Abstract
Escherichia coli O157:H7 is an endemic pathogen causing a variety of human diseases including mild diarrhea, hemorrhagic colitis, hemolytic-uremic syndrome, and thrombotic thrombocytopenic purpura. This study concerns the exploitation of bacteriophages as biocontrol agents to eliminate the pathogen E. coli O157:H7. Two distinct lytic phages (e11/2 and e4/1c) isolated against a human strain of E. coli O157:H7, a previously isolated lytic phage (pp01), and a cocktail of all three phages were evaluated for their ability to lyse the bacterium in vivo and in vitro. Phage e11/2, pp01, and the cocktail of all three virulent phages resulted in a 5-log-unit reduction of pathogen numbers in 1 h at 37°C. However, bacteriophage-insensitive mutants (BIMs) emerged following the challenge. All tested BIMs had a growth rate which approximated that of the parental O157 strain, although many of these BIMs had a smaller, more coccoid cellular morphology. The frequency of BIM formation (10−6 CFU) was similar for e11/2, pp01, and the phage cocktail, while BIMs insensitive to e4/1c occurred at the higher frequency (10−4 CFU). In addition, BIMs commonly reverted to phage sensitivity within 50 generations. In an initial meat trial experiment, the phage cocktail completely eliminated E. coli O157:H7 from the beef meat surface in seven of nine cases. Given that the frequency of BIM formation is low (10−6 CFU) for two of the phages, allied to the propensity of these mutants to revert to phage sensitivity, we expect that BIM formation should not hinder the use of these phages as biocontrol agents, particularly since low levels of the pathogen are typically encountered in the environment.
Since Escherichia coli O157:H7 was identified in 1983 (30), it has emerged as an important human pathogen. The infective dose can be as low as 10 cells, and symptoms of infection include diarrhea, hemorrhageic colitis, hemolytic-uremic syndrome and thrombotic thrombocytopenic purpura (39). Cattle are known to be asymptomatic carriers of E. coli O157:H7 (6) and indeed are major reservoirs for this bacterium (11, 12, 44). Outbreaks have been attributed to food, water, and person-to-person and direct fecal contact (9). A recent study in the United Kingdom found the prevalence rate of E. coli O157 in cattle feces presented for slaughter to be 7.5% (28). E. coli is a robust bacterium, and studies have shown that E. coli O157:H7 survives in bovine feces for up to 56 days at 22°C and up to 70 days at 5°C (41) and has a high resistance to acid (14, 18). One important route of food contamination by O157 is the transmission of the bacterium from bovine feces onto meat during slaughter (10).
The exploitation of bacteriophages as a realistic approach to the control of pathogens has attracted considerable interest in recent years (24, 37) because of the emergence of antibiotic-resistant bacteria. Phage therapy has been used successfully since the early 1920s, but this has been largely confined to regions within the former Soviet Union (5). Many of the research results reported in the past have attributed variable efficiencies to the phage therapy applied. This is due to a number of factors including poor understanding of bacterial pathogenesis, lack of knowledge of phage-host interactions, and the possible use of impure phage preparations in some studies (1).
Previous studies demonstrating phage-mediated biocontrol of pathogenic E. coli in animals have generated very promising results. For example, calves and piglets with diarrhea due to experimentally administered pathogenic E. coli were cured within 8 h following phage administration (33). Further studies found that phage could act very successfully as a prophylactic. Experimentally induced diarrhea could be prevented by spraying the litter in the calf rooms with aqueous phage suspensions or by keeping calves in uncleaned rooms previously occupied by calves whose E. coli infection had been treated by phage administration (35). Recent results of phage therapy against other bacterial pathogens have shown considerable potential. For example, it has been shown that intraperitoneal injections of bacteria (Staphlococcus aureus or vancomycin-resistant Enterococcus faeicum) eventually cause death in mice whereas the administration of an intraperitoneal injection of phage following the initial injection significantly reduces the lethality of the bacteria (4, 23).
Phages specific for E. coli O157 have previously been isolated from natural environments and characterized (4, 19, 26, 31). These could have considerable potential for biocontrol of the pathogen either as animal feed inoculants or as decontaminants. In one study (19), phage infection and plaque formation on O157 were influenced by the nature of the host cell O157 lipopolysaccharide (LPS). Strains that did not express the O157 antigen or that expressed a truncated LPS were not susceptible to plaque formation or lysis by phage. In addition, strains that expressed abundant intermediate-molecular-weight LPS did not support plaque formation but were lysed in liquid culture. This study suggested that O157 antigen-specific phages could be applied for biocontrol of E. coli O157:H7 in animals and fresh foods without compromising the viability of other members of the normal flora or food quality. In another study, O157 phage-host interactions were monitored both in wastewater treatment plants (38) and in a chemostat continuous culture (25). In the latter, where E. coli O157:H7 and phage pp01 were combined, a rapid decrease in viable-cell count was observed, followed by the emergence of phage-resistant bacteria. These resistant bacteria were found to have altered OmpC expression, suggesting the involvement of this protein in phage attachment. A previous study also found OmpC to be the receptor used by phage during E. coli O157:H7 attachment (42). However, phage mutants with an altered host range, which could overcome the phage-resistant bacteria, were also identified (25). Thus, coevolution of phage and E. coli O157:H7 proceeded mutually in a chemostat continuous culture (25).
In this study, lytic phages against a human isolate of E. coli O157:H7 were isolated and evaluated for their ability to lyse the bacterium in vivo and in vitro. During in vivo work, the anti-O157 phage cocktail was assessed for its ability to eliminate small numbers of E. coli O157:H7 on meat surfaces and was found to control the pathogen in seven of nine cases. However, during the in vitro challenge, bacteriophage-insensitive mutants (BIMs) emerged at a similar frequency for phage e11/2, pp01, and the phage cocktail whereas a higher frequency was obtained for phage e4/1c. The BIMs tested generally had a growth rate which approximated that of the parent O157, and all eventually reverted to the phage- sensitive phenotype.
MATERIALS AND METHODS
Bacterial strains.
Nontoxigenic E. coli O157:H7 strain P1432, as described previously (22), was used as a representative O157:H7 strain for phage isolation, propagation, and evaluation. This strain does not produce Shiga toxin 1 or 2 because it lacks the genes encoding these factors. A rifampin-resistant derivative of P1432 was also employed. All strains (Table 1) were grown at 30 or 37°C in brain heart infusion (BHI) broth (Merck). Solid media contained 1% (wt/vol) bacteriological agar (Oxoid, UK). The strains were stocked in BHI broth containing 40% glycerol and stored at −80°C. Cultures were stored at 4°C and transferred periodically.
TABLE 1.
Bacteriophage host range
| Bacterial isolate | Plaque formationa
|
Originb | ||
|---|---|---|---|---|
| φ el1/2 | φ pp01 | φ e4/1c | ||
| Nontoxigenic E. coli O157:H7 | ||||
| P1432 | +++ | +++ | +++ | DPRC |
| AR12900 | +++ | ++ | +++ | DPRC |
| Toxigenic E. coli O157:H7 | ||||
| ATCC 43895 | + | + | + | NFC |
| Ent C9490 | + | + | + | NFC |
| 380-94 | + | + | + | NFC |
| BFB2OY | + | ++ | + | NFC |
| BP1S2SO | ++ | ++ | +++ | NFC |
| BBS3LK | + | + | + | NFC |
| MS3MO | + | + | +++ | NFC |
| BPS2D | + | ++ | + | NFC |
| BPS3DL | + | + | + | NFC |
| MP2S3CN | + | + | + | NFC |
| MP1S3KK | − | − | − | NFC |
| MP2S2DL | − | − | − | NFC |
| Collection of gram-negative bacteria | ||||
| E. coli Qβ | − | − | − | DPRC |
| E. coli NC1061 | − | − | − | DPRC |
| E. coli wt555 | − | − | − | DPRC |
| E. coli JM109 (K-12) | − | − | − | DPRC |
| Salmonella enterica serovar Typhimurium DT104-25 | − | − | − | DPRC |
| Salmonella enterica serovar Typhimurium LT2 | − | − | − | DPRC |
| Salmonella enterica serovar Typhimurium BAA-185 | − | − | − | DPRC |
| Salmonella enterica serovar Derby | − | − | − | DPRC |
| Pseudomonas fluorescens | − | − | − | DPRC |
+++, EOP 1 to 0.5; ++, EOP 0.5 to 0.2; +, EOP 0.2 to 0.001; −, bacterial strain was not susceptible to phage attack.
DPRC, Dairy Products Research Centre, Moorepark, Fermoy, Co. Cork, Ireland; NFC, National Food Centre, Castleknock, Co. Dublin, Ireland.
Bacteriophage enrichment and isolation.
Fecal material was screened as a source of E. coli O157:H7-specific phage. Human fecal samples were obtained from patients hospitalized due to gastrointestinal illness, and bovine fecal samples were obtained from dairy farms, a cattle mart, and a slaughterhouse over a 4-month period. Portions (5 ml or 5 g) were added to 20-ml volumes of early-log-phase E. coli O157:H7 strain P1342 (106 CFU/ml grown at 37°C) containing 10 mM CaCl2. These cultures were incubated overnight at 37°C with shaking (200 rpm). Samples (1.5 ml) were then withdrawn and centrifuged at 20,800 × g in a microcentrifuge for 5 min. The supernatant was removed, filter sterilized (pore size, 0.45 μm), and subjected to a plaque assay (see below) using E. coli O157:H7 strain P1432 as the host. Plaques appearing on lawns of the host bacterium were propagated as described by Sambrook and Russell (32).
Bacteriophage propagation and bacteriophage assays.
All three phages were routinely propagated on strain P1432. A 1% inoculum of the propagating strain (P1432) was grown in BHI broth (1 liter) containing 2% 1 M CaCl2 and incubated at 37°C for 2 h with shaking (100 rpm). The sample was split into two, phage was added to one sample, and both were incubated for a further 3 h at 37°C. The samples were pooled and incubated for a further 2 h at 37°C with shaking. Then 1 M NaCl was added, and the sample was stirred for 1 h at 4 °C. The sample was centrifuged at 6,000 × g for 1 h, and the supernatant (BHI lysate) was filter sterilized (pore size, 0.45 μm). Concentrated phage preparations were obtained by polyethylene glycol 8000 precipitation of BHI lysates followed by CsCl density gradient centrifugation. Methods were used as described by Sambrook and Russell (32). Plaque assays were performed as described previously (29), except that BHI agar and BHI overlays (0.7% agar) were employed for fresh overnight cultures. Plates were incubated overnight at 37°C.
Electron microscopy.
Pure, high-titer phage stocks were prepared from CsCl gradients to achieve titers in excess of 109 PFU/ml. Each sample was stained negatively with 1% uranyl acetate, and electron micrographs were taken at various magnifications. Based on the morphology of the phage, the three coliphages were classified into their respective family on the basis of the guidelines of the International Committee on Taxonomy of Viruses (27).
Bacteriophage DNA isolation and restriction.
Contaminating bacterial DNA was removed from the concentrated phage solution (1 ml) by adding 5 μl of 1,000-U DNase I (Roche Diagnostics) and incubating at 37°C for 30 min. Nucleic acids were extracted by adjusting the mixture with 1% sodium dodecyl sulfate and 6 μl of 0.5 M EDTA and subsequently digested with 6 μl of proteinase K (10 mg/ml) (Sigma-Aldrich) for 1 h at 37°C. Proteinaceous material was removed by using phenol-chloroform-isoamyl alcohol (25:24:1) (Sigma-Aldrich). The phage DNA was extracted by treatment with Tris- saturated phenol and chloroform. The DNA solution was then ethanol precipitated. Phage DNA was digested with EcoRV (New England Biolabs) and electrophoresed on 0.7% horizontal agarose (32).
Bacteriophage host range.
All three phages were assessed for their ability to form plaques on a range of gram-negative bacteria (Table 1). If a phage plaqued on a strain, its efficiency of plaquing (EOP) was determined using E. coli O157:H7 strain P1432 as the reference strain. The EOP was determined by expressing the phage titer of the susceptible strain relative to the phage titer of the reference strain.
Bacterial challenge tests.
Challenge tests were performed with bacteria in BHI broth in the presence of 10 mM CaCl2. The medium (500 ml) was inoculated (1%) with a fresh overnight bacterial culture and then incubated at 12, 30, or 37°C with shaking. Samples were taken every hour and plated in triplicate. After 4 h of incubation, the 500-ml culture was split into five 100-ml volumes and phage was added, maintaining a multiplicity of infection (MOI) of greater than 1 (generally 100). The cultures were reincubated at the appropriate temperature. Samples were taken every hour, and BIMs were enumerated by plate counting in triplicate. Standard deviations were determined.
Contaminated-meat trials.
Steak meat was cut into equal portions, and these portions were placed on a petri dish equidistant from each other. The petri dish containing the meat was incubated at 37°C for 3 h. E. coli O157:H7 (rifampin-resistant strain P1432) was grown to an optical density of 0.3 in BHI broth at 37°C with shaking. The cells were harvested and washed three times in sterile distilled H2O. This culture was then diluted to give a titer of approximately 2 × 103 CFU/ml. A 100-μl portion was spotted onto the surface of each piece of meat. The meat containing the bacteria was incubated at 37°C for a further 1 h. A 1-ml volume of a phage cocktail (e11/2, e4/1c, and pp01) at a titer of 2 × 108 PFU/ml (multiplicity of infection = 1 million) was spotted onto the meat surfaces in the petri dish. The control (meat and E. coli O157:H7) and test meat samples were incubated for a further 1 h. Following this, each piece of meat was individually enriched for E. coli by being placed in 40 ml of BHI broth and incubated at 37°C for 2 h. Following enrichment, the BHI samples were assayed for E. coli by performing a plate count on violet red bile agar containing rifampin. The plates were incubated overnight at 37°C.
BIM observation, selection, growth, and phage sensitivity.
BIMs were isolated as distinct colonies on agar plates following bacterial challenge tests. BIMs and the parental E. coli O157:H7 strain P1432 cells were observed using 63× objective oil immersion interfence contrast (Zeiss Axioplan Transmitted Light Microscope). Colonies with different morphologies were inoculated into tubes containing 5 ml of BHI broth and incubated at 37°C overnight. Ninety-six-well microtiter plates were incubated statically and used to determine the growth rate for P1432 (parental strain) and the BIMs. The total volume in each well was 200 μl (BHI broth). Each culture was grown in triplicate from a 2% inoculum of overnight cultures. The plates also contained a number of blank wells (medium only) and a number of control wells (parental strain P1432). The optical density at 600 nm was recorded every hour for 9 h (Anthos 2001; O.D. 600 nm Anthos Labtec Instruments). The plates were incubated at 37°C. Triplicate readings were averaged, and values of blanks (medium only) were subtracted from these readings. Standard deviations were determined. A selection of BIMs were propagated through 50 generations at 37°C. Each propagated BIM was then checked for phage susceptibility by a plaque assay.
Determination of the frequency of emergence of BIMs.
Plaque assays were performed using an overnight culture of strain P1432 containing known cell numbers and phages of known titer. The MOI was always greater than 1 (generally 10). The plates were incubated overnight at 37°C. All resulting colonies were counted, and the BIM frequency (number of surviving colonies/original bacterial titer) was determined. All experiments were performed in triplicate, and standard deviations were determined.
RESULTS
Characterization of phages e11/2, pp01, and e4/1c.
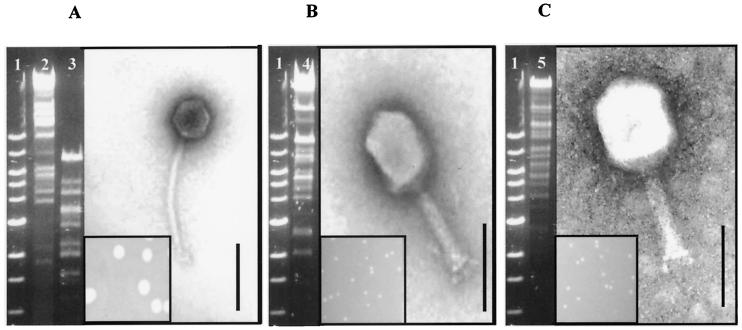
This study initially involved the isolation of two distinct phages (e11/2 and e4/1c) from bovine farmyard slurry samples. In addition, a third phage, pp01, was obtained from the Tokyo Institute of Technology. All three phages plaqued on E. coli O157:H7 strain P1432. Phages e11/2 and pp01 formed pinpoint plaques (0.5 mm) on P1432, whereas e4/1c formed medium-sized plaques (3 mm) (Fig. 1). High-titer phage stocks prepared for each phage contained phage particles at concentrations between 109 and 1012 PFU/ml.
FIG. 1.
Electron micrographs of phages negatively stained with 1% uranyl acetate; restriction digest of phage DNA restricted with EcoRV and phage plaque morphology. (A) Phage e4/1c and associated plaque. (B) Phage e11/2 and associated plaque. (C) Phage pp01 and associated plaque. Bar, 100 nm. Lanes: 1, 1 kb; 2, T4 DNA; 3, e4/1c DNA; 4, e11/2 DNA; 5, pp01 DNA. The DNA was run on a 0.7% agarose gel.
Morphological analysis of phages e11/2, pp01, and e4/1c by electron microscopy (Fig. 1) allowed each to be classified into its respective viral family and order. Phages e11/2 and pp01 had a contractile, long, rigid, and relatively thick tail. The tail length of phage e11/2 is 117 ± 4 nm, whereas that of pp01 is 112 ± 2 nm. It is clear from the electron micrographs that both of these phages contain a baseplate with spikes. Both phage also have the characteristic elongated head. The head diameter of phage e11/2 is 112 ± 2 nm, and that of pp01 is 118 ± 4 nm. Phages e11/2 and pp01 thus have the A2 morphotype and can be classified into the Myoviridae family. Phage e4/1c has a relatively thin, long, noncontractile, and flexible tail with an isometric head. The tail length of phage e4/1c was determined to be 201 ± 1 nm, and its head diameter was 64 ± 1 nm. This phage can thus be classified into the Siphoviridae family. All three lytic phages (e11/2, pp01 and e4/1c) belong to the order Caudovirales.
The bacteriophage DNA was digested with EcoRV, and the fragments were analyzed by agarose gel electrophoresis (0.7% agarose) (Fig. 1). Although the phage DNA may not be totally digested in each case, it is clear that both e11/2 and pp01 showed a greater resemblance to T4 than did e4/1c. This result was not unexpected since all three phages (T4, e11/2, and pp01) belong to the Myoviridae family. Most endonucleases do not normally digest T4 DNA because the DNA contains glucosylated hydroxymethyl cytosine instead of cytosine (20). We think that this is true for phage e11/2 and pp01 since since most endonucleases used in this study could not restrict the DNA.
Bacteriophage host range.
All three phages were assessed for their ability to plaque on a range of gram-negative bacteria including 12 distinct toxigenic E. coli O157:H7 and 2 nontoxigenic E. coli O157:H7 strains. All toxigenic E. coli O157:H7 strains were shown by multiplex PCR to contain virulence factors such as vt1, vt2, eaeA, hlyA, P0157, and FliCH7 (G. Duffy, personal communication). Three of the toxigenic E. coli O157:H7 strains were involved in major outbreaks in the United States (ATCC 43895, Ent C9490, and 380- 94) and were obtained from culture collections. The other nine toxigenic strains were isolated from various minced-meat samples obtained from retailers and butchers in Ireland. All three phages displayed similar lytic spectra. Only two toxigenic E. coli O157:H7 strains (MP1S3KK and MP2S2DL) showed resistance to all three phages. The phages were capable of lysing 12 of the 14 E. coli O157:H7 strains shown in Table 1. In addition to the lytic activities, the EOP was determined for all the phages against all the strains. These values were determined using E. coli O157:H7 strain P1432 as the reference strain, and all results are shown in Table 1.
Bacterial challenge tests.
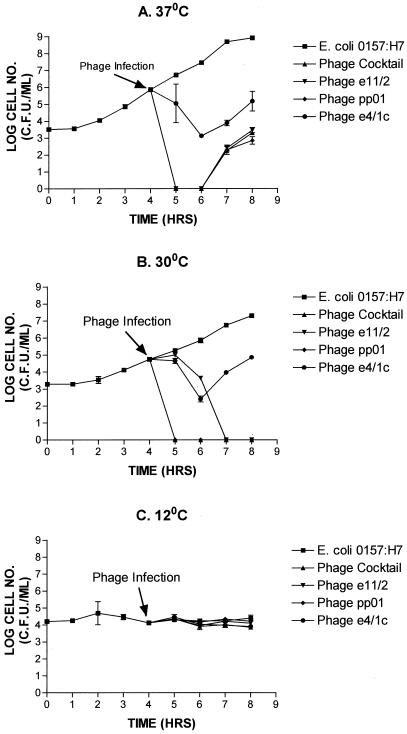
To investigate the relative ability of each phage (either alone or in combination) to lyse E. coli O157:H7 (P1432) in vitro, challenge trails were performed which involved the addition of phage (109 PFU/ml) to a mid-exponential-phase culture at 37, 30 and 12°C (Fig. 2). At both 37 and 30°C with the cocktail, e11/2, and pp01 treatments, a reduction in viable numbers of the culture to below detectable levels was observed. Similarly, phage e4/1c caused a 3-log-unit reduction in the number of viable cells within 2 h at 37°C, and a 3-log-unit reduction in the number of viable cells after treatment with e4/1c at 30°C was also evident. In contrast, a 5-log-unit reduction was seen in 1 or 3 h after treatment with the cocktail or phage pp01 at 30°C. However, the culture started to grow again within 2 to 3 h when incubated at 37°C for all phages when they were applied individually or as a cocktail. This regrowth was not observed for incubation at 30°C, except for cultures treated with phage e4/1c. The bacterial viable numbers of E. coli O157:H7 strain P1432 during the 12°C challenge test were not reduced by the phage cocktail or individual phage addition because the culture did not grow during this period (Fig. 2C). It was determined that all three phages could form plaques on a lawn of E. coli O157:H7 strain P1432 at 12°C following sufficient incubation (results not shown).
FIG. 2.
Comparison of the lytic ability of phage and phage cocktails to lyse E. coli O157:H7 strain P1432 at different temperatures in vitro. Challenge tests were carried out at 37°C (A), 30°C (B), and 12°C (C).
Contaminated-meat trials.
Given that meat is commonly contaminated by E. coli O157 during slaughter, this assay was performed to determine whether phage cocktails could be exploited to eliminate the bacterium or reduce its incidence on meat carcasses. For this experiment, 18 pieces of meat were inoculated with a rifampin-resistant derivative of E. coli O157:H7 strain P1432. A phage cocktail composed of phages e11/2, e4/1c, and pp01 was pipetted evenly onto nine pieces of meat. No phages were pipetted onto another nine pieces, which acted as controls. The enrichment step was included to permit the detection of any surviving E. coli cells. As expected, all nine control pieces of meat were positive, exhibiting counts of E. coli O157:H7 of ∼105 CFU/ml. In the samples where phage cocktails had been added, seven of the nine samples were completely free of E. coli O157:H7 as determined by viable plate count after enrichment. The two positive samples had E. coli O157:H7 counts of less than 10 CFU/ml.
BIMs.
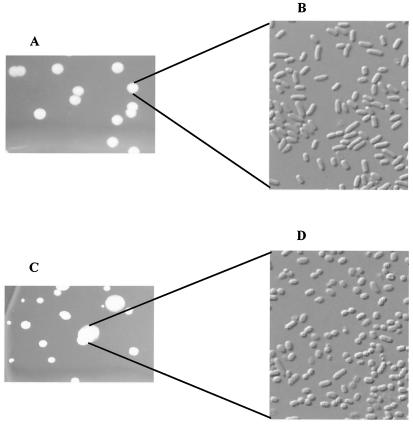
BIMs which had emerged during the in vitro challenge tests at 37°C were selected for further analysis. Mutant colony morphology differed significantly from that of the parental strain, P1432. Strain P1432 appeared rod shaped, whereas the BIM's appeared coccoid and were smaller (Fig. 3). A selection of BIM colonies were inoculated and grown overnight at 37°C. Following overnight growth, each culture was inoculated (2%) and grown at 37°C in triplicate to determine a growth rate. The growth rate for strain P1432 was 1.07 ± 0.01 h−1. The growth rate of 10 BIM isolates following challenge with the phage cocktail did not differ significantly from that of the parent and was recorded as 1.03 ± 0.07 h−1. Ten BIMs which had emerged from the challenge with phage e11/2 alone had a growth rate of 1.13 ± 0.15 h−1, 10 BIMs associated with the phage pp01 challenge had a growth rate of 0.86 ± 0.06 h−1, and 10 BIMs from the e4/1c challenge had a growth rate of 1.03 ± 0.05 h−1. Interestingly, none of the BIMs in this study could reach the turbidity of the parent strain, but they could reach an equivalent cell number where tested (results not shown). Each BIM was checked for susceptibility to all three phages (e11/2, pp01, and e4/1c) after being passed through 50 generations. All BIMs which could form a lawn on a plate were found to be susceptible to each phage (Table 2) to various degrees.
FIG. 3.
Comparison of BIM cells to the parental E. coli O157:H7 strain cells. P1432 (A) Parental E. coli O157:H7 colonies; (B) parental E. coli O157:H7 cells; (C) phage cocktail BIM colonies; (D) phage cocktail BIM cells.
TABLE 2.
BIM susceptibility after 50 generations of growth
| Phage cocktail BIMs | Phage e11/2 BIMs
|
Phage pp01 BIMs
|
Phage e4/1c BIMs
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BIMa | E11/2 | PP01 | E4/1C | BIM | E11/2 | PP01 | E4/1C | BIM | E11/2 | PP01 | E4/1C | BIM | E11/2 | PP01 | E4/1C |
| BIM1 | +++ | + | +++ | BIM1 | ++ | + | + | BIM1 | +++ | +++ | ++ | BIM1 | + | ++ | ++ |
| BIM2 | ++ | + | +++ | BIM2 | ++ | + | + | BIM2 | +++ | +++ | ++ | BIM2 | + | + | + |
| BIM3 | ++ | + | + | BIM3 | ++ | + | + | BIM3 | +++ | +++ | ++ | BIM3 | ++ | ++ | + |
| BIM4 | + | + | + | BIM4 | + | + | + | BIM4 | +++ | +++ | ++ | BIM4 | ++ | ++ | ++ |
| BIM5 | ++ | ++ | + | BIM5 | + | + | ++ | BIM5 | +++ | +++ | ++ | BIM5 | +++ | +++ | +++ |
| BIM6 | ++ | + | +++ | BIM6 | INSb | INS | INS | BIM6 | +++ | +++ | ++ | BIM6 | +++ | +++ | +++ |
| BIM7 | ++ | + | +++ | BIM7 | ++ | ++ | ++ | BIM7 | +++ | +++ | ++ | BIM7 | +++ | +++ | +++ |
| BIM8 | ++ | + | +++ | BIM8 | + | + | ++ | BIM8 | +++ | +++ | ++ | BIM8 | ++ | ++ | ++ |
| BIM9 | ++ | + | +++ | BIM9 | + | + | + | BIM9 | +++ | +++ | ++ | BIM9 | +++ | +++ | +++ |
| BIM10 | + | + | + | BIM10 | INS | INS | INS | BIM10 | +++ | +++ | ++ | BIM10 | INS | INS | INS |
BIM1 to BIM10: BIMs which emerged from strain P1432 when exposed to different phage combinations.
INS, insufficient growth to form a lawn.
Frequency of BIM isolation.
Assays were performed in vitro to determine the frequency at which BIMs emerged after phage challenge at 37°C using a single phage or phage combinations. BIM frequencies were similar for all phage combinations except when phage e4/1c was applied alone (Table 3). Phage e4/1c caused a greater than 100-fold increase in BIM frequency (Table 3) relative to the other two phage (e11/2 and pp01). This result suggests differences in the mechanism of infection between the Siphoviridae phage (e4/1c) and the two Myoviridae phage (e11/2 and pp01). Such differences could be due to different receptors for phage adsorption. However, the BIMs derived from all three phages exhibited resistance to subsequent infection, suggesting that another mechanism is involved.
TABLE 3.
BIM frequency at 37°C
| Phage combination | BIM frequency (mean ± SD)a |
|---|---|
| e11/2 | 1.2 × 10−6 ± 4.9 × 10−7 |
| e4/1c | 3.3 × 10−4 ± 1.1 × 10−4 |
| pp01 | 1.9 × 10−6 ± 5.7 × 10−7 |
| e11/2 + pp01 | 1.2 × 10−6 ± 5.0 × 10−7 |
| e4/1c + pp01 | 1.5 × 10−6 ± 6.0 × 10−7 |
| e11/2 + e4/1c | 1.2 × 10−6 ± 4.2 × 10−7 |
| e11/2 + pp01 + e4/1c | 1.1 × 10−6 ± 4.2 × 10−7 |
SD, standard deviation. Results were obtained from triplicate experiments.
DISCUSSION
It has been shown that E. coli O157:H7 is prevalent in domestic animals, especially cattle (11, 12, 40, 44). Previous studies evaluated the reduction of E. coli O157:H7 in cattle and sheep prior to slaughter when using microbial feeds (21, 43) and a change of diet (14). It is widely believed that phage therapy may have potential in the reduction of harmful bacteria in animals (17). Research involving the application of phage therapies in animal models has produced very promising results (2, 3, 15, 16, 21, 23, 33-36, 43). In this work we evaluated phages as biocontrol agents in vitro and in vivo (on the meat surface). Two anti-O157:H7 phages (e11/2 and e4/1c) were isolated from environmental samples in this study. The third phage (pp01) was taken from a previous collection. Phages were characterized by plaque morphology, electron microscopy, and restriction analysis. In the course of in vitro challenge tests, all phages caused a significant decrease in E. coli O157 cell numbers. BIM formation as reported in the literature (13) is associated with point mutations in genes encoding receptor molecules on the bacterial cell surface. Reversal is therefore assumed to be due to a reversion of the mutation within the population or acquistion of a compensatory mutation. The application of the phage cocktail to meat illustrates its potential role in the elimination of E. coli O157:H7 from the surface.
Phages were relatively easily isolated from the environment following the enrichment step with overnight incubation. The majority of the phages isolated at the outset gave cloudy plaques on the plaquing host. These were discarded on the basis that they were likely to be temperate, while clear-plaque formers were retained since they were most likely to be lytic on the plaquing host (21, 31). All phages isolated from human and bovine fecal samples appeared to be temperate phages, which correlates with a previous study (7). Two lytic phages (e11/2 and e4/1c) were isolated from bovine farmyard slurry samples.
Previous studies characterized phages, based on plaque size, host range (19), and electron micrographs (31). Characterization of all three lytic phages involved a number of criteria. The initial determinant was based on plaque size. Phages e11/2 and pp01 formed pinpoint plaques whereas phage e4/1c formed medium-size plaques on the E. coli O157:H7 host. Electron microscopy classified all phages into their respective families. Even though two phages (e11/2 and pp01) belong to the Myoviridae family, they are unusual in that they have an elongated head much like that of phage T4. In contrast, the third phage (e4/1c) is typical of the Siphoviridae family in that it exhibits a common morphotype. It is not surprising, therefore, that the restriction digests of the two Myoviridae phage appear similar while the pattern for phage e4/1c was completely different, with a smaller genome of approximately 40 kb.
All three phages were tested for their ability to form plaques on a range of gram-negative bacteria. Each phage was shown to be specific for E. coli O157:H7 in that other E. coli strains and other gram-negative bacteria were not lysed. The susceptibility of bacterial strains to phage attack differed, and this may be due to variation of receptor molecules (adsorption blocking), restriction modification systems in the host, or other phage-resistant systems such as abortive infection (8). All toxigenic E. coli O157:H7 isolates were shown by pulsed-field gel electrophoresis (4a) to be distinct. These results suggest that the normal bacterial flora would not be affected by the application of these phages for treatment of E. coli O157:H7. PCR analysis was carried out to evaluate whether any of the three phages (e11/2, pp01, and e4/1c) contained the toxin genes vt1 and vt2 or the virulence factors eaeA and hlyA. While the results were negative for vt1, vt2, and hlyA, we found that phage e4/1c contained the virulence factor gene eaeA (results not shown), which was probably picked up from the host at some point.
The ability of the phage cocktail to control E. coli O157:H7 on the meat surface gave favorable results. The initial E. coli O157:H7 cell numbers (103 CFU/g) correlate with levels observed in bovine feces, which could potentially contaminate the meat carcass (28). The phage titer used represents a typical titer of phage lysates, obtained from phage propagation. Even though the MOI used in these experiments was up to 106-fold, it is worthwhile noting that this typical ratio of phage to cell number would be encountered if the phage lysate was incorporated into a wash step during the slaughter process as part of an HACCP system. In our study, the bacterium was completely eliminated in seven of nine cases when the phage cocktail was applied. This suggests that phage therapy could be introduced as a control measure to eliminate E. coli O157:H7 from the carcass surface during slaughter. Another possible application of phage therapy may involve the addition of the phage cocktail to cattle hides. These observations favor the exploitation of phages for biocontrol of E. coli O157:H7.
When the in vitro E. coli O157:H7 challenges were performed by using individual phages or phage cocktails at different temperatures, significant decreases in bacterial cell numbers were obtained at 30 and 37°C. This lytic ability was greatly reduced at 12°C because the culture was not growing, even though phages were capable of plaque formation at this temperature. The prevalence of BIM formation at 37°C was not surprising because the cell numbers are larger due to a higher growth rate. The correlation with growth rate is possibly temperature associated; while the BIM frequency may be the same, a larger cell number is present at 37°C due to an increased growth rate. However, the prevalence of BIM formation for phage e4/1c was surprising because the rate of BIM formation seems extraordinarily high.
A selection of BIMs of the E. coli O157:H7 strain P1432 were assayed and compared to the parental strain. Pulsed-field gel electrophoresis was carried out on a number of BIMs whose pattern showed that they were derived from the original strain, P1432 (results not shown). BIM colony morphology differed greatly among the different BIMs and also differed from that of the parent strain, an observation which was also made recently in another study (25). No BIM could reach a turbidity equivalent to that of the parent strain (P1432). This is possibly associated with the observation that the cells changed from being rod shaped to coccoid, which reduced their overall size.
E. coli O157:H7 is a prevalent pathogen with a low infectious dose. This study indicates that phage therapy could be a viable method of controlling this pathogen. The frequency of BIM formation is low, and all mutants revert to phage sensitivity; therefore, we expect that BIMs should not hinder the use of these phages as biocontrol agents of the pathogen E. coli O157:H7.
Acknowledgments
We thank Geraldine Duffy of the National Food Centre, Castleknock, Co. Dublin, for access to the toxigenic strains and category 3 laboratory. We also thank Horst Neve of the Federal Research Centre of Nutrition and Food, Institute for Microbiology, Kiel, Germany, for electron microscopy of phages.
This research was funded by the Irish Government under the FIRM Programme as part of National Development Plan 2000-2006 and by The Science Foundation of Ireland. Gary O'Flynn is in receipt of a Teagasc Walsh Fellowship.
REFERENCES
- 1.Barrow, P. 2001. The use of bacteriophages for treatment and prevention of bacterial disease in animals and animal models of human infection. J. Chem. Technol. Biotechnol. 76:677-682. [Google Scholar]
- 2.Barrow, P., M. Lovell, and A. Berchieri, Jr. 1998. Use of lytic bacteriophage for control of experimental Escherichia coli septicemia and meningitis in chickens and calves. Clin. Diagn. Lab. Immunol. 5:294-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berchieri, A., Jr., M. A. Lovell, and P. A. Barrow. 1991. The activity in the chicken alimentary tract of bacteriophages lytic for Salmonella typhimurium. Res. Microbiol. 142:541-549. [DOI] [PubMed] [Google Scholar]
- 4.Biswas, B., S. Adhya, P. Washart, B. Paul, A. N. Trostel, B. Powell, R. Carlton, and C. R. Merril. 2002. Bacteriophage therapy rescues mice bacteremic from a clinical isolate of vancomycin-resistant Enterococcus faecium. Infect. Immun. 70:204-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4a.Cagney, C., H. Crowley, G. Duffy, J. J. Sheridan, S. O’Brien, E. Carney, W. A. Anderson, D. A. McDowell, and I. S. Blair. 2004. Prevalence and numbers of Escherichia coli O157:H7 in minced beef and beef burgers from butcher shops and supermarkets in the Republic of Ireland. Food Microbiol. 21:203-212. [Google Scholar]
- 5.Chanishvili, N., T. Chanishvili, M. Tediashvili, and P. A. Barrow. 2001. Phages and their application against drug-resistant bacteria. J. Chem. Technol. Biotechnol. 76:689-699. [Google Scholar]
- 6.Cray, W. C., Jr., and H. W. Moon. 1995. Experimental infection of calves and adult cattle with Escherichia coli O157:H7. Appl. Environ. Microbiol. 61:1586-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dhillon, T. S., E. K. Dhillon, H. C. Chau, W. K. Li, and A. H. Tsang. 1976. Studies on bacteriophage distribution: virulent and temperate bacteriophage content of mammalian feces. Appl. Environ. Microbiol. 32:68-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duckworth, D. H., J. Glenn, and D. J. McCorquodale. 1981. Inhibition of bacteriophage replication by extrachromosomal genetic elements. Microbiol. Rev. 45:52-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duffy, G. 2003. Verocytoxigenic Escherichia coli in animal faeces, manures and slurries. J. Appl. Microbiol. 94(Suppl. 1):94-103. [DOI] [PubMed] [Google Scholar]
- 10.Elder, R. O., J. E. Keen, G. R. Siragusa, G. A. Barkocy-Gallagher, M. Koohmaraie, and W. W. Laegreid. 2000. Correlation of enterohemorrhagic Escherichia coli O157 prevalence in feces, hides, and carcasses of beef cattle during processing. Proc. Natl. Acad. Sci. USA 97:2999-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faith, N. G., J. A. Shere, R. Brosch, K. W. Arnold, S. E. Ansay, M. S. Lee, J. B. Luchansky, and C. W. Kaspar. 1996. Prevalence and clonal nature of Escherichia coli O157:H7 on dairy farms in Wisconsin. Appl. Environ. Microbiol. 62:1519-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heuvelink, A. E., F. L. van den Biggelaar, E. de Boer, R. G. Herbes, W. J. Melchers, J. H. Huis in't Veld, and L. A. Monnens. 1998. Isolation and characterization of verocytotoxin-producing Escherichia coli O157 strains from Dutch cattle and sheep. J. Clin. Microbiol. 36:878-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hill, C. 1993. Bacteriophage and bacteriophage resistance in lactic acid bacteria. FEMS Microbiol. Rev. 12:87-108. [Google Scholar]
- 14.Hovde, C. J., P. R. Austin, K. A. Cloud, C. J. Williams, and C. W. Hunt. 1999. Effect of cattle diet on Escherichia coli O157:H7 acid resistance. Appl. Environ. Microbiol. 65:3233-3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huff, W. E., G. R. Huff, N. C. Rath, J. M. Balog, and A. M. Donoghue. 2002. Prevention of Escherichia coli infection in broiler chickens with a bacteriophage aerosol spray. Poult. Sci. 81:1486-1491. [DOI] [PubMed] [Google Scholar]
- 16.Huff, W. E., G. R. Huff, N. C. Rath, J. M. Balog, H. Xie, P. A. Moore, Jr., and A. M. Donoghue. 2002. Prevention of Escherichia coli respiratory infection in broiler chickens with bacteriophage (SPR02). Poult. Sci. 81:437-441. [DOI] [PubMed] [Google Scholar]
- 17.Joerger, R. D. 2003. Alternatives to antibiotics: bacteriocins, antimicrobial peptides and bacteriophages. Poult. Sci. 82:640-647. [DOI] [PubMed] [Google Scholar]
- 18.Jordan, K. N., L. Oxford, and C. P. O'Byrne. 1999. Survival of low-pH stress by Escherichia coli O157:H7: correlation between alterations in the cell envelope and increased acid tolerance. Appl. Environ. Microbiol. 65:3048-3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kudva, I. T., S. Jelacic, P. I. Tarr, P. Youderian, and C. J. Hovde. 1999. Biocontrol of Escherichia coli O157 with O157-specific bacteriophages. Appl. Environ. Microbiol. 65:3767-3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kutter, E., T. Stidham, B. Guttman, E. Kutter, D. Batts, S. Peterson, T. Djavakhishvili, F. Arisaka, V. Mesyanzhinov, W. Ruger, and G. Mosig. 1994. Genomic map of bacteriophage T4, p. 491-519. In J. D. Karam (ed.), Molecular biology of bacteriophage T4. ASM Press, Washington, D.C.
- 21.Lema, M., L. Williams, and D. R. Rao. 2001. Reduction of fecal shedding of enterohemorrhagic Escherichia coli O157:H7 in lambs by feeding microbial feed supplement. Small Rumin. Res. 39:31-39. [DOI] [PubMed] [Google Scholar]
- 22.Maher, M. M., K. N. Jordan, M. E. Upton, and A. Coffey. 2001. Growth and survival of E. coli O157:H7 during the manufacture and ripening of a smear- ripened cheese produced from raw milk. J. Appl. Microbiol. 90:201-207. [DOI] [PubMed] [Google Scholar]
- 23.Matsuzaki, S., M. Yasuda, H. Nishikawa, M. Kuroda, T. Ujihara, T. Shuin, Y. Shen, Z. Jin, S. Fujimoto, M. D. Nasimuzzaman, H. Wakiguchi, S. Sugihara, T. Sugiura, S. Koda, A. Muraoka, and S. Imai. 2003. Experimental protection of mice against lethal Staphylococcus aureus infection by novel bacteriophage phi MR11. J. Infect. Dis. 187:613-624. [DOI] [PubMed] [Google Scholar]
- 24.Merril, C. R., D. Scholl, and S. L. Adhya. 2003. The prospect for bacteriophage therapy in Western medicine. Nat. Rev. Drug. Discov. 2:489-497. [DOI] [PubMed] [Google Scholar]
- 25.Mizoguchi, K., M. Morita, C. R. Fischer, M. Yoichi, Y. Tanji, and H. Unno. 2003. Coevolution of bacteriophage PP01 and Escherichia coli O157:H7 in continuous culture. Appl. Environ. Microbiol. 69:170-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morita, M., Y. Tanji, K. Mizoguchi, T. Akitsu, N. Kijima, and H. Unno. 2002. Characterization of a virulent bacteriophage specific for Escherichia coli O157:H7 and analysis of its cellular receptor and two tail fiber genes. FEMS Microbiol. Lett. 211:77-83. [DOI] [PubMed] [Google Scholar]
- 27.Murphy, F. A., C. M. Fauquet, D. H. L. Bishop, S. A. Ghabrial, A. W. Jarvis, G. P. Martelli, M. A. Mayo, and M. D. Summers. 1995. Virus taxonomy: sixth report of the International Committee on Taxonomy of Viruses. Springer-Verlag, Vienna, Austria.
- 28.Omisakin, F., M. MacRae, I. D. Ogden, and N. J. Strachan. 2003. Concentration and prevalence of Escherichia coli O157 in cattle feces at slaughter. Appl. Environ. Microbiol. 69:2444-2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Sullivan, D., D. P. Twomey, A. Coffey, C. Hill, G. F. Fitzgerald, and R. P. Ross. 2000. Novel type I restriction specificities through domain shuffling of HsdS subunits in Lactococcus lactis. Mol. Microbiol. 36:866-875. [DOI] [PubMed] [Google Scholar]
- 30.Riley, L. W., R. S. Remis, S. D. Helgerson, H. B. McGee, J. G. Wells, B. R. Davis, R. J. Hebert, E. S. Olcott, L. M. Johnson, N. T. Hargrett, P. A. Blake, and M. L. Cohen. 1983. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N. Engl. J. Med. 308:681-685. [DOI] [PubMed] [Google Scholar]
- 31.Ronner, A. B., and D. O. Cliver. 1990. Isolation and characterization of a coliphage specific for Escherichia coli O157:H7. J. Food Prot. 53:944-947. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Smith, H. W., and M. B. Huggins. 1983. Effectiveness of phages in treating experimental Escherichia coli diarrhoea in calves, piglets and lambs. J. Gen. Microbiol. 129:2659-2675. [DOI] [PubMed] [Google Scholar]
- 34.Smith, H. W., and M. B. Huggins. 1982. Successful treatment of experimental Escherichia coli infections in mice using phage: its general superiority over antibiotics. J. Gen. Microbiol. 128:307-318. [DOI] [PubMed] [Google Scholar]
- 35.Smith, H. W., M. B. Huggins, and K. M. Shaw. 1987. The control of experimental Escherichia coli diarrhoea in calves by means of bacteriophages. J. Gen. Microbiol. 133:1111-1126. [DOI] [PubMed] [Google Scholar]
- 36.Smith, H. W., M. B. Huggins, and K. M. Shaw. 1987. Factors influencing the survival and multiplication of bacteriophages in calves and in their environment. J. Gen. Microbiol. 133:1127-1135. [DOI] [PubMed] [Google Scholar]
- 37.Sulakvelidze, A., Z. Alavidze, and J. G. Morris, Jr. 2001. Bacteriophage therapy. Antimicrob. Agents Chemother. 45:649-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tanji, Y., K. Mizoguchi, M. Yoichi, M. Morita, N. Kijima, H. Kator, and H. Unno. 2003. Seasonal change and fate of coliphages infected to Escherichia coli O157:H7 in a wastewater treatment plant. Water Res. 37:1136-1142. [DOI] [PubMed] [Google Scholar]
- 39.Tarr, P. I. 1995. Escherichia coli O157:H7: clinical, diagnostic, and epidemiological aspects of human infection. Clin. Infect. Dis. 20:1-8; quiz, 9-10. [DOI] [PubMed] [Google Scholar]
- 40.Vuddhakul, V., N. Patararungrong, P. Pungrasamee, S. Jitsurong, T. Morigaki, N. Asai, and M. Nishibuchi. 2000. Isolation and characterization of Escherichia coli O157 from retail beef and bovine feces in Thailand. FEMS Microbiol. Lett. 182:343-347. [DOI] [PubMed] [Google Scholar]
- 41.Wang, G., T. Zhao, and M. P. Doyle. 1996. Fate of enterohemorrhagic Escherichia coli O157:H7 in bovine feces. Appl. Environ. Microbiol. 62:2567-2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu, S. L., K. L. Ko, C. S. Chen, Y. C. Chang, and W. J. Syu. 2000. Characterization of the distal tail fiber locus and determination of the receptor for phage AR1, which specifically infects Escherichia coli O157:H7. J. Bacteriol. 182:5962-5968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao, T., M. P. Doyle, B. G. Harmon, C. A. Brown, P. O. Mueller, and A. H. Parks. 1998. Reduction of carriage of enterohemorrhagic Escherichia coli O157:H7 in cattle by inoculation with probiotic bacteria. J. Clin. Microbiol. 36:641-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao, T., M. P. Doyle, J. Shere, and L. Garber. 1995. Prevalence of enterohemorrhagic Escherichia coli O157:H7 in a survey of dairy herds. Appl. Environ. Microbiol. 61:1290-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]