Abstract
Giardia cysts in 131 raw wastewater samples from Milwaukee, Wis., were genotyped by sequence analysis of the triosephosphate isomerase gene which showed the presence of two distinct genotypes (assemblages A and B) of Giardia duodenalis. Of the 131 samples, 111 belonged to assemblage A, and the remaining samples belonged to assemblage B. A high degree of genetic polymorphism was evident within the assemblage B cluster, with 10 distinct subgenotypes identified, eight of which have not been reported before.
Giardiasis is the most common parasitic disease in the United States. A recent surveillance study of 6 years (January 1992 through December 1997) revealed occurrences of transmission in all major geographic areas of United States, with an estimated incidence of 2.5 million cases annually (12). Giardiasis is also a common cause of diarrheal disease in other mammals (35). Based on host origins, 41 species of Giardia have been named (5, 33). However, the taxonomy of Giardia at the species level is unresolved because only limited morphological differences are present between different species. Thus far, only six species in the genus are considered valid: Giardia duodenalis (synonym G. lamblia or G. intestinalis) in a wide range of mammals, Giardia agilis in amphibians, Giardia muris in rodents, Giardia ardeae and Giardia psittaci in birds, and Giardia microti in muskrats and voles (7, 8, 9, 11, 32).
Waterborne outbreaks of giardiasis are a major public health problem in many industrialized nations, including the United Kingdom, Sweden, Canada, and the United States (20, 26). Human sewage has been considered a source of Giardia cyst contamination in water. In Canada and Italy, a high prevalence (73 to 100%) of Giardia cysts was reported in raw sewage samples (4, 14, 34). The public health importance and contamination sources of Giardia cysts found in water, however, are largely unclear, because very few studies have been carried out to genetically characterize the Giardia cysts in water. Nevertheless, G. duodenalis cysts of assemblage A have been identified in a few clams collected from the Rhode River, a Chesapeake Bay subestuary in Maryland (13).
Recently, we described the development of a Giardia genotyping tool based on a sequence characterization of the triosephosphate isomerase (TPI) gene of G. duodenalis from humans, domestic animals, and wild mammals (29). In this communication, we have used this molecular tool in the detection and differentiation of Giardia spp. in raw wastewater samples from Milwaukee, Wis.
Wastewater samples.
A total of 237 raw wastewater samples were obtained from the Jones Island wastewater treatment plant in Milwaukee between November 1999 and February 2003 (with an average of two to four samples per week and no more than one sample per day). Each sample was a composite from three separate siphons that delivered influent to the treatment plant: domestic sewage, combined domestic and industrial sewage, and diluted domestic sewage overflow. For each siphon, a small quantity (10 to 15 ml), based on the proportioned diurnal flow, was automatically drawn every 15 min and deposited in a 15-liter refrigerated tank on a diurnal flow-proportioned basis. After 24 h, each tank was brought to the laboratory and manually mixed in proportions to the volumes delivered by the three systems. Only 50 ml of the 24-h composite was analyzed for each sample. The 50-ml subsample was concentrated by centrifugation at 1,000 × g for 10 min. Giardia cysts in pellets were isolated by immunomagnetic separation (IMS), with magnetic beads coated with an anti-Giardia monoclonal antibody (Dynal, Inc., Lake Success, N.Y.). The purified Giardia cysts (attached to magnetic beads) were used in DNA extraction.
Some samples were also processed for microscopic detection of Giardia cysts. A portion of each sample (50 ml) was centrifuged at 1,800 × g for 15 min. The supernatant was discarded and the packed pellet volume (≤0.5 ml) was resuspended. Procedures described in the U.S. Environmental Protection Agency method 1623 (30) were used to purify one-half (5 ml) of the resuspended pellet by IMS using magnetic beads (Dynal, Inc.), stained with fluorescein isothiocyanate-labeled anti-Cryptosporidium and anti-Giardia monoclonal antibodies (Merifluor; Meridian Bioscience, Inc.), and examined under a fluorescent microscope. Giardia cysts were identified by fluorescence characteristics, size, and shape and then enumerated. Any discrepancy was resolved by differential interference contrast microscopy.
DNA extraction.
To extract DNA from concentrates of wastewater, the IMS beads with bound Giardia cysts were subjected to five freeze-thaw cycles, incubated at 56°C with 1 mg of proteinase K (Sigma, St. Louis, Mo.) per ml for at least 1 h, and diluted with an equal volume of ACS grade ethanol. DNA was extracted by passing the cyst-ethanol suspension through QIAamp DNA Mini isolation columns (QIAGEN, Valencia, Calif.).
PCR and sequencing analyses.
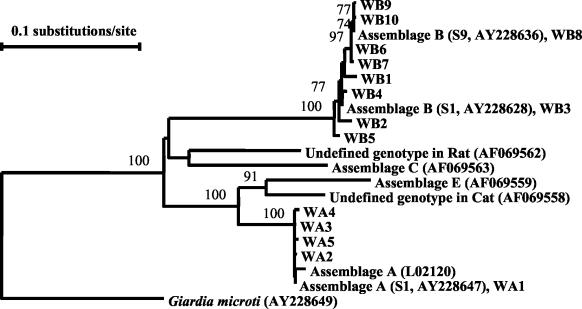
Giardia cysts in samples were identified to the species and genotype (assemblage) level using a previously described nested PCR protocol (29), which amplified a 605-bp fragment of the TPI gene in the primary PCR and a 532-bp fragment in the secondary PCR. The secondary PCR products were purified using Microcon PCR centrifugal filter devices (MILLIPORE, Bedford, Mass.) and sequenced using an ABI 3100 automated sequencer (Perkin Elmer). Sequence accuracy was confirmed by two-directional sequencing of two separate PCR products from each sample. Multiple alignments of the nucleotide sequences were performed using the PILEUP program in the Wisconsin Package version 9.0 programs (Genetics Computers Group, Madison, Wis.). To assess the extent of genetic diversity of Giardia species in samples and their evolutionary relationships to other Giardia species and assemblages, a phylogenetic analysis was carried out. In the analysis, published Giardia TPI nucleotide sequences (GenBank accession no. L02116, L02120, U57897, AF06957 to AF069565, and AY228628 to AY228649) were aligned with the TPI sequences from the samples obtained in this study. A neighbor-joining tree was constructed based on the evolutionary distances calculated by Kimura-2-parameter analysis using the program TreeconW (http://rrna.uia.ac.be/dcse/help/treecon.html). Subsequently, because only assemblages A and B of G. duodenalis were found in the samples, G. microti (GenBank accession no. AY228649) was used as the outgroup because of its genetic relatedness to G. duodenalis (Fig. 1). The reliability of these trees was assessed using the bootstrapping method (10) with 1,000 pseudoreplicates.
FIG. 1.
Formation of two major clusters (assemblages A and B) by G. duodenalis in wastewater in a neighbor-joining analysis of the TPI nucleotide sequences. Assemblage A subgenotypes in wastewater are labeled as WA1 to WA5, and assemblage B subgenotypes in wastewater are labeled as WB1 to WB10. Numbers on branches are bootstrapping values (in percentages) using 1,000 replicates; only values of >70% are shown.
Giardia genotypes in wastewater.
In this study, a total of 237 samples were examined for Giardia spp. by TPI-based diagnostic PCR protocols. Of these, 131 samples (55%) were PCR positive. Fifty-two samples were also processed by Environmental Protection Agency method 1623, all of which were positive by immunofluorescence microscopy. The average number of cysts per 25 ml was 95.3 (range, 13 to 223). All microscopy-positive samples were amplified by PCR. Nucleotide sequences (532 bp) were generated from all PCR-amplified TPI fragments. The extent of genetic diversity was assessed by multiple alignments of the TPI sequences with sequences of known Giardia species and genotypes. This analysis revealed the presence of two major genotypes of G. duodenalis in the samples examined (Table 1).
TABLE 1.
Distribution of Giardia genotypes in wastewater samples from Milwaukee
| Sample collection date | Total no. of samples | No. of positive samples | Assemblage(s) (genotypes) |
|---|---|---|---|
| November 1999 | 3 | 1 | B (1) |
| December 1999 | 2 | 1 | A (1) |
| April 2000 | 6 | 1 | B (1) |
| May 2000 | 10 | 4 | A (4) |
| June 2000 | 12 | 5 | A (4), B (1) |
| July 2000 | 9 | 2 | A (1), B (1) |
| August 2000 | 8 | 1 | A (1) |
| September 2000 | 12 | 5 | A (4), B (1) |
| October 2000 | 15 | 5 | A (3), B (2) |
| November 2000 | 12 | 3 | A (2), B (1) |
| December 2000 | 13 | 11 | A (9), B (2) |
| January 2001 | 14 | 11 | A (9), B (2) |
| February 2001 | 12 | 1 | A (1) |
| March 2001 | 11 | 5 | A (5) |
| April 2001 | 13 | 7 | A (6), B (1) |
| May 2001 | 12 | 4 | A (4) |
| July 2001 | 15 | 10 | A (10) |
| March 2002 | 15 | 2 | A (1), B (1) |
| October 2002a | 5 | 5 | A (5) |
| November 2002a | 12 | 12 | A (8), B (4) |
| December 2002a | 9 | 9 | A (7), B (2) |
| January 2003a | 18 | 18 | A (18) |
| February 2003a | 8 | 8 | A (8) |
These samples were also analyzed by Environmental Protection Agency method 1623 (30) and were identified as positive for Giardia spp. by microscopy.
To confirm the existence of two of the Giardia genotypes in wastewater, a neighbor-joining tree was constructed with published TPI sequences and sequences from this study, with a G. ardeae sequence (GenBank accession no. AF069564) as the outgroup in an initial analysis and a G. microti sequence (GenBank accession no. AY228649) in the secondary analysis. Phylogenetic analysis revealed that all TPI sequences from wastewater were placed in two large clusters, assemblage A and assemblage B, with 111 samples belonging to assemblage A and 20 samples belonging to assemblage B (Fig. 1).
Intragenotypic variations.
Genetic variations in the TPI nucleotide sequences from the samples were evident within both assemblages A and B. By using the GenBank sequence L02120 as the reference sequence, the samples that grouped in assemblage A had 11 single nucleotide polymorphisms (SNPs) (Table 2). Eight of the 11 SNPs occurred at the third position of the codons and were synonymous in nature. Altogether, five distinct types of TPI sequences of assemblage A were evident in the samples, of which four were new subgenotypes. These five types of sequences were named subgenotypes WA1 to WA5, to reflect intragenotypic variations in assemblage A. Of the 111 assemblage A samples, the majority of them (107, subgenotype WA1) were identical to a TPI sequence (GenBank accession no. AY228647) that was obtained from six human samples from Peru (29). The remaining four subgenotypes (WA2, WA3, WA4, and WA5) were represented each by one sample (Table 3).
TABLE 2.
Variation in the TPI nucleotide sequences of subgenotypes of the G. duodenalis assemblage A
| Subgenotypeb | Nucleotide at positiona:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 15 | 27 | 79 | 86 | 96 | 129 | 254 | 363 | 399 | 510 | 546 | |
| L02120 | C | C | G | C | G | T | A | G | C | G | G |
| W1 | T | G | G | C | G | C | A | G | T | G | A |
| W2 | T | G | G | T | G | C | A | G | T | G | A |
| W3 | T | G | G | C | G | C | G | G | T | G | A |
| W4 | T | G | G | C | G | C | A | A | T | A | A |
| W5 | T | G | A | C | A | C | A | G | T | G | A |
Nucleotide position numbers according to L02120, with the beginning of the coding region as position no. 1. Underlined numbers indicate the third codon positions.
WA1, samples 1503, 2030, 2032, 2033, 2038, 2080, 2099, 2106, 2107, 2110, 2234, 2236, 2238, 2242, 2424, 2437, 2467, 2469, 2474, 2593, 2598, 2624, 2633, 2635, 2636, 2680, 2681, 2682, 2683, 2684, 2685, 2822, 2823, 2834, 2835, 3191, 3192, 3193, 3443, 3446, 3447, 3649, 3902, 3903, 3908, 3916, 3917, 3919, 3992, 4010, 4059, 4060, 4062, 4221, 4222, 4223, 4233, 4234, 4240, 4241, 5407, 7090, 7091, 7092, 7093, 7094, 7109, 7111, 7113, 7114, 7158, 7159, 7160, 7326, 7328, 7330, 7331, 7332, 7333, 7334, 7335, 7384, 7385, 7386, 7387, 7388, 7389, 7390, 7391, 7392, 7393, 7394, 7395, 7396, 7397, 7483, 7484, 7485, 7486, 7487, 7488, 7489, 7506, 7507, 7508, 7509, and 7510; WA2, sample 3906; WA3, sample 4220; WA4, sample 4218; WA5, sample 4230.
TABLE 3.
Distribution of subgenotypes of assemblages A and B in the 131 Giardia-positive wastewater samples in Milwaukee
| Assemblage and subgenotype | Total no. | Percentage |
|---|---|---|
| Assemblage A | 111 | 84.7 |
| WA1 | 107 | 81.7 |
| WA2 | 1 | 0.8 |
| WA3 | 1 | 0.8 |
| WA4 | 1 | 0.8 |
| WA5 | 1 | 0.8 |
| Assemblage B | 20 | 15.3 |
| WB1 | 2 | 1.5 |
| WB2 | 1 | 0.8 |
| WB3 | 1 | 0.8 |
| WB4 | 1 | 0.8 |
| WB5 | 1 | 0.8 |
| WB6 | 3 | 2.3 |
| WB7 | 2 | 1.5 |
| WB8 | 5 | 3.8 |
| WB9 | 1 | 0.8 |
| WB10 | 3 | 2.3 |
A very high degree of genetic polymorphism was seen among the limited number of G. duodenalis assemblage B samples. By using the GenBank sequence L02116 as the reference sequence, 26 SNPs were evident within the 20 samples that clustered in assemblage B (Table 4). Twenty-one of the SNPs occurred at the third position of the codons, but only one of the SNPs was nonsynonymous (a change of a glutamic acid to an asparagine at nucleotide position 105). Altogether, 10 types of sequences were obtained and were named subgenotypes WB1 to WB10 to reflect intragenotypic variations in assemblage B. Eight of the subgenotypes were new. Five of the subgenotypes (WB2, WB3, WB4, WB5, and WB9) were represented each by a single sample; two subgenotypes (WB1 and WB7) were represented by two samples each, two subgenotypes (WB6 and B10) had three samples each, and one subgenotype (WB8) had five samples (Table 3). Two of the subgenotypes (WB3 and WB8) were identical to S1 and S9 subgenotypes of assemblage B recently reported for humans (India, Peru, and the United States) and wild mammals (beavers and muskrats) in Maryland, respectively (29).
TABLE 4.
Variation in the TPI nucleotide sequences of subgenotypes of the G. duodenalis assemblage B
| Subgenotypeb | Nucleotide at positiona:
|
|||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 27 | 39 | 45 | 91 | 105 | 111 | 165 | 168 | 198 | 210 | 216 | 240 | 263 | 271 | 280 | 297 | 304 | 333 | 342 | 393 | 402 | 429 | 438 | 471 | 483 | 534 | |
| L02116 | T | A | T | T | G | G | T | T | C | A | C | C | G | C | A | A | A | C | G | C | A | G | T | A | A | T |
| WB1 | G | G | C | T | G | G | C | T | C | G | C | C | G | C | G | G | A | T | T | C | A | G | T | A | A | C |
| WB2 | G | G | C | C | G | A | C | C | C | G | T | C | G | T | A | A | A | C | G | C | A | G | T | C | G | C |
| WB3 | G | G | T | C | G | G | C | C | C | G | C | C | G | C | A | A | A | C | G | C | A | G | T | A | A | C |
| WB4 | G | G | T | C | G | G | C | C | C | G | C | C | G | C | A | A | G | C | G | T | A | G | T | A | A | C |
| WB5 | G | G | T | C | G | G | T | C | T | G | C | T | G | C | A | A | A | C | G | C | G | G | C | A | G | C |
| WB6 | G | A | T | T | G | G | T | T | C | G | C | C | G | C | A | A | A | C | G | C | A | G | T | A | A | C |
| WB7 | G | A | T | T | T | G | T | T | C | G | C | C | G | C | A | A | A | C | G | C | A | G | T | A | A | C |
| WB8 | G | A | T | T | G | G | T | T | C | A | C | C | G | C | A | A | A | C | G | C | A | G | T | A | A | C |
| WB9 | G | A | T | T | G | G | T | T | C | A | C | C | A | C | A | A | A | C | G | C | A | G | T | A | A | C |
| WB10 | G | A | T | T | G | G | T | T | C | A | C | C | G | C | A | A | A | C | G | C | A | A | T | A | A | C |
Nucleotide position numbers according to L02116, with the beginning of the coding region as position no. 1. Underlined numbers indicate the third codon positions.
WB1, samples 2623 and 2634; WB2, sample 2436; WB3, sample 1794; WB4, sample 2434; WB5, sample 3920; WB6, samples 7115, 7116, and 7117; WB7, samples 7327 and 7329; WB8, samples 1418, 2109, 2476, 2824, and 2833; WB9, sample 2100; WB10, samples 2426, 5409, and 7110.
Public health significance.
Two major groups of G. duodenalis have been recognized in humans, assemblages A and B (Table 5). In addition to these two assemblages, phylogenetic analysis of glutamate dehydrogenase, elongation factor 1α, TPI, and small subunit (SSU) rRNA nucleotide sequences have identified three to five other host-adapted lineages of G. duodenalis in cattle, dogs, cats, and rodents (23, 24, 25, 27). Only the two human-pathogenic assemblages were found in wastewater in this study, even though rodents were expected to be present in the wastewater distribution system, indicating that rodents were not significant contributors to Giardia genotypes found in this study. The TPI gene was reported to display the highest degree of polymorphism in G. duodenalis, which allows it to differentiate G. duodenalis at both genotype and intragenotype levels (23, 29). It has a further advantage over the SSU rRNA gene in being able to be easily amplified without the use of special buffers designed for the PCR of GC-rich targets (29). One potential problem of the technique is its limited ability to detect mixed genotypes in environmental samples.
TABLE 5.
Distribution of assemblages A and B in humans in some studies
| Location | No. of positive samples analyzed | Assemblage of samplesa
|
Reference | ||
|---|---|---|---|---|---|
| A | B | Mixed | |||
| Australia | 13 | 13 (1 I, 12 II) | 0 | 0 | 17 |
| 11 | 4 (2 I, 2 II) | 7 (1 III, 6 IV) | 0 | 2 | |
| 4 | 2 (1 I, 1 II) | 2 (1 III, 1 IV) | 0 | 23 | |
| Australia, Cambodia, China, United States | 9 | 5 (3 I, 2 II) | 4 (4 III) | 0 | 21 |
| China | 3 | 0 | 2 (III) | 1 (I + III) | 22 |
| 8 | 4 | 4 | 0 | 36 | |
| Germany | 12 | 11 (5 I, 6 II) | 1 (1 III) | 0 | 18 |
| India | 10 | 0 | 10 | 0 | 29 |
| Italy | 30 | 24 | 6 | 0 | 3 |
| Korea | 7 | 7 | 0 | 0 | 36 |
| Mexico | 22 | 22 (22 II) | 0 | 0 | 28 |
| 26 | 26 (26 I) | 0 | 0 | 6 | |
| New Zealand | 5 | 5 (1 I, 4 II) | 0 | 0 | 19 |
| Peru | 25 | 6 | 19 | 0 | 29 |
| The Netherlands | 18 | 9 | 9 | 0 | 16 |
| 24 | 12 (Polish) | 12 (Belgian) | 0 | 15 | |
| United Kingdom | 33 | 9 | 21 | 3 (A + B) | 1 |
| United States | 2 | 0 | 2 | 0 | 29 |
For assemblage A, the numbers of group I, group II, and Polish samples are given in parentheses. For assemblage B, the numbers of group III, group IV, and Belgian samples are given.
The two human-pathogenic genotypes (assemblages A and B) of G. duodenalis are distributed worldwide (Table 5). The distribution of each genotype in humans, however, differs according to the studies and geographic locations. For example, in a study in Mexico, all 22 samples and derived clones from humans belonged to the restriction fragment length polymorphism (RFLP) subtype II of the assemblage A, with a complete lack of assemblage B. Biological or geographic factors were attributed to the predominance of one subtype in the study area, but the tool used divides assemblage A only to two subtypes (28). Another study conducted in Mexico had also shown that all 26 homogeneous isolates from humans belonged to assemblage A (6). Similarly, all seven human isolates in Korea characterized at the SSU rRNA locus were from assemblage A (36). However, several studies conducted in India, Peru, and the United Kingdom showed that assemblage B was responsible for more human infections than assemblage A (Table 5).
In recent years, several attempts have been made to detect and differentiate Giardia spp. in environmental samples by using PCR techniques (4, 31, 34). The distribution of Giardia spp. in environmental samples is likely dependent on human, agricultural, and wildlife activities. Two human-pathogenic genotypes of G. duodenalis were identified in 14 environmental samples (assemblage A in 10 and assemblage B in 4) from Canada by using a SSU rRNA-based PCR-RFLP protocol (31, 34). In Italy, 16 samples from four wastewater plants were analyzed by a beta-giardin-based PCR-RFLP method; assemblage A was found in eight of the samples, and both assemblages A and B were found in the remaining eight samples (4). The predominance of assemblage A in wastewater in Italy is in agreement with the genotype distribution in humans in the same country (3) (Table 5).
In the present study with a much larger sample size, both human-pathogenic genotypes (assemblages A and B) were found in 131 wastewater samples. The majority (84.7%) of the samples (111 samples) belonged to G. duodenalis assemblage A, which had five distinct subgenotypes (WA1 to WA5). However, one subgenotype (WA1) accounted for most of the assemblage A samples (107 of 111), indicating that humans in Milwaukee were infected with subgenotype WA1. This subgenotype was identical to a sequence previously obtained from a cultured isolate from humans (assemblage A, group II; GenBank accession no. AF069557) in Australia (23) and from six fecal samples from humans (GenBank accession no. AY228647) in Peru (29). The significance of a predominant subgenotype in Milwaukee is not clear. It is tempting to conclude a common source of human infection was responsible for the wide occurrence of subgenotype WA1. In contrast, 20 assemblage B samples had a high genetic diversity (with 10 distinct subgenotypes), indicating that it is unlikely that the transmission of Giardia infection in Milwaukee is restricted to one source. It is likely that assemblage A parasites are more conserved and evolve more slowly than assemblage B parasites. It is also possible that subgenotype WA1 of assemblage A is more infectious to humans than other Giardia parasites. Recently, it has been reported that assemblage B was seen in patients with persistent diarrhea, whereas assemblage A was seen mostly in patients with intermittent diarrhea (16).
Nucleotide sequence accession numbers.
Distinct nucleotide sequences of the TPI gene of G. duodenalis in wastewater were deposited in the GenBank database under accession no. AY368157 to AY368171.
Acknowledgments
This study was supported in part by a research grant from the AWWA Research Foundation.
We thank Birhane Dashew, Jeff MacDonald, and Sanjib Bhattacharya for their assistance in sample collection and Clem Ng for microscopic examination.
REFERENCES
- 1.Amar, C. F., P. H. Dear, S. Pedraza-Diaz, N. Looker, E. Linnane, and J. McLauchlin. 2002. Sensitive PCR-restriction fragment length polymorphism assay for detection and genotyping of Giardia duodenalis in human feces. J. Clin. Microbiol. 40:446-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrews, R. H., P. T. Monis, P. L. Ey, and G. Mayrhofer. 1998. Comparison of the levels of intra-specific genetic variation within Giardia muris and Giardia intestinalis. Int. J. Parasitol. 8:1179-1185. [DOI] [PubMed] [Google Scholar]
- 3.Caccio, S. M., M. De Giacomo, and E. Pozio. 2002. Sequence analysis of the beta-giardin gene and development of a polymerase chain reaction-restriction fragment length polymorphism assay to genotype Giardia duodenalis cysts from human fecal samples. Int. J. Parasitol. 32:1023-1030. [DOI] [PubMed] [Google Scholar]
- 4.Caccio, S. M., M. De Giacomo, F. A. Aulicino, and E. Pozio. 2003. Giardia cysts in wastewater treatment plants in Italy. Appl. Environ. Microbiol. 69:3393-3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell, S. R., H. van Keulen, S. L. Erlandsen, J. B. Senturia, and J. L. Jarrol. 1990. Giardia sp.: comparison of electrophoretic karyotypes. Exp. Parasitol. 71:470-482. [DOI] [PubMed] [Google Scholar]
- 6.Cedillo-Rivera, R., J. M. Darby, J. A. Enciso-Moreno, G. Ortega-Pierres, and P. L. Ey. 2003. Genetic homogeneity of axenic isolates of Giardia intestinalis derived from acute and chronically infected individuals in Mexico. Parasitol. Res. 90:119-123. [DOI] [PubMed] [Google Scholar]
- 7.Erlandsen, S. L., and W. L. Bemrick. 1987. SEM evidences for a new species Giardia psittaci. J. Parasitol. 73:623-629. [PubMed] [Google Scholar]
- 8.Erlandsen, S. L., W. J. Bemrick, C. L. Wellis, D. E. Feely, L. Kundson, S. R. Cambell, H. van Keulen, and E. L. Jarrol. 1990. Axenic culture and characterization of Giardia ardeae from the great blue heron (Ardea herodias). J. Parasitol. 76:717-724. [PubMed] [Google Scholar]
- 9.Feely, D. E. 1988. Morphology of the cyst of Giardia microti by light and electron microscopy. J. Protozool. 35:52-54. [DOI] [PubMed] [Google Scholar]
- 10.Felsenstein, J. 1985. Confidence limits on the phylogenies: an approach using bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 11.Filice, F. P. 1952. Studies on the cytology and life history of a Giardia from the laboratory rat. Univ. Calif. Publ. Zool. 57:53-146. [Google Scholar]
- 12.Furness, B. W., M. J. Beach, and J. M. Roberts. 2000. Giardiasis surveillance—United States, 1992-1997. Morb. Mortal. Wkly. Rep. 49:1-13. [PubMed] [Google Scholar]
- 13.Graczyk, T. K., R. C. Thompson, R. Fayer, P. Adams, U. M. Morgan, and E. J. Lewis. 1999. Giardia duodenalis cysts of genotype A recovered from clams in the Chesapeake Bay subestuary, Rhode River. Am. J. Trop. Med. Hyg. 61:526-529. [DOI] [PubMed] [Google Scholar]
- 14.Heitman, T. L., L. M. Frederick, J. R. Viste, N. J. Guselle, U. M. Morgan, R. C. Thompson, and M. E. Olson. 2002. Prevalence of Giardia and Cryptosporidium and characterization of Cryptosporidium spp. isolated from wildlife, human, and agricultural sources in the North Saskatchewan River Basin in Alberta, Canada. Can. J. Microbiol. 48:530-541. [DOI] [PubMed] [Google Scholar]
- 15.Homan, W. L., M. Gilsing, H. Bentala, L. Limper, and F. van Knapen. 1998. Characterization of Giardia duodenalis by polymerase-chain-reaction fingerprinting. Parasitol. Res. 84:707-714. [DOI] [PubMed] [Google Scholar]
- 16.Homan, W. L., and T. G. Mank. 2001. Human giardiasis: genotype linked differences in clinical symptomatology. Int. J. Parasitol. 31:822-826. [DOI] [PubMed] [Google Scholar]
- 17.Hopkins, R. M., B. P. Meloni, D. M. Groth, J. D. Wetherall, J. A. Reynoldson, and R. C. Thompson. 1997. Ribosomal RNA sequencing reveals differences between the genotypes of Giardia isolates recovered from humans and dogs living in the same locality. J. Parasitol. 83:44-51. [PubMed] [Google Scholar]
- 18.Karanis, P., and P. L. Ey. 1998. Characterization of axenic isolates of Giardia intestinalis established from humans and animals in Germany. Parasitol. Res. 84:442-449. [DOI] [PubMed] [Google Scholar]
- 19.Learmonth, J. J., G. Ionas, A. B. Pita, and R. S. Cowie. 2003. Identification and genetic characterisation of Giardia and Cryptosporidium strains in humans and dairy cattle in the Waikato region of New Zealand. Water Sci. Technol. 47:21-26. [PubMed] [Google Scholar]
- 20.Ljungstrom, I., and B. Castor. 1992. Immune response to Giardia lamblia in a water-borne outbreak of giardiasis in Sweden. J. Med. Microbiol. 36:347-352. [DOI] [PubMed] [Google Scholar]
- 21.Lu, S., J. Wen, J. Li, and F. Wang. 2002. DNA sequence analysis of the triose phosphate isomerase gene from isolates of Giardia lamblia. Chin. Med. J. 115:99-102. [PubMed] [Google Scholar]
- 22.Lu, S. Q., A. C. Baruch, and R. D. Adam. 1998. Molecular comparison of Giardia lamblia isolates. Int. J. Parasitol. 28:1341-1345. [DOI] [PubMed] [Google Scholar]
- 23.Monis, P. T., R. H. Andrews, G. Mayrhofer, and P. L. Ey. 1999. Molecular systematics of the parasitic protozoan Giardia intestinalis. Mol. Biol. Evol. 16:1135-1144. [DOI] [PubMed] [Google Scholar]
- 24.Monis, P. T., R. H. Andrews, G. Mayrhofer, J. Kulda, J. L. Isaac-Renton, and P. L. Ey. 1998. Novel lineages of Giardia intestinalis identified by genetic analysis of organisms isolated from dogs in Australia. Parasitology 116:7-19. [DOI] [PubMed] [Google Scholar]
- 25.Monis, P. T., G. Mayrhofer, R. H. Andrews, W. L. Homan, L. Limper, and P. L. Ey. 1996. Molecular genetic analysis of Giardia intestinalis isolates at the glutamate dehydrogenase locus. Parasitology 112:1-12. [DOI] [PubMed] [Google Scholar]
- 26.Moore, A. C., B. L. Herwaldt, G. F. Craun, R. L. Calderon, A. K. Highsmith, and D. D. Juranek. 1993. Surveillance for waterborne disease outbreaks—United States, 1991-1992. Morb. Mortal. Wkly. Rep. CDC Surveill. Summ. 42:1-22. [PubMed] [Google Scholar]
- 27.Mowatt, M. R., E. C. Weinbach, T. C. Howard, and T. T. Nash. 1994. Complementation of Escherichia coli glycolysis mutant by Giardia lamblia triosephosphate isomerase. Exp. Parasitol. 78:85-92. [DOI] [PubMed] [Google Scholar]
- 28.Ponce-Macotela, M., M. N. Martinez-Gordillo, R. M. Bermudez-Cruz, P. M. Salazar-Schettino, G. Ortega-Pierres, and P. L. Ey. 2002. Unusual prevalence of the Giardia intestinalis A-II subtype amongst isolates from humans and domestic animals in Mexico. Int. J. Parasitol. 32:1201-1202. [DOI] [PubMed] [Google Scholar]
- 29.Sulaiman, I. M., R. Fayer, C. Bern, R. H. Gilman, J. M. Trout, P. M. Schantz, P. Das, A. A. Lal, and L. Xiao. Triosephosphate isomerase gene characterization and potential zoonotic transmission of multispecies Giardia duodenalis. Emerg. Infect. Dis. 9:1444-1452. [DOI] [PMC free article] [PubMed]
- 30.U.S. Environmental Protection Agency. April 2001. Method 1623: Cryptosporidium and Giardia in water by IFA/IMS/FA. EPA 821-R-01-025. Office of Water, U.S. Environmental Protection Agency, Washington, D.C.
- 31.van Keulen, H., P. T. Macechko, S. Wade, S. Schaaf, P. M. Wallis, and S. L. Erlandsen. 2002. Presence of human Giardia in domestic, farm and wild animals, and environmental samples suggests a zoonotic potential for giardiasis. Vet. Parasitol. 108:97-107. [DOI] [PubMed] [Google Scholar]
- 32.van Keulen, H., D. E. Feely, T. Macechko, E. L. Jarrol, and S. L. Erlandsen. 1998. The sequence of Giardia small subunit rRNA shows that voles and muskrats are parasitized by a unique species Giardia microti. J. Parasitol. 84:294-300. [PubMed] [Google Scholar]
- 33.van Keulen, H., R. Gutell, M. Gates, S. Campbell, S. L. Erlandsen, E. L. Jarrol, J. Kulda, and E. A. Meyer. 1993. Unique phylogenetic position of Diplomonadida based on the complete small subunit ribosomal RNA sequence of Giardia ardeae, Giardia muris, Giardia duodenalis and Hexamita sp. FASEB J. 7:223-231. [DOI] [PubMed] [Google Scholar]
- 34.Wallis, P. M., S. L. Erlandsen, J. L. Isaac-Renton, M. E. Olson, W. J. Robertson, and H. van Keulen. 1996. Prevalence of Giardia cysts and Cryptosporidium oocysts and characterization of Giardia spp. isolated from drinking water in Canada. Appl. Environ. Microbiol. 62:2789-2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiao, L. 1994. Giardia infection in farm animals. Parasitol. Today 10:436-438. [DOI] [PubMed] [Google Scholar]
- 36.Yong, T. S., S. J. Park, U. W. Hwang, H. W. Yang, K. W. Lee, D. Y. Min, H. J. Rim, Y. Wang, and F. Zheng. 2000. Genotyping of Giardia lamblia isolates from humans in China and Korea using ribosomal DNA sequences. J. Parasitol. 86:887-891. [DOI] [PubMed] [Google Scholar]