Abstract
Chronic periodontitis is an inflammatory disease of the periodontium affecting nearly 65 million adults in the United States. Changes in subgingival microbiota have long been associated with chronic periodontitis. Recent culture-independent molecular studies have revealed the immense richness and complexity of oral microbial communities. However, data sets across studies have not been directly compared, and whether the observed microbial variations are consistent across different studies is not known. Here, we used 16S rRNA sequencing to survey the subgingival microbiota in 25 subjects with chronic periodontal disease and 25 healthy controls and compared our data sets with those of three previously reported microbiome studies. Consistent with data from previous studies, our results demonstrate a significantly altered microbial community structure with decreased heterogeneity in periodontal disease. Comparison with data from three previously reported studies revealed that subgingival microbiota clustered by study. However, differences between periodontal health and disease were larger than the technical variations across studies. Using a prediction score and applying five different distance metrics, we observed two predominant clusters. One cluster was driven by Fusobacterium and Porphyromonas and was associated with clinically apparent periodontitis, and the second cluster was dominated by Rothia and Streptococcus in the majority of healthy sites. The predicted functional capabilities of the periodontitis microbiome were significantly altered. Genes involved in bacterial motility, energy metabolism, and lipopolysaccharide biosynthesis were overrepresented in periodontal disease, whereas genes associated with transporters, the phosphotransferase system, transcription factors, amino acid biosynthesis, and glycolysis/gluconeogenesis were enriched in healthy controls. These results demonstrate significant alterations in microbial composition and function in periodontitis and suggest genes and metabolic pathways associated with periodontal disease.
INTRODUCTION
Periodontal disease is a common disease affecting many adults in the United States. If left untreated, chronic periodontitis (CP) can lead to serious problems such as tooth loss. Indeed, periodontal disease is the number one cause of tooth loss in the United States, accounting for half of all tooth loss in U.S. adults (1). There is considerable evidence that the clinical impact of periodontal disease extends beyond the oral cavity (2). Strong associations have been found between periodontitis and a number of systemic conditions, including cardiovascular disease (3, 4), preterm delivery and low-birth-weight babies (5), diabetes mellitus (6–8), and rheumatoid arthritis (9–11).
The activity of periodontal disease is determined by a complex interplay between the immune system and periodontal pathogens (12, 13). Alterations in subgingival microbiota have long been associated with the development and progression of periodontitis (14). In susceptible individuals, perturbations in host homeostasis are induced by changes in the polymicrobial community, with several microorganisms frequently being associated with periodontal lesions. These microorganisms include the red complex, consisting of Tannerella forsythia, Porphyromonas gingivalis, and Treponema denticola, and the orange complex, consisting of Fusobacterium nucleatum subspecies, Fusobacterium periodonticum subspecies, Prevotella intermedia, Prevotella nigrescens, and Micromonas micros (15). While the presence of these putative periodontal pathogens in periodontal pockets correlates with clinical measures of periodontal disease, the extremely complex microbial populations in the oral cavity, which include many uncultivable or unnamed species, suggest that additional organisms may be involved (16). Indeed, thanks to recent advances in high-throughput sequencing technologies, >700 oral bacterial species have now been identified (17). Many of these bacteria are uncultivable, and their impact on periodontal health remains unexplored.
Over the past 3 decades, numerous studies have investigated the polymicrobial communities in subgingival pockets in an attempt to elucidate the role of microbiota in periodontal disease (15, 17–20). Early studies using culture-based techniques were labor-intensive (21), and many organisms remained uncultivable despite improvements in culturing techniques (22). Subsequent molecular approaches based on DNA-DNA hybridization and microarray techniques substantially improved our understanding of these microbial communities (23–27). However, since knowledge of nucleic acid signatures or hybridization probes are required a priori, these methods precluded detailed analyses of entire microbial communities. Recently, culture-independent molecular methods targeting small-subunit rRNA sequences (16S rRNA) in combination with high-throughput DNA sequencing have emerged as a popular approach to profile microbial flora from diverse sites of the human body. By amplification and deep sequencing of 16S rRNA gene segments, this approach has provided an unprecedented view of human-associated microbial communities and has uncovered novel or previously unrecognized microbes associated with health and disease (16, 28–31). The Human Microbiome Project (HMP) data set is currently the largest reference data set (143 individuals) for the subgingival microbiome (32). However, because this data set was generated from individuals with minimal or no periodontal disease, the entire spectrum of the subgingival microbiota was not captured. Several groups have since used the 16S rRNA and metagenomic approaches to survey subgingival microbiota from subjects recruited in the community (30, 33). These “community” data sets have demonstrated a difference between periodontitis-associated and health-associated microbiota in both the overall microbial community structure (30, 31) and the predicted functional capabilities (34, 35). A number of bacterial taxa and genes were found to be differentially abundant between health and disease. However, data sets across studies have not been directly compared, and whether the observed microbial variations are consistent across different studies is not known.
The goal of this study was to compare the subgingival microbiomes between subjects with chronic periodontitis and healthy controls (HCs) and to consolidate our data with data from previous studies to determine whether differences in subgingival microbiota between health and disease are conserved across studies. We used 16S rRNA deep sequencing to analyze subgingival microbiota from 25 subjects with chronic periodontal disease and 25 periodontally healthy controls and compared our data set with previously reported data sets from the HMP, Griffen et al. (30), and Abusleme et al. (31). In addition, we used PICRUSt (phylogenetic investigation of communities by reconstruction of unobserved states) (36), a bioinformatics tool designed to predict metagenome functional content from marker gene surveys and full genomes, to predict functional compositions of subgingival metagenomes in healthy and diseased individuals.
MATERIALS AND METHODS
Subjects.
A total of 50 subjects were entered into the study, 25 with chronic periodontitis and 25 without evidence of periodontal disease (healthy controls). Study participants were recruited from the Periodontal Disease Research Center at the University of Florida, Gainesville, FL, over a 6-month period between August 2010 and January 2011. Clinical evaluation and data, including pocket depths, were collected by a single periodontist. Exclusion criteria included a history of systemic diseases that could interfere with clinical characteristics, incidence, or progression of periodontal disease; periodontal treatment within the previous 6 months; and chronic treatment with any medication known to affect periodontal status within the previous 3 months (i.e., antibiotics, nonsteroidal anti-inflammatory drugs [NSAIDs], and contraceptives). Clinical diagnosis and selection of subjects were based on clinical and radiographic criteria proposed by the American Academy of Periodontology (37). Two sites were sampled for each of the 50 subjects. Samples from the healthy group had a clinical attachment loss (CAL) of ≤3 mm, whereas samples from the CP group had a CAL of ≥5 mm. Pertinent information concerning the study protocol was explained to each subject, and informed consent was obtained from all participants as required by the study protocol approved by the University of Florida Institutional Review Board.
Sample collection.
Subgingival microbiota was collected from two different sites in each subject by inserting a sterile absorbent paper point to the depth of the sulcus and moving it laterally along the surface of the tooth and the sulcular epithelial lining. The paper point sample was placed directly into a bead-beating tube containing 750 μl buffer (PowerSoil DNA extraction kit; Mobio, Carlsbad, CA) and stored at −20°C until further processing. The probing depths of subgingival pockets were 6.8 ± 1.1 mm for the 50 diseased sites (24 sites at 6 mm, 18 sites at 7 mm, and 8 sites at ≥8 mm) and ≤3 mm for all 50 healthy sites.
Sample preparation and PCR amplification.
Genomic DNA was extracted from each sample by using the Mobio (Carlsbad, CA) PowerSoil DNA extraction kit according to the manufacturer's instructions. For each sample, the bacterial 16S rRNA V1-V3 gene segment was amplified in quadruplicates by using composite primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 534R (5′-ATTACCGCGGCTGCTGG-3′). The forward and reverse primers contained a Lib-A unidirectional Titanium tag sequence required for Roche/454 sequencing, and each reverse primer also included a unique barcode to allow multiplex deep sequencing. Each 20-μl PCR mixture contained 2 μl of the purified DNA template, 1× Accuprime PCR buffer II, 5 μM the forward primer, 5 μM the reverse primer, and 1 U of Accuprime Taq High Fidelity polymerase. PCR amplification was performed as follows: a denaturation step at 95°C for 30 s followed by 25 cycles of denaturation at 95°C for 30 s, annealing at 56°C for 30 s, and extension at 68°C for 5 min.
Sample pooling and pyrosequencing.
Four replicates of barcoded PCR products for each sample were pooled and analyzed on a 1% SYBR Safe (Invitrogen, Carlsbad, CA) agarose gel. Gel slices containing amplicons of the expected size (∼600 bp) were excised and purified by using the Qiagen gel extraction kit (Qiagen, Valencia, CA). Purified PCR products were quantified by using a Qubit HS DNA quantification kit (Invitrogen, Carlsbad, CA) and pooled with an equal molar concentration. The use of barcodes allowed multiplexing and sequencing on the Roche/454 pyrosequencing platform in five pools, generating a total of 406,502 raw reads. After trimming of primer and barcode sequences and quality control (see the supplemental material), 352,682 V1-V3 16S rRNA sequence reads were available for analysis (averaging 3,343 ± 1,925 reads per sample and an amplicon length of 495 nucleotides [nt]). Good's coverage estimates were >0.95 for all samples (0.9905 ± 0.0081). Rarefaction analysis indicates that the sampling effort was sufficient for the majority of the samples, and further sampling would yield few additional operational taxonomic units (OTUs) (see Fig. S1 in the supplemental material).
Sequence analysis.
Pyrosequence reads were demultiplexed based on barcode and reverse primer 534R sequences by using custom scripts. Barcode and primer sequences were removed and trimmed to a minimum length of 423 bp. Trimmed reads were compared against the Human Oral Microbiome Database (HOMD) (38) by using USEARCH (39) with ≥97% sequence identity and ≥80% alignable query criteria. OTUs were formed by collapsing hits by their human oral taxon (HOT) identification. Additional statistical, alpha diversity, and beta diversity analyses were performed by using linear discriminant analysis coupled with effect size measurements (LEfSe) (40), R, and Qiime (41). Enterotype analysis was implemented in R. Prediction of metagenome functional content was performed based on 16S rRNA sequences by using the PICRUSt software package (36).
Comparison of microbiome data sets.
Raw sequencing reads were obtained from three other studies: the HMP (42), Griffen et al. (30), and Abusleme et al. (31). The reads were downloaded from the HMP DACC website and the Sequence Read Archive (SRA) (SRA accession number SRP009299 for data reported by Griffen et al. and SRA accession number SRP012422 for data reported by Abusleme et al.). Because of the differences in amplified 16S rRNA regions and read lengths across studies, all raw sequence reads were reprocessed. The common region V1-V2 (27F-342R) shared among the four studies was used for direct comparison. The filtering step was adjusted by using the following criteria: average per-base quality score of 25, no ambiguous bases, and a minimum length of 200 bp. Reprocessed reads were then integrated into our existing pipeline.
Nucleotide sequence accession number.
The reads are available in the SRA database under BioProject accession number PRJNA269205.
RESULTS
Microbial community structure.
A total of 457 operational taxonomic units (OTUs) were identified from 100 subgingival microbial communities (50 communities in the CP group and 50 communities in the control group), which belonged to 122 different genera in 10 different phyla. Subgingival microbial communities between sites within subjects were more similar to one another than sites between subjects (Jaccard similarity index) (see Fig. S2 in the supplemental material). In the following analyses, microbial communities in different sites within each subject were treated independently. Analysis of microbial diversity, composition, and structure using pooled data sets yielded similar results.
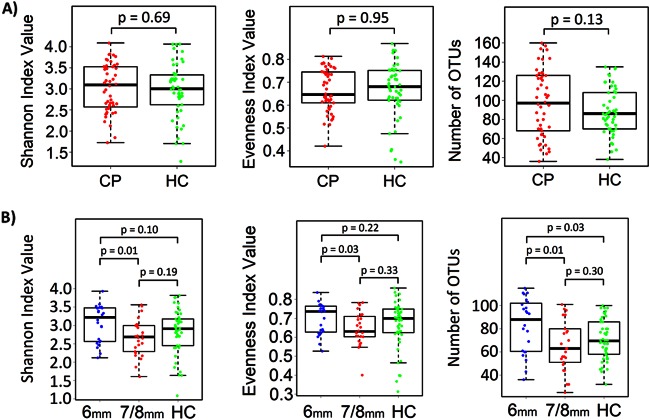
We first compared microbial diversity, evenness, and species richness of subgingival microbial communities between CP and healthy control (HC) subjects. Microbial diversity (determined by the Shannon diversity index) was not significantly different between the diseased and control sites (P = 0.69 by Student's t test) (Fig. 1A). Similarly, no significant difference in evenness or species richness (number of OTUs) was observed. When CP sites were stratified by pocket depth, the Shannon diversity index, species richness, and evenness values were significantly higher in 6-mm pockets than in 7-mm/8-mm pockets (P ≤ 0.05) (Fig. 1B). Compared to healthy sites, species richness but not diversity was significantly high in 6-mm pockets (P = 0.03). No significant difference in species richness, diversity, or evenness was observed between 7-mm/8-mm pockets and healthy sites (P > 0.05).
FIG 1.
Microbial diversity, evenness, and richness in subgingival microbiota in subjects with chronic periodontitis and healthy controls, shown in box plots. (A) The Shannon diversity index was used to estimate microbial diversity for each group. The species evenness index was calculated by using the formula J′ = H′/H′max, where H′ is the Shannon diversity index and H′max is the maximal value of H′ (i.e., ln S, where S is the total number of species in the community). Species richness was defined as the number of OTUs identified in each sample. Each point represents an individual subgingival sample. (B) CP sites were stratified by pocket depths (6 mm or 7 mm/8 mm). P values (Student's t test) are shown above the bars for each comparison.
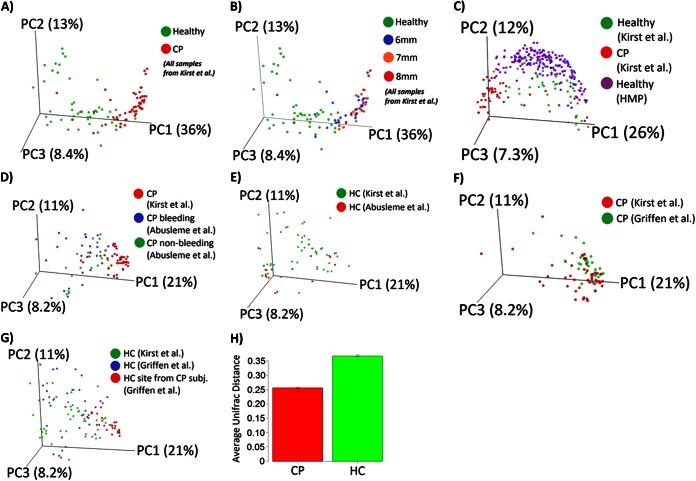
Next, we applied the UniFrac method to compare the degree of phylogenetic overlap in the overall microbial community structures between the CP and HC groups. In both weighted and unweighted UniFrac analyses, subgingival microbial communities in the CP group clustered separately from those in the HC group (Fig. 2A). The separation between samples along the first principal coordinate (PC1) explains ∼36% of the microbiome variations, likely reflecting differences in clinical phenotype. Compared to deep sites (7 mm and ≥8 mm), the shallow sites (6-mm pockets) clustered closely with the HC group (Fig. 2B). This separation according to clinical phenotype was also observed when data sets reported by the HMP (healthy subjects only), Abusleme et al. (31), and Griffen et al. (30) (both healthy and periodontitis groups) were included in the analysis (Fig. 2C to G). The average weighted, normalized UniFrac distance between pairs of samples within the CP group was significantly lower than that in the HC group (Fig. 2H), indicating a decrease in microbial heterogeneity in subjects with chronic periodontitis compared to HCs.
FIG 2.
Comparison of subgingival microbial community composition. Weighted UniFrac analysis was used to generate distances among different samples. Scattered plots were then generated by using principal coordinate analysis. The percentage of variation explained by each principal coordinate (PC) is indicated on the axes. Each point represents a microbial community. (A) Microbial communities in subjects with chronic periodontitis versus healthy controls. (B) Microbial communities in subjects with chronic periodontitis at pocket depths of 6 mm, 7 mm, and 8 mm versus healthy controls. (C) Subgingival microbial communities in this study versus the subgingival data set from the Human Microbiome Project. (D) Bleeding sites and nonbleeding sites from the study by Abusleme et al. (31) versus sites from subjects with chronic periodontitis in this study. (E) Healthy sites from the study by Abusleme et al. versus healthy sites from this study. (F) Chronic periodontitis sites from the study by Griffen et al. (30) versus CP sites from this study. (G) Healthy sites from subjects with chronic periodontitis and healthy sites from healthy controls from the study by Griffen et al. versus healthy sites from this study. (H) Average UniFrac distance between pairs of samples within each group, indicating lower heterogeneity in subgingival microbial communities in the chronic periodontitis group. Error bars indicate standard errors of the means.
Subgingival microbial composition.
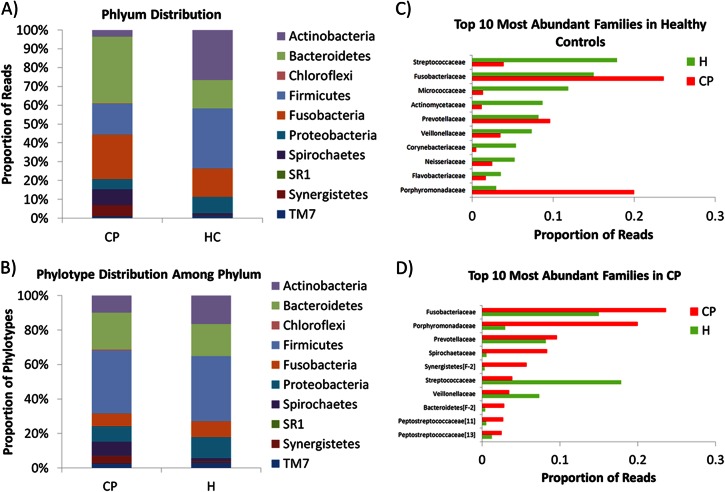
The aggregate subgingival microbiota in healthy sites was dominated by Actinobacteria (26.6%) and Firmicutes (31.9%), while Bacteroidetes (15.1%), Fusobacteria (15.0%), and Proteobacteria (8.8%) were less abundant (Fig. 3A). Members of the Spirochaetes, TM7, Synergistetes, and SR1 phyla were minor constituents of the healthy subgingival microbiota (each <1.6%), and Chloroflexi sequences were completely absent. In the chronic periodontitis group, Bacteroidetes (35.4%) and Synergistetes (6.0%) were more abundant (P < 0.0001 by multivariate analysis). In contrast, the relative proportions of bacterial phylotypes for most phyla did not differ significantly (P > 0.3) (Fig. 3B). At the family level, Streptococcaceae, Fusobacteriaceae, Micrococcaceae, and Actinomycetaceae were dominant members of the healthy subgingival microbiota (17.9%, 15.0%, 11.9%, and 8.7%, respectively), constituting a combined average of 53.5% among healthy samples (Fig. 3C). In the chronic periodontitis group, 61.6% of reads belonged to members of the Fusobacteriaceae, Porphyromonadaceae, Prevotellaceae, and Spirochaetaceae (Fig. 3D). Of the 457 total OTUs identified, 79 OTUs (17.3%) were unique to CP sites, 34 OTUs (7.4%) were unique to healthy sites, and 344 (75.3%) were shared between the two groups. The majority of OTUs unique to healthy sites (22 of 34, or 64.7%) were members of the Proteobacteria, while the remaining unique OTUs belonged to the Firmicutes, Bacteroidetes, and Actinobacteria (8, 3, and 1 OTUs, respectively). In contrast, 18 of 79 OTUs (22.8%) unique to chronic periodontitis sites belonged to the Spirochaetes phylum, and 27 and 18 OTUs were members of the Firmicutes and Bacteroidetes, respectively.
FIG 3.
Relative abundance of bacterial taxa at the phylum level in subjects with chronic periodontitis (CP) and healthy controls (HC). (A) Relative proportion of sequence reads for each phylum. (B) Relative proportions of bacterial phylotypes for each phylum. (C) Top 10 most abundant families in HCs. (D) Top 10 most abundant families in subjects with CP.
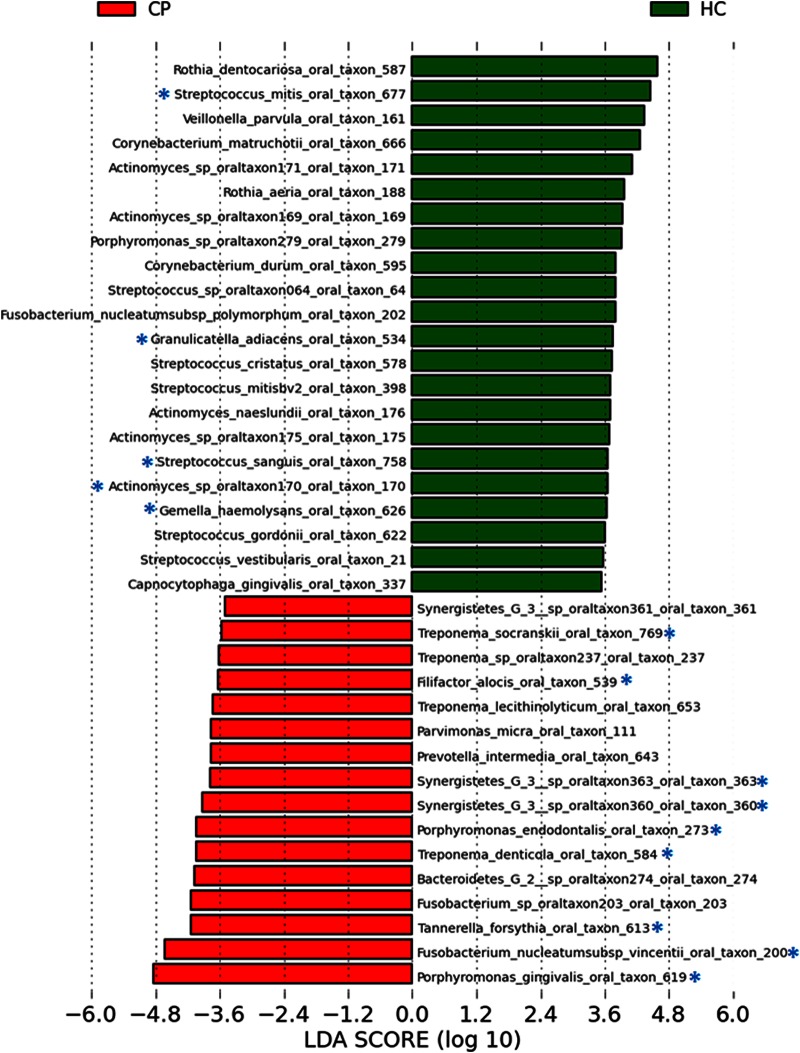
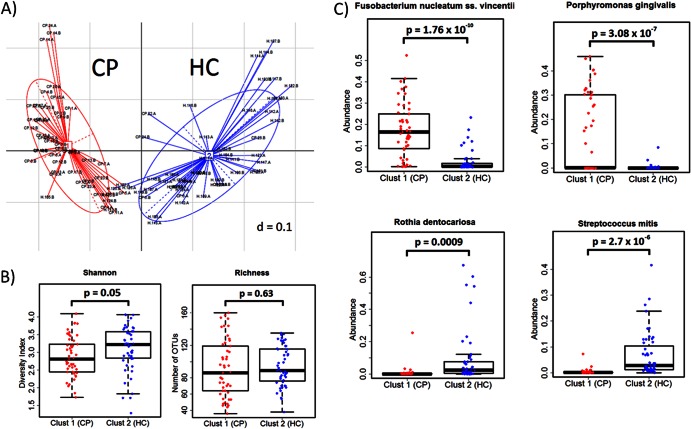
To identify bacterial taxa that were differentially abundant between the two groups, we used LEfSe (linear discriminant analysis coupled with effect size measurements) and a multivariate approach. LEfSe identified 36 OTUs (of 457 total OTUs [7.9%]) differentially enriched in the chronic periodontitis group and 50 OTUs (10.9%) enriched in the HC group. The multivariate approach (43) identified 19 OTUs as being significantly more abundant in periodontitis patients and 5 OTUs as being significantly more abundant in healthy controls (Table 1). Eighteen OTUs were identified by both methods as being significantly abundant in chronic periodontitis, and five OTUs (Actinomyces oral taxon 170, Streptococcus mitis, Streptococcus sanguis, Gemella haemolysans, and Granulicatella adjacens) were strongly associated with periodontal health (Fig. 4). These five taxa combined represented 10.3% of the total reads in HCs (∼76 to 100% of sites) but only 1.5% of the total reads in the CP group (ranging from 16 to 72% of CP sites).
TABLE 1.
Subgingival phylotypes significantly enriched in subjects with chronic periodontitis and healthy controls
| Phylotype associated with group |
|---|
| Healthy controls |
| Actinomyces oral taxon 170 |
| Gemella haemolysans |
| Granulicatella adiacens |
| Streptococcus mitis |
| Streptococcus sanguis |
| Chronic periodontitis |
| Porphyromonas endodontalis |
| Porphyromonas gingivalisa |
| Tannerella forsythiaa |
| Prevotella oral taxon 526 |
| Streptococcus constellatus |
| Eubacterium saphenum |
| Eubacterium minutum |
| Filifactor alocis |
| Mogibacterium timidum |
| Peptostreptococcaceae oral taxon 103 |
| Fusobacterium nucleatum subsp. vincentiia |
| Desulfobulbus oral taxon 041 |
| Treponema denticolaa |
| Treponema maltophilum |
| Treponema parvum |
| Treponema socranskii |
| Synergistetes oral taxon 360 |
| Synergistetes oral taxon 363 |
| Synergistetes oral taxon 452 |
Organism in the orange or red complex, as defined by Socransky et al. (15).
FIG 4.
Differentially abundant bacterial phylotypes identified by linear discriminant analysis (LDA) coupled with effect size measurements (LEfSe). Bacterial taxa enriched in healthy sites are indicated with positive linear discriminant analysis scores, and taxa enriched in periodontitis sites are indicated with negative linear discriminant analysis scores. Only taxa that met the significant linear discriminant analysis threshold of 3.5 are shown. Phylotypes that were also significantly different between the two groups by multivariate analysis are indicated by an asterisk. The oral taxon numbers are derived from the Human Oral Microbiome Database.
Of the 18 most differentially enriched taxa in periodontitis (Fig. 4), 3 belonged to the red complex, as defined by Socransky et al. (15) (Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia), and 1 was a member of the orange complex (Fusobacterium nucleatum subsp. vincentii). Members of the red and orange complexes (19 OTUs, or 4.2% of the 457 OTUs identified in this study) represented 44.9% of pyrosequence reads in the CP group and 17.5% in the control group. In total, members of the five major subgingival microbial complexes (43 OTUs), as defined by Socransky et al. (15), accounted for 49.4% of all sequence reads in the CP group and 39.3% in the control group. Two additional taxa, Filifactor alocis and Fretibacterium sp. (Synergistetes G-3 oral taxon 361), which are not part of the microbial complexes, were significantly enriched in sites of subjects with chronic periodontitis (86% and 3%, respectively). Interindividual variations in the relative proportions of dominant taxa were observed in both groups (see Fig. S3 in the supplemental material).
Clustering of subgingival microbial communities using distance metrics.
The gut microbiome of an individual can be classified into one of three “enterotypes” or clusters based on the abundance of key bacterial taxa (Bacteroides, Prevotella, and Ruminococcus) (44). More recent studies, however, suggest that most HMP and published “community” microbiome data sets fall into gradients rather than discrete clusterings of microbial communities (45). To examine whether distinct clusters of subgingival communities, or “periodontotypes,” are present in our data set, we utilized the same analytical approach as the one implemented previously by Arumugam et al. (44) with our combined healthy and chronic periodontitis data sets. By applying five different distance metrics (weighted and unweighted UniFrac distances, Jensen-Shannon divergence, root Jensen-Shannon divergence, and Bray-Curtis distance) (see Materials and Methods) to the combined data set, the optimal number of clusters was found to be 2 according to both the Calinski-Harabasz (CH) index and prediction score (Fig. 5A and data not shown). The prediction score indicated strong support for independent clusters (ranging from 0.81 to 0.92 for all distance metrics used). The species diversity and richness estimates between the two clusters were similar (Fig. 5B). The two clusters were dominated by different taxa, with one cluster being driven by Fusobacterium and Porphyromonas and the second cluster being dominated by Rothia and Streptococcus (Fig. 5C). Clinically apparent periodontal disease was significantly correlated with Fusobacterium and Porphyromonas belonging to the first cluster (88%, or 43 of 49 samples; P < 0.001), whereas healthy sites were predominant in the second cluster (86%, or 44 of 51 samples; P < 0.001).
FIG 5.
Clustering for subgingival microbial communities. (A) Prediction scores based on weighted UniFrac distances showing strong support for two independent clusters. One cluster is dominated by periodontitis sites (red), and the second cluster consists of mostly healthy microbial communities (blue). (B) Measures of microbial diversity and species richness in the two clusters (P > 0.5). (C) Relative proportions of sequence reads according to the taxa indicated.
Functional profiling of subgingival communities.
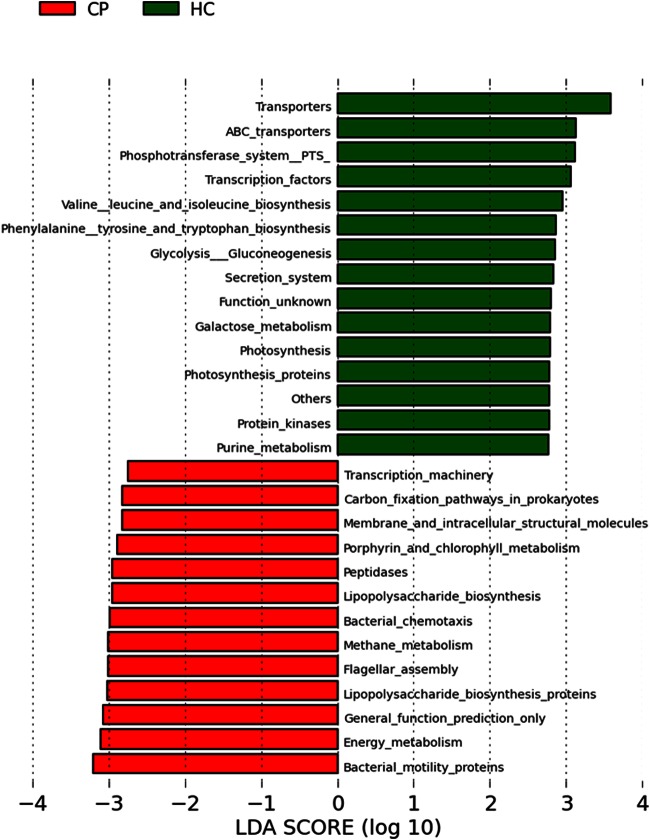
To identify biological functions differentially enriched in the subgingival metagenomes of subjects with chronic periodontitis, we applied PICRUSt (phylogenetic investigation of communities by reconstruction of unobserved states) (36), a computational algorithm that predicts functions based on 16S rRNA sequence information. Analysis using PICRUSt revealed that genes involved with bacterial motility, energy metabolism, lipopolysaccharide (LPS) biosynthesis, flagellar assembly, methane metabolism, bacterial chemotaxis, and peptidases were significantly more abundant in the subgingival metagenome of subjects with chronic periodontitis. In healthy controls, genes involved with transporters, the phosphotransferase system, transcription factors, amino acid biosynthesis, and glycolysis/gluconeogenesis were overrepresented in the microbiome (Fig. 6).
FIG 6.
Differentially abundant gene functions in subjects with chronic periodontitis and healthy controls. Functional categories of genes of the subgingival metagenome were predicted by using PICRUSt, and differentially abundant functions were then identified by using linear discriminant analysis (LDA) coupled with effect size measurements (LEfSe). Gene functions enriched in healthy sites are indicated with positive linear discriminant analysis scores, and functions differentially enriched in periodontitis sites are indicated with negative linear discriminant analysis scores. Only gene functions that have a linear discriminant analysis score threshold of 2.75 are shown.
DISCUSSION
A detailed analysis of the subgingival microbiome is critical for understanding the role of subgingival microbial communities in chronic periodontitis. Recent work in the microbiome field has revealed the immense richness and complexity of the oral microbiome. In this study, we used 16S rRNA sequencing to determine the composition and structure of the subgingival microbiomes in subjects with chronic periodontitis and healthy controls and compared our data set with data reported previously by the HMP, Griffen et al. (30), and Abusleme et al. (31). Comparison of the four data sets revealed that subgingival microbiota clustered by study, but differences in subgingival microbiomes between periodontal health and disease were larger than the technical variations across the four studies. Using an algorithm that predicts gene functions based on 16S rRNA sequence information, we identified several gene categories that were highly abundant in chronic periodontitis. Furthermore, we identified two major clusters strongly correlated with clinical measures of periodontal disease. These two clusters were distinguishable by species composition, with one cluster driven by Fusobacterium and Porphyromonas and the second cluster dominated by Rothia and Streptococcus. Our data also indicate that subgingival microbial communities in subjects with chronic periodontitis are more homogeneous than those in healthy controls, suggesting that a limited repertoire of species or genes is shared, which may play a role in the etiology and progression of periodontal disease.
Previous studies by Socransky et al. (46) and Griffen et al. (30) reported greater microbial diversity and species richness in sites with chronic periodontitis compared to healthy controls. In this study, we observed no significant difference in microbial diversity between healthy and periodontitis sites, although species richness (i.e., number of OTUs) was slightly higher in diseased sites (Fig. 1A). Interestingly, when deep and shallow sites were analyzed separately, species richness and diversity were significantly higher in shallow sites (i.e., 6-mm pockets) than in healthy control sites. In deeper sites (≥7-mm pockets), richness and diversity measures were similar or lower than in healthy control sites (Fig. 1B). This difference in microbial diversity and richness according to pocket depths may explain the difference observed between our data set and data from previous studies. However, these results also support a model of microbial succession, in which members of disease-associated bacteria initially invade the healthy microbiota, resulting in a diverse community consisting of both health- and disease-associated microbiota. As disease progresses, this transitional microbiota is then replaced by predominantly disease-associated organisms, leading to a more homogenous microbiota (Fig. 2).
Culture-independent 16S rRNA approaches have been utilized to characterize subgingival microbiota in healthy and diseased subjects in several studies (30, 31, 42). However, data sets from these studies could not be readily compared because amplification and sequencing strategies (e.g., 16S rRNA gene segments targeted) were different across studies. The Human Microbiome Project was the largest of the four data sets, including ours, consisting of 183 samples from 143 subjects with no or minimal periodontal disease (pocket size of <4 mm; some were collected from the same subjects at different times). The community data set of Griffen et al. (30) included a total of 87 samples from 29 subjects with chronic periodontitis (one diseased site and one healthy site from each subject) and 29 healthy controls (one sample each). The community data set of Abusleme et al. surveyed 61 samples from 22 subjects with periodontitis (one bleeding site and one nonbleeding site) and 17 healthy controls (multiple samples from some subjects). The present study evaluated 25 subjects with chronic periodontitis and 25 healthy controls (2 sites per subject). In order to compare these four data sets, we obtained raw sequence reads from published studies, trimmed the reads to identical lengths, and reprocessed them by using the same analytical pipeline (see the supplemental material). By using these procedures, we observed modest differences in microbial composition and community structure across the four data sets (Fig. 2C to G). These results suggest that technical procedures used in different studies, such as amplified 16S rRNA regions (V1-V3 versus V1-V2), sequencing chemistry (FLX versus Titanium chemistry), read lengths (∼250 bp versus ∼400 bp), and sampling techniques (paper point versus scaling), likely account for the observed differences across the four studies. This observation is also consistent with a recent meta-analysis of human microbiome studies (47), in which the authors concluded that technical variability across different studies likely explains the observed variations in the composition of microbiota. Alternatively, our data may reflect genuine differences in subgingival microbial communities across data sets. Because the origins of samples were different for the four studies (Europe, North America, and South America), host genetics, diet, or other environmental factors may have contributed to the differences in microbiota composition. We note that all three community data sets (30, 31; this study) display sufficiently large and systematic microbial shifts in chronic periodontitis compared to healthy control sites. This observation implicates a functional role for dysbiosis of the subgingival microbiome in chronic periodontitis.
As in previous studies (30, 31, 48), a difference in microbial composition was apparent between diseased and control sites at all taxonomic levels from phylum to species. Of 457 bacterial taxa, 50 were identified as being significantly enriched in healthy sites by using the LEfSe approach. Among these 50 taxa, 5 were significantly more abundant in healthy sites based on a conservative multivariate analysis (43). Most of the health-associated organisms identified in our study belong to the genus Streptococcus, and several are oral commensals, such as Rothia. In periodontitis, 36 of 457 taxa were significantly enriched based on LEfSe analysis, 18 of which were significantly more abundant, as determined by multivariate analysis (43). Among the 18 disease-associated taxa, most were members of the red and orange complexes, as defined by Socranski et al. (15) (Table 1). Two bacterial phylotypes, Filifactor alocis and Synergistetes G-3, which were recently linked to periodontitis (30, 31), were also identified in our data set. Filifactor alocis is a fastidious, Gram-positive anaerobic rod found to be highly prevalent and abundant in patients with generalized aggressive periodontitis and chronic periodontitis (49). This organism encodes several virulence factors, which may play a role in the pathogenesis of periodontal disease by enabling the organism to persist in periodontal pockets (50). Members of the phylum Synergistetes, a newly recognized phylum of Gram-negative anaerobic bacteria, were recently implicated in periodontal disease (51).
Recent gut microbiome surveys suggest that individuals can be classified into clusters or “enterotypes” based on the abundance of key bacterial species in the gut microbiota (44). Significant changes in gut enterotypes have been associated with long-term dietary changes (52). We examined whether subgingival microbiota could also be classified into different clusters or “periodontotypes” based on abundances of key bacterial genera. By using approaches described previously by Arumugam et al. (44) and Wu et al. (52), our subgingival data sets clustered into two periodontotypes distinguished primarily by levels of Fusobacterium/Porphyromonas and Rothia/Streptococcus bacteria. The majority of samples from subjects with chronic periodontitis belonged to one cluster, characterized by high levels of Fusobacterium and Porphyromonas bacteria. The second cluster consists of predominantly healthy sites, distinguishable by high levels of Rothia and Streptococcus bacteria. Thus, these data support an association between periodontotype and clinical measures of periodontal disease. Recently, Koren et al. (45) examined the HMP data sets from healthy volunteers who had minimal or no periodontal disease but failed to identify discrete clustering of subgingival microbiota in this population. Instead, those authors observed a continuous gradient according to the relative abundances of Prevotella, Fusobacterium, and Treponema. We note that in the HMP data set, subjects with periodontitis were not included, and although both healthy and diseased sites were analyzed in the present study, subjects with mild-to-moderate periodontal disease (i.e., subgingival pocket depths of between 3 and 6 mm) were not included. Thus, while our data support a model for periodontotypes, we could not exclude the possibility of a continuous abundance gradient of key bacteria within the human population. A larger number of samples encompassing the entire spectrum of periodontal health is warranted in order to confirm our conclusions.
Our 16S rRNA analysis demonstrates significant interindividual variations in taxonomic profiles within both health- and disease-associated microbiota (see Fig. S3 in the supplemental material). In contrast, predicted functions encoded by the subgingival metagenome were less variable within groups. For example, no taxa were universally present in all subjects, although several OTUs (e.g., Streptococcus and Rothia) were prevalent in healthy sites. In contrast, unlike microbial taxa, most functional categories or metabolic pathways were evenly distributed, and several were ubiquitous among individuals. The most abundant gene categories were transporters, DNA repair and recombination proteins, ribosomes, purine and pyrimidine metabolism proteins, and peptidases, which likely reflect the basic requirements of microbial life in the subgingival habitat. Despite the conservation of core functions, the relative abundances of some gene functions differed significantly between subjects with periodontitis and healthy controls. Notably, functions related to bacterial motility, energy metabolism, lipopolysaccharide (LPS) biosynthesis, flagellar assembly, methane metabolism, bacterial chemotaxis, and peptidases were abundant in chronic periodontitis cases. These results are consistent with recent metagenomic data (34, 53). Going forward, elucidating the role of key bacteria encoding these functions and understanding the basis of individual variations in microbiomes and metagenomes will be essential in future studies.
Supplementary Material
ACKNOWLEDGMENTS
We thank Zenith Maddipatla for enterotype analysis and members of the Wang laboratory for technical support and helpful discussions.
This work was supported by NIH/NIDCR grant R56 DE02556, James & Esther King Biomedical Research Program of Florida Department of Health grant 2KN08, the University of Florida Research Opportunity fund, NIH/NIAID grant K08 AI077713, and the University of Florida Department of Medicine.
We declare that we have no conflict of interest.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02712-14.
REFERENCES
- 1.Albandar JM, Brunelle JA, Kingman A. 1999. Destructive periodontal disease in adults 30 years of age and older in the United States, 1988-1994. J Periodontol 70:13–29. doi: 10.1902/jop.1999.70.1.13. [DOI] [PubMed] [Google Scholar]
- 2.Seymour GJ, Ford PJ, Cullinan MP, Leishman S, Yamazaki K. 2007. Relationship between periodontal infections and systemic disease. Clin Microbiol Infect 13(Suppl 4):S3–S10. doi: 10.1111/j.1469-0691.2007.01798.x. [DOI] [PubMed] [Google Scholar]
- 3.Mattila KJ, Nieminen MS, Valtonen VV, Rasi VP, Kesaniemi YA, Syrjala SL, Jungell PS, Isoluoma M, Hietaniemi K, Jokinen MJ. 1989. Association between dental health and acute myocardial infarction. BMJ 298:779–781. doi: 10.1136/bmj.298.6676.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reyes L, Herrera D, Kozarov E, Rolda S, Progulske-Fox A. 2013. Periodontal bacterial invasion and infection: contribution to atherosclerotic pathology. J Periodontol 84(4 Suppl):S30–S50. doi: 10.1902/jop.2013.1340012. [DOI] [PubMed] [Google Scholar]
- 5.Offenbacher S, Katz V, Fertik G, Collins J, Boyd D, Maynor G, McKaig R, Beck J. 1996. Periodontal infection as a possible risk factor for preterm low birth weight. J Periodontol 67(10 Suppl):1103–1113. [DOI] [PubMed] [Google Scholar]
- 6.Ryan ME, Carnu O, Kamer A. 2003. The influence of diabetes on the periodontal tissues. J Am Dent Assoc 134(Spec No):34S–40S. doi: 10.14219/jada.archive.2003.0370. [DOI] [PubMed] [Google Scholar]
- 7.Lalla E, Papapanou PN. 2011. Diabetes mellitus and periodontitis: a tale of two common interrelated diseases. Nat Rev Endocrinol 7:738–748. doi: 10.1038/nrendo.2011.106. [DOI] [PubMed] [Google Scholar]
- 8.Bullon P, Newman HN, Battino M. 2014. Obesity, diabetes mellitus, atherosclerosis and chronic periodontitis: a shared pathology via oxidative stress and mitochondrial dysfunction? Periodontol 2000 64:139–153. doi: 10.1111/j.1600-0757.2012.00455.x. [DOI] [PubMed] [Google Scholar]
- 9.Mercado FB, Marshall RI, Klestov AC, Bartold PM. 2001. Relationship between rheumatoid arthritis and periodontitis. J Periodontol 72:779–787. doi: 10.1902/jop.2001.72.6.779. [DOI] [PubMed] [Google Scholar]
- 10.Bartold PM, Marshall RI, Haynes DR. 2005. Periodontitis and rheumatoid arthritis: a review. J Periodontol 76(11 Suppl):2066–2074. doi: 10.1902/jop.2005.76.11-S.2066. [DOI] [PubMed] [Google Scholar]
- 11.Mikuls TR, Payne JB, Yu F, Thiele GM, Reynolds RJ, Cannon GW, Markt J, McGowan D, Kerr GS, Redman RS, Reimold A, Griffiths G, Beatty M, Gonzalez SM, Bergman DA, Hamilton BC III, Erickson AR, Sokolove J, Robinson WH, Walker C, Chandad F, O'Dell JR. 2014. Periodontitis and Porphyromonas gingivalis in patients with rheumatoid arthritis. Arthritis Rheumatol 66:1090–1100. doi: 10.1002/art.38348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Teles R, Teles F, Frias-Lopez J, Paster B, Haffajee A. 2013. Lessons learned and unlearned in periodontal microbiology. Periodontol 2000 62:95–162. doi: 10.1111/prd.12010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hajishengallis G. 2010. Complement and periodontitis. Biochem Pharmacol 80:1992–2001. doi: 10.1016/j.bcp.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Socransky SS, Haffajee AD. 2005. Periodontal microbial ecology. Periodontol 2000 38:135–187. doi: 10.1111/j.1600-0757.2005.00107.x. [DOI] [PubMed] [Google Scholar]
- 15.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL Jr. 1998. Microbial complexes in subgingival plaque. J Clin Periodontol 25:134–144. doi: 10.1111/j.1600-051X.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 16.Kumar PS, Griffen AL, Barton JA, Paster BJ, Moeschberger ML, Leys EJ. 2003. New bacterial species associated with chronic periodontitis. J Dent Res 82:338–344. doi: 10.1177/154405910308200503. [DOI] [PubMed] [Google Scholar]
- 17.Paster BJ, Olsen I, Aas JA, Dewhirst FE. 2006. The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontol 2000 42:80–87. doi: 10.1111/j.1600-0757.2006.00174.x. [DOI] [PubMed] [Google Scholar]
- 18.Tanner AC, Goodson JM. 1986. Sampling of microorganisms associated with periodontal disease. Oral Microbiol Immunol 1:15–22. doi: 10.1111/j.1399-302X.1986.tb00310.x. [DOI] [PubMed] [Google Scholar]
- 19.Moore WE, Moore LV. 1994. The bacteria of periodontal diseases. Periodontol 2000 5:66–77. doi: 10.1111/j.1600-0757.1994.tb00019.x. [DOI] [PubMed] [Google Scholar]
- 20.Paster BJ, Boches SK, Galvin JL, Ericson RE, Lau CN, Levanos VA, Sahasrabudhe A, Dewhirst FE. 2001. Bacterial diversity in human subgingival plaque. J Bacteriol 183:3770–3783. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanner AC, Haffer C, Bratthall GT, Visconti RA, Socransky SS. 1979. A study of the bacteria associated with advancing periodontitis in man. J Clin Periodontol 6:278–307. doi: 10.1111/j.1600-051X.1979.tb01931.x. [DOI] [PubMed] [Google Scholar]
- 22.Kroes I, Lepp PW, Relman DA. 1999. Bacterial diversity within the human subgingival crevice. Proc Natl Acad Sci U S A 96:14547–14552. doi: 10.1073/pnas.96.25.14547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Socransky SS, Smith C, Martin L, Paster BJ, Dewhirst FE, Levin AE. 1994. “Checkerboard” DNA-DNA hybridization. Biotechniques 17:788–792. [PubMed] [Google Scholar]
- 24.Olson JC, Cuff CF, Lukomski S, Lukomska E, Canizales Y, Wu B, Crout RJ, Thomas JG, McNeil DW, Weyant RJ, Marazita ML, Paster BJ, Elliott T. 2011. Use of 16S ribosomal RNA gene analyses to characterize the bacterial signature associated with poor oral health in West Virginia. BMC Oral Health 11:7. doi: 10.1186/1472-6831-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Socransky SS, Haffajee AD, Smith C, Martin L, Haffajee JA, Uzel NG, Goodson JM. 2004. Use of checkerboard DNA-DNA hybridization to study complex microbial ecosystems. Oral Microbiol Immunol 19:352–362. doi: 10.1111/j.1399-302x.2004.00168.x. [DOI] [PubMed] [Google Scholar]
- 26.Colombo AP, Boches SK, Cotton SL, Goodson JM, Kent R, Haffajee AD, Socransky SS, Hasturk H, Van Dyke TE, Dewhirst F, Paster BJ. 2009. Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the human oral microbe identification microarray. J Periodontol 80:1421–1432. doi: 10.1902/jop.2009.090185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shaddox LM, Alfant B, Tobler J, Walker C. 2010. Perpetuation of subgingival biofilms in an in vitro model. Mol Oral Microbiol 25:81–87. doi: 10.1111/j.2041-1014.2009.00549.x. [DOI] [PubMed] [Google Scholar]
- 28.Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, Lakshmanan A, Wade WG. 2010. The human oral microbiome. J Bacteriol 192:5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar PS, Griffen AL, Moeschberger ML, Leys EJ. 2005. Identification of candidate periodontal pathogens and beneficial species by quantitative 16S clonal analysis. J Clin Microbiol 43:3944–3955. doi: 10.1128/JCM.43.8.3944-3955.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Griffen AL, Beall CJ, Campbell JH, Firestone ND, Kumar PS, Yang ZK, Podar M, Leys EJ. 2012. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J 6:1176–1185. doi: 10.1038/ismej.2011.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abusleme L, Dupuy AK, Dutzan N, Silva N, Burleson JA, Strausbaugh LD, Gamonal J, Diaz PI. 2013. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME J 7:1016–1025. doi: 10.1038/ismej.2012.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. 2007. The human microbiome project. Nature 449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bizzarro S, Loos BG, Laine ML, Crielaard W, Zaura E. 2013. Subgingival microbiome in smokers and non-smokers in periodontitis: an exploratory study using traditional targeted techniques and a next-generation sequencing. J Clin Periodontol 40:483–492. doi: 10.1111/jcpe.12087. [DOI] [PubMed] [Google Scholar]
- 34.Wang J, Qi J, Zhao H, He S, Zhang Y, Wei S, Zhao F. 2013. Metagenomic sequencing reveals microbiota and its functional potential associated with periodontal disease. Sci Rep 3:1843. doi: 10.1038/srep01843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu B, Faller LL, Klitgord N, Mazumdar V, Ghodsi M, Sommer DD, Gibbons TR, Treangen TJ, Chang Y, Li S, Stine OC, Hasturk H, Kasif S, Segre D, Pop M, Amar S. 2012. Deep sequencing of the oral microbiome reveals signatures of periodontal disease. PLoS One 7:e37919. doi: 10.1371/journal.pone.0037919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, Beiko RG, Huttenhower C. 2013. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Armitage GC. 1999. Development of a classification system for periodontal diseases and conditions. Ann Periodontol 4:1–6. doi: 10.1902/annals.1999.4.1.1. [DOI] [PubMed] [Google Scholar]
- 38.Chen T, Yu WH, Izard J, Baranova OV, Lakshmanan A, Dewhirst FE. 2010. The Human Oral Microbiome Database: a Web accessible resource for investigating oral microbe taxonomic and genomic information. Database (Oxford) 2010:baq013. doi: 10.1093/database/baq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 40.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. 2011. Metagenomic biomarker discovery and explanation. Genome Biol 12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Methé BA, Nelson KE, Pop M, Creasy HH, Giglio MG, Huttenhower C, Gevers D, Petrosino JF, Abubucker S, Badger JH, Chinwalla AT, Earl AM, FitzGerald MG, Fulton RS, Hallsworth-Pepin K, Lobos EA, Madupu R, Magrini V, Martin JC, Mitreva M, Muzny DM, Sodergren EJ, Versalovic J, Wollam AM, Worley KC, Wortman JR, Young SK, Zeng Q, Aagaard KM, Abolude OO, Allen-Vercoe E, Alm EJ, Alvarado L, Andersen GL, Anderson S, Appelbaum E, Arachchi HM, Armitage G, Arze CA, Ayvaz T, Baker CC, Begg L, Belachew T, Bhonagiri V, Bihan M, Blaser MJ, Bloom T, Bonazzi VR, Brooks P, Buck GA, et al. . 2012. A framework for human microbiome research. Nature 486:215–221. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chi YY, Gribbin M, Lamers Y, Gregory JF III, Muller KE. 2012. Global hypothesis testing for high-dimensional repeated measures outcomes. Stat Med 31:724–742. doi: 10.1002/sim.4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, Bertalan M, Borruel N, Casellas F, Fernandez L, Gautier L, Hansen T, Hattori M, Hayashi T, Kleerebezem M, Kurokawa K, Leclerc M, Levenez F, Manichanh C, Nielsen HB, Nielsen T, Pons N, Poulain J, Qin J, Sicheritz-Ponten T, Tims S, Torrents D, Ugarte E, Zoetendal EG, Wang J, Guarner F, Pedersen O, de Vos WM, Brunak S, Doré J, MetaHIT Consortium, Antolín M, Artiguenave F, Blottiere HM, Almeida M, Brechot C, Cara C, Chervaux C, Cultrone A, Delorme C, Denariaz G. 2011. Enterotypes of the human gut microbiome. Nature 473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koren O, Knights D, Gonzalez A, Waldron L, Segata N, Knight R, Huttenhower C, Ley RE. 2013. A guide to enterotypes across the human body: meta-analysis of microbial community structures in human microbiome datasets. PLoS Comput Biol 9:e1002863. doi: 10.1371/journal.pcbi.1002863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Socransky SS, Haffajee AD, Smith C, Dibart S. 1991. Relation of counts of microbial species to clinical status at the sampled site. J Clin Periodontol 18:766–775. doi: 10.1111/j.1600-051X.1991.tb00070.x. [DOI] [PubMed] [Google Scholar]
- 47.Lozupone CA, Stombaugh J, Gonzalez A, Ackermann G, Wendel D, Vazquez-Baeza Y, Jansson JK, Gordon JI, Knight R. 2013. Meta-analyses of studies of the human microbiota. Genome Res 23:1704–1714. doi: 10.1101/gr.151803.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumar PS, Leys EJ, Bryk JM, Martinez FJ, Moeschberger ML, Griffen AL. 2006. Changes in periodontal health status are associated with bacterial community shifts as assessed by quantitative 16S cloning and sequencing. J Clin Microbiol 44:3665–3673. doi: 10.1128/JCM.00317-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schlafer S, Riep B, Griffen AL, Petrich A, Hubner J, Berning M, Friedman A, Gobel UB, Moter A. 2010. Filifactor alocis—involvement in periodontal biofilms. BMC Microbiol 10:66. doi: 10.1186/1471-2180-10-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aruni AW, Roy F, Fletcher HM. 2011. Filifactor alocis has virulence attributes that can enhance its persistence under oxidative stress conditions and mediate invasion of epithelial cells by Porphyromonas gingivalis. Infect Immun 79:3872–3886. doi: 10.1128/IAI.05631-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vartoukian SR, Palmer RM, Wade WG. 2009. Diversity and morphology of members of the phylum “Synergistetes” in periodontal health and disease. Appl Environ Microbiol 75:3777–3786. doi: 10.1128/AEM.02763-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, Sinha R, Gilroy E, Gupta K, Baldassano R, Nessel L, Li H, Bushman FD, Lewis JD. 2011. Linking long-term dietary patterns with gut microbial enterotypes. Science 334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Frias-Lopez J, Duran-Pinedo A. 2012. Effect of periodontal pathogens on the metatranscriptome of a healthy multispecies biofilm model. J Bacteriol 194:2082–2095. doi: 10.1128/JB.06328-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.