those sites containing T to C conversion, the presence of false positives was minimized (Mukherjee et al., 2011). Overall, we annotated 3,140 sites in the 3′UTR, which contain the most (~58%) PAR-CLIP sites. PAR-CLIP sites were also found in coding regions (~17%), introns (5%) and intergenic sites (17%). The roles of such sites are not clear at present. In addition, our dataset did not contain significant number of G-rich sequences, which could interact with Ago2 in a miRNA-independent manner (Leung et al., 2011). This work complements previous characterization of genome-wide miRNA binding sites in HEK293, HeLa cells and mouse and human brain tissues (Boudreau et al., 2013; Hafner et al., 2010a; Lebedeva et al., 2011; Mukherjee et al., 2011). However, transcriptome-wide modulation of miRNA/ mRNA interactions by an RBP has not been described.
ELAVL1/ miRNA interactions on the transcriptome
To accurately assign miRNA sites on the BMDM transcriptome and to determine those that are regulated by ELAVL1, we determined the expression patterns and abundance of miRNA and mRNA species by RNAseq analysis of purified RNA from BMDM. For miRNA expression characterization, we compared the datasets from total small RNA preparations as well as from the PAR-CLIP experiments.
RNAseq analysis indicated that WT and Elavl1 KO BMDM contains largely similar transcript profiles. This was surprising given that ELAVL1 is a major RNA stabilizing protein that interacts with hundreds to thousands of targets (Lebedeva et al., 2011; Mukherjee et al., 2011). Indeed, approximately 16% of transcripts in the genome contain potential ELAVL1 binding sites and PAR-CLIP analysis using Anti-ELAVL1 antibodies have identified > 26,000 sites in human cells (Lebedeva et al., 2011; Mukherjee et al., 2011). In our experiments using primary BMDM, the Elavl1 gene is deleted by the LysM-Cre nuclease, which is activated early in the differentiation of the myeloid lineage cells(Chang et al., 2013). Thus, under steady-state conditions, even though ELAVL1-mediated RNA stabilization mechanisms were absent, compensatory mechanisms may have been activated to balance the transcriptome. We uncovered one such mechanism as described below.
We considered transcripts which show strong targeting by miRNAs, as defined by those sites with ≥ 100 reads in the PAR-CLIP dataset. Among the 3,033 sites, significant fraction show increased expression in Elavl1 KO cells when miRNA and ELAVL1 binding sites. Previous studies, which examined ELAVL1 interaction with select mRNAs, suggested that RBP binding to mRNAs can have both positive and negative effects on miRNA-mediated gene suppression. Indeed, many of the miRNA binding sites which show alterations in response to ELAVL1 levels contain proximal binding sites for the RBP (see examples such as c-Myc). Interestingly, the fluctuations in miRNA binding to these sites were much higher in magnitude than the corresponding changes in expression of the corresponding mRNAs. Nevertheless, increased miRNA binding is associated generally with decreased expression of mRNAs. Thus, ELAVL1 antagonizes miRNA-mediated suppression of macrophage gene expression. Our dataset has been implemented in the genome browser in a searchable format (http://genome.ucsc.edu/cgi-bin/hgTracks?hgS_doOtherUser=submit&hgS_otherUserName=Yi%2DChien%20Lu&hgS_otherUserSessionName=Ago2%20PARLIP%20and%20mRNAseq), which provides a resource and should facilitate further studies on post-transcriptional gene regulation in macrophages.
Macrophage genes involved in vascular development and angiogenesis are targeted by miRNAs and modulated by ELAVL1
Significant numbers of BMDM mRNAs that show high miRNA binding regulate the process of vascular development/ angiogenesis. These processes are involved in key biological processes such as wound healing as well as in disease processes such as tumor angiogenesis and chronic inflammatory diseases. Indeed, macrophages are critical for vascular network formation during development because they chaperone filopodia-containing “tip” cells from vascular sprouts and allow them to fuse together to form a primary vascular network (Fantin et al., 2013). Our data suggest that miRNA-mediated gene regulation is an important mechanism regulating macrophage regulation of blood vessel formation and morphogenesis, consistent with recent studies (Chang et al., 2013; Zhang et al., 2012). Since macrophage functions have been implicated in angiogenic switch of tumors (Ruffell et al., 2012), tumor resistance to anti-angiogenic drugs and metastasis (De Palma and Lewis, 2013), molecular targets identified in this work can be tested for their requirement in these processes.
Further analysis of transcripts targeted by miRNAs and the subset that shows sensitivity to ELAVL1 levels revealed coordinate regulation of several cellular processes such as chemotaxis/ cell migration, paracrine interaction with vascular cells, and signal transduction and gene transcription. This work provides a firm foundation to further investigate how the miRNA network is regulated in complex processes such as angiogenesis and inflammation. Previous work has shown that macrophage phenotype switch, for example the so called “M2 phenotype”, is important in angiogenic switch and metastasis(De Palma and Lewis, 2013; Ruffell et al., 2012). It will be of interest to determine if the miRNAs identified in this work are altered during the phenotype switch of macrophages.
Identification of a miRNA-regulated synchronized expression of ELAVL1 and ZFP36
We identified a major miRNA-targeted site in the 3′UTR of ZFP36 (a.k.a. tristeraspolin/ TTP), a major RNA binding protein that induces destabilization of ARE-containing mRNAs (Blackshear et al., 2003). This site was predicted as the target of miR-27 and ELAVL1, suggesting the competitive interaction of ELAVL1 and miR-27 to regulate Zfp36 mRNA. Both ELAVL1 and ZFP36 are ARE-binding proteins that interact with similar cis-acting sequences on mRNAs. However, in contrast to ELAVL1, which is located primarily in the nucleus, ZFP36 is cytoplasmic. Indeed, the inducible prostaglandin synthase (Ptgs2 mRNA) and the oncogene (c-Myc mRNA) are both reciprocally regulated by ZFP36 and ELAVL1(Rounbehler et al., 2012). Our data reveal a previously unrecognized miRNA-mediated regulatory system, whereby the levels of RNA stability and instability factors are synchronized. When ELAVL1 levels are reduced, miR-27-mediated suppression of ZFP36 is enhanced, thereby reducing the levels of this RNA instability factor. This mechanism may account, at least in part, our observation that under steady-state conditions, the transcriptome of WT and Elavl1 KO cells are largely similar.
In summary, this work reports the comprehensive mapping of miRNA binding sites in BMDM, an important innate immune cell type. Further, our dataset also describes the miRNA binding sites that are modulated by ELAVL1, a major RBP that regulates gene expression by interacting with ARE and UREs. Moreover, we provide evidence for coordinated miRNA targeting of transcripts that regulate blood vessel development and angiogenesis. In addition, analysis of this dataset identified miRNA-mediated coordination of expression of ELAVL1 and ZFP36, two major RBPs that induce or suppress gene expression, respectively. This resource should allow further interrogation of macrophage post-transcriptional gene regulatory mechanisms in vascular biology, immunology and tumor biology.
MATERIALS AND METHODS
Animals
Myeloid specific deletion of Elavl1 in C57Bl/6 mice was generated by crossing Elavl1 floxed mice (Ghosh et al., 2009) with mice expressing Cre recombinase driven by the lysozyme M promoter. All animal care and experimental protocols were conducted following the guidelines of the institutional care and use committee of Weill Cornell Medical College.
Cell Culture
HEK293T cell lines were maintained in DMEM growth medium containing high glucose (4.5 g/L) (Sigma), L-glutamine, 10% fetal bovine serum (Invitrogen). Cells were cultured at 37C in a humidified atmosphere containing 5% CO2.
BMDM culture and PAR-CLIP
Animals were sacrificed using carbon dioxide and bone marrow cells from mouse femur and tibia were collected by flushing through PBS with a 23-guage needle. Bone marrow cells were cultured in one 150mm Petri dishes with complete DMEM and 20% of L929 cell culture medium for 6 days (Weischenfeldt and Porse, 2008). BMDM for transfection and sequencing library preparation were then washed twice with PBS and cultured in complete DMEM with 20% of L929 cell culture medium over night and harvested with 0.2 mM EDTA in PBS.
PAR-CLIP protocol for BMDM was described previously (Hafner et al., 2010a, b) with several minor described in supplemental experimental procedures.
PARalyzer annotation and miRNA target sites predication
For WT and ELAVL1 KO PAR-CLIP library, PARalyzer was used to identify binding sites as described previously (Corcoran et al., 2011). Detail analysis methods are described in supplemental experimental procedure. Clusters that overlapped with predicted miRNA binding site from Target Scan database (TargetScan 6.2, mouse non-conserved and conserved predictions) were identified using custom scripts. Customized miRNA prediction procedure and signal-to-noise calculation method on figure 3A are further described in supplemental information.
Total RNA isolation, miRNA isolation and high-throughput sequencing for mRNA or miRNA
Total RNA and small RNAs were isolated as detailed in the manufacturer’s protocols (Clonetech) and RNA quality was checked by an Agilent 2100 bioanalyzer (Agilent technologies). Small RNA and mRNA libraries preparation were followed as manufacturer’s protocols (Illumina Small RNA v1.5 Sample Preparation Kit and Illumina mRNA sequencing Sample Preparation Kit, Illumina). All libraries were sequenced for single-reads, 42 cycles on the Illumina Genome Analyzer IIx (Illumina). Detailed protocol for sequencing data analysis is further described in supplemental information.
qRT-PCR analysis for miRNAs and mRNAs
Total miRNA was subject to poly(A) tailing reaction and reverse transcribed with oligo-dT adaptor primer (qScript miRNA cDNA synthesis kit, Quanta) and followed by qPCR amplification with the specific miRNA sequence primer and oligo-dT adaptor sequence primer (PerfeCTa® SYBR® Green system, Quanta). Expression of specific miRNAs was normalized to the value of U6 snRNA. To validate mRNA expression by qRT-PCR, RNA was reverse transcribed with MMLV reverse transcriptase (Invitrogen) and cDNAs were amplified with specific primers (primer bank) using SYBR green master mix (PerfeCTa® SYBR® Green system, Quanta).
GO Analysis
Genes with high PAR-CLIP signals at 3′ UTR (≥ 200 reads in WT or Elavl1 KO) were performed for gene ontology analysis by using DAVID Bioinformatics program (Huang da et al., 2009a, b). The expressed genes from mRNA sequencing dataset (≥ 0.1 RPKM) were used as a background for this analysis.
Luciferase miRNA target reporter assay
Zfp36 3′UTR fragment (762 bp) was cloned into the pmirGLO vector (Promega). Mutations were made in the seed region of miR-27b binding site, ARE1 binding site, and ARE2 binding site. Detail primer sequences are described in supplemental information
HEK293T cells were co-transfected with 50ng/ml pmirGLO vector containing Zfp36 3′UTR (WT or mutant) miR-27b mimic (0 nM to 40 nM) (Thermo Scientific) using Lipofectamine-2000 (Invitrogen). After 40 hr, the cells were lysed and luciferase activity was measured using a luminometer. Firefly Luciferase values were normalized with Renilla luciferase values.
Supplementary Material
Figure S1. Mouse AGO2 protein was immuno-precipitated by AGO2 antibody in PAR-CLIP procedure. Related to Figure 1A.
(A) Autoradiogram image of an SDS-gel resolving radio-labeled RNA cross-linked to Ago2 IPs. Autoradiogram of SDS PAGE gels of RISC complexes are 32P-labeled, cross-linked and immunoprecipitated (IP) with Ago2 IgG but not by non-specific mouse IgG. Protein-RNA complexes on SDS gels were cut from 100 kDa to130 kDa range for library preparation and sequencing. (B) AGO2 expression in Elavl1 KO BMDM. Western blot analysis of Ago2 protein in lysates of BMDM used in the PAR-CLIP experiments.
Figure S2. miRNAseq results from BMDM. Related to Figure 2A.
miRNA profiles from 2 WT and 2 Elavl1 KO miRNA-seq data. The color code represents log2 scale of relative frequencies fold changes determined by sequencing. Only 5 out of 211 highly expressed mature miRNAs (Reads comprise > 0.001% of total aligned miRNA reads) have more than 2 fold change in expression level (4 increase and 1 decrease after Elavl1 KO).
Figure S3. Identification of high confidence Ago2 PAR-CLIP sites from BMDM. Related to Figure 3A.
Total number of Ago2 PAR-CLIP sites from WT and Elavl1 KO BMDM were sorted based on the number of reads (Right Y axis; blue line). The left Y axis represents % overlap in PAR-CLIP sites between WT and Elavl1 KO samples (black line). Note that high read Ago2 PAR-CLIP sites have high degree of overlap, which is an index of reproducibility between independent replicates. Selected sites that have > 100 combined reads (3,033 sites) were marked with a red box for further analysis.
Figure S4. miRNAs binding is regulated by Elavl1. Related to Figure 5.
UCSC genome browser views PAR-CLIP data and mRNA-seq data on cell surface receptor genes Tgfbr1 (B), Itgav (C) and Tgfbr2 (J), secreted angiogenic factor genes Pdgfa (F) and Vegfa (G), transcription factor genes Hif1a (D) and E2f7(E), and intracellular signal transducing protein genes Map3k7(A), Smad5 (H) and Smad7 (I). Listed miRNAs below PAR-CLIP signals indicate predicted miRNAs. The color code represents log10 scale of average miRNA reads from miRNA-seq data. Green bars represents potential Elavl1 binding sequences that contain UUUNUUU or less than 2 A base in 7 poly U region (ex. UUAUUAU or UAUUUAU). Red-labeled miRNAs are the most-likely candidates for a given binding site.
Figure S5. Zfp36 gene expression is not significantly affected in Elavl1 KO BMDM. Related to Figure 7E.
(A)Quantitative RT-PCR result of Zfp36 mRNA levels in BMDM after LPS stimulation. Experiments were repeated 3 times in different sets of WT and Elavl1 KO BMDM. (B) Mir-27a/b expression level is not significantly affected in Elavl1 KO BMDM. Quantitative RT-PCR of miR-27b expression level in BMDM after LPS stimulation. (N=2). MiR-27a and miR-27b expression was amplified by Qunata qScript™ microRNA Quantification System and normalized by U6 small nuclear RNA. There is only one nucleotide difference between miR-27a and miR-27b (C to U at 19 nt) and hard to completely distinguish by qRT-PCR results. We used combined miR-27a and miR-27b to represent the overall miR-27 expression level in BMDM.
Figure S6. Elavl1 shRNA suppressed ~90% of Elavl1 polypeptide. Related to Figure 7H.
Western blot analysis detected the Elavl1 expression level in HEK293T cell transfected with control or Elavl1 shRNA. All error bars indicate the standard deviation. P value were determined by Student’s t-test. ***P < 0.001.
Bowtie alignment results shows 211 miRNAs were detected from the miRNAseq data of 2 WT and 2 Elavl1 KO BMDM, relate to figure 2A.
Goby analysis result shows a list of mRNAs was expressed in BMDM (average RPKM > 0.1), relate to figure 2B. Input mRNAseq data were from 4 WT and 4 Elavl1 KO BMDM.
The list of predicted miRNA/mRNA pairs for PAR-CLIP sites. Relate to figure 2B.
PAR-CLIP analysis result shows a list of angiogenic related genes that are regulated by miRNAs. Relate to figure 5.
Acknowledgments
This work is supported by NIH grants (RO1-HL49094 and U54-HL117798) to TH. We thank Dr. Fabien Campagne, Weill Cornell Medical College for help with the Goby software and RNAseq analysis.
Footnotes
Author Contributions: YCL designed and conducted experiments and wrote the manuscript, SHC did the GO analysis, MH assisted in the PAR-CLIP experiments and PARalyzer analysis, XL helped with luciferase reporter assays, TT assisted with PAR-CLIP experimental design and overall research strategy, OE conducted the the bioinformatic analysis of PAR-CLIP, RNAseq and miRNA prediction studies, and TH helped write the manuscript and overall supervision of the work. All authors read and edited the manuscript.
Data submission to public databases: RNAseq of mRNA and miRNAs, and PAR-CLIP clusters from WT and Elavl1 KO BMDM have been submitted to the GEO database with the following accession numbers:
The miRNA binding sites can be accessed at the following site: http://genome.ucsc.edu/cgi-bin/hgTracks?hgS_doOtherUser=submit&hgS_otherUserName=Yi%2DChien%20Lu&hgS_otherUserSessionName=Ago2%20PARLIP%20and%20mRNAseq
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdelmohsen K, Gorospe M. Posttranscriptional regulation of cancer traits by HuR. Wiley interdisciplinary reviews. RNA. 2010;1:214–229. doi: 10.1002/wrna.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelmohsen K, Lal A, Kim HH, Gorospe M. Posttranscriptional orchestration of an anti-apoptotic program by HuR. Cell cycle. 2007;6:1288–1292. doi: 10.4161/cc.6.11.4299. [DOI] [PubMed] [Google Scholar]
- Al-Ahmadi W, Al-Ghamdi M, Al-Haj L, Al-Saif M, Khabar KS. Alternative polyadenylation variants of the RNA binding protein, HuR: abundance, role of AU-rich elements and auto-Regulation. Nucleic acids research. 2009;37:3612–3624. doi: 10.1093/nar/gkp223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhandari A, Gordon W, Dizon D, Hopkin AS, Gordon E, Yu Z, Andersen B. The Grainyhead transcription factor Grhl3/Get1 suppresses miR-21 expression and tumorigenesis in skin: modulation of the miR-21 target MSH2 by RNA-binding protein DND1. Oncogene. 2013;32:1497–1507. doi: 10.1038/onc.2012.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- Blackshear PJ, Lai WS, Kennington EA, Brewer G, Wilson GM, Guan X, Zhou P. Characteristics of the interaction of a synthetic human tristetraprolin tandem zinc finger peptide with AU-rich element-containing RNA substrates. The Journal of biological chemistry. 2003;278:19947–19955. doi: 10.1074/jbc.M301290200. [DOI] [PubMed] [Google Scholar]
- Boudreau RL, Jiang P, Gilmore BL, Spengler RM, Tirabassi R, Nelson JA, Ross CA, Xing Y, Davidson BL. Transcriptome-wide Discovery of microRNA Binding Sites in Human Brain. Neuron. 2013 doi: 10.1016/j.neuron.2013.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger K, Muhl B, Kellner M, Rohrmoser M, Gruber-Eber A, Windhager L, Friedel CC, Dolken L, Eick D. 4-thiouridine inhibits rRNA synthesis and causes a nucleolar stress response. RNA Biol. 2013;10 doi: 10.4161/rna.26214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang SH, Hla T. Gene regulation by RNA binding proteins and microRNAs in angiogenesis. Trends Mol Med. 2011;17:650–658. doi: 10.1016/j.molmed.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang SH, Lu YC, Li X, Hsieh WY, Xiong Y, Ghosh M, Evans T, Elemento O, Hla T. Antagonistic function of the RNA-binding protein HuR and miR-200b in post-transcriptional regulation of vascular endothelial growth factor-A expression and angiogenesis. The Journal of biological chemistry. 2013;288:4908–4921. doi: 10.1074/jbc.M112.423871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekulaeva M, Filipowicz W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr Opin Cell Biol. 2009;21:452–460. doi: 10.1016/j.ceb.2009.04.009. [DOI] [PubMed] [Google Scholar]
- Chinchilla A, Lozano E, Daimi H, Esteban FJ, Crist C, Aranega AE, Franco D. MicroRNA profiling during mouse ventricular maturation: a role for miR-27 modulating Mef2c expression. Cardiovasc Res. 2011;89:98–108. doi: 10.1093/cvr/cvq264. [DOI] [PubMed] [Google Scholar]
- Ciafre SA, Galardi S. microRNAs and RNA-binding proteins: a complex network of interactions and reciprocal regulations in cancer. RNA Biol. 2013;10:935–942. doi: 10.4161/rna.24641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran DL, Georgiev S, Mukherjee N, Gottwein E, Skalsky RL, Keene JD, Ohler U. PARalyzer: definition of RNA binding sites from PAR-CLIP short-read sequence data. Genome biology. 2011;12:R79. doi: 10.1186/gb-2011-12-8-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Palma M, Lewis CE. Macrophage regulation of tumor responses to anticancer therapies. Cancer Cell. 2013;23:277–286. doi: 10.1016/j.ccr.2013.02.013. [DOI] [PubMed] [Google Scholar]
- Denkert C, Koch I, von Keyserlingk N, Noske A, Niesporek S, Dietel M, Weichert W. Expression of the ELAV-like protein HuR in human colon cancer: association with tumor stage and cyclooxygenase-2. Mod Pathol. 2006;19:1261–1269. doi: 10.1038/modpathol.3800645. [DOI] [PubMed] [Google Scholar]
- Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annual review of biochemistry. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- Fantin A, Vieira JM, Plein A, Denti L, Fruttiger M, Pollard JW, Ruhrberg C. NRP1 acts cell autonomously in endothelium to promote tip cell function during sprouting angiogenesis. Blood. 2013;121:2352–2362. doi: 10.1182/blood-2012-05-424713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farazi TA, Spitzer JI, Morozov P, Tuschl T. miRNAs in human cancer. J Pathol. 2011;223:102–115. doi: 10.1002/path.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh M, Aguila HL, Michaud J, Ai Y, Wu MT, Hemmes A, Ristimaki A, Guo C, Furneaux H, Hla T. Essential role of the RNA-binding protein HuR in progenitor cell survival in mice. J Clin Invest. 2009;119:3530–3543. doi: 10.1172/JCI38263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannice R, Erreni M, Allavena P, Buscaglia M, Tozzi R. Chemokines mRNA expression in relation to the Macrophage Migration Inhibitory Factor (MIF) mRNA and Vascular Endothelial Growth Factor (VEGF) mRNA expression in the microenvironment of endometrial cancer tissue and normal endometrium: a pilot study. Cytokine. 2013;64:509–515. doi: 10.1016/j.cyto.2013.07.024. [DOI] [PubMed] [Google Scholar]
- Gorospe M, Tominaga K, Wu X, Fahling M, Ivan M. Post-Transcriptional Control of the Hypoxic Response by RNA-Binding Proteins and MicroRNAs. Front Mol Neurosci. 2011;4:7. doi: 10.3389/fnmol.2011.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grande-Garcia A, del Pozo MA. Caveolin-1 in cell polarization and directional migration. Eur J Cell Biol. 2008;87:641–647. doi: 10.1016/j.ejcb.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr, Jungkamp AC, Munschauer M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010a;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jungkamp AC, Munschauer M, et al. PAR-CliP--a method to identify transcriptome-wide the binding sites of RNA binding proteins. J Vis Exp. 2010b doi: 10.3791/2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic acids research. 2009a;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009b;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Humtsoe JO, Liu M, Malik AB, Wary KK. Lipid phosphate phosphatase 3 stabilization of beta-catenin induces endothelial cell migration and formation of branching point structures. Mol Cell Biol. 2010;30:1593–1606. doi: 10.1128/MCB.00038-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsanou V, Milatos S, Yiakouvaki A, Sgantzis N, Kotsoni A, Alexiou M, Harokopos V, Aidinis V, Hemberger M, Kontoyiannis DL. The RNA-binding protein Elavl1/HuR is essential for placental branching morphogenesis and embryonic development. Mol Cell Biol. 2009;29:2762–2776. doi: 10.1128/MCB.01393-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedde M, Strasser MJ, Boldajipour B, Oude Vrielink JA, Slanchev K, le Sage C, Nagel R, Voorhoeve PM, van Duijse J, Orom UA, et al. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell. 2007;131:1273–1286. doi: 10.1016/j.cell.2007.11.034. [DOI] [PubMed] [Google Scholar]
- Kedde M, van Kouwenhove M, Zwart W, Oude Vrielink JA, Elkon R, Agami R. A Pumilio-induced RNA structure switch in p27-3′ UTR controls miR-221 and miR-222 accessibility. Nature cell biology. 2010;12:1014–1020. doi: 10.1038/ncb2105. [DOI] [PubMed] [Google Scholar]
- Keene JD. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M. HuR recruits let-7/RISC to repress c-Myc expression. Genes & development. 2009;23:1743–1748. doi: 10.1101/gad.1812509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koide N, Odkhuu E, Naiki Y, Tsolmongyn B, Ito K, Komatsu T, Yoshida T, Yokochi T. Augmentation of LPS-induced vascular endothelial cell growth factor production in macrophages by transforming growth factor-beta1. Innate Immun. 2013 doi: 10.1177/1753425913509291. [DOI] [PubMed] [Google Scholar]
- Lebedeva S, Jens M, Theil K, Schwanhausser B, Selbach M, Landthaler M, Rajewsky N. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Molecular cell. 2011;43:340–352. doi: 10.1016/j.molcel.2011.06.008. [DOI] [PubMed] [Google Scholar]
- Leung AK, Young AG, Bhutkar A, Zheng GX, Bosson AD, Nielsen CB, Sharp PA. Genome-wide identification of Ago2 binding sites from mouse embryonic stem cells with and without mature microRNAs. Nature structural & molecular biology. 2011;18:237–244. doi: 10.1038/nsmb.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levadoux-Martin M, Gouble A, Jegou B, Vallet-Erdtmann V, Auriol J, Mercier P, Morello D. Impaired gametogenesis in mice that overexpress the RNA-binding protein HuR. EMBO Rep. 2003;4:394–399. doi: 10.1038/sj.embor.embor803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lue H, Dewor M, Leng L, Bucala R, Bernhagen J. Activation of the JNK signalling pathway by macrophage migration inhibitory factor (MIF) and dependence on CXCR4 and CD74. Cell Signal. 2011;23:135–144. doi: 10.1016/j.cellsig.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai A, Veltel S, Pellinen T, Padzik A, Coffey E, Marjomaki V, Ivaska J. Competitive binding of Rab21 and p120RasGAP to integrins regulates receptor traffic and migration. The Journal of cell biology. 2011;194:291–306. doi: 10.1083/jcb.201012126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J, Wang D, Mataleena P, He B, Niu D, Katayama K, Xu X, Ojala JR, Wang W, Shu Q, et al. Myo1e impairment results in actin reorganization, podocyte dysfunction, and proteinuria in zebrafish and cultured podocytes. PLoS One. 2013;8:e72750. doi: 10.1371/journal.pone.0072750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee N, Corcoran DL, Nusbaum JD, Reid DW, Georgiev S, Hafner M, Ascano M, Jr, Tuschl T, Ohler U, Keene JD. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Molecular cell. 2011;43:327–339. doi: 10.1016/j.molcel.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasheed SA, Teo CR, Beillard EJ, Voorhoeve PM, Casey PJ. MicroRNA-182 and microRNA-200a control G-protein subunit alpha-13 (GNA13) expression and cell invasion synergistically in prostate cancer cells. The Journal of biological chemistry. 2013;288:7986–7995. doi: 10.1074/jbc.M112.437749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rounbehler RJ, Fallahi M, Yang C, Steeves MA, Li W, Doherty JR, Schaub FX, Sanduja S, Dixon DA, Blackshear PJ, et al. Tristetraprolin impairs myc-induced lymphoma and abolishes the malignant state. Cell. 2012;150:563–574. doi: 10.1016/j.cell.2012.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffell B, Affara NI, Coussens LM. Differential macrophage programming in the tumor microenvironment. Trends Immunol. 2012;33:119–126. doi: 10.1016/j.it.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y, Suzuki H, Tsugawa H, Imaeda H, Matsuzaki J, Hirata K, Hosoe N, Nakamura M, Mukai M, Saito H, et al. Overexpression of miR-142-5p and miR-155 in gastric mucosa-associated lymphoid tissue (MALT) lymphoma resistant to Helicobacter pylori eradication. PLoS One. 2012;7:e47396. doi: 10.1371/journal.pone.0047396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonda N, Simonato F, Peranzoni E, Cali B, Bortoluzzi S, Bisognin A, Wang E, Marincola FM, Naldini L, Gentner B, et al. miR-142-3p prevents macrophage differentiation during cancer-induced myelopoiesis. Immunity. 2013;38:1236–1249. doi: 10.1016/j.immuni.2013.06.004. [DOI] [PubMed] [Google Scholar]
- Valluru M, Staton CA, Reed MW, Brown NJ. Transforming Growth Factor-beta and Endoglin Signaling Orchestrate Wound Healing. Front Physiol. 2011;2:89. doi: 10.3389/fphys.2011.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo DT, Abdelmohsen K, Martindale JL, Qiao M, Tominaga K, Burton TL, Gelfond JA, Brenner AJ, Patel V, Trageser D, et al. The oncogenic RNA-binding protein Musashi1 is regulated by HuR via mRNA translation and stability in glioblastoma cells. Mol Cancer Res. 2012;10:143–155. doi: 10.1158/1541-7786.MCR-11-0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Guo Y, Chu H, Guan Y, Bi J, Wang B. Multiple Functions of the RNA-Binding Protein HuR in Cancer Progression, Treatment Responses and Prognosis. Int J Mol Sci. 2013;14:10015–10041. doi: 10.3390/ijms140510015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijts BG, Bakker WJ, Cornelissen PW, Liang KH, Schaftenaar FH, Westendorp B, de Wolf CA, Paciejewska M, Scheele CL, Kent L, et al. E2F7 and E2F8 promote angiogenesis through transcriptional activation of VEGFA in cooperation with HIF1. EMBO J. 2012;31:3871–3884. doi: 10.1038/emboj.2012.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weischenfeldt J, Porse B. Bone Marrow-Derived Macrophages (BMM): Isolation and Applications. CSH Protoc. 2008 doi: 10.1101/pdb.prot5080. pdb prot5080. [DOI] [PubMed] [Google Scholar]
- Wheeler AP, Ridley AJ. RhoB affects macrophage adhesion, integrin expression and migration. Exp Cell Res. 2007;313:3505–3516. doi: 10.1016/j.yexcr.2007.07.014. [DOI] [PubMed] [Google Scholar]
- Yablonka-Reuveni Z, Rivera AJ. Influence of PDGF-BB on proliferation and transition through the MyoD-myogenin-MEF2A expression program during myogenesis in mouse C2 myoblasts. Growth Factors. 1997;15:1–27. doi: 10.3109/08977199709002109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yester JW, Tizazu E, Harikumar KB, Kordula T. Extracellular and intracellular sphingosine-1-phosphate in cancer. Cancer Metastasis Rev. 2011;30:577–597. doi: 10.1007/s10555-011-9305-0. [DOI] [PubMed] [Google Scholar]
- Zhang J, Modi Y, Yarovinsky T, Yu J, Collinge M, Kyriakides T, Zhu Y, Sessa WC, Pardi R, Bender JR. Macrophage beta2 integrin-mediated, HuR-dependent stabilization of angiogenic factor-encoding mRNAs in inflammatory angiogenesis. Am J Pathol. 2012;180:1751–1760. doi: 10.1016/j.ajpath.2011.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Mouse AGO2 protein was immuno-precipitated by AGO2 antibody in PAR-CLIP procedure. Related to Figure 1A.
(A) Autoradiogram image of an SDS-gel resolving radio-labeled RNA cross-linked to Ago2 IPs. Autoradiogram of SDS PAGE gels of RISC complexes are 32P-labeled, cross-linked and immunoprecipitated (IP) with Ago2 IgG but not by non-specific mouse IgG. Protein-RNA complexes on SDS gels were cut from 100 kDa to130 kDa range for library preparation and sequencing. (B) AGO2 expression in Elavl1 KO BMDM. Western blot analysis of Ago2 protein in lysates of BMDM used in the PAR-CLIP experiments.
Figure S2. miRNAseq results from BMDM. Related to Figure 2A.
miRNA profiles from 2 WT and 2 Elavl1 KO miRNA-seq data. The color code represents log2 scale of relative frequencies fold changes determined by sequencing. Only 5 out of 211 highly expressed mature miRNAs (Reads comprise > 0.001% of total aligned miRNA reads) have more than 2 fold change in expression level (4 increase and 1 decrease after Elavl1 KO).
Figure S3. Identification of high confidence Ago2 PAR-CLIP sites from BMDM. Related to Figure 3A.
Total number of Ago2 PAR-CLIP sites from WT and Elavl1 KO BMDM were sorted based on the number of reads (Right Y axis; blue line). The left Y axis represents % overlap in PAR-CLIP sites between WT and Elavl1 KO samples (black line). Note that high read Ago2 PAR-CLIP sites have high degree of overlap, which is an index of reproducibility between independent replicates. Selected sites that have > 100 combined reads (3,033 sites) were marked with a red box for further analysis.
Figure S4. miRNAs binding is regulated by Elavl1. Related to Figure 5.
UCSC genome browser views PAR-CLIP data and mRNA-seq data on cell surface receptor genes Tgfbr1 (B), Itgav (C) and Tgfbr2 (J), secreted angiogenic factor genes Pdgfa (F) and Vegfa (G), transcription factor genes Hif1a (D) and E2f7(E), and intracellular signal transducing protein genes Map3k7(A), Smad5 (H) and Smad7 (I). Listed miRNAs below PAR-CLIP signals indicate predicted miRNAs. The color code represents log10 scale of average miRNA reads from miRNA-seq data. Green bars represents potential Elavl1 binding sequences that contain UUUNUUU or less than 2 A base in 7 poly U region (ex. UUAUUAU or UAUUUAU). Red-labeled miRNAs are the most-likely candidates for a given binding site.
Figure S5. Zfp36 gene expression is not significantly affected in Elavl1 KO BMDM. Related to Figure 7E.
(A)Quantitative RT-PCR result of Zfp36 mRNA levels in BMDM after LPS stimulation. Experiments were repeated 3 times in different sets of WT and Elavl1 KO BMDM. (B) Mir-27a/b expression level is not significantly affected in Elavl1 KO BMDM. Quantitative RT-PCR of miR-27b expression level in BMDM after LPS stimulation. (N=2). MiR-27a and miR-27b expression was amplified by Qunata qScript™ microRNA Quantification System and normalized by U6 small nuclear RNA. There is only one nucleotide difference between miR-27a and miR-27b (C to U at 19 nt) and hard to completely distinguish by qRT-PCR results. We used combined miR-27a and miR-27b to represent the overall miR-27 expression level in BMDM.
Figure S6. Elavl1 shRNA suppressed ~90% of Elavl1 polypeptide. Related to Figure 7H.
Western blot analysis detected the Elavl1 expression level in HEK293T cell transfected with control or Elavl1 shRNA. All error bars indicate the standard deviation. P value were determined by Student’s t-test. ***P < 0.001.
Bowtie alignment results shows 211 miRNAs were detected from the miRNAseq data of 2 WT and 2 Elavl1 KO BMDM, relate to figure 2A.
Goby analysis result shows a list of mRNAs was expressed in BMDM (average RPKM > 0.1), relate to figure 2B. Input mRNAseq data were from 4 WT and 4 Elavl1 KO BMDM.
The list of predicted miRNA/mRNA pairs for PAR-CLIP sites. Relate to figure 2B.
PAR-CLIP analysis result shows a list of angiogenic related genes that are regulated by miRNAs. Relate to figure 5.