Abstract
Ser/Thr- and Tyr-Protein kinases constitute a key switch underlying the dynamic nature and graded regulation of signal transduction and pathway activities in cellular organization. Here we describe the identification and characterization of Dusty, a single-copy gene that arose in metazoan evolution and encodes a putative dual Ser/Thr and Tyr protein kinase with unique structural features. Dusty is widely expressed in vertebrates, broadly distributed in the central nervous system, and deregulated in certain human cancers. Confocal imaging of transiently expressed human Dusty-GFP fusion proteins showed a cytoplasmic distribution. Dusty proteins from lower to higher species display an increasing degree of sequence conservation from the N-terminal non-catalytic domain to C-terminal catalytic domain. The non-catalytic region has eight conserved cysteine residues, multiple potential kinasedocking motifs and phosphorylation sites, whereas the catalytic domain is divergent and about equally distant of Ser/Thr and Tyr protein kinases. Homology analyses identified the essential catalytic residues, suggesting that Dusty homologues all possess the enzymatic activity of a protein kinase. Taken together, Dusty is a unique evolutionarily selected group of divergent protein kinases that may play important functional roles in the brain and other tissues of vertebrates.
Keywords: Dusty protein kinase, Tissue specificity, Protein structure, Gene evolution, Vertebrate animals, Human cancer
1. Introduction
Protein-serine/threonine and protein-tyrosine kinases (PSK and PTK), which transfer the γ-phosphate from ATP to the −OH group of Ser/Thr and Tyr in substrate proteins, comprise two major classes of protein kinases [1]. They provide a key switch dictating the dynamic nature and graded regulation of signal transduction pathway activities in cellular organization [2, 3]. In the course of evolution from simple unicellular organism to complex multicellular metazoan natural selection has heavily invested on this switch by repeatedly exploiting two tactics for functional and regulatory innovations. In the one, the catalytic cores have been diversified to expand their repertoire of protein substrates [4]. The complement of PKs of the human genome or the kinome totals more than 500 genes [5] and a comparable set in mice is evident [6]. In the other, such cores have been joined to a range of novel domains that modulate reaction specificity and network assembly [7, 8]. Accordingly, these kinases have acquired a multitude of spatiotemporal processes to physiologically fine-tune their enzymatic and non-enzymatic activities.
PSK and PTK have largely preserved their specificity for S/T-OH and Y-OH without cross reactivity, despite a dozen dual-specificity PKs known to intramolecularly autophosphorylate either amino acid [9, 10]. The dominance of PSK in yeast [11], the early evolution of the tyrosine kinase-like group and associated tyrosine phosphorylation [12], and later increase of PTK in metazoans [13-16] imply the origin of PTK from PSK by gene duplication and diversification. In this process two mechanisms, not necessarily mutually exclusive, could redirect catalytic subfunctionalization. One entails a creation of hybrid protein kinase domains generated of different PSK as intermediates, which were then diversified to PTK by fast evolution [9, 16]. The other may involve a process of many small steps in the form of nonsynonymous substitutions in a PSK ancestor or its duplicate, leading to the origin of PTK and ultimately tyrosine-based catalysis. Due to a deep rooting and long history of PK evolution [17], which would cause mutation saturation in catalytic domains, it is difficult to identify those intermediate products that are derived from the above-mentioned selection events.
In this paper we report detailed analyses of Dusty PK, a putative and more recently evolved dual Ser/Thr and Tyr kinase in a variety of species, notably vertebrates. Dusty was named SgK496 by genome scan as a novel member of human kinome [5], when we deposited six Dusty genes from zebrafish to human (AY208850 to AY208855). Our deposited Dusty was renamed as the receptor interaction protein family member 5 (RIP5) [18], but this view has been challenged by the unifying features of the RIP PK family including the newly identified members [19]. Here we describe the primary structure, gene evolution, tissue specific expression and unique divergent features of the catalytic domain of Dusty proteins. We provide also evidence that Dusty is not RIP5 and more importantly show that Dusty is a divergent protein kinase selected to play some potentially important functional roles in the brain and other tissues.
2. Materials and methods
2.1. Cloning of Dusty protein kinases
RNA samples were isolated from various species using Trizol reagent (Invitrogen) or obtained from colleagues or commercial sources (Clontech, Novagen and Seegene). Cloning methods followed standard protocols [20]. Full-length Dusty cDNAs were assembled from sequencing of overlapping PCR products using RACE kits (Gibco) and gene specific primers (GSP). The open reading frame (ORF) of Dusty was PCR-amplified by GSPs that cover the initiation and stop codons, respectively (supplementary table 1), subcloned in pCRII-TOPO vector and sequenced to completion. These plasmids served as master templates for construction of expression vectors.
2.2. Screening and characterization of mouse Dusty gene
A mouse BAC (bacterial artificial chromosome) library CitbCJ7 of 129Sv ES cell origin [21] was PCR-screened using GSP-mE3F and GSP-mE3R (supplementary table 1). The 664-bp product of exon 3 indicated positive clones. BAC plasmids were isolated using the alkaline lysis method and analyzed by pulsed gel electrophoresis, fingerprinted by exon-specific PCR or mapped by Southern blot. Exons or introns were amplified from BAC clones, and their junctions were defined by alignment with the cDNA sequence. The promoter region of Dusty genes was amplified by PCR from human or mouse genomic libraries using adaptor primers and exon 1 GSPs. The fragments were gel-purified and sequenced. The cis-acting transcription factor binding motifs were searched according to the published method [22] (http://cic.cs.wustl.edu/wordspy).
2.3. Bioinformatics and phylogenetic analyses
Dusty sequences were used for database search by BLAST [23]. Short sequence motifs were identified using Scansite [24] and Minimotif Miner [25]. Secondary structures and hydrophobic profiles were analyzed with the DNASTAR Protean software. Protein sequences were aligned using Muscle (version 3.52) [26] from which the matching codon sequences were derived. Phylogenetic trees were reconstructed using the maximal likelihood (ML) method as implemented in PhyML (version 2004) [27] under the JTT (Jones-Taylor-Thornton) + 4G (four categories of Gamma substitution rates) + I (invariant sites) model [28]. Adaptive evolution of Dusty genes was assessed using maximum likelihood methods [29, 30]. For classification and homology analyses, the retrieved ePK domains (E value<1e-4) of known 3D structures were aligned with Dusty using both multiple [26] and pair-wise sequence alignment. A phylogram was obtained with Mega (version 3.0) [31].
2.4. Northern blot analysis and RNA FISH
Northern blots of human and animals and human Cancer Profiling Array were purchased (Clontech and Seegene) and hybridized with Dusty cDNA probes. Dusty probes are species-specific for exon 3 or ePK domain (supplementary table 2). Blots were washed under high stringency. RNA fluorescent in situ hybridization (FISH) to mouse embryo and adult tissue sections was done as described [32]. Two probes were made for labeling: P-I contained Dusty exon 3 (657-bp) cloned in pCRScript Sk(+) vector (Stratagene) and P-II covered the ePK coding sequence (861-bp) cloned in pCRII-TOPO vector (supplementary table 2). Antisense and sense P-I or P-II probes were generated from Not I-digested Dusty plasmids via in vitro transcription by T7/T3 and Sp6/T7 RNA polymerases, respectively.
2.5. Polyclonal antibody production
To produce antibodies for Dusty, the sequence of human Dusty254-458 was PCR-amplified with primers Dusty254F and Dusty458R (supplementary table 1). The product was digested with EcoRI and XhoI, and cloned into compatible sites upstream of 6xHis-tag in pET28b vector (Novagen). The Dusty-6xHis plasmid was transformed into E. coli BL21 cells and induced with 0.3 mM of IPTG (30°C, 4 h) for protein expression. After sonication and cell lysis, the Dusty-6xHis protein was purified on a Ni-NTA column (Qiagen). About 300μg of protein in total was emulsified with adjuvant and injected into rabbits, as described [33].
2.6. Vector construction, cell culture, transfection, and confocal microscopy
Expression vectors were constructed using high fidelity pfu DNA polymerase (Stratagene) and verified free of mutations by sequencing. To tag green fluorescence protein (GFP), human Dusty and its truncated versions were amplified with various GSP (supplementary table 1). The product was digested with restriction enzymes and subcloned into pEGFP-C1 or -N3 vector (Clontech). Human (HEK293, K562, HeLa, HL60) and nonhuman (COS7) cells (ATCC) were cultured at 37°C in DMEM containing 10% FCS (Invitrogen) in a 5% CO2 incubator. Cells for transient expression were grown in 6-well plate for 24h and added with plasmid and lipofectamine (Invitrogen). In imaging analysis, 3×105 HEK293 cells were plated on 35 mm coverglass (MatTek), transfected with 1 μg of Dusty-pEGFP or pEGFP-Dusty plasmid plus lipofectamine. After 24 h, GFP was excited at 488nm with an argon laser and fluorescent cells were viewed under a Zeiss LSM510 confocal microscope. The images captured were processed using the image browser software.
2.7. Western blot analysis
Western blot analysis of Dusty protein was performed using cellular extracts from mouse native tissues (Biochain), human cell lines or Dusty-transfected nonhuman cells. The blots were probed with the rabbit primary antiserum against Dusty254-458 (1:1,000) and then stained with HRP-linked donkey anti-rabbit IgG (1:5,000) (Amersham). Proteins were visualized with a chemiluminescent kit (Pierce).
3. Results
3.1. Organismal distribution, RNA structure and genomic organization
16 full-length Dusty cDNAs were cloned from 14 vertebrates and 2 invertebrates. Database analyses showed: 1) Dusty is of single-copy in all 16 species; 2) Dusty is ubiquitous in vertebrates, but absent from unicellular organisms and has a patchy distribution in invertebrates: it is found in early metazoans such as Cnidarians (Hydra and Nematostella), arthropods (Apis, Tribolium, Bombyx) and echinoderms (sea urchin: Strongylocentrotus purpuratus), but has apparently been secondarily lost from both Drosophila and C. elegans genomes as well as from Ciona, as is seen with other kinase families throughout evolution [12].
Of vertebrates from zebrafish to human, Dusty mRNA encodes a protein of 885-931 amino acids (molecular mass of 100-105 kDa). Fig.1A shows the mRNA and genomic structure of human Dusty as a model. Human Dusty mRNA is 7,898-base long with a 5' atypical Kozak sequence and a long 3'-UTR sequence with multiple polyadenylation signals. Likewise, Dusty mRNA form is of similar size in other mammals and chicken (see below).
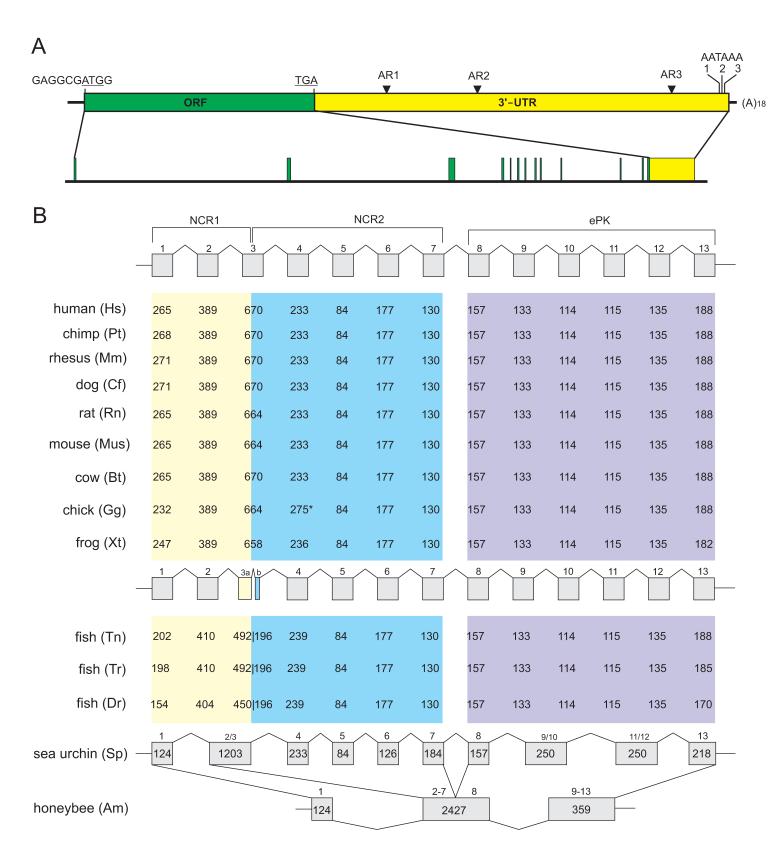
Fig. 1.
Dusty gene structure and organization. (A) The mRNA structure and genomic organization of human Dusty as a model. The 7,898-base mRNA is divided into ORF, 3'UTR and poly(A) tail, where initiation and stop codons, Alu repeats (AR) and three polyadenylation signals (AATAAA) are shown. Division of mRNA into 13 exons is drawn to scale. (B) Exon-intron structure of Dusty in vertebrates. Exons (in bp) in correspondence to the protein sequence regions are denoted. Chicken exon 4 has an insertion (star-denoted) and fish exon 3 is split (3a and 3b). The organization of sea urchin Dusty is similar to that of vertebrates with fewer exon splits but is quite different from honeybee Dusty (bottom).
Next we screened a mouse BAC library and detected 5 Dusty clones. Characterization of the clones together with database analysis revealed an exon-intron structure that corresponds to protein domain organization from sea urchin to human (Fig. 1B). Dusty is divided into 13 exons from frog to human and 14 exons in fish where exon 3 is split in two smaller ones. Other shared features of vertebrate Dusty genes include a G/C-rich promoter (not shown), conforming to their expression in many tissues (see below), and a preserved size of equivalent exons, except for the 5' and 3' exons (Fig. 2B). However, the exon number is reduced to 10 in sea urchin and 3 in honeybee. These data suggest that during metazoan evolution Dusty gene had subject to a profound organizational change prior to its ubiquitous spread in vertebrates.
Fig. 2.
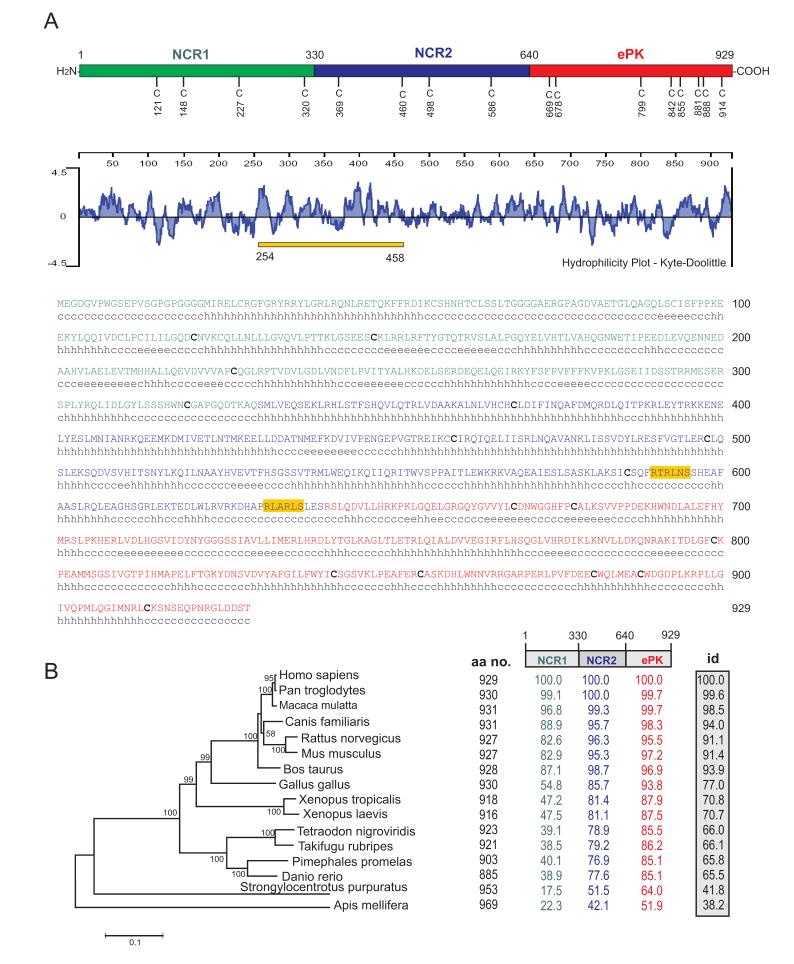
Dusty protein structure and evolution. (A) Human Dusty is shown as a model; it is divided in NCR1 (1-330), NCR2 (331-640) and ePK (641-929) domains, and contains 16 conserved Cys residues (whose positions are numbered) in vertebrates. The hydropathy plot, primary sequence and secondary structural elements (c, coiled-coil; e, extended strand; h, helix) are shown. The secondary structure shown for human Dusty is representative of other mammals, including chimp, rhesus monkey, cow, dog, rat and mouse. The bar denotes the peptide 254-458 for antibody production. Potential phosphorylation sites conserved in vertebrates are shaded (Akt/PKB: RTRLNS; and Pak: RLARLS). Conserved Cys residues are bolded. (B) The phylogram and pair-wise identity of Dusty proteins. Species names and bootstrap values are given (left). A scale bar denotes 0.2 substitutions per amino acid site. Shown also are the polypeptide size (total amino acids), domain identity (color-coded) and overall identity (shaded box) of each protein, as referred to human Dusty. Note the divergence of NCR1 among species.
3.2. Sequence organization and structural features
Comparison of 16 Dusty PKs led to a protein model of three-domain organization: two non-catalytic regions (NCR1 and NCR2) and ePK domain (Fig. 2A) (supplementary data 1). The protein structure and hydrophilicity are shown for human Dusty as a model. The absolute conservation of 16 cysteines across vertebrates is notable (4 in NCR1, 4 in NCR2, and 8 in ePK). NCR1 and NCR2 share no homology to known conserved domains, but harbor some putative phosphorylation sites (Fig. 2A, two examples are given). NCR1 and NCR2 also contain multiple short stretches of hydrophobic sequence (Fig. 2A, middle) but no apparent membrane-spanning segment.
To decipher the relationship of Dusty proteins, a phylogram was obtained. The tree topology is congruent with the known order of animal species, where vertebrate proteins form a cogent cluster most likely derived from invertebrate ones (Fig. 2B, left). To explore diversification of Dusty, pairwise sequence identities based on the three-region model were computed. As referred to human, the overall identity is steadily increased from honeybee to chimpanzee (Fig. 2B, right). A region-wise division further reveals that this increase is characterized by a gradient of conservation from NCR1 (least conserved) to NCR2 (more conserved) to ePK (most conserved) (Fig. 2B). This asymmetric pattern of sequence divergence points to a faster evolution of NCR1 vs. a slower evolution of NCR2 and ePK. Indeed, we found that only NCR1 has been subject to adaptive evolution in a few codon sites among the mammalian group including human, chimp, monkey, cow, dog, rat and mouse (supplementary table 3).
3.3. Tissue specific expression in vertebrates
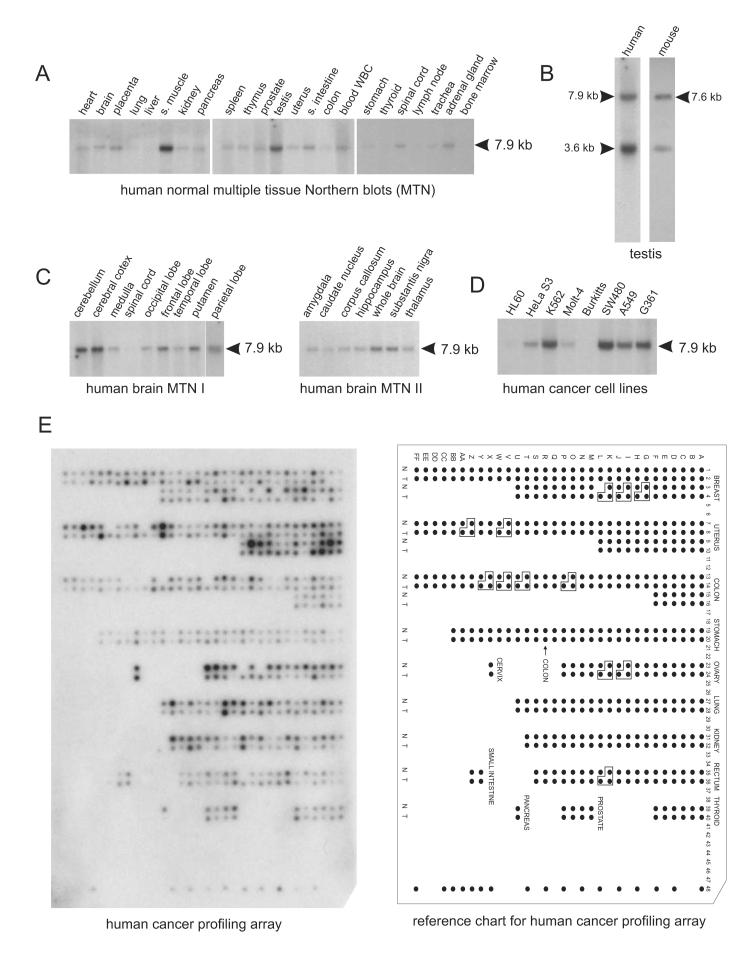
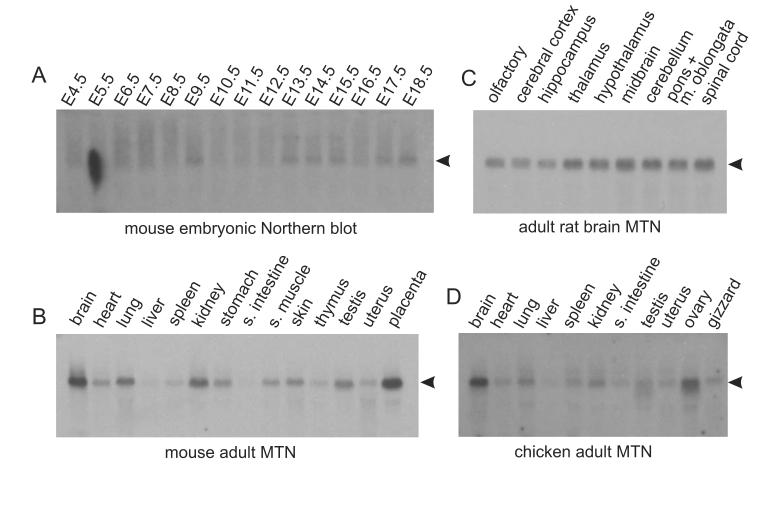
To assess the abundance and location of Dusty expression, RNA blot analysis was done on four vertebrates. In human, Dusty was expressed at variable levels as a single form in multiple tissues (brain, heart, kidney, lung or muscle) (Fig. 3A) and two forms in testis (Fig. 3B). The pattern of Dusty expression in human testis was independently shown [18]. Further analysis revealed its broad distribution in various CNS regions (Fig. 3C). Human Dusty was apparently deregulated in certain cancer cell lines (erythroleukemic K562 and colon cancer SW480, but not cancer cells of myeloid or lymphoid origin) (Fig. 3D). As shown by cancer profiling array, human Dusty was notably down-regulated in tumors of breast, ovary, lung and pancreas (Fig. 3E). Dusty expression in animals showed a comparable pattern. In mouse, Dusty was expressed at a basal level in almost all stages during embryonic development with peaks at E9.5, E13.5 and onward (Fig. 4A), and variably expressed in multiple adult tissues (Fig. 4B), including two transcripts specifically expressed in the testis tissue (Fig. 3B). Similarly, Dusty is expressed in many regions of adult rat brain (Fig. 4C) and in chicken adult tissues (Fig. 4D).
Fig. 3.
Northern blot analysis of human Dusty. (A) Northern blots of 23 human tissues probed with Dusty-NB1 (exon 3-specific). The 7.9-kb band is denoted. (B) Northern blots of Dusty expressed in human (left) and mouse testis (right). Note a short mRNA form is seen in both species. (C) Northern blots of Dusty in human CNS regions. (D) Northern blots of Dusty in human cancer cell lines. In blots A to D (all from Clontech), each lane was loaded with 2 μg poly(A)+ RNA. Hybridization of actin probe was quite uniform and is not shown for brevity. (E) Human Cancer Profiling Array. The array (Clontech Cat#7841-1) was hybridized with the Dusty-CPA probe encoding the ePK domain (see supplementary table 2). N: normal tissue; and T: tumor tissue. The orientation grid for the array is shown at right. The identity and information of tumor tissue samples can be accessed on Clontech website or available from the authors upon request. Hybridization of the array with human ubiquitin cDNA was quite uniform and is not shown for brevity. Abbreviations: s. muscle, skeletal muscle; and s. intestine, small intestine.
Fig. 4.
Northern blot analysis of animal Dusty. (A) Northern blots of mouse embryonic development (E4.5 to E18.5) probed with Dusty-NB2. Note a basal expression in all stages and an elevation at E13.5 and onward. (B) Northern blots of mouse adult tissues. (C) Northern blots of rat adult brain probed with Dusty-NB3. Note Dusty is present in spinal cord. (D) Northern blots of adult chicken tissues probed with Dusty-NB4. All probes are exon 3 and species-specific. In blots A to D (all from SeeGene), each lane was loaded with 20 μg of total RNA and the loading was monitored with 28S and 18S rRNA bands. Hybridization of the blots with the actin or ubiquitin probe was quite uniform and is not shown for brevity.
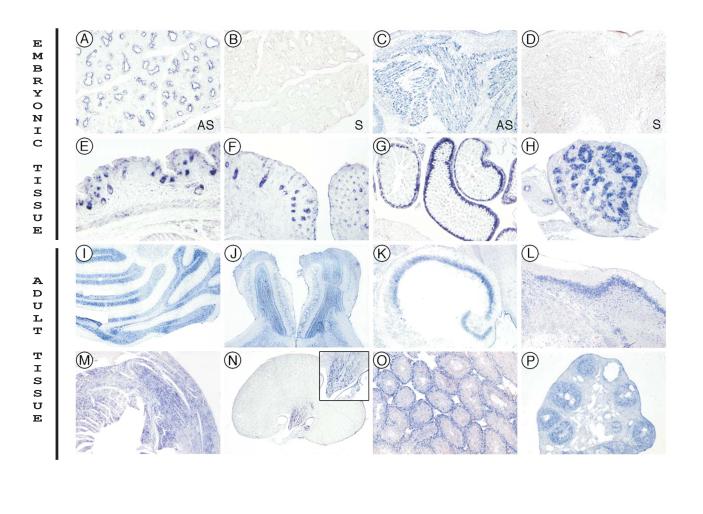
To refine the expression sites of Dusty, FISH was done in mouse as a major animal model. The results matched that of RNA blots (Fig. 3) and revealed further details of the sites of expression in embryonic and adult tissues. In mouse embryo at E14.5 (Fig. 5), Dusty was already expressed in lung (5A) and muscle (5C), and at E18.5 in skin (5E), whisker (5F), gut (5G), and testis (5H), which are from the three embryonic layers, respectively. In adult mouse, Dusty was expressed highly in brain tissues including cerebellum, olfactory, hippocampus, and cerebral cortex (Fig. 5I to L) as well as other tissues like heart, kidney, ovary and testis (Fig. 5M to 5P). Taken together, the tissue distribution of Dusty is a conserved feature at least in mammals and birds.
Fig. 5.
Dusty RNA FISH in mouse embryonic and adult tissues. All the images were obtained with the P-I probe. A—H are for mouse embryonic tissues. A and B, E14.5 lung. C and D, E14.5 skeletal muscle. E —H, E18.5: E, skin; F, whisker; G, intestine; H, testis. A, C, E, F, G and H are obtained with the antisense (AS) probe, while B and D are controls of the sense (S) probe. I —P are for mouse adult tissues with the antisense probe. I—L are for the brain: I, cerebellum; J, olfactory; K, hippocampus; L, cerebral cortex. M, heart; N, kidney (inset: the high magnification of renal collecting tubules); O, testis; and P, ovary. Magnifications are 80x: A, B, C, D, E, H, O, P; 50x: F, N-inset; 40x: G, K, L, M; 20x: I, J; 8x: N.
3.4. Protein expression and subcellular localization
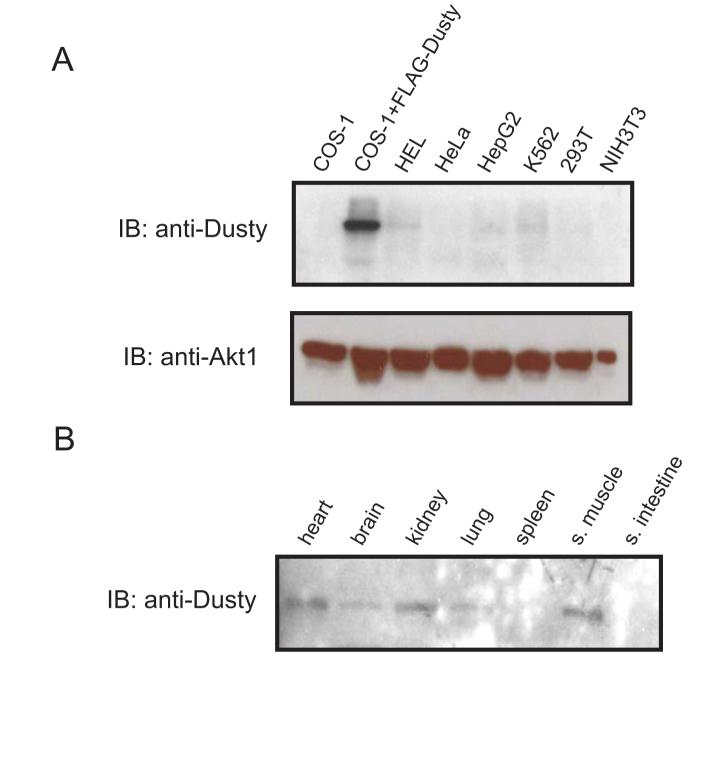
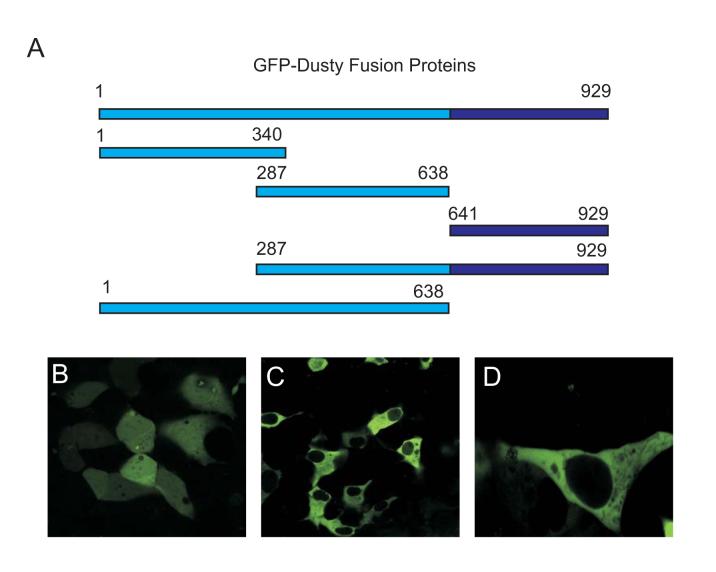
To establish Dusty expression at a protein level, a polyclonal antibody was raised for His-tagged human Dusty254-458. With this antibody, Dusty protein was demonstrated by western blot analysis of extracts from human cell lines and transfected cells, and mouse native tissues. In human cells, a protein band >100 kDa was seen (Fig. 6A), conforming to the predicted size of Dusty. Mouse Dusty protein was detected in brain, heart, muscle, kidney and lung (Fig. 6B). To define subcellular location, various GFP-Dusty forms (Fig. 7A) were expressed in cells followed by confocal imaging. In cells transfected with pEGFP alone, the fluorescence signal was evenly distributed in the cytoplasm and nucleus (Fig. 7B). In the cells expressing either full-length GFP-Dusty or its truncated version, the signal also displayed a relatively uniform pattern in the cytoplasm (Fig. 7B and C), with exclusion from certain subcellular organelles.
Fig. 6.
Western blot analysis. The blots were probed with a polyclonal antibody against human Dusty254-458. (A) Endogenous Dusty detected in different cell lines with Dusty-specific antibody. The transiently expressed Flag-Dusty was used as positive control and Akt/PKBα as a loading control. (B) Mouse Dusty on multiple normal tissue blots detected with the human Dusty antibody.
Fig. 7.
Subcellular localization of GFP-tagged human Dusty. (A) The full-length GFP-Dusty and truncated forms are depicted, whose positions refer to human Dusty. (B) An image of pEGFP vector expressed in HEK293 cells as control was captured by confocal microscopy. (C) An image of full-length GFP-Dusty indicates the cytoplasmic location. (D) A representative image of GFP-Dusty truncated versions indicates the similar subcellular location, as shown above.
3.5. Classification and conservation pattern of Dusty ePK domains
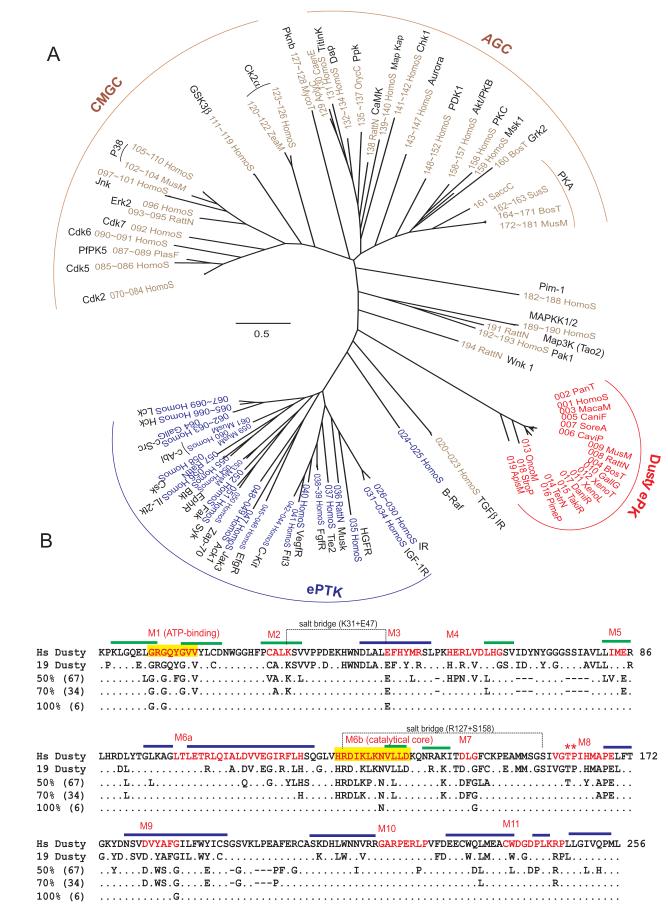
Having shown ePK to be the most conserved part of Dusty, we explored their classification, conservation pattern and evolutionary relationship with known PSK and PTK. The ML optimal tree shows that Dusty ePKs form a specific branch, almost equally distant from PSK and PTK major clusters (Fig. 8A). This approximate equal distance may reflect the unique feature of Dusty ePK, which might stem from ePK exchanges through gene conversion. Alternatively, Dusty may be a very divergent protein kinase and so it is distant from all known kinases. Expanded identification and analysis of Dusty orthologues and their ePKs among invertebrates should yield clues to these hypotheses. Comparison of Dusty ePKs revealed conservation and variation patterns, providing insight into which residues are crucial for catalytic function and thus are constrained in evolution (Fig. 8B). Importantly, Dusty ePKs are not only conserved but also retain all essential catalytic motifs (Fig. 8B) previously identified [4,16], suggesting that Dusty proteins possess a kinase catalytic activity. Notably, the Thr-Pro dipeptide in the activation loop is also conserved in all Dusty ePKs (Fig. 8B), which may be a substrate of a CMGC (proline-directed) kinase or subject to autophosphrylation [34].
Fig. 8.
Phylogram and conservation pattern of Dusty ePK domains. (A) The ML optimal tree was reconstructed from 194 ePK (19 Dusty and 175 PSK+PTK) using JTT+4G+I model. The tree is not rooted. The ePK taxa of PSK, PTK and Dusty are marked with brown, blue and red, respectively. Except Dusty, other ePKs have known 3D structures. The ID and accession numbers of the total dataset are listed in supplemental table 4. The bar denotes 0.5 substitutions per amino acid site. The name and division of major protein kinase groups are denoted. (B) Functional motifs of ePK gleaned from the multiple sequence alignment between 19 Dusty and 175 PSK/PTK. The first row is human Dusty (1-256 equals to 651-906aa in Fig. 2A), the second row conserved residues among 19 Dusty (consensus), and third to fifth rows conserved residues among 194 ePK on 50, 70, and 100% levels. Specific motifs (M1 through M11) are marked in red. The ATP binding motif (M1) and catalytic core (M6a) are shaded in yellow. The conserved TP residues in the activation loop (M8) are denoted by stars. The α-helix and β-sheet are denoted with blue and green bars, respectively, while the remaining are loops.
4. Discussion
Here we characterized Dusty, a unique set of protein kinases present in invertebrates to vertebrates. We showed that Dusty PK is composed of a novel N-terminal non-catalytic region (NCR1 and NCR2) that is connected to a composite but highly conserved C-terminal ePK domain. Phylogenetic analysis provides evidence for the origin of Dusty during metazoan evolution and demonstrates a fast (adaptive) evolution of NCR1 domain in mammals. Of vertebrate Dusty PKs, NCR1 and NCR2 share no homology to any known conserved protein domain, but harbor eight conserved Cys residues and putative phosphorylation sites. A detailed expression analysis showed the broad distribution of Dusty PKs in vertebrate tissues including various CNS regions. These data, together with the fact that Dusty is a single-copy gene establish the orthologous relationship among Dusty PKs, indicating that they perform the same functional roles in different organisms.
The recognition of Dusty PKs as a unique protein kinase disputes the classification of human Dusty as RIP5, a premise largely based on the alignment of ePK domains [18]. The RIP PKs are essential sensors and integrators of cellular stress signals [19]. They play crucial regulatory roles in cell death or cell survival by binding cell-surface receptors (e.g., TNF receptors) [35] and then triggering the activation of transcription factors [36]. All known and recently identified RIP1-7 are organized similarly: their N-termini contain ePK and C-termini other domains. The uncharacterized human RIP5 (SgK288) has multiple C-terminal ankrin repeats and has previously been annotated as a RIPK [5], and totally differs from the domain structure of Dusty here described. Moreover, the Dusty ePK phylogram does not cover RIPs due to low identities. Together, these results preclude a family membership between Dusty and RIP, implying that even Dusty may participate in stress response as a novel signal transducer, it is unlikely involved in RIP-mediated pathway activities.
Although there is a dramatic difference in exon-intron organization between honeybee and sea urchin, the organization of Dusty genes is conserved and so is their protein region, particularly with regard to the NCR2 and ePK domain. Despite a fast evolution of NCR1, which may impart a species-specific function, the expression profiles as observed in four vertebrates suggest strongly that Dusty PKs may play important functional roles in both brain and other tissues. Significantly, mouse Dusty mimics human Dusty in chromosomal location, protein structure, CNS distribution and tissue-specificity, thereby providing a useful animal model for future studies. Although preliminary, the deregulation of Dusty in certain human cancers warrants a more detailed investigation, given the observation that Dusty induced apoptosis when over-expressed [18].
The finding that Dusty proteins contain an ePK domain and putative phosphorylation sites raises the possibility that they are both an enzyme and a substrate protein of phosphorylation. The conservation of the activation loop in Dusty ePK domains also points to their possible occurrence as a substrate of a CMGC kinase and capability of autophosphorylation [34]. Our bioinformatics analyses suggest that Dusty PKs intrinsically retain a catalytic activity, given that the amino acid residues essential for catalysis of phosphate transfer are absolutely conserved. In keeping with this theme, our preliminary studies indeed demonstrated the ability of human Dusty PK to phosphorylate model substrates and that human Dusty is a target of Akt/PKB phosphorylation (Dong and Huang, unpublished data). In future studies, a major undertaking should be directed to identify the physiologic substrates of Dusty PKs and to probe the mechanistic aspect of their actions.
The novelty of Dusty PKs lies in both their non-catalytic region as discussed above, and their catalytic domain as presented below. The Dusty ePK domain is composite in nature, as revealed by the following observations. 1) Database analyses showed that Dusty ePK shares almost the same degree of sequence identity with that of PSK and PTK. 2) The ePK phylogram demonstrated that the Dusty branch is almost in equal evolutionary distances to PSK and PTK clusters. 3) The consensus sequence of Dusty ePK is characterized by a patched conservation of essential catalytic residues pertinent to both ePK groups. 4) The 3D model for Dusty ePK unveiled some fine topologic details that mingle the features of known PSK or PTK 3D structures (data not shown). These molecular and topologic facets imply a novel catalytic phosphate-transfer mechanism set in Dusty PKs, which necessitates a thorough structural scrutiny. Nonetheless, the occurrence of a divergent ePK domain in Dusty proteins does raise a fundamental question related to its evolutionary origin and natural selection. Could Dusty PK act to phosphorylate both protein-serine/threonine and protein-tyrosine via intermolecular catalysis or serve to provide a regulatory platform for a crosstalk between PSK and PTK? Given the diversity in the mode and regulation of various protein kinases [37-40], answers to these challenging questions should yield informative insights into the structure-function relationships of Dusty PKs and their connection with other signaling pathway activities.
Supplementary Material
Acknowledgments
We appreciate Tian Ye for his expertise in fusion protein expression, antibody production and microscopic immunofluorescence staining. We thank Michel Tarnawski for confocal imaging and staff members of the Laboratory of Microchemistry for DNA sequencing.
Abbreviations
- ePK
eukaryotic protein kinase catalytic domain
- BAC
bacterial artificial chromosome
- bp
basepair
- EST
expressed sequence tags
- FISH
fluorescence in situ hybridization
- GFP
green fluorescence protein
- GSP
gene-specific primer
- kb
kilobase
- nt
nucleotide
- ORF
open reading frame
- PCR
polymerase chain reaction
- PSK
protein-serine/threonine kinase
- PTK
protein-tyrosine kinase
- RACE
rapid amplification of cDNA ends
- RT-PCR
reverse transcriptase PCR
- UTR
untranslated region
Footnotes
This work was supported in part by NIH Grant DK67274 and institutional funds. The nucleotide sequences described here have the following GenBank accession numbers: AY208850, AY208851, AY208852, AY208853, AY208854, AY208855, AY364232, AY364233, AY429674, AY429675, AY429676, AY429677, AY429678, AY429679, AY429680, AY429681, AY641092, AY641093, AY641094, AY641095, AY641096, DQ013067, DQ013068, DQ054504, DQ341264, DQ419945, and DQ419946.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kennelly PJ, Krebs EG. Consensus sequences as substrate specificity determinants for protein kinases and protein phosphatases. J. Biol. Chem. 1991;266:15555–15558. [PubMed] [Google Scholar]
- 2.Hunter T. Protein kinases and phosphatases: The Yin and Yang of protein phosphorylation and signaling. Cell. 1995;80:2 25–236. doi: 10.1016/0092-8674(95)90405-0. [DOI] [PubMed] [Google Scholar]
- 3.Pawson T, Scott JD. Protein phosphorylation in signaling – 50 years and counting. Trends Biochem. Sci. 2005;30:286–290. doi: 10.1016/j.tibs.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 4.Hanks SK, Quinn A-M, Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- 5.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 6.Caenepeel S, Charydczak G, Sudarsanam S, Hunter T, Manning G. The mouse kinome: Discovery and comparative genomics of all mouse protein kinases. Proc. Natl. Acad. Sci. U. S. A. 2004;101:11707–11712. doi: 10.1073/pnas.0306880101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445–452. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- 8.Kholodenko BN. Cell-signalling dynamics in time and space. Nat. Rev. Mol. Cell Biol. 2006;7:165–176. doi: 10.1038/nrm1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lindberg RA, Quinn M-A, Hunter T. Dual-specificity protein kinases: will any hydroxyl do? Trends Biochem. Sci. 1992;17:114–119. doi: 10.1016/0968-0004(92)90248-8. [DOI] [PubMed] [Google Scholar]
- 10. http://www.brenda.uni-koeln.de/php/result_flat.php4?ecno=2.7.1.1.112.DUA.
- 11.Hunter T, Plowman GD. The protein kinases of budding yeast: six score and more. Trends Biochem. Sci. 1997;22:18–22. doi: 10.1016/s0968-0004(96)10068-2. [DOI] [PubMed] [Google Scholar]
- 12.Goldberg JM, Manning G, Liu A, Fey P, Pilcher KE, Xu Y, Smith JL. The Dictyostelium kinome – Analysis of the protein kinases from a simple model organism. PloS Genet. 2006;2:291–303. doi: 10.1371/journal.pgen.0020038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Plowman GD, Sudarsanam S, Bingham J, Whyte D, Hunter T. The protein kinases of Caenorhabditis elegans: A model for signal transduction in multicellular organisms. Proc. Natl. Acad. Sci. U. S. A. 1999;96:13603–13610. doi: 10.1073/pnas.96.24.13603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morrison DK, Murakami MS, Cleghon V. Protein kinases and phosphatases in the Drosophila genome. J. Cell Biol. 2000;150:F57–62. doi: 10.1083/jcb.150.2.f57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manning G. Genomic overview of protein kinases. In: WormBook, editor. The C. elegans Research Community. WormBook; 2005. doi/10.1895/wormbook.1.60.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanks SK. Genomic analysis of the eukaryotic protein kinase superfamily: a perspective. Genome Biol. 2003;4:111.1–111.7. doi: 10.1186/gb-2003-4-5-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leonard CL, Aravind L, Koonon EV. Novel families of putative protein kinases in bacteria and archaea: evolution of the “eukaryotic” protein kinase superfamily. Genome Res. 1998;8:1038–1047. doi: 10.1101/gr.8.10.1038. [DOI] [PubMed] [Google Scholar]
- 18.Zha J, Zhou Q, Xu L-G, Chen D, Li L, Zhai Z, Shu H-B. RIP5 is a RIP-homologous inducer of cell death. Biochem. Biophys. Res. Commun. 2004;319:298–303. doi: 10.1016/j.bbrc.2004.04.194. [DOI] [PubMed] [Google Scholar]
- 19.Meylan E, Tschopp J. The RIP kinases: crucial integrators of cellular stress. Trends Biochem. Sci. 2005;30:151–159. doi: 10.1016/j.tibs.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook J, Russel M. Molecular Cloning: A Laboratory Manual. 3rd Ed. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 2001. [Google Scholar]
- 21.Shizuya H, Birren B, Kim U-J, Mancino V, Slepak T, Tachiiri Y, Simon MI. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc. Natl. Acad. Sci. U. S. A. 1992;89:8794–8797. doi: 10.1073/pnas.89.18.8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang G, Tao Y, Zhang W. WordSpy: identifying transcription factor binding motifs by building a dictionary and learning a grammar. Nucleic Acids Res. 2005;33:W412–416. doi: 10.1093/nar/gki492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Obenauer JC, Cantley LC, Yaffe MB. Scansite 2.0: Proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;31:3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Balla S, Thapar V, Verma S, Luong T, Faghri T, Huang C-H, Rajasekaran S, del Campo JJ, Shinn JH, Mohler WA, Maciejewski MW, Gryk MR, Piccirillo B, Schiller SR, Schiller MR. Minimotif Miner: a tool for investigating protein function. Nat. Methods. 2006;3:175–177. doi: 10.1038/nmeth856. [DOI] [PubMed] [Google Scholar]
- 26.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:694–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 28.Felsenstein J. Inferring phylogenies from protein sequences by parsimony, distance, and likelihood methods. Methods Enzymol. 1996;266:418–427. doi: 10.1016/s0076-6879(96)66026-1. [DOI] [PubMed] [Google Scholar]
- 29.Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 30.Yang Z, Bielawski JP. Statistical methods for detecting molecular adaptation. Trends Ecol. Evol. 2000;15:496–503. doi: 10.1016/S0169-5347(00)01994-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar S, Tamura K, Jakobsen IB, Nei M. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics. 2001;17:1244–1245. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- 32.Hui C.-c, Joyner AL. A mouse model of Greig cephalo–polysyndactyly syndrome: the extra–toesJ mutation contains an intragenic deletion of the Gli3 gene. Nat. Genet. 1993;3:241–246. doi: 10.1038/ng0393-241. [DOI] [PubMed] [Google Scholar]
- 33.Harlow E, Lane D. Antibodies: A Laboratory Manual. 1st Ed. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1988. [Google Scholar]
- 34.Kannan N, Newwald AF. Evolutionary constraints associated with functional specificity of the CMGC protein kinases MAPK, CDK, GSK, SRPK, DYRK, and CK2alpha. Protein Sci. 2004;2004;13:2059–2077. doi: 10.1110/ps.04637904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stanger BZ, Leder P, Lee T-H, Kim E, Seed B. RIP: A novel protein containing a death domain that interacts with Fas/APO-1 (CD95) in yeast and causes cell death. Cell. 1995;81:513–523. doi: 10.1016/0092-8674(95)90072-1. [DOI] [PubMed] [Google Scholar]
- 36.Micheau O, Tschopp J. Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell. 2003;114:181–190. doi: 10.1016/s0092-8674(03)00521-x. [DOI] [PubMed] [Google Scholar]
- 37.Kim C, Xuong NH, Taylor SS. Crystal structure of a complex between the catalytic and regulatory (RIα) subunits of PKA. Science. 2005;307:690–696. doi: 10.1126/science.1104607. [DOI] [PubMed] [Google Scholar]
- 38.Lochhead PA, Sibbet G, Morrice N, Cleghon V. Activation-Loop Autophosphorylation is mediated by a novel transitional intermediate form of DYRKs. Cell. 2005;121:925–936. doi: 10.1016/j.cell.2005.03.034. [DOI] [PubMed] [Google Scholar]
- 39.Dar AC, Dever TE, Sicheri F. Higher-order substrate recognition of eIF2α by the RNA-dependent protein kinase PKR. Cell. 2005;122:887–900. doi: 10.1016/j.cell.2005.06.044. [DOI] [PubMed] [Google Scholar]
- 40.Furdui CM, Lew ED, Schlessinger J, Anderson KS. Autophosphorylation of FGFR1 kinase is mediated by a sequential and precisely ordered reaction. Mol. Cell. 2006;21:711–717. doi: 10.1016/j.molcel.2006.01.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.