Abstract
Bacterial and fungal populations associated with the rhizosphere of healthy black spruce (Picea mariana) seedlings and seedlings with symptoms of root rot were characterized by cloned rRNA gene sequence analysis. Triplicate bacterial and fungal rRNA gene libraries were constructed, and 600 clones were analyzed by amplified ribosomal DNA restriction analysis and grouped into operational taxonomical units (OTUs). A total of 84 different bacterial and 31 different fungal OTUs were obtained and sequenced. Phylogenetic analyses indicated that the different OTUs belonged to a wide range of bacterial and fungal taxa. For both groups, pairwise comparisons revealed that there was greater similarity between replicate libraries from each treatment than between libraries from different treatments. Significant differences between pooled triplicate samples from libraries of genes from healthy seedlings and pooled triplicate samples from libraries of genes from diseased seedlings were also obtained for both bacteria and fungi, clearly indicating that the rhizosphere-associated bacterial and fungal communities of healthy and diseased P. mariana seedlings were different. The communities associated with healthy and diseased seedlings also showed distinct ecological parameters as indicated by the calculated diversity, dominance, and evenness indices. Among the main differences observed at the community level, there was a higher proportion of Acidobacteria, Gammaproteobacteria, and Homobasidiomycetes clones associated with healthy seedlings, while the diseased-seedling rhizosphere harbored a higher proportion of Actinobacteria, Sordariomycetes, and environmental clones. The methodological approach described in this study appears promising for targeting potential rhizosphere-competent biological control agents against root rot diseases occurring in conifer nurseries.
The rhizosphere is defined as the soil environment directly under the influence of living roots (18). It is a niche where complex microbial communities are supported by nutrients released by root exudates, mucilage, and sloughed-off root cells (6). It has been demonstrated that a high proportion of rhizosphere- and other soil-inhabiting microorganisms cannot be cultured on synthetic media at this time (16, 18, 34). Culture-independent methods, mainly supported by the development of PCR-based technologies, have improved detection, identification, and characterization of microorganisms directly from complex substrates such as soil, overcoming the limitations of cultivation-based approaches (14, 32). Many primers and probes having different specificities, ranging from universal to specific (2, 4, 9, 26), are now routinely used for molecular detection and identification of microorganisms. Using these taxonomical markers, increasing numbers of culture-independent studies aimed at better defining the microbial biodiversity of the rhizosphere are now reported each year using denaturing or temperature gradient gel electrophoresis or amplified ribosomal DNA restriction analysis (ARDRA) (5, 7, 8, 10, 17, 20, 29, 38). The development and improvement of these technologies have accelerated the discovery of new microbial species, refining our classical view of soil microbial diversity and generating new questions about the roles and functions of these organisms (18).
Some rhizosphere-inhabiting microbes are known to participate in detrimental interactions with the plants that lead to the development of root diseases, while others are clearly beneficial to the plants by stimulating their growth, enhancing nutrient uptake, or playing a significant role in controlling soilborne pathogens and the diseases they cause (36). This biological approach to the control of root diseases, known as biological control, relies on the idea that rhizosphere communities can be manipulated to promote natural disease suppression by mechanisms such as competition, production of antibiotics, parasitism, and induced resistance (3, 12, 39). However, very little is known about which rhizosphere microbial taxa play a significant role in controlling soilborne plant pathogens and root diseases, as well as how soilborne plant pathogens and disease development may alter rhizosphere microbial communities and the ecology of the rhizosphere under natural or controlled environmental conditions. We have clearly not yet reached the full potential concerning the use and management of disease-suppressive microbial communities to control root diseases. Therefore, more information is needed to better understand the role and implication that different rhizosphere-inhabiting microorganisms might play in biological control.
Root rot diseases, mainly caused by fungal pathogens, are common in conifer nurseries (13). The impact of these diseases on nursery seedling production is reflected by economic losses. Root rot diseases may not only cause mortality at nursery sites but may also affect survival rates after outplanting to reforestation sites. Mortality rates as high as 40% for the first year after outplanting have been reported for Picea mariana (25). An important factor is that symptoms are not always visible on infected seedlings, as the disease can remain latent and develop only after transplantation (11). The main pathogens that cause root diseases of conifers under nursery conditions belong to the fungal genera Cylindrocladium, Rhizoctonia, and Fusarium and the Oomycota genera Pythium and Phytophthora (13). To date, very few efficient biologically based control strategies have been developed to control these pathogens under conifer nursery conditions (23). A better understanding of the rhizosphere microbial ecology and diversity associated with conifer seedlings grown in tree nurseries would most certainly benefit the development of efficient biological control strategies.
Therefore, the aim of this study was to use a molecular culture-independent approach to (i) profile the main bacterial and fungal rhizosphere taxa associated with healthy black spruce (Picea mariana) seedlings and seedlings with symptoms of root rot grown in a conifer nursery and (ii) contrast the differences in terms of rhizosphere microbial community composition with respect to the health status of the seedlings.
MATERIALS AND METHODS
Sampling site.
Soil samples were collected in September 2001 from a governmental tree nursery located in the eastern region of Canada (Saint-Modeste, Québec, Canada) where a high incidence of root rot diseases has been reported (L. Innes, personal communication). In this nursery, black spruce propagation is routinely obtained by cuttings and rooting in compartmentalized production trays for 1 year under greenhouse conditions, followed by 2 years of growth outside before being shipped for reforestation. Six soil samples (50 to 100 g) were randomly collected from the rhizosphere of 3-year-old black spruce seedlings grown in one production plot. Soil samples were taken from the rhizosphere of three healthy seedlings and three seedlings with symptoms of root rot. Black spruce seedlings presumed to be healthy were carefully examined under a dissecting microscope to confirm the absence of disease symptoms. Rhizosphere soil samples were obtained by vigorously shaking the P. mariana seedling and collecting the soil portion in close contact with the root system. Soil samples were maintained on ice and transported back to the laboratory where they were rapidly lyophilized and kept at −80°C prior to soil genomic DNA extraction.
DNA extraction.
Total genomic DNA was extracted from 250 mg of a freeze-dried soil subsample using the MoBio UltraClean Soil DNA Isolation kit (MoBio Laboratories, Inc., Solana Beach, Calif.) according to the manufacturer's protocol with the following modifications: a FastPrep apparatus (Thermo Savant, Holbrook, N.Y.) was used at speed level 4 for 15 s instead of vortexing the soil samples for initial cell disruption. The purified DNA was resuspended in 50 μl of solution S5 (MoBio Laboratories, Inc.) and stored at −20°C until PCR amplification.
Bacterial and fungal rRNA gene amplification.
PCR amplification targeting bacterial partial 16S rRNA gene was performed using the 968f and 1401r primers (24), while the NS1 and NS2 primers (37) were used for fungal partial 18S rRNA gene amplification. Five microliters of 10× PCR buffer (Qiagen, Mississauga, Ontario, Canada), 5 μl of a 5 μM concentration of each bacterial or fungal primer, 1 μl of a 10 mM concentration of each deoxyribonucleoside triphosphate, 2 μl of a 1/20 dilution of soil genomic DNA or 2 μl of double-distilled H2O (negative control), 1.25 U of Taq DNA polymerase (Qiagen), and double-distilled H2O were combined in a final volume of 50-μl PCR mixtures. Reaction mixtures were overlaid with mineral oil prior to PCR amplification. PCR amplification was performed using a MJ Research PT-100 thermal cycler and the following steps: (i) an initial denaturation step (3 min at 94°C); (ii) 35 cycles, with 1 cycle consisting of denaturation (1 min at 94°C), annealing (1 min at 62°C [primers 968f and 1401r] or 55°C [primers NS1 and NS2]), and elongation (2 min at 72°C); and (iii) a final extension step (10 min at 72°C). Products were visualized by 1.5% agarose gel electrophoresis in 1× Tris-acetate-EDTA (TAE) buffer and stained with ethidium bromide (1% [wt/vol]). Bands were excised using a sterile scalpel, and DNA was purified from gel slices by using the QIAquick gel extraction kit (Qiagen).
Shotgun cloning and construction of bacterial and fungal rRNA gene libraries.
Each replicate library (six libraries of genes from the rhizosphere of three healthy seedlings and three diseased seedlings) was produced using different soil samples. PCR amplicons obtained from each of the three samples of the rhizosphere from three healthy or diseased seedlings using the 968f-1401r or NS1-NS2 primer set (total of 12 different samples) were each ligated into pCR4-TOPO vectors and transformed into One Shot TOP10 chemically competent Escherichia coli following the manufacturer's protocol (Invitrogen, Carlsbad, Calif.). Seventy randomly picked colonies per sample were each grown overnight at 37°C in 5 ml of Luria-Bertani broth containing 100 μl of ampicillin/ml and stored in 25% glycerol at −80°C. Plasmid DNA extraction was performed using the QIAprep Spin Miniprep kit (Qiagen), and the size of the insert was determined by both EcoRI restriction digest analysis and PCR amplification using the 968f-1401r or NS1-NS2 primer set. For each cloning reaction (total of 12), 50 clones containing an insert of the expected size (approximately 433 bp using 968f-1401r or 555 bp using NS1-NS2) were used for ARDRA.
ARDRA and sequencing.
After PCR amplification of cloned rRNA genes using the 968f-1401r or NS1-NS2 primer set, 10 μl of each amplicon (600 amplicons in total) were digested for 2 h at 37°C with 2.5 U of HinfI, 2.5 U of HaeIII, and 2.5 U of RsaI (New England Biolabs, Mississauga, Ontario, Canada) in 15-μl reaction mixtures. Restriction patterns were visualized by 3% agarose gel electrophoresis in 1× TAE buffer and stained with ethidium bromide (1% [wt/vol]). Gel images were digitalized, and restriction patterns were analyzed using the Quantity One software (Bio-Rad, Mississauga, Ontario, Canada). Each different restriction pattern was defined as an operational taxonomical unit (OTU). Plasmid DNA from one of each different representative OTU was commercially sequenced (DNA Landmarks, St-Jean-sur-Richelieu, Québec, Canada) on an ABI Prism 3700 DNA analyzer (Applied Biosystems, Foster City, Calif.) using T3 and T7 primers.
Sequence analysis.
DNA sequences were edited, and consensus sequences were obtained using DNAMAN version 4.13 (Lynnon BioSoft.). All sequenced clones were analyzed for the presence of chimeras using the CHIMERA CHECK program (version 2.7) from the Ribosomal Database Project (RDP) (http://rdp.cme.msu.edu/html/). Sequences suspected of being chimeric were not included in further analysis. All remaining sequences were screened against those in the GenBank database using BLASTn (1). On the basis of BLASTn similarity searches, sequences that were suspected of not being of bacterial or fungal origin were discarded. The most homologous sequences found in the GenBank database were used to construct a multiple-sequence alignment using ClustalX (version 1.81) (33) for both the bacterial and fungal clone libraries. A neighbor-joining analysis was performed using PAUP (version 4.0b10) (31) with 1,000 bootstrap replicates. Phylogenetic trees were edited using Treeview (version 1.6.6).
Diversity indices.
Clones having the same OTU were grouped, and the frequency of each OTU was determined. A number of diversity indices were calculated for each of the bacterial and fungal libraries. These indices included three different species richness indicators: (i) the total number of OTUs per library; (ii) the Shannon diversity index, a general diversity index that considers both species richness and evenness; and (iii) the Margalef index, which measures richness by using a ratio between the number of OTUs and the ln function of the number of clones analyzed (15). Three different indices were also calculated for evenness and dominance: (i) the Shannon evenness index, which is the ratio of the Shannon diversity index to the maximum possible value that could theoretically be obtained with the observed number of OTUs; (ii) the Simpson's index, which gives the probability that two clones chosen at random will be from the same OTU; and (iii) the Berger-Parker index, which is the relative abundance of the most abundant OTU (15). To determine whether the rRNA gene sequences obtained for each library of genes from the rhizosphere of healthy and diseased seedlings were different, homologous coverage curves, Cx(D), and heterologous coverage curves, Cxy(D) were calculated by a Cramér-von Mises test statistic and compared by a Monte Carlo test procedure (28). Similarity coefficients, which reflected the proportions of shared OTUs, were calculated by performing pairwise comparisons of the libraries of genes from the rhizosphere of healthy and diseased seedlings (21).
Nucleotide sequence accession numbers.
The nucleotide sequences determined in this study have been deposited in the National Center for Biotechnology Information (NCBI) database under accession numbers AY321198 to AY321281 (bacterial clones) and AY321682 to AY321712 (fungal clones).
RESULTS
High-molecular-weight DNA recovered from every soil sample was successfully amplified using both 968f-1401r and NS1-NS2 primer sets. Preliminary experiments, using ARDRA on 16S and 18S rRNA gene clones obtained from replicate rhizosphere DNAs revealed high similarity of the OTU patterns, suggesting a low degree of variability caused by sampling, DNA extraction, PCR amplification, cloning, and ARDRA (data not shown).
To confirm that the grouping of the different OTUs using 16S and 18S partial rRNA gene digestion with HinfI, HaeIII, and RsaI was reproducible and reliable for discriminating between the different bacterial and fungal clones, three replicates of three different bacterial and fungal OTUs were randomly selected and sequenced. The results revealed no important difference in sequence composition between the replicates of each of these different OTUs (data not shown), suggesting that the OTU grouping based on restriction digestion patterns was efficient for discriminating between bacterial and fungal rRNA gene clones analyzed in this study.
Of the partial bacterial 16S rRNA gene fragments amplified from the rhizosphere of healthy and diseased P. mariana seedlings and shotgun cloned, 300 different inserts (150 from healthy and 150 from diseased seedling treatments) were analyzed by ARDRA. ARDRA profiles revealed 84 different OTUs, and each OTU was sequenced. Thirty-four OTUs were found to be associated only with the rhizosphere of healthy seedlings, 26 OTUs were associated only with the rhizosphere of diseased seedlings, and 24 were found associated with the rhizosphere of both healthy and diseased seedlings. Phylogenetic analysis of the different OTUs and their closest relatives in the GenBank database was used to assign each environmental clone to a major bacterial group. The results of the sequence analysis of these OTUs, their phylogenetic affiliation, and their incidence among the rhizosphere of healthy and diseased seedlings are shown in Table 1.
TABLE 1.
Bacterial sequences from the GenBank database with the highest similarity to each OTU and distribution of OTUs in the rhizosphere of healthy and diseased P. mariana seedlings
| OTU | BLAST match | Accession no. | Similarity (%) | % Clonesa
|
|
|---|---|---|---|---|---|
| Healthy | Diseased | ||||
| 01 | Neochlamydia hartmannellae | AF177275 | 96 | 3 | |
| 02 | Uncultured gammaproteobacterium clone JG36-TzT-191 | AJ534626 | 93 | 3 | |
| 03 | Parachlamydia acanthamoebae | Y07556 | 93 | 3 | |
| 04 | Uncultured Acidobacteria bacterium clone VC47 | AY211077 | 94 | 3 | |
| 05 | Uncultured bacterium TM7LH15 | AF269005 | 96 | 3 | |
| 06 | Aquaspirillum autotrophicum | AB074524 | 99 | 3 | |
| 07 | Uncultured gammaproteobacterium YNPRH65B | AF465652 | 99 | 3 | |
| 08 | Uncultured bacterium clone a13152 | AY102338 | 98 | 3 | |
| 09 | Uncultured soil bacterium clone C06 | AF507679 | 94 | 3 | |
| 10 | Uncultured gammaproteobacterium YNPRH65B | AF465652 | 99 | 3 | |
| 11 | Uncultured soil bacterium clone C065 | AF507697 | 91 | 3 | |
| 12 | Bacillus sp. | AF548880 | 98 | 3 | |
| 13 | Uncultured Holophaga sp. | AJ536864 | 96 | 2 | |
| 14 | Paenibacillus alginolyticus | AB073362 | 99 | 2 | |
| 15 | Uncultured candidate division TM7 clone NoosaAW57 | AF269013 | 94 | 2 | |
| 16 | Sporichthya polymorpha | AB025317 | 97 | 2 | |
| 17 | Uncultured bacterium clone CR99-7-30 | AF429064 | 89 | 2 | |
| 18 | Uncultured bacterium TM7LH20 | AF269006 | 95 | 2 | |
| 19 | Polyangium sp. | M94280 | 93 | 2 | |
| 20 | Uncultured Acidobacteria bacterium isolate OF20 | AY177758 | 98 | 2 | |
| 21 | Uncultured candidate division BD bacterium clone BDfull1 | AF545649 | 95 | 1 | |
| 22 | Uncultured Acidobacteriaceae bacterium clone Rs-M39 | AB089126 | 97 | 1 | |
| 23 | Mycobacterium sp. | AF494537 | 99 | 1 | |
| 24 | Oxalobacter formigenes | U49755 | 93 | 1 | |
| 25 | Frateuria aurentia | AJ010481 | 97 | 1 | |
| 26 | Pseudonocardia chlorethenivorans | AF454510 | 97 | 1 | |
| 27 | Microthrix parvicella | X82546 | 91 | 1 | |
| 28 | Pseudomonas sp. | AF105381 | 99 | 1 | |
| 29 | Uncultured bacterium clone JG30a-KF-32 | AJ536876 | 91 | 1 | |
| 30 | Uncultured soil bacterium clone C1105 | AF507688 | 95 | 1 | |
| 31 | Saccharothrix tangerinus | AB020031 | 99 | 1 | |
| 32 | Nitrosospira sp. | AF359341 | 99 | 1 | |
| 33 | Uncultured bacterium clone CCM6b | AY221067 | 96 | 1 | |
| 34 | Oxalobacter formigenes | U49755 | 93 | 1 | |
| 35 | Uncultured Rubrobacteridae bacterium 0649-1G9 | AF234119 | 93 | 2 | 1 |
| 36 | Uncultured alphaproteobacterium clone JG34-KF-258 | AJ532705 | 96 | 2 | 1 |
| 37 | Uncultured gammaproteobacterium YNPRH65B | AF465652 | 98 | 3 | 1 |
| 38 | Unidentified bacterial species clone 32-10 | Z95710 | 97 | 3 | 2 |
| 39 | Uncultured Acidobacteria bacterium isolate OF23 | AY177759 | 97 | 3 | 2 |
| 40 | Uncultured soil bacterium clone C1220 | AF507649 | 96 | 3 | 2 |
| 41 | Parachlamydia acanthamoebae | Y07556 | 93 | 2 | 2 |
| 42 | Uncultured bacterium DGGEb band YNPRH-B18 | AF465676 | 94 | 2 | 2 |
| 43 | Uncultured Acidobacteria bacterium isolate OF23 | AY177759 | 95 | 2 | 2 |
| 44 | Uncultured soil bacterium clone C1220 | AF507649 | 97 | 1 | 1 |
| 45 | Parachlamydia acanthamoebae | Y07556 | 92 | 1 | 1 |
| 46 | Parachlamydia acanthamoebae | Y07556 | 93 | 1 | 1 |
| 47 | Uncultured gammaproteobacterium YNPRH65B | AF465652 | 95 | 1 | 1 |
| 48 | Uncultured bacterium clone NOS7.157WL | AF432607 | 97 | 1 | 1 |
| 49 | Parachlamydia acanthamoebae | Y07556 | 94 | 1 | 1 |
| 50 | Uncultured bacterium DGGE band YNPRH-B18 | AF465676 | 94 | 1 | 1 |
| 51 | Bacterial clone RB30 | Z95720 | 99 | 1 | 3 |
| 52 | Uncultured eubacterium WD260 | AJ292673 | 99 | 1 | 3 |
| 53 | Uncultivated soil bacterium clone S023 | AF013550 | 98 | 1 | 2 |
| 54 | Thiobacillus prosperus | AY034139 | 90 | 1 | 2 |
| 55 | Uncultured gammaproteobacterium clone JG36-TzT-191 | AJ534626 | 96 | 1 | 2 |
| 56 | Gordonia jacobaea | AF251791 | 90 | 1 | 2 |
| 57 | Uncultured actinobacterium clone OBPB53 | AY193072 | 90 | 1 | 2 |
| 58 | Uncultured bacterium NoosaAW57 | AF269013 | 95 | 1 | 2 |
| 59 | Conexibacter woesei | AJ440237 | 97 | 1 | |
| 60 | Marmoricola aurantiacus | Y18629 | 96 | 1 | |
| 61 | Uncultured sludge bacterium S41 | AF234734 | 97 | 1 | |
| 62 | Unidentified betaproteobacterium | AB006750 | 94 | 1 | |
| 63 | Burkholderia glathei | AY154379 | 99 | 1 | |
| 64 | Rathayibacter tritici | X77438 | 98 | 1 | |
| 65 | Uncultured Verrucomicrobium sp. clone V97 | AY143745 | 97 | 1 | |
| 66 | Kitasatospora sp. | AB022877 | 93 | 1 | |
| 67 | Bacillus benzoevorans | Y14693 | 98 | 1 | |
| 68 | Uncultured Acidobacteria bacterium isolate OF20 | AY177758 | 96 | 1 | |
| 69 | Ammoniphilus oxalaticus | Y14579 | 99 | 1 | |
| 70 | Neochlamydia hartmannellae | AF177275 | 92 | 1 | |
| 71 | Uncultured actinobacterium ML602J-44 | AF486498 | 93 | 1 | |
| 72 | Neochlamydia hartmannellae | AF177275 | 95 | 2 | |
| 73 | Actinomycete clone Ep T1.148 | Z73370 | 98 | 2 | |
| 74 | Nitrosospira sp. | AY123805 | 95 | 2 | |
| 75 | Uncultured betaproteobacterium clone ccs4 | AY133078 | 95 | 2 | |
| 76 | Uncultured verrucomicrobium DEV031 | AJ401123 | 94 | 2 | |
| 77 | Parachlamydia acanthamoebae | Y07556 | 94 | 2 | |
| 78 | Uncultured bacterium clone JG30a-KF-32 | AJ536876 | 92 | 2 | |
| 79 | Nitrosospira sp. | AY123808 | 94 | 3 | |
| 80 | Burkholderia sp. | AY134849 | 99 | 3 | |
| 81 | Uncultured bacterium clone JG30a-KF-32 | AJ536876 | 91 | 5 | |
| 82 | Nocardiopsis compostus KS9 | AF360734 | 91 | 5 | |
| 83 | Uncultured bacterium DGGE band YNPRH-B18 | AF465676 | 94 | 5 | |
| 84 | Nitrospira sp. | AJ224042 | 97 | 5 | |
Percentage of clones showing the same OTU from the rhizosphere of healthy or diseased seedlings.
DGGE, denaturing gradient gel electrophoresis.
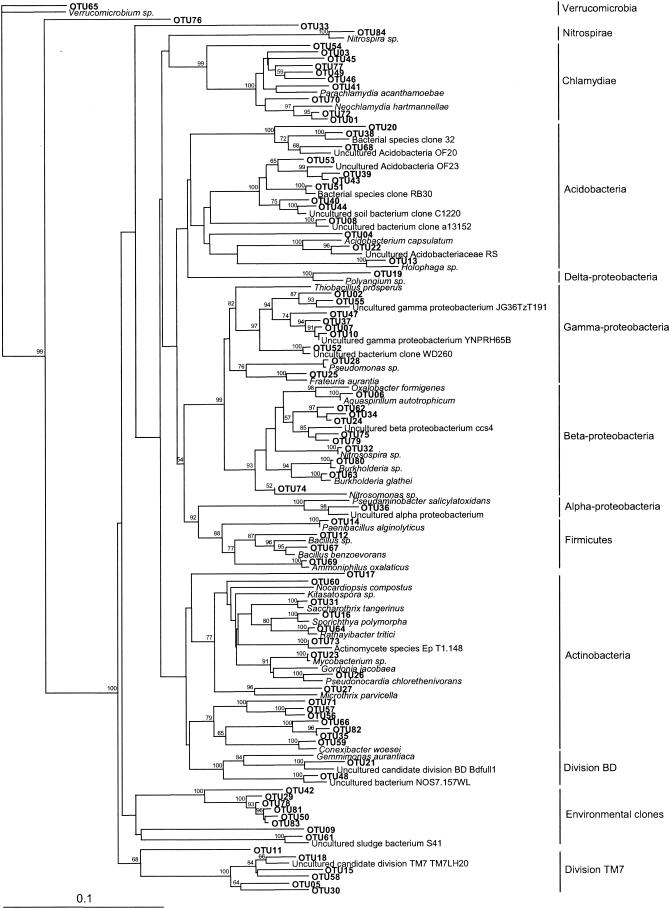
The phylogenetic analysis indicated that the bacterial DNAs amplified from the rhizosphere of P. mariana are taxonomically diverse and can be grouped into 13 different bacterial groups (Fig. 1). From this analysis, we determined that the majority of 16S rRNA gene clones obtained from the rhizosphere of healthy seedlings grouped with the Proteobacteria (27%) and Acidobacteria (25%) (Table 2). The analysis also revealed that the 16S rRNA gene clones obtained from the rhizosphere of diseased seedlings were not associated with the same main groups and in the same proportions (Proteobacteria [22%], Actinobacteria [18%], environmental clones [17%], and Acidobacteria [16%]) as those ones obtained from the rhizosphere of healthy seedlings (Table 2). Environmental clones represent an arbitrary grouping of clones that have never been found in cultivated organisms and that have been retrieved from environmental conditions only by culture-independent studies. Clones affiliated with the Deltaproteobacteria were detected from the rhizosphere of only one healthy seedling, while clones affiliated with the Nitrospirae and Verrucomicrobia were detected only from the rhizosphere of diseased seedlings.
FIG. 1.
Unrooted neighbor-joining tree representing the phylogenetic relationships of 84 bacterial OTU sequences from the rhizosphere of healthy and diseased P. mariana seedlings to the most closely related sequences obtained from BLAST searches. The numbers at the nodes of the tree are bootstrap values for each node with 1,000 bootstrap resamplings (values below 50 are not shown). The bar represents 0.1 substitution per site.
TABLE 2.
Relative abundance of clones with respect to different bacterial and fungal taxa from triplicate soil samples obtained from the rhizosphere of healthy and diseased P. mariana seedlings
| Phylogenetic group | Relative bacterial and fungal clone abundance (%)
|
|||||||
|---|---|---|---|---|---|---|---|---|
| rRNA gene library from healthy seedlings
|
rRNA gene library from diseased seedlings
|
|||||||
| Rep Aa | Rep B | Rep C | All | Rep A | Rep B | Rep C | All | |
| Bacteria | ||||||||
| Acidobacteria | 36 | 10 | 31 | 25 | 15 | 13 | 19 | 16 |
| Actinobacteria | 11 | 10 | 9 | 10 | 19 | 22 | 12 | 18 |
| Firmicutes | 3 | 9 | 4 | 6 | 2 | |||
| Proteobacteria | 32 | 38 | 14 | 27 | 8 | 30 | 30 | 22 |
| Alphaproteobacteria | 3 | 1 | 3 | 1 | ||||
| Betaproteobacteria | 11 | 13 | 8 | 4 | 15 | 18 | 13 | |
| Deltaproteobacteria | 5 | 2 | ||||||
| Gammaproteobacteria | 21 | 18 | 11 | 16 | 4 | 15 | 9 | 7 |
| Chlamydiae | 7 | 10 | 11 | 10 | 12 | 19 | 3 | 13 |
| Nitrospirae | 8 | 3 | 3 | 4 | ||||
| Verrucomicrobia | 4 | 6 | 3 | |||||
| Candidate division BD | 7 | 2 | 4 | 1 | ||||
| Candidate division TM7 | 4 | 13 | 11 | 10 | 4 | 3 | 2 | |
| Environmental clones | 10 | 9 | 7 | 22 | 13 | 15 | 17 | |
| Unidentified bacteria | 4 | 8 | 6 | 6 | 4 | 3 | 2 | |
| Fungi | ||||||||
| Ascomycota | 13 | 20 | 28 | 20 | 68 | 48 | 33 | 49 |
| Sordariomycetes | 20 | 14 | 12 | 32 | 38 | 18 | 29 | |
| Dothideomycetes | 18 | 4 | 9 | 10 | ||||
| Eurothiomycetes | 14 | 4 | ||||||
| Pezizomycetes | 13 | 4 | ||||||
| Mitosporic Ascomycota | 18 | 6 | 6 | 10 | ||||
| Basidiomycota | 87 | 70 | 72 | 76 | 22 | 47 | 53 | 41 |
| Homobasidiomycetes | 87 | 70 | 72 | 76 | 18 | 47 | 47 | 38 |
| Heterobasidiomycetes | 4 | 6 | 3 | |||||
| Unidentified fungi | 10 | 4 | 10 | 6 | 15 | 10 | ||
Rep A, replicate A.
Three hundred different partial fungal 18S rRNA gene fragments (150 from healthy and 150 from diseased seedling treatments) were also amplified from the rhizosphere of healthy and diseased P. mariana seedlings and shotgun cloned. ARDRA of these inserts revealed 31 different OTUs, and each of these OTUs was sequenced. Eight OTUs were found to be associated only with the rhizosphere of healthy seedlings, 20 OTUs were associated only with the rhizosphere of diseased seedlings, and 3 were found associated with the rhizosphere of both healthy and diseased seedlings.
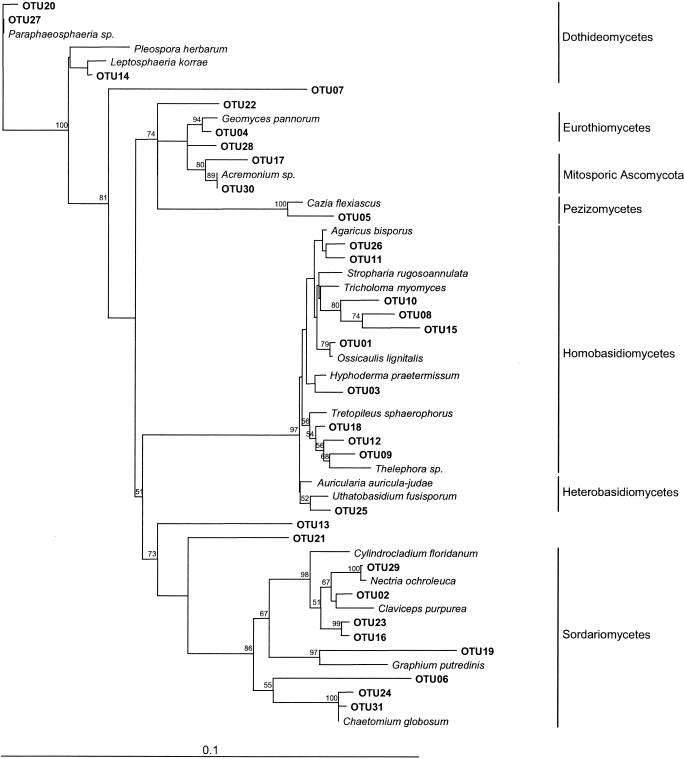
Phylogenetic analysis of the different OTUs and their closest relatives in the GenBank database was performed to assign each environmental clone to a major fungal group. The results of the sequence analysis of these OTUs, their phylogenetic affiliation, and their incidence among the rhizosphere of healthy and diseased seedlings are shown in Table 3. Phylogenetic analysis indicated that the fungal DNAs amplified from the rhizosphere of P. mariana are taxonomically diverse and can be grouped into seven different groups belonging to the Ascomycota and Basidiomycota (Fig. 2). Healthy rhizosphere-associated fungal 18S rRNA gene clones were not associated with the same main groups and in the same proportions as the ones associated with diseased seedlings (Table 2). The majority of the clones found in the rhizosphere of healthy seedlings grouped with the Homobasidiomycetes (76%), while the ones found in the rhizosphere of diseased seedlings mainly grouped with the Homobasidiomycetes (38%) and Soradiomycetes (29%). Clones affiliated with the Eurothiomycetes and Pezizomycetes were detected from the rhizosphere of only one healthy seedling, while clones affiliated with the Dothideomycetes, mitosporic Ascomycota, and Heterobasidiomycetes were detected only from the rhizosphere of diseased seedlings.
TABLE 3.
Fungal sequences from the GenBank database with highest similarity to each OTU and distribution of OTUs in the rhizosphere of healthy and diseased P. mariana seedlings
| OTU | BLAST match | Accession no. | % Similarity | % Clonesa
|
|
|---|---|---|---|---|---|
| Healthy | Diseased | ||||
| 01 | Ossicaulis lignatilis | AF334923 | 99 | 24 | |
| 02 | Geosmithia putterillii | AB032069 | 99 | 8 | |
| 03 | Hyphoderma praetermissum | AF518580 | 99 | 4 | |
| 04 | Geomyces pannorum | AB016174 | 99 | 4 | |
| 05 | Cazia flexiascus | U42666 | 98 | 4 | |
| 06 | Chaetomium globosum | AB048285 | 94 | 4 | |
| 07 | Acremonium sp. | AJ278754 | 97 | 4 | |
| 08 | Ossicaulis lignatilis | AF334923 | 97 | 4 | |
| 09 | Thelephora sp. | AF026627 | 98 | 24 | 2 |
| 10 | Tricholoma myomyces | AF287841 | 98 | 12 | 22 |
| 11 | Hyphoderma praetermissum | AF518580 | 99 | 8 | 4 |
| 12 | Tretopileus sphaerophorus | AB006005 | 98 | 1 | |
| 13 | Chaetomium globosum | AB048285 | 93 | 1 | |
| 14 | Leptosphaeria korrae | AF486626 | 99 | 1 | |
| 15 | Tricholoma myomyces | AF287841 | 96 | 1 | |
| 16 | Cylindrocladium floridanum | AY321197 | 99 | 1 | |
| 17 | Acremonium sp. | AJ278754 | 98 | 1 | |
| 18 | Tretopileus sphaerophorus | AB006005 | 99 | 1 | |
| 19 | Graphium putredinis | AB007683 | 94 | 1 | |
| 20 | Paraphaeosphaeria sp. | AB096264 | 99 | 1 | |
| 21 | Chaetomium globosum | AB048285 | 95 | 1 | |
| 22 | Pleospora herbarum | U43458 | 93 | 2 | |
| 23 | Cylindrocladium floridanum | AY321197 | 99 | 4 | |
| 24 | Chaetomium globosum | AB048285 | 99 | 2 | |
| 25 | Uthatobasidium fusisporum | AF518593 | 99 | 3 | |
| 26 | Agaricus bisporus | L36658 | 99 | 6 | |
| 27 | Paraphaeosphaeria sp. | AB096264 | 100 | 7 | |
| 28 | Acremonium sp. | AJ278754 | 98 | 7 | |
| 29 | Nectria ochroleuca | AB003950 | 99 | 7 | |
| 30 | Acremonium sp. | AJ278754 | 100 | 9 | |
| 31 | Chaetomium globosum | AB048285 | 99 | 13 | |
Percentage of clones showing the same OTU from the rhizosphere of healthy or diseased seedlings.
FIG. 2.
Unrooted neighbor-joining tree representing the phylogenetic relationships of 31 fungal OTU sequences from the rhizosphere of healthy and diseased P. mariana seedlings to the most closely related sequences obtained from BLAST searches. The numbers at the nodes of the tree indicate bootstrap values for each node with 1,000 bootstrap resamplings (values below 50 are not shown). The bar represents 0.1 substitution per site.
The Cramér-von Mises test statistic and Monte Carlo comparison test procedure indicated that the bacterial libraries associated with the rhizosphere of healthy and diseased P. mariana seedlings were significantly different (P < 0.05). For the fungal libraries, as the number of different OTUs obtained was low (11 for healthy and 23 for diseased seedlings), this statistical approach was not used, since a larger number of sequences, usually more than 20 different sequences per library, is required to give a realistic comparison (28). However, the distribution of OTUs shown in Table 3 clearly suggests that the fungal communities associated with healthy and diseased seedlings are different.
Similarity coefficients were calculated by performing pairwise comparisons of the different replicates of bacterial and fungal libraries (Table 4). The similarity coefficients obtained between healthy-healthy and diseased-diseased seedling replicates for bacterial or fungal libraries were all higher than the ones obtained for comparisons between healthy-diseased seedling treatments, indicating that all replicates from libraries of genes from the rhizosphere of healthy or diseased seedlings were more similar to each other than to different treatments.
TABLE 4.
Pairwise comparisons of bacterial and fungal rRNA gene library compositions obtained from triplicate rhizosphere soil samples associated with healthy and diseased P. mariana seedlings
| Phylogenetic group and rhizosphere sample | Librarya | Similarity coefficientb
|
||||
|---|---|---|---|---|---|---|
| rRNA gene libraries from healthy seedlings
|
rRNA gene libraries from diseased seedlings
|
|||||
| Rep A | Rep B | Rep C | Rep A | Rep B | ||
| Bacteria | ||||||
| Healthy seedlings | Healthy (Rep B) | 0.259 | ||||
| Healthy (Rep C) | 0.255 | 0.364 | ||||
| Diseased seedlings | Diseased (Rep A) | 0.233 | 0.118 | 0.078 | ||
| Diseased (Rep B) | 0.118 | 0.203 | 0.231 | 0.292 | ||
| Diseased (Rep C) | 0.078 | 0.237 | 0.231 | 0.375 | 0.536 | |
| Fungi | ||||||
| Healthy seedlings | Healthy (Rep B) | 0.500 | ||||
| Healthy (Rep C) | 0.500 | 0.489 | ||||
| Diseased seedlings | Diseased (Rep A) | 0.211 | 0.095 | 0.190 | ||
| Diseased (Rep B) | 0.250 | 0.222 | 0.222 | 0.400 | ||
| Diseased (Rep C) | 0.222 | 0.100 | 0.200 | 0.667 | 0.500 | |
Library of rRNA genes from triplicate rhizosphere soil samples associated with healthy and diseased P. mariana seedlings. Rep, replicate.
Calculated by the method of McCaig et al. (21).
Diversity indices were calculated using ARDRA data obtained from either bacterial or fungal clones and from each library of genes from the rhizosphere of healthy and diseased seedlings (Table 5). For bacteria, a higher richness was associated with healthy seedlings; this was reflected by the higher values obtained with the three different species richness indices used. The same indices were applied to fungal libraries and revealed that a higher richness was associated with diseased seedlings. In the case of evenness and dominance indices, no important differences were obtained when comparing the libraries of bacterial or fungal genes from the rhizosphere of healthy and diseased seedlings. However, overall, the indices suggest a slightly higher dominance of some bacterial species associated with diseased seedlings, while for fungi, slightly higher values were associated with fungi associated with healthy seedlings, suggesting that some fungal species were more dominant in healthy than diseased seedling treatments.
TABLE 5.
Diversity indices for bacterial and fungal OTUs associated with the rhizosphere of healthy and diseased P. mariana seedlings
| Phylogenetic group and sourcea | Species richness index
|
Evenness or dominance index
|
||||
|---|---|---|---|---|---|---|
| No. of recorded OTUs | Shannon index (H′) | Margalef index (Dmg) | Shannon evenness index (E) | Simpson's index (D) | Berger-Parker index (d) | |
| Bacteria | ||||||
| Healthy seedlings | 58 | 4.036 | 12.298 | 0.994 | 0.012 | 0.039 |
| Diseased seedlings | 50 | 3.566 | 10.916 | 0.911 | 0.014 | 0.045 |
| Fungi | ||||||
| Healthy seedlings | 11 | 2.116 | 2.316 | 0.882 | 0.141 | 0.240 |
| Diseased seedlings | 23 | 2.600 | 4.831 | 0.829 | 0.090 | 0.221 |
Source of bacteria or fungi from the rhizosphere of healthy and diseased P. mariana seedlings.
DISCUSSION
The aim of this study was to characterize the diversity and community structure of bacteria and fungi associated with the rhizosphere of P. mariana seedlings grown under nursery conditions using a highly discriminant culture-independent approach and assess differences in community structure with respect to the health status of the seedlings. Although several other rRNA gene analysis-based approaches were available to perform more rapid analysis (e.g., denaturing or temperature gradient gel electrophoresis), the ARDRA approach used in this study provided a high level of resolution and the generation of additive and retrievable data which might also be used to generate other taxonomical probes and primers (e.g., to target smaller groups of microorganisms) for use in further studies.
To our knowledge, the present work is the first reported culture-independent study aimed at characterizing the microbial diversity that exists under conifer nursery conditions. As a small number of samples was studied, caution has to be taken when drawing conclusions from such a limited set of data. Nevertheless, the results obtained clearly indicated that the bacterial and fungal diversity associated with the rhizosphere of P. mariana seedlings grown under nursery conditions was considerable. Different diversity and phylogenetic analyses (see Results) supported the hypothesis that the bacterial and fungal communities associated with the rhizosphere of healthy and diseased P. mariana seedlings are different. Similarity coefficients, calculated by performing pairwise comparisons to assess the variability associated with the sampling strategy, clearly indicated that the triplicate rRNA gene libraries used for healthy and diseased seedling-associated treatments were more similar within than among treatments). These analyses supported the view that our sampling was sufficient to provide a realistic portrait of the microbial diversity in this system and that the variability obtained, always inherent with any such studies, did not compromise the interpretation of the results.
Our results clearly support the hypothesis that the bacterial and fungal communities were altered in relation to the health status of P. mariana seedlings. As reflected by a higher number of different OTUs, healthy P. mariana seedlings showed a higher rhizosphere-associated bacterial diversity than diseased seedlings, while the opposite trend was observed for fungi. The high frequency of some fungal OTUs suggested that very few fungal species dominated the rhizosphere of P. mariana seedlings (mainly organisms closely related to Ossicaulis lignitalis, Thelephora sp., and Tricholoma myomyces), while for bacteria, no OTU was found at high frequency. Among the most important differences observed between the bacterial and fungal communities associated with the rhizosphere of healthy and diseased seedlings, clones related to the Nitrospirae, Verrucomicrobia, Dothideomycetes, mitosporic Ascomycota, and Heterobasidiomycetes were found associated only with some diseased seedlings, while clones related to the Deltaproteobacteria, Eurothiomycetes, and Pezizomycetes were found associated only with healthy seedlings. Although it remains difficult to interpret these differences ecologically and functionally due to the important biodiversity detected, these results, which were clearly correlated with the health status of P. mariana seedlings, certainly are of interest and may help to narrow searches using PCR primer sets targeting smaller bacterial and fungal groups.
Very few culture-independent studies have looked at the differences that may exist between the rhizosphere microbial communities associated with healthy and diseased plants and how these changes may influence disease development and/or resistance (22, 39). Yang et al. (39) detected significant changes in bacterial communities associated with healthy and Phytophthora-infected avocado roots, a conclusion also reached by McSpadden Gardener et al. (22) while investigating changes in populations of rhizosphere bacteria associated with take-all disease of wheat caused by Gaeumannomyces graminis var. tritici. Although it was hypothesized in both studies that Pseudomonas populations might have been involved in biological control against those two different fungal soilborne pathogens, clear evidence supporting this hypothesis was not found.
The bacterial and fungal sequences amplified from the rhizosphere of P. mariana belonged to different phylogenetic groups that have also been reported in other culture-independent studies as important rhizosphere-inhabiting organisms. For example, Smalla et al. (29) reported that gram-positive bacteria with a high G+C content (or Actinobacteria) were among the most dominant bacteria in the rhizosphere of strawberry (Fragaria ananassa), oilseed rape (Brassica napus), and potato (Solanum tuberosum) grown under field conditions. McCaig et al. (21) reported that the most important bacterial groups found in a grassland dominated by grass species, such as Agrostis capillaries, Festuca ovina, Trifolium repens, and Trifolium perenne, belonged to the Proteobacteria (mainly the α-group) and Actinobacteria, while Chow et al. (5) reported that the Proteobacteria (mainly the α- and β-groups) and Acidobacteria dominated the rhizosphere of lodgepole pine (Pinus contorta) under forest conditions. The main bacterial groups identified in the present study, mainly the Proteobacteria, Acidobacteria, and Actinobacteria, were the same as those identified in other studies, even if the plant species and the ecosystems studied were completely different. It is interesting that members of different bacterial groups for which no or only a very limited number of species have been successfully cultured so far, such as the Verrucomicrobia, Acidobacteria, candidate division BD, candidate division TM7, and environmental clones, were detected at high levels (up to 44% of the bacterial rRNA gene sequences amplified) in this study. Because different molecular markers and methodological approaches have been used so far to detect and identify fungi and because very few culture-independent studies have been performed in this field (19, 27, 30), it is not yet possible to make reliable comparisons and obtain a general culture-independent portrait of the most dominant fungal groups commonly associated with the rhizosphere of plants. In the present study, Homobasidiomycetes, Sordaryomycetes, Dothideomycetes, and mitosporic Ascomycota, all belonging to the Ascomycota or Basidiomycota, were the groups most often found associated with the rhizosphere of P. mariana seedlings.
The determination of the causal agent(s) was not the objective of this study and since only the rhizosphere soil and not the root system was investigated for the presence of microorganisms, we cannot yet clearly conclude on which main root rot-causing pathogen(s) was involved. In light of the results obtained, a better hypothesis would be that organisms related to Fusarium (Nectria ochroleuca, 99% similarity) and Cylindrocladium (Cylindrocladium floridanum, 99% similarity) might have contributed to the disease symptoms clearly observed in P. mariana seedlings with symptoms of root rot, a hypothesis also supported by the fact that these organisms were found only in the rhizosphere of diseased seedling. It is interesting that other microbial members that are generally known for their beneficial impact on plants and biological control potential against soilborne plant pathogens (35), such as species related to Pseudomonas (99% similarity with Pseudomonas sp.), Bacillus (98% similarity with Bacillus sp.), and Paenibacillus (99% similarity with Paenibacillus alginolyticus), were detected only in the rhizosphere of healthy seedlings. However, because the specificity range of the PCR primers used was large and only a limited number of samples was studied, these results should mainly be considered as interesting avenues for further investigations, where selected group-specific PCR primers could be used to reveal a more precise taxonomical identification.
As most studies performed so far to identify potential biocontrol agents against given soilborne plant pathogens were conducted using isolation techniques and culture-based approaches, only a few potential rhizosphere competent biocontrol agents have been sampled (18). Hence, it appears reasonable to believe that the discovery of new rhizosphere competent biocontrol agents will most likely benefit from culture-independent studies and that we might eventually identify yet uncultured microorganisms that may play a significant role in biological control of soilborne pathogens. Better knowledge of these organisms may also promote the development of new approaches to culture them and/or manage their population.
We have not yet been able to clearly demonstrate whether the communities associated with healthy roots in fact promote disease suppression or whether their presence is simply a direct consequence of the absence of the pathogen. Nevertheless, the clear demonstration that the communities in the rhizosphere of diseased seedlings are different (as seen by the distinct rRNA gene amplification profiles obtained in this study) certainly represents a good starting point for further analyses. The next steps will involve determining the ecological role of microbial communities associated with selected healthy seedlings and clearly demonstrating the mode of action of biological control.
Acknowledgments
This work was supported in part by a NSERC-Biocontrol Network grant to M. St-Arnaud, L. Bernier, and R. C. Hamelin.
We are indebted to Vladimir Vujanovic for providing biological material and expertise. We thank Etienne Yergeau for technical assistance.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, I. C., C. D. Campbell, and J. I. Prosser. 2003. Potential bias of fungal 18S rDNA and internal transcribed spacer polymerase chain reaction primers for estimating fungal biodiversity in soil. Environ. Microbiol. 5:36-47. [DOI] [PubMed] [Google Scholar]
- 3.Boland, G. J., and L. D. Kuykendall (ed.). 1998. Plant-microbe interactions and biological control. Marcel Dekker, Inc., New York, N.Y.
- 4.Borneman, J., and R. J. Hartin. 2000. PCR primers that amplify fungal rRNA genes from environmental samples. Appl. Environ. Microbiol. 66:4356-4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chow, M. L., C. C. Radomski, J. M. McDermott, J. Davies, and P. E. Axelrood. 2002. Molecular characterization of bacterial diversity in lodgepole pine (Pinus contorta) rhizosphere soils from British Columbia forest soils differing in disturbance and geographic source. FEMS Microbiol. Ecol. 42:347-357. [DOI] [PubMed] [Google Scholar]
- 6.Curl, E. A., and B. Truelove. 1986. The rhizosphere, vol. 15. Springer-Verlag, Giessen, Germany.
- 7.Duineveld, B. M., G. A. Kowalchuk, A. Keijzer, J. D. van Elsas, and J. A. van Veen. 2001. Analysis of bacterial communities in the rhizosphere of chrysanthemum via denaturing gradient gel electrophoresis of PCR-amplified 16S rRNA as well as DNA fragments coding for 16S rRNA. Appl. Environ. Microbiol. 67:172-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duineveld, B. M., A. S. Rosado, J. D. van Elsas, and J. A. van Veen. 1998. Analysis of the dynamics of bacterial communities in the rhizosphere of the chrysanthemum via denaturing gradient gel electrophoresis and substrate utilization patterns. Appl. Environ. Microbiol. 64:4950-4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gardes, M., and T. D. Bruns. 1993. ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol. Ecol. 2:113-118. [DOI] [PubMed] [Google Scholar]
- 10.Gomes, N. C. M., H. Heuer, J. Schonfeld, R. Costa, L. Mendonca-Hagler, and K. Smalla. 2001. Bacterial diversity of the rhizosphere of maize (Zea mays) grown in tropical soil studied by temperature gradient gel electrophoresis. Plant Soil 232:167-180. [Google Scholar]
- 11.Hamelin, R. C., P. Bérubé, M. Gignac, and M. Bourassa. 1996. Identification of root rot fungi in nursery seedlings by nested multiplex PCR. Appl. Environ. Microbiol. 62:4026-4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Handelsman, J., and E. V. Stabb. 1996. Biocontrol of soilborne plant pathogens. Plant Cell 8:1855-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hansen, E. M., and K. J. Lewis. 1997. Compendium of conifer diseases. APS Press, St. Paul, Minn.
- 14.Head, I. M., J. R. Saunders, and R. W. Pickup. 1998. Microbial evolution, diversity, and ecology: a decade of ribosomal RNA analysis of uncultivated microorganisms. Microb. Ecol. 35:1-21. [DOI] [PubMed] [Google Scholar]
- 15.Hill, C. J., K. A. Walsh, J. A. Harris, and B. F. Moffett. 2003. Using ecological diversity measures with bacterial communities. FEMS Microbiol. Ecol. 43:1-11. [DOI] [PubMed] [Google Scholar]
- 16.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kandeler, E., P. Marschner, D. Tscherko, T. S. Gahoonia, and N. E. Nielsen. 2002. Microbial community composition and functional diversity in the rhizosphere of maize. Plant Soil 238:301-312. [Google Scholar]
- 18.Kent, A. D., and E. W. Triplett. 2002. Microbial communities and their interactions in soil and rhizosphere ecosystems. Annu. Rev. Microbiol. 56:211-236. [DOI] [PubMed] [Google Scholar]
- 19.Kowalchuk, G. A., S. Gerards, and J. W. Woldendorp. 1997. Detection and characterization of fungal infections of Ammophila arenaria (Marram grass) roots by denaturing gradient gel electrophoresis of specifically amplified 18S rDNA. Appl. Environ. Microbiol. 63:3858-3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marilley, L., G. Vogt, M. Blanc, and M. Aragno. 1998. Bacterial diversity in the bulk soil and rhizosphere fractions of Lolium perenne and Trifolium repens as revealed by PCR restriction analysis of 16S rDNA. Plant Soil 198:219-224. [Google Scholar]
- 21.McCaig, A. E., L. A. Glover, and J. I. Prosser. 1999. Molecular analysis of bacterial community structure and diversity in unimproved and improved upland grass pastures. Appl. Environ. Microbiol. 65:1721-1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McSpadden Gardener, B. B., and D. M. Weller. 2001. Changes in populations of rhizosphere bacteria associated with take-all disease of wheat. Appl. Environ. Microbiol. 67:4414-4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morin, C., J. Samson, and M. Dessureault. 1999. Protection of black spruce seedlings against Cylindrocladium root rot with ectomycorrhizal fungi. Can. J. Bot. 77:169-174. [Google Scholar]
- 24.Nübel, U., B. Engelen, A. Felske, J. Snaidr, A. Wieshuber, R. I. Amann, W. Ludwig, and H. Backhaus. 1996. Sequence heterogeneities of genes encoding 16S rRNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J. Bacteriol. 178:5636-5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saunders, J. E., J. Juzwik, and R. Hutchison. 1992. Outplanting survival of Cylindrocladium root rot affected black spruce seedlings. Can. J. For. Res. 22:1204-1207. [Google Scholar]
- 26.Schmalenberger, A., F. Schwieger, and C. C. Tebbe. 2001. Effect of primers hybridizing to different evolutionarily conserved regions of the small-subunit rRNA gene in PCR-based microbial community analyses and genetic profiling. Appl. Environ. Microbiol. 67:3557-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sigler, W. V., and R. F. Turco. 2002. The impact of chlorothalonil application on soil bacterial and fungal populations as assessed by denaturing gradient gel electrophoresis. Appl. Soil Ecol. 21:107-118. [Google Scholar]
- 28.Singleton, D. R., M. A. Furlong, S. L. Rathbun, and W. B. Whitman. 2001. Quantitative comparisons of 16S rRNA gene sequence libraries from environmental samples. Appl. Environ. Microbiol. 67:4374-4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smalla, K., G. Wieland, A. Buchner, A. Zock, J. Parzy, S. Kaiser, N. Roskot, H. Heuer, and G. Berg. 2001. Bulk and rhizosphere soil bacterial communities studied by denaturing gradient gel electrophoresis: plant-dependent enrichment and seasonal shifts revealed. Appl. Environ. Microbiol. 67:4742-4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smit, E., P. Leeflang, B. Glandorf, J. D. van Elsas, and K. Wernars. 1999. Analysis of fungal diversity in the wheat rhizosphere by sequencing of cloned PCR-amplified genes encoding 18S rRNA and temperature gradient gel electrophoresis. Appl. Environ. Microbiol. 65:2614-2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Swofford, D. L. 2002. PAUP*: phylogenetic analysis using parsimony (and other methods), 4.0 Beta. Sinauer Associates, Sunderland, Mass.
- 32.Theron, J., and T. E. Cloete. 2000. Molecular techniques for determining microbial diversity and community structure in natural environments. Crit. Rev. Microbiol. 26:37-57. [DOI] [PubMed] [Google Scholar]
- 33.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ward, D. M., R. Weller, and M. M. Bateson. 1990. 16S rRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature 345:63-65. [DOI] [PubMed] [Google Scholar]
- 35.Weller, D. M. 1988. Biological control of soilborne plant pathogens in the rhizosphere with bacteria. Annu. Rev. Phytopathol. 26:379-407. [Google Scholar]
- 36.Whipps, J. M. 2001. Microbial interactions and biocontrol in the rhizosphere. J. Exp. Bot. 52:487-511. [DOI] [PubMed] [Google Scholar]
- 37.White, T. J., T. Bruns, S. Lee, and J. Taylor. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315-322. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press, Inc., San Diego, Calif.
- 38.Yang, C. H., and D. E. Crowley. 2000. Rhizosphere microbial community structure in relation to root location and plant iron nutritional status. Appl. Environ. Microbiol. 66:345-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang, C. H., D. E. Crowley, and J. A. Menge. 2001. 16S rDNA fingerprinting of rhizosphere bacterial communities associated with healthy and Phytophthora infected avocado roots. FEMS Microbiol. Ecol. 35:129-136. [DOI] [PubMed] [Google Scholar]