Abstract
Partial sequences of ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) (EC 4.1.1.39) genes were retrieved from samples taken along a redox gradient in alkaline, hypersaline Mono Lake, Calif. The form I gene (cbbL) was found in all samples, whereas form II (cbbM) was not retrieved from any of the samples. None of the RuBisCO sequences we obtained were closely related (nucleotide similarity, <90%) to sequences in the database. Some could be attributed to organisms isolated from the lake (Cyanobium) or appearing in enrichment cultures. Most (52%) of the sequences fell into in one clade, containing sequences that were identical to sequences retrieved from an enrichment culture grown with nitrate and sulfide, and another clade contained sequences identical to those retrieved from an arsenate-reducing, sulfide-oxidizing enrichment.
Mono Lake is a closed-basin, alkaline, hypersaline lake located at the western edge of the Great Basin in eastern California. Saline lakes and similar alkaline, hypersaline environments are widespread today and have been important features of the hydrosphere since the earliest stages of the formation of the Earth's oceans (15). At ∼85-g liter−1 salinity and pH 9.8, Mono Lake is a midpoint on the gradient of saline lakes, and as such, its chemistry and the general features of its ecology are similar to those of numerous other saline lakes and similar environments in the western United States and elsewhere in the world (21).
Summer thermal stratification and the high rates of primary production characteristic of many temperate saline lakes result in seasonal bottom water anoxia. Saline lakes are also prone to longer periods of anoxia during meromixis (persistent chemical stratification). During the past two decades, two extended periods (>5 year) of meromixis were initiated at Mono Lake by unusually high runoff. Reduced species, especially sulfide, ammonia, and methane, accumulate to high concentrations beneath the chemocline (14, 17). Because of the strong redox gradients that are generated when the lake is stratified, chemolithoautotrophy may be an important trophic mode in Mono Lake, particularly during meromixis.
A common carbon fixation pathway for chemolithoautotrophic microorganisms is via the reductive pentose phosphate or Calvin-Benson-Bassham (CBB) cycle. One of the key enzymes in the CBB pathway, ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) (EC 4.1.1.39), catalyzes two competing reactions: the carboxylation or the oxygenolysis of ribulose-1,5-bisphosphate. RuBisCO exists in two forms. The form I enzyme is made up of eight large catalytic subunits and eight small subunits, with the cbbL (rbcL in plants) gene encoding the large subunit and the cbbS (rbcS in plants) gene encoding the small subunit (16). This structural form is typically found in all terrestrial plants, algae, cyanobacteria, and most photo- and chemoautotrophic bacteria (26).
Form II RuBisCO is composed of two large catalytic subunits that are encoded by the cbbM gene and arranged in a homodimeric structure. Although the active site residues are conserved in the two forms, the overall level of similarity between the forms I and II large subunits is only 25 to 30% (24). The cbbM gene has been found in several photosynthetic bacteria (27), aerobic and facultatively anaerobic chemoautotrophic bacteria (4, 11, 16, 22, 25), and dinoflagellates (18).
Due to its relatively large size, degree of conservation, widespread distribution, functional significance, and an increasing number of published sequences from chemoautotrophs, the RuBisCO gene is particularly useful for studying chemoautotrophic populations, in addition to and for comparison with other genes, such as the rRNA gene (28). We investigated the use of RuBisCO as an indicator of the diversity of carbon-fixing microorganisms in the water column of Mono Lake. In this study, we analyzed the distribution and phylogenetic richness of the cbbL and cbbM genes in four samples selected to represent the range of water column redox conditions in Mono Lake.
MATERIALS AND METHODS
Sample collection.
Water samples were collected on 20 July 2000 at Mono Lake Station 6 (37°57.822′N, 119°01.305′W) as described previously (13). Vertical profiles of temperature, pressure, conductivity, photosynthetically active radiation, beam attenuation, and fluorescence were obtained with a SeaBird SeaCat profiler equipped with a Licor 2π PAR sensor, a WetLabs 10-cm-path-length transmissometer, and a WetLabs WetStar fluorometer. Oxygen profiles were taken with a YSI polarographic oxygen sensor equipped with a Clark-type electrode. Water samples were collected at discrete depths using a Niskin water sampler. Samples were screened as necessary to remove brine shrimp, placed in dark plastic bottles, and held in coolers until they were processed (within 4 h of collection).
DNA collection and extraction.
Methods of DNA collection, extraction, and purification were as described previously (9, 13). Water samples (500 to 1,000 ml) were pressure filtered (∼50 kPa) through Millipore Sterivex filter cartridges (0.22-μm pore size) to collect microbial biomass for subsequent DNA extraction. Excess water was expelled, and then the cartridges were filled with extraction buffer, capped, frozen on dry ice, shipped to the laboratory, and stored at −70°C until processed. Blanks were prepared with each set of samples using Sterivex cartridges through which no water had been filtered.
PCR and cloning.
Samples from 2 m (mixolimnion), 17.5 m (base of the oxycline, also corresponding to a fluorescence maximum/transparency minimum), 23 m (top of the chemocline), and 35 m (monimolimnion) were chosen for analysis. These depths were chosen because they represent the range of water column redox conditions (see Fig. 1). Although only four samples taken from one station on one day were analyzed in detail, extensive analysis of the distribution of Mono Lake bacteria ribotypes using survey techniques (PCR and denaturing gradient gel electrophoresis) indicates that the composition of these samples is representative of other depths, stations and times (12; J. T. Hollibaugh, N. Bano, S. Humayoun, and G. LeCleir, unpublished data). These are also the same samples that were used in an analysis of the diversity of 16S rRNA genes of Mono Lake bacteria (13).
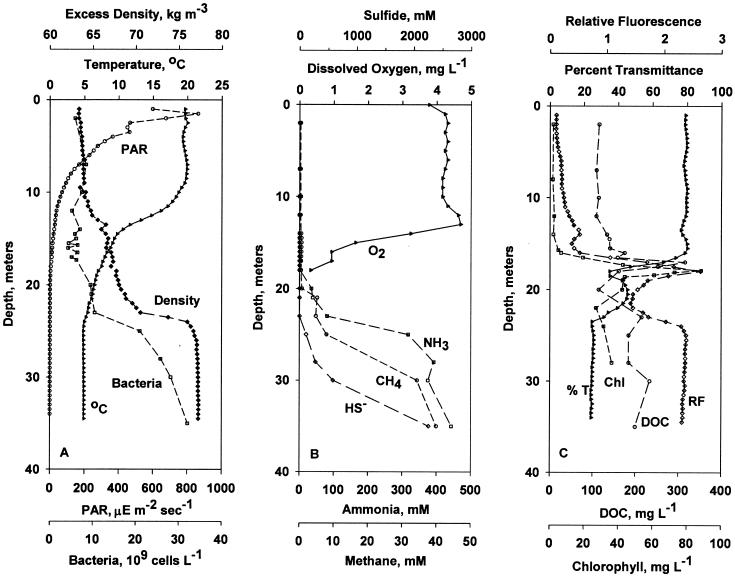
FIG. 1.
Vertical distributions of physical and biological variables at Station 6 in Mono Lake on 20 July 2000. (A) Temperature (°C), irradiance (PAR), excess density (Density), and abundance of bacteria (Bacteria); (B) dissolved oxygen (O2), ammonia (NH3), sulfide (S−2), and methane (CH4); (C) relative fluorescence (RF) (arbitrary units), transparency (% T), chlorophyll (Chl), and dissolved organic carbon (DOC). The chlorophyll profile was constructed from a low-resolution cast made on 17 July 2000 and a high-resolution cast to define the chlorophyll maximum taken on 20 July 2000. mgL −1, milligrams per liter; μE m−2 sec−1, micro-Einsteins per square meter per second.
RuBisCO form I cbbL genes were amplified with primers 595f (forward; 5′-GACTTCACCAAAGACGACGA-3′) and 1387r (reverse; 5′-TCGAACTTGATTTCTTTCCA-3′), corresponding to positions 595 to 615 and 1387 to 1405, respectively, of the cbbL gene from Anabaena strain 7120 (7). These primers are reported to amplify a wide range of green-like form 1A and 1B genes but not red-like form 1B or 1C or Archaea genes (7). Form II cbbM genes were amplified with primers cbbMf (5′-ATCATCAARCCSAARCTSGGCCTGCGTCCC-3′) and cbbMr (5′-MGAGGTGACSGCRCCGTGRCCRGCMCGRTG-3′), which were designed from alignments of gene sequences from the Riftia pachyptila endosymbiont and Rhodospirillum rubrum (7). Primers were synthesized either by Operon Technologies (Oakland, Calif.) or by the University of Georgia Molecular Genetics Instrumentation Facility. PCR mixtures were prepared in a total volume of 100 μl containing 1× PCR buffer, 2.5 mM MgCl2, 200 μM (each) deoxyribonucleotide triphosphate, 1 μM (each) primer, and 20 to 100 ng of template DNA. PCR was performed using the following reaction conditions: initial denaturation at 95°C for 5 min, pause at 82°C to add Taq DNA polymerase (2.5 U; Promega), and then 25 cycles consisting of denaturation (1 min at 94°C), annealing (1 min at 52°C), extension (1 min at 72°C), and a final extension at 72°C for 45 min. Reactions were run in triplicate, with replicates combined and purified using Wizard PCR preparation kits (Promega). The purified PCR amplicons were cloned as described previously (13).
Phylogenetic analysis.
All sequences were obtained from an automatic sequencer operated by Molecular Genetics Instrumentation Facility. A total of 57 clones (11 from 2 m, 17 from 17.5 m, 15 from 23 m, and 14 from 35 m) were selected randomly; DNA was extracted from them, purified, and sequenced using the plasmid primer SP6, T7, or both. Sequence reads were ∼650 bp. Most clones were sequenced in only one direction; however, a subset of clones representing major clades were sequenced from both ends to obtain complete sequences of the inserts. Nucleotide sequences were aligned and converted to inferred amino acid sequences by using programs in the Wisconsin package (version 10.0) Genetics Computer Group Inc. Sequences (nucleotide and amino acid) were compared to known sequences using BLAST (2). Phylogenetic trees of RuBisCO genes were constructed using the ARB program. The topology of the nucleotide tree was established using full-length sequences (772 bp after primer sequences were removed) and the neighbor-joining method. Bootstrap analysis (5,000 replicates) was performed, and then partial sequences (including some of the reference sequences) were inserted into the consensus tree. The inferred amino acid tree used the neighbor-joining method and evolutionary (Kimura) distances and was constructed with PHYLIP (8), using 228 positions. Bootstrap values for the amino acid tree are based on 100 replicates. The final trees were unrooted, with a Chlorella-like algal form I RuBisCO large subunit sequence used as the outgroup.
Nucleotide sequence accession numbers.
Sequences have been deposited in GenBank under accession numbers AY291478 to AY291533 and AY293400 as indicated on the figures.
RESULTS AND DISCUSSION
Limnological characteristics.
Vertical profiles of physical and biological variables in Mono Lake on the sampling day are shown in Fig. 1. Figure 1A shows that the water column was stably stratified with a prominent density discontinuity at 23 m. Water temperature declined from 20°C at 9 m to 8°C at 14 m and 5°C at 25 m. Irradiance decreased to 1% of the surface irradiance at a depth of 16.5 m. Oxygen concentration, shown in Fig. 1B, decreased below 12 m to the limit of detection at 17 m. Ammonia was first detected at a depth of 20 m; ammonia concentration increased rapidly to >400 μM at 27 m. Sulfide was first detected at 23 m; sulfide concentration increased rapidly (to >2,000 μM) in the monimolimnion. Methane was first detected at 20 m and increased to >40 μM in the monimolimnion. Chlorophyll concentration, dissolved organic carbon (DOC) concentration, and relative fluorescence exhibited maxima at a depth of 16 to 18 m, while transparency exhibited a minimum at this same depth (Fig. 1C). The unicellular green alga Picocystis salinarum was a major component of this “plate” (23). The abundance of bacteria increased from 5 × 109 cells liter−1 in the surface layer to 30 × 109 cells liter−1 in the monimolimnion (Fig. 1A). We also noted changes in the morphologies and sizes of cells over the vertical profile, with small, single cells dominant in the mixolimnion and large cells and many chains abundant in anoxic waters (data not shown). Bacterial abundance did not exhibit a maximum at the depth of the fluorescence maximum and transparency minimum (16 to 18 m), consistent with previous reports that a plate of photosynthetic bacteria does not form at the base of the oxycline in Mono Lake, unlike many other saline lakes (5, 6).
Phylogenetic affiliations of RuBisCO genes.
Amplicons of the form I RuBisCO gene (cbbL) were obtained from all samples. The diversity of cbbL genes we retrieved from our Mono Lake samples was lower than we expected. Coverage estimates (10) were 82, 83, 93, and 93% for libraries from 2, 17.5, 23, and 35 m, respectively. Fifty-two of the fifty-seven sequences we obtained are classified as form IA RuBisCO, and five form 1B sequences were from cyanobacteria. Nucleotide (Fig. 2) and inferred amino acid (Fig. 3) tree topologies were generally congruent, with one exception (discussed below). The nucleotide sequences fell into nine distinct clades, with the largest clade containing 42% of the sequences. One clade of four closely related sequences merged with the largest clade when inferred amino acid sequences were used for phylogenetic analysis, leaving only eight distinct clades, with the largest containing 52% of the sequences. Most of the sequences we obtained are not closely affiliated (nucleotide similarities, <90%) with RuBisCO sequences in the databases; however, some of them were closely related to sequences obtained from Mono Lake isolates or enrichments.
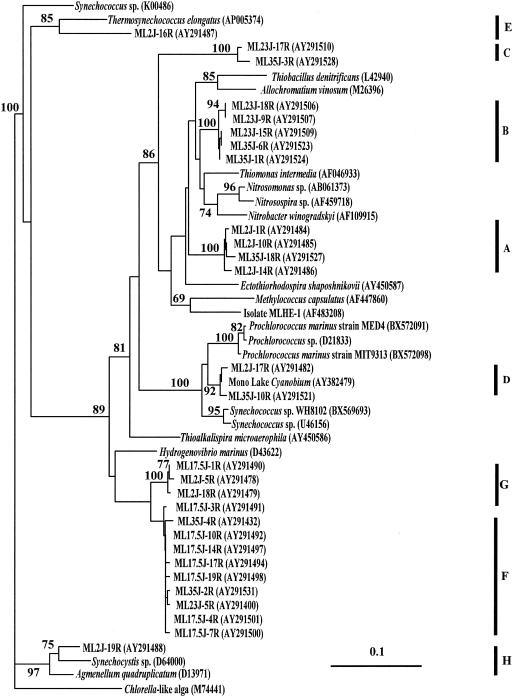
FIG. 2.
Phylogenetic relationships of cbbL genes retrieved from Mono Lake. The tree was constructed with the ARB program using full-length (772-bp) sequences, bootstrap analysis was performed (values of >50% are shown), and then shorter sequences were inserted. Sampling depth is indicated as the first set of numbers in the clone identifier; for example, for clone ML2J-16R, ML = Mono Lake; 2 = sampling depth in meters; J = month sampled (July); 16R = clone number from that library. GenBank accession numbers and lengths of partial sequences are given in parentheses following the clone identifier.
FIG. 3.
Phylogenetic relationships of cbbL genes retrieved from Mono Lake based on an analysis of amino acid sequences (234 positions) inferred from the longest nucleotide sequences used in Fig. 2. This is a neighbor-joining tree based on Kimura distances. Bootstrap values of >70% are shown. GenBank accession numbers are given in parentheses following the clone identifier.
Clades A, B, C, and D grouped together on a major branch of the Mono Lake RuBisCO tree that included reference sequences from a Mono Lake Cyanobium, Thiobacillus denitrificans, Allochromatium vinosum (Chromatium vinosum), Methylococcus capsulatus, Nitrobacter winogradskyi, Thiomonas intermedia (Thiobacillus intermedius), and an arsenite-oxidizing member of the Ectothiorhodospiraceae isolated from Mono Lake, strain MLHE-1 (20). The assignment of sequences to various clades in this branch was strongly supported by bootstrap analysis; however, the overall branch topology was not.
Clade A contains three sequences from the oxic mixolimnion and one from the monimolimnion that were most similar to RuBisCO sequences from T. denitrificans, A. vinosum, and M. capsulatus.
Clade B is dominated by sequences retrieved from chemocline and monimolimnion samples (Fig. 2). Sequences in Clade B were most similar to RuBisCO sequences from Nitrosomonas sp., Nitrosospira sp., N. winogradskyi, and T. intermedia (T. intermedius).
Clade C sequences were found in chemocline and monimolimnion samples and were most similar to those of Mono Lake isolate strain MLHE-1 (20) and to clade B (nucleotide sequence, Fig. 2). We recently retrieved RuBisCO sequences with >99% similarity (431 of 435 bp) to those of clade C from an arsenate-reducing, sulfide-oxidizing enrichment culture derived from a Mono Lake sample.
Clade D contains form 1B sequences that are >99% similar to the RuBisCO sequence from a Cyanobium-like unicellular cyanobacterium isolated from Mono Lake (C. Budinoff and J. T. Hollibaugh, unpublished data). The Cyanobium is abundant in subchemocline waters (∼108 cells liter−1; Hollibaugh, unpublished data), and although it grows photoautotrophically in the laboratory (C. Budinoff, unpublished data), it has not yet been observed in surface water samples from Mono Lake. It is unclear whether the monimolimnion population is actively growing, represents a relict population from the last time the lake completely mixed, or is maintained by a sinking flux (for example, brine shrimp fecal pellets).
Branches E and H each contain one sequence retrieved from the mixolimnion sample. These sequences were most closely related to cyanobacterial genes (nucleotide similarities of 77 and 82%, respectively).
Clades F and G grouped together on the second major branch of the Mono Lake RuBisCO tree along with the reference sequence from Hydrogenovibrio marinus (nucleotide similarities of 89 and 87%, respectively). Clades F1 and F2 account for more than half of the sequences we retrieved. Their nucleotide sequences are >95% similar; however, their amino acid sequences are >99% similar, because differences in the nucleotide sequences correspond to variability in the third base pair of codons for the same amino acids. No Clade F sequences were retrieved from the surface water sample; most were retrieved from the micro-oxic or anoxic region between the oxycline and the chemocline (17.5- and 23-m samples), where gradients of reduced substrates were strongest (Fig. 1). Sequences that were identical to those in clade F1 were retrieved from dilution cultures of Mono Lake water amended with nitrate and sulfide (Hollibaugh et al., unpublished). Clade G contains sequences from the mixolimnion and the oxycline.
No PCR products were obtained from any samples when primers specific for the form II RuBisCO gene, cbbM, were used for amplification, despite successful amplification of the gene from the positive control (R. rubrum, ATCC 9791). We also failed to obtain RuBisCO sequences when samples were amplified with RuBisCO primers used in a study of chemoautotrophs in groundwater (1).
Given the variety and strength of geochemical gradients in the Mono Lake water column, we expected a greater diversity of chemoautotrophs and thus of form 1a and form 2 RuBisCO genes in our samples. The low diversity of RuBisCO sequences may reflect a low diversity of organisms; low diversity of RuBisCO genes due to physicochemical conditions in Mono Lake; inefficient extraction of DNA from some groups of organisms, for example, methanotrophs; or restricted specificity of the primer set.
Elsaied and Naganuma (7) report identifying a total of 29 different cbbL sequences (operational taxonomic units [OTUs])from 300 cloned genes. Their samples covered a range of habitats associated with a hydrothermal vent site, including sediment, water, and symbionts. The richness of cbbL genes they detected (0.10 OTU clone−1) is comparable to the richness we observed (0.12 OTU clone−1), despite the differences in habitat diversity. However, they detected cbbM genes in their samples (24 OTUs in 250 cbbM clones) where we detected no cbbM genes in Mono Lake samples. Alfreider et al. (1) recovered 24 cbbL OTUs in 100 clones (0.24 OTU clone−1) and 6 cbbM OTUs in 40 clones (0.15 OTU clone−1) in groundwater samples. Nanba et al. (19) retrieved at least 7 different OTUs from 10 clones of a microbial mat sample and at least 23 different OTUs from 110 clones retrieved from volcanic soil samples. These comparisons are not entirely satisfactory, because each study used a different primer set. Furthermore, the heterogeneity of these environments is greater than that of the Mono Lake water column, but there are as yet no other comparable studies from other aquatic environments.
RuBisCO gene diversity might be low because Mono Lake presents a very stable environment with regard to RuBisCO substrates. Because of the elevated pH (9.8) and high dissolved inorganic carbon concentrations of Mono Lake (360 mM; W.-J. Cai and Y. Wang, unpublished data), Mono Lake partial CO2 pressure and alkalinity are elevated (512 and 593 μatm and 661 and 634 mM, respectively, measured in two samples by the procedure described in reference 3 [Cai and Wang, unpublished data]) relative to those of offshore surface seawater in equilibrium with the atmosphere (∼365 μatm and 2.2 mM). Oxygen concentrations are low and constant (∼0) anywhere in the lake below the oxycline. Thus, the Mono Lake environment may be less variable with regard to the conditions under which RuBisCO functions than other aquatic environments, which may be reflected in lower diversity of RuBisCO genes. Alternatively, chemoautotrophic populations may be diverse, reflecting the diversity of physiochemical conditions in the lake's highly stratified water column, but Mono Lake chemoautotrophs may rely on other pathways for carbon fixation or the RuBisCO primer set we used may not adequately sample this diversity.
Another possible reason for the apparent low richness of RuBisCO genes in Mono Lake samples may be that the primers we used recognize a subset of cbbL sequences that are not well represented in Mono Lake chemoautotrophs. The composition of the Mono Lake chemoautotroph population may be significantly different from those of populations of chemoautotrophs in the other environments that have been studied. This speculation is supported by the observation that none of the sequences we obtained was closely related (nucleotide similarities of <90%) to environmental sequences obtained by Alfreider et al. (1) or Elsaied and Naganuma (7).
Acknowledgments
This work was supported by National Science Foundation grant MCB 99-77886 to J.T.H. and S. B. Joye.
We are grateful to G. LeCleir for help with sample collection and for providing cell count and DOC data and to C. R. Budinoff for his help with data analysis. S. B. Joye and S. Carini supplied sulfide, ammonia, and methane data. Field laboratory facilities, limnological data processing, and other survey and logistical support were provided by R. Jellison, S. Roll, and the Sierra Nevada Aquatic Research Laboratory with support from the National Science Foundation (MCB-9977901). We are also grateful to two anonymous reviewers for comments that greatly helped improve the manuscript.
REFERENCES
- 1.Alfreider, A., C. Vogt, D. Hoffmann, and W. Babel. 2003. Diversity of ribulose-1,5-bisphosphate carboxylase/oxygenase large-subunit genes from groundwater and aquifer microorganisms. Microb. Ecol. 45:317-328. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Cai, W.-J., W. J. Wiebe, Y. Wang, and J. E. Sheldon. 2000. Intertidal marsh as a source of dissolved inorganic carbon and a sink of nitrate in the Satilla River-estuarine complex in the southeastern U.S. Limnol. Oceanogr. 45:1743-1752. [Google Scholar]
- 4.Chung, S. Y., T. Yaguchi, H. Nishihara, Y. Igarashi, and T. Kodama. 1993. Purification of the L2 RuBisCO from a marine obligately autotrophic hydrogen-oxidizing bacterium Hydrogenovibrio marinus strain MH-110. FEMS Microb. Lett. 109:49-54. [DOI] [PubMed] [Google Scholar]
- 5.Cloern, J. E., B. E. Cole, and R. S. Oremland. 1983. Autotrophic processes in meromictic Big Soda Lake, Nevada. Limnol. Oceanogr. 28:1049-1061. [Google Scholar]
- 6.Cloern, J. E., B. E. Cole, and R. S. Oremland. 1983. Seasonal changes in the chemistry and biology of a meromictic lake (Big Soda Lake, Nevada, USA). Hydrobiology 105:195-206. [Google Scholar]
- 7.Elsaied, H., and T. Naganuma. 2001. Phylogenetic diversity of ribulose-1,5-bisphosphate carboxylase/oxygenase large-subunit genes from deep-sea microorganisms. Appl. Environ. Microbiol. 67:1751-1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Felsenstein, J. 1993. PHYLIP (Phylogeny Inference Package), v. 3.5 ed. University of Washington, Seattle, Wash.
- 9.Ferrari, V. C., and J. T. Hollibaugh. 1999. Distribution of microbial assemblages in the central Arctic Ocean basin studied by PCR/DGGE: analysis of a large data set. Hydrobiology 401:55-68. [Google Scholar]
- 10.Good, I. J. 1953. The population frequencies of species and the estimation of the population parameters. Biometrika 40:237-264. [Google Scholar]
- 11.Hernandez, J. M., S. H. Baker, S. C. Lorbach, J. M. Shively, and F. R. Tabita. 1996. Deduced amino acid sequence, functional expression, and unique enzymatic properties of the form I and form II ribulose bisphosphate carboxylase/oxygenase from the chemoautotrophic bacterium Thiobacillus denitrificans. J. Bacteriol. 178:347-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hollibaugh, J. T., P. S. Wong, N. Bano, S. K. Pak, E. M. Prager, and C. Orrego. 2001. Stratification of microbial assemblages in Mono Lake, California, and response to a mixing event. Hydrobiology 446:45-60. [Google Scholar]
- 13.Humayoun, S. B., N. Bano, and J. T. Hollibaugh. 2003. Depth distribution of microbial diversity in Mono Lake, a meromictic soda lake in California. Appl. Environ. Microbiol. 69:1030-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jellison, R., and J. M. Melack. 1993. Meromixis in hypersaline Mono Lake, California. 1. Stratification and vertical mixing during the onset, persistence, and breakdown of meromixis. Limnol. Oceanogr. 38:1008-1019. [Google Scholar]
- 15.Kempe, S., and E. T. Degens. 1985. An early soda ocean? Chem. Geol. 53:95-108. [Google Scholar]
- 16.Kobayashi, H., A. M. Viale, T. Takabe, T. Akazawa, K. Wada, J. Shinozaki, K. Kobayashi, and M. Sugiura. 1991. Sequence and expression of genes encoding the large and small subunits of ribulose-1,5-bisphosphate carboxylase/oxygenase from Chromatium vinosum. Gene 97:55-62. [DOI] [PubMed] [Google Scholar]
- 17.Miller, L. G., R. Jellison, R. S. Oremland, and C. W. Culbertson. 1993. Meromixis in hypersaline Mono Lake, California. 3. Biogeochemical response to stratification and overturn. Limnol. Oceanogr. 38:1040-1051. [Google Scholar]
- 18.Morse, D., P. Salois, P. Markovic, and J. W. Hastings. 1995. A nuclear-encoded form II RuBisCO in dinoflagellates. Science 268:1622-1624. [DOI] [PubMed] [Google Scholar]
- 19.Nanba, K., G. King, and K. Dunfield. 2004. Analysis of facultative lithotroph distribution and diversity on volcanic deposits using the large sub-unit of ribulose 1,5-biphosphate carboxylase/oxygenase. Appl. Environ. Microbiol., in press. [DOI] [PMC free article] [PubMed]
- 20.Oremland, R. S., S. E. Hoeft, N. Bano, R. A. Hollibaugh, and J. T. Hollibaugh. 2002. Anaerobic oxidation of arsenite in Mono Lake water and by a facultative chemoautotroph, strain MLHE-1. Appl. Environ. Microbiol. 68:4795-4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oren, A. 2002. Halophilic Microorganisms and their environments, vol. 5. Kluwer Academic Publishers, Dordrecht, The Netherlands..
- 22.Robinson, J. J., J. L. Stein, and C. M. Cavanaugh. 1998. Cloning and sequencing of a form II ribulose-1,5-bisphosphate carboxylase/oxygenase from the bacterial symbiont of the hydrothermal vent tubeworm Riftia pachyptila. J. Bacteriol. 180:1596-1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roesler, C. S., C. W. Culbertson, S. M. Etheridge, R. Goericke, R. P. Kiene, L. G. Miller, and R. S. Oremland. 2002. Distribution, production, and ecophysiology of Picocystis strain ML in Mono Lake, California. Limnol. Oceanogr. 47:440-452. [Google Scholar]
- 24.Shively, J. M., G. van Keulen, and W. G. Meijer. 1998. Something from almost nothing: carbon dioxide fixation in chemoautotrophs. Annu. Rev. Microbiol. 52:191-230. [DOI] [PubMed] [Google Scholar]
- 25.Stoner, M. T., and J. M. Shively. 1993. Cloning and expression of the D-ribulose-1,5-bisphosphate carboxylase/oxygenase form II gene from Thiobacillus intermedius in Escherichia coli. FEMS Microb. Lett. 107:287-292. [DOI] [PubMed] [Google Scholar]
- 26.Tabita, F. R. 1999. Microbial ribulose-1,5-bisphosphate carboxylase/oxygenase: a different perspective. Photosyn. Res. 60:1-28. [Google Scholar]
- 27.Tabita, F. R. 1988. Molecular and cellular regulation of autotrophic carbon dioxide fixation in microorganisms. Microbiol. Rev. 52:155-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watson, G. M. F., and F. R. Tabita. 1997. Microbial ribulose-1,5-bisphosphate carboxylase/oxygenase: a molecule for phylogenetic and enzymological investigation. FEMS Microb. Lett. 146:13-22. [DOI] [PubMed] [Google Scholar]