Abstract
We report engineering Neurospora crassa to improve the yield of cellobiose and cellobionate from cellulose. A previously engineered strain of N. crassa (F5) with six of seven β-glucosidase (bgl) genes knocked out was shown to produce cellobiose and cellobionate directly from cellulose without the addition of exogenous cellulases. In this study, the F5 strain was further modified to improve the yield of cellobiose and cellobionate from cellulose by increasing cellulase production and decreasing product consumption. The effects of two catabolite repression genes, cre-1 and ace-1, on cellulase production were investigated. The F5 Δace-1 mutant showed no improvement over the wild type. The F5 Δcre-1 and F5 Δace-1 Δcre-1 strains showed improved cellobiose dehydrogenase and exoglucanase expression. However, this improvement in cellulase expression did not lead to an improvement in cellobiose or cellobionate production. The cellobionate phosphorylase gene (ndvB) was deleted from the genome of F5 Δace-1 Δcre-1 to prevent the consumption of cellobiose and cellobionate. Despite a slightly reduced hydrolysis rate, the F5 Δace-1 Δcre-1 ΔndvB strain converted 75% of the cellulose consumed to the desired products, cellobiose and cellobionate, compared to 18% converted by the strain F5 Δace-1 Δcre-1.
INTRODUCTION
Cellulosic biomass is an attractive resource for the development of biofuels and chemicals due to its widespread abundance, low cost, and distinction from food crops (1, 2). The recalcitrance of cellulosic biomass (converting the biomass into fermentable sugars) is the predominant obstacle to commercialization of this technology, as the associated processing steps (pretreatment, cellulase production, and enzymatic hydrolysis) comprise about 40% of the overall production cost (3, 4). Consolidating the process into fewer processing steps is one way to improve the overall economics (5–7). Utilizing lignocellulolytic microorganisms to directly hydrolyze the cellulose into reactive intermediates such as sugar or sugar-like intermediates for subsequent conversion to fuels or chemicals is one such way to consolidate the process (8). The major limitation of such a process is carbon loss due to the consumption of the hydrolysis products by the lignocellulolytic organism. A metabolic engineering strategy can be employed to direct the carbon flow toward products, thereby preserving those products for the subsequent conversion to fuels and chemicals.
Neurospora crassa is a model microorganism, and its genetics, biochemistry, and fungal biology have been extensively studied for many years (9). The tools for genetic manipulation are readily available (9). The sequenced genome and functional sexual crossing made it possible to construct strains with multiple mutations in a relatively short time (10, 11). N. crassa is also a proficient plant cell wall degrader, and it produces a wide spectrum of cellulases and hemicellulases. Additionally, it produces cellobiose dehydrogenase (CDH) under cellulolytic conditions, which oxidizes cellobiose to its aldonic acid, cellobionic acid (12, 13). Efficient hydrolysis of cellulose requires several cellulase enzymes, including endoglucanases (EGs) (EC 3.2.1.4) and cellobiohydrolases (CBHs) (EC 3.2.1.91). Endoglucanases hydrolyze cellulose internally, while exoglucanases hydrolyze cellulose at the reducing and nonreducing ends. Cellobiose is the primary product of this hydrolysis, which can be further hydrolyzed by β-glucosidase (BGL) (EC 3.2.1.21) to form glucose (14). In previous studies, we engineered N. crassa for direct cellobiose and cellobionate production from cellulose (8, 15). By knocking out six of the seven bgl genes in N. crassa (designated strain F5), more than 7 g/liter of cellobiose was produced as the primary cellulose hydrolysis product without any cellulase addition in 96 h. A small amount (less than 1 g/liter) of cellobionate was produced along with cellobiose due to cellobiose oxidation by CDH. The hydrolysis products of cellobionate are glucose and gluconate, both of which could be substrates for the production of fuels and chemicals (15). Of the hydrolyzed cellulose, about 50% was directed toward the production of cellobiose and cellobionate in the F5 strain (8, 15). The unmodified wild-type strain consumed all the cellulose for cell growth without any cellobiose or cellobionate accumulation. The deletion of six of seven bgl genes did not negatively affect the rate of cellulose hydrolysis, and the cellulose conversion achieved by the F5 strain increased compared to that of the wild type. The level of cellulase production was comparable to that of the wild type (8).
In this study, we aimed to further modify the N. crassa F5 strain to improve the yield of cellobiose and cellobionate from cellulose by promoting cellulose hydrolysis via improved cellulase production and decreasing product consumption. Specifically, we aimed to improve cellulase production by knocking out a carbon catabolite repression (CCR) gene, cre-1, and a cellulase repressor gene, ace-1. The zinc finger transcription factor CRE1/CRE-1/CreA is a known carbon catabolite repressor for many plant-degrading fungi, including N. crassa, aspergilli, and Trichoderma reesei (16–22). Deletion of the cre-1 (NCU08807) gene in N. crassa and the cre1 gene in T. reesei led to improved cellulase production (16–18). Additionally, another zinc finger transcription factor, encoded by ace1, in T. reesei was found to repress cellulases and hemicellulose gene expression (23). This gene alone is subjected to CRE-1-dependent CCR (24). An ace1 orthologue (NCU09333) has been identified in N. crassa, and the effects of such a mutation on the homologous gene in N. crassa are currently unknown (25). Since ace-1 derepresses some of the same cellulases as cre-1 in other filamentous fungi, this opens the possibility of derepressing cellulase expression in N. crassa by knocking out the ace-1 gene and possibly obtaining a synergistic effect with a double knockout of the ace-1 and cre-1 genes. Furthermore, the ndvB gene (NCU09425), encoding a cellobionate phosphorylase enzyme, was previously shown to phosphorylate cellobionic acid to α-d-glucose 1-phosphate and d-gluconic acid, both of which can be metabolized by N. crassa (26). In this study, we also analyzed the effects of an ndvB knockout strain on the hydrolysis of cellulose for the production of cellobiose and cellobionate.
MATERIALS AND METHODS
Fungal strains.
The N. crassa strains used in this study and their sources are given in Table 1. The F5 Δcre-1, F5 Δcre-1 Δace-1, and F5 Δcre-1 Δace-1 ΔndvB strains were constructed in this study.
TABLE 1.
Strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| F5 | bgl-1::hph bgl-2::hph bgl-3::hph bgl-4::hph bgl-6::hph bgl-7::hph matA | 8 |
| F5-25a | bgl-1::hph bgl-2::hph bgl-3::hph bgl-4::hph bgl-6::hph bgl-7::hph mata | 8 |
| F5 Δace-1 | F5 ace-1::six | This study |
| F5 Δmus-51 Δcre-1 | F5 mus-51::six cre-1::six | 27 |
| F5 Δcre-1 | F5 cre-1::six | 27 |
| F5 Δace-1 Δcre-1 | F5 ace-1::six cre-1::six | This study |
| F5 Δmus-51 Δace-1 Δcre-1 | F5 mus-51::six ace-1::six cre-1::six | This study |
| F5 Δace-1 Δcre-1 ΔndvB | F5 mus-51::ace-1::six cre-1::six | This study |
Construction of the F5 Δace-1 and F5 Δcre-1 Δace-1 strains.
To construct the F5 Δace-1 and F5 Δcre-1 Δace-1 strains, 1.5-kb-long 5′ and 3′ flanking regions of the ace-1 gene (NCU09333) were amplified from the wild-type genomic DNA using Phusion high-fidelity DNA polymerase (ThermoScientific, Waltham, MA) and primer pair ES034JF and ES035JF or ES036JF and ES037JF, respectively (see Fig. S1A in the supplemental material). Unique DraI restriction sites were introduced at the ends of the deletion construct in the primers ES034JF and ES037JF (shown in lowercase in Table 2) to facilitate release of the fragment from the pUC19 vector. Using the GeneArt Seamless Cloning & Assembly kit (Invitrogen), a xylP(p)β-rec/six(bar) recyclable marker cassette was incorporated to create a deletion cassette, as previously described (27). The ace-1 deletion cassette was released from the plasmid using DraI and was used to transform the F5 Δmus-51 Δcre-1 using the standard transformation method (27). Twelve transformants resistant to phosphinothricin were screened by diagnostic PCR using primer pairs ES038JF-Sv739 and ES039JF-Sv740. Ten transformants showed correct ace-1 replacement (see Fig. S1B in the supplemental material). Lack of the ace-1 gene was confirmed with primers ES056JF and ES057JF (see Fig. S1C). The marker cassette was removed as described before (27), and marker excision was confirmed by PCR using primer pair ES058JF and ES059JF (see Fig. S1D). One of the resulting transformants was crossed with strain F5-25a to obtain the homokaryotic F5 Δcre-1, F5 Δace-1, and F5 Δcre-1 Δace-1strains.
TABLE 2.
Primers used in strain construction and verification
| Primer | Sequencea |
|---|---|
| ES034JF | AATTCGAGCTCGGTACtttaaaGAAGTCGACTGCATCAGG |
| ES035JF | GGACCTGAGTGAgatGTTTGCTGAGTTGTGTGGAG |
| ES036JF | TGGTCCATCTAGTgatGACAAGTTGGGGAGAACGC |
| ES037JF | GCCAAGCTTGCATGCCtttaaaGGCAGATTCAATAACGACC |
| ES038JF | AGCGTTTGTTGTCGAACCC |
| ES039JF | GATGAGGAAGCAAGCAGAGG |
| ES056JF | CTCTCCCACTACTCCAGCC |
| ES057JF | AGATGTCCTGAGATGATGGC |
| ES058JF | CCAAGCCTGAATACCAACCC |
| ES059JF | AAATGAACAATATCAGCAAGGG |
| ES113JF | AATTCGAGCTCGGTACgaattcTATTTAGGATACAGTAGCAGCG |
| ES114JF | GGACCTGAGTGAtttTTGGTTGTGTGTGAAGTTGAG |
| ES115JF | TGGTCCATCTAGTtttAACCTTACAGTGACTATTCCG |
| ES116JF | GCCAAGCTTGCATGCCgaattcGGATGTTGAGCACCTTGACG |
| ES117JF | ACTACGTTCTTCGATAGTAGG |
| ES118JF | TTCTTGACCACCGTGTGAC |
| ES119JF | GCTACGAAATCACCAACCC |
| ES120JF | GATGTCCTTGACGTGAGGC |
| Sv739 | ACAAATAAGTATACTCTATTGACC |
| Sv740 | AGAGTAGGTCATTTAAGTTGAGC |
Lowercase indicates a restriction site (see Materials and Methods).
Construction of the F5 Δcre-1 Δace-1 ΔndvB strain.
To construct the F5 Δcre-1 Δace-1 ΔndvB strain, 1-kb-long 5′ and 3′ flanking regions of the ndvB gene (NCU09425) were amplified from wild-type genomic DNA using Phusion high-fidelity DNA polymerase and primer pair ES113JF and ES114JF or ES115JF and ES116JF, respectively (see Fig. S2A in the supplemental material). Unique EcoRI restriction sites were introduced at the ends of the deletion construct in primers ES113JF and ES116JF (shown in lowercase in Table 2) to facilitate release of the fragment from the pUC19 vector. Using the GeneArt Seamless Cloning & Assembly kit, a gh10-2(p)β-rec/six(bar-tk) recyclable marker cassette was incorporated to create a deletion cassette, as described by Szewczyk et al. (28). The 3′ flanking region of the ndvB gene incorporated in the cassette was sequenced to ensure correct amplification of the hypothetical gene NCU09424 located only 559 bp downstream of ndvB. The ndvB deletion cassette was released from the plasmid using EcoRI restriction enzyme digestion. The cassette was then used to transform the F5 Δmus-51 Δcre-1 Δace-1 strain. The positive transformants resistant to phosphinothricin were screened by diagnostic PCR using primer pairs ES117JF-Sv739 and ES118JF-Sv740 (see Fig. S2A in the supplemental material). Twenty of 24 screened ndvB transformants showed correct ndvB replacement (see Fig. S2B). Lack of the ndvB gene was confirmed by PCR with primer pair ES119JF and ES120JF (see Fig. S2C). The marker cassette was removed as described before (18), and marker excision was confirmed by PCR using primer pair ES117JF and ES118JF (see Fig. S1D).
Fermentation experiments.
N. crassa strains were grown on agar with 1× Vogel's salts and 1.5% sucrose in an incubator at 30°C with light (29). After 3 days, flasks were removed from the incubator, and the strains were grown for 7 days at room temperature. After a total of 10 days of growth, the conidia were harvested in water and filtered through eight layers of cheesecloth. Fermentation experiments were conducted in 250-ml Erlenmeyer flasks with a 50-ml working volume, 1× Vogel's salts medium, and 20 g/liter Avicel unless otherwise noted. At the beginning of each fermentation, 0.5 g/liter of glucose was added to initiate cell growth. Conidia were inoculated at a volume to yield a final optical density at 420 nm (OD420) of 0.15. Flasks were incubated at 28°C in a rotary shaker at 200 rpm with light. Samples were taken at various time intervals for enzyme activity analysis and compositional analysis.
Sample analysis.
Concentrations of sugars (glucose, cellobiose, and cellobionate) were analyzed using a Shimadzu high-pressure liquid chromatograph (HPLC) equipped with a refraction index detector, photodiode array (PDA) detector, and CARBOSep Coregel-87C (Transgenomic, San Jose, CA, USA) column. Four millimolar calcium chloride at a flow rate of 0.6 ml/min was used as the mobile phase.
Enzyme assays.
CDH activity was determined by following the decrease in absorbance of 2,6-dichlorophenolindophenol (DCPIP) at 520 nm according to previously established methods, using cellobiose as the substrate (30).
Exoglucanase activity was measured similarly, with the following modifications. One hundred microliters of 1 mg/ml p-nitrophenyl-β-d-lactopyranoside in 50 mM citric acid buffer (pH 5.0) was incubated at 37°C for 15 min. Eighty microliters of sample was added to the substrate and incubated for 15 min at 37°C. The reaction was stopped with 120 μl of 1 M NaOH. Absorbance was immediately measured at 405 nm and compared to that of p-nitrophenol standards (0 to 10 mM) in citric acid buffer (pH 5.0). One unit of enzyme activity is defined as the amount of enzyme required to liberate 1 μmol of p-nitrophenol per minute under the given assay conditions.
Endoglucanase activity was measured as described by King et al. in a 96-well plate with minor modifications (31). One hundred microliters of 2% carboxymethyl cellulose in 50 mM sodium citrate buffer (pH 5.0) was incubated at 37°C for 15 min. Eighty microliters of sample was added to the substrate and incubated for an additional 15 min at 37°C, after which the reaction was stopped by adding 120 μl of 3,5-dinitrosalicylic acid (DNS) reagent (0.01 g DNS, 3 g sodium potassium tartrate, 4 ml 1 M NaOH, with a total volume of 10 μl made with deionized [DI] water). The reaction mixture was transferred to PCR plates and incubated in a thermocycler at 95°C for 5 min, followed by 1 min at 4°C. The absorbance was measured at 540 nm and compared to that of glucose standards (0 to 1.0 g/liter). One unit of activity corresponds to the release of 1 μmol of reducing sugar per minute.
Mycelial biomass measurements.
The dry weight of mycelia contained in the fermentation samples was measured by extracting ergosterol from the mycelia and measuring the amount by HPLC (32). The fermentation residue was collected by filtration through a 0.8-μm membrane. All the residue, including the mycelia, was harvested and frozen in liquid nitrogen for 1 h, and ethanol (6 ml) was added to the frozen sample and incubated at 37°C for 2 h with shaking. An aliquot of KOH solution (60% [wt/vol], 0.8 ml) was added to the mixture, which was then heated to 97°C for 20 min. This sample was cooled and neutralized with HCl (36.5%, ∼0.7 ml). The solution was extracted three times with 5 ml hexane, the hexane fractions were combined, and air was used to evaporate the solvent. The residue was dissolved in ethanol (1 ml), filtered through a 0.22-μm membrane filter, analyzed by HPLC with a PDA detector on a reverse-phase column (Zorbax Eclipse Plus C18 [4.6 by 250 mm, 5-μm particle size]; Agilent), and eluted at 1.0 ml/min with methanol-water (97:3, vol/vol). The amount of biomass was quantified using a standard curve prepared with known N. crassa dry biomass.
RESULTS
Effects of cre-1 and ace-1 gene deletions on enzyme production.
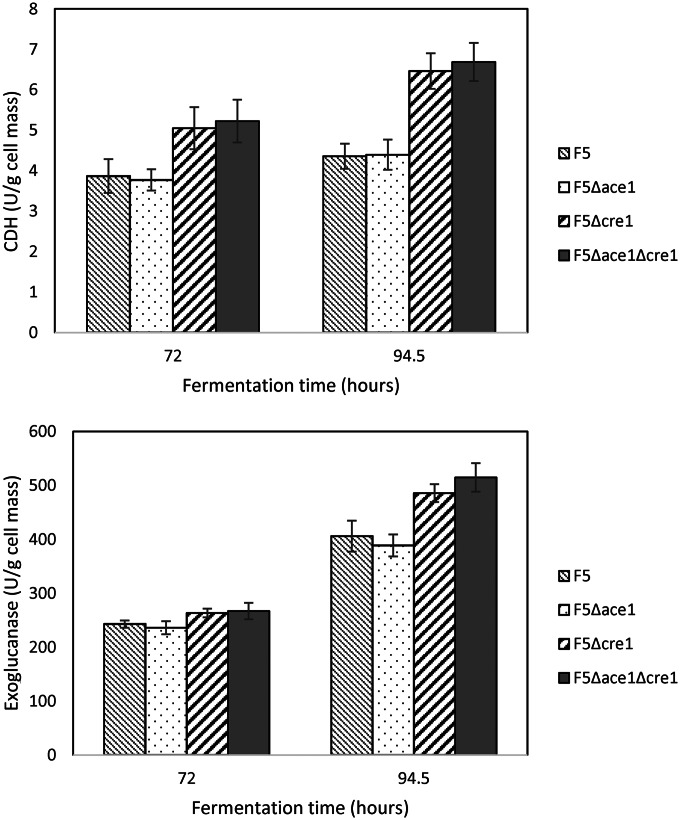
Strains with single and double knockouts of the ace-1 and cre-1 genes were evaluated for cellulase production in shake flask fermentations with 20 g/liter Avicel. Exoglucanase, endoglucanase, and CDH activities were analyzed at 3 and 4 days of culture. The enzyme activity data normalized to mycelial biomass are shown in Fig. 1. Production of exoglucanase and CDH was improved for strains containing the cre-1 deletion. While the F5 Δace-1 single-knockout strain was indistinguishable from the parent strain, F5 Δace-1 Δcre-1 had exoglucanase production that was improved by 26% and CDH production that was improved by 50% compared to those of the F5 parent strain. All of the strains showed similar endoglucanase activity (see Fig. S3 in the supplemental material) and rate of biomass production (see Fig. S4 in the supplemental material).
FIG 1.
CDH and exoglucanase activities for ace-1 and/or cre-1 knockout strains at selected time points. The values shown are the means for biological triplicates. Error bars are the standard deviations.
Effects of cre-1 and ace-1 gene deletions on cellobiose and cellobionate production.
Shake flask experiments in Vogel's medium with 20 g/liter Avicel were analyzed for the production of cellobiose. No difference in cellobiose production was observed for the ace-1 and cre-1 knockout strains (see Fig. S5 in the supplemental material). Interestingly, despite the presence of a high level of CDH, no cellobionate was detected over the time course of the fermentation. This prompted the investigation of the gene(s) responsible for cellobionate and/or cellobiose metabolism in N. crassa, as discussed below.
Effects of the ndvB gene deletion on cellobiose, cellobionate, and cell mass production.
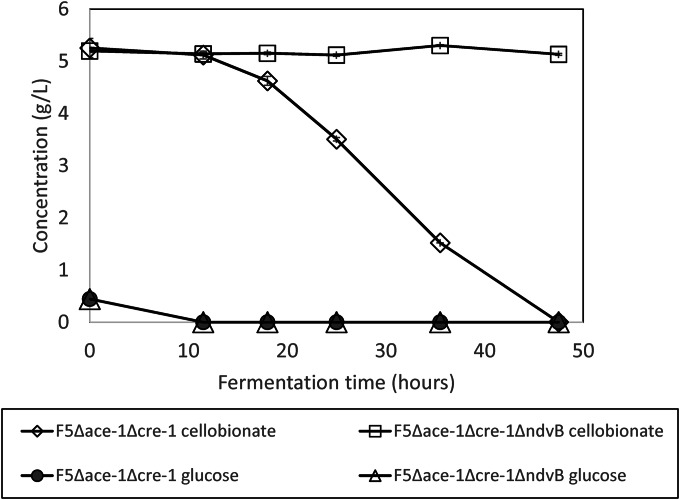
The F5 Δace-1 Δcre-1 ΔndvB knockout strain was compared to the parent strain (F5 Δace-1 Δcre-1) in a shake flask fermentation experiment in Vogel's medium with 0.5 g/liter glucose to initiate cell growth and 5 g/liter cellobionate to evaluate cellobionate consumption. The results are shown in Fig. 2. Glucose was completely utilized within 11.5 h for both strains. For the F5 Δace-1 Δcre-1 strain, cellobionate consumption began after glucose was utilized, and all cellobionate was completely used up in 48 h. In contrast, no cellobionate was consumed for the F5 Δace-1 Δcre-1 ΔndvB knockout strain, confirming that the ndvB gene was responsible for cellobionate metabolism.
FIG 2.
Cellobionate consumption by the ndvB knockout F5 Δace-1 Δcre-1 ΔndvB strain compared to the parent F5 Δace-1 Δcre-1 strain. The values shown are the means for biological triplicates for each strain. Error bars are the standard deviations for the biological triplicates.
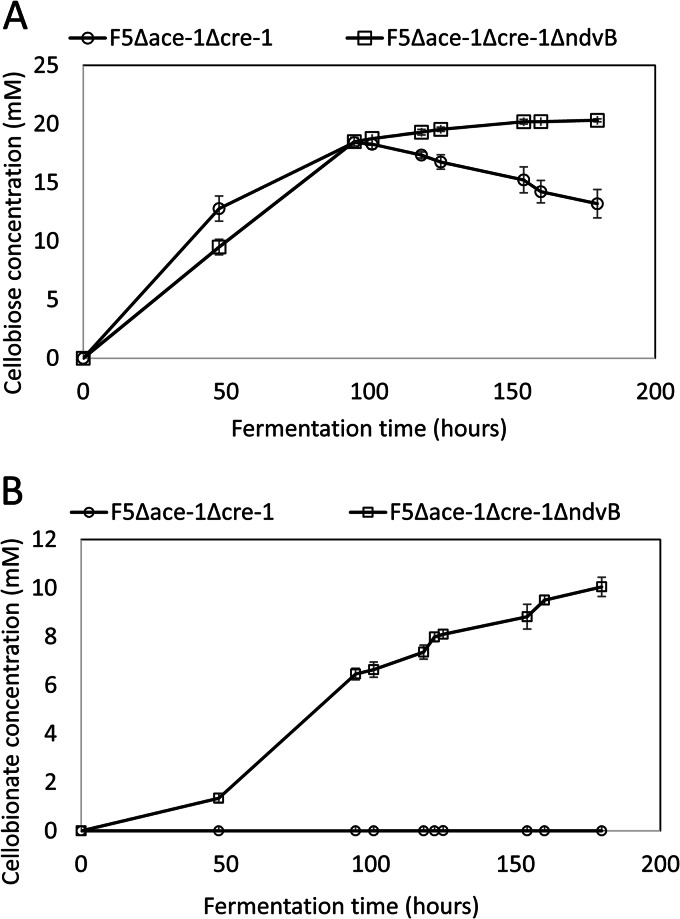
The F5 Δace-1 Δcre-1 and F5 Δace-1 Δcre-1 ΔndvB strains were subsequently evaluated in a shake flask fermentation in Vogel's medium with 20 g/liter Avicel. The concentrations of cellobiose and cellobionate production along the time course are presented in Fig. 3. Cellobiose production increased over the course of 167 h of fermentation time for the F5 Δace-1 Δcre-1 ΔndvB strain, with a maximum concentration of just over 20 mM, whereas the parent strain reached a similar maximum of 18.4 mM cellobiose at 95 h, followed by a steady decrease in concentration over the remainder of the fermentation. There was no detectable cellobionate production for the F5 Δace-1 Δcre-1 strain, whereas the F5 Δace-1 Δcre-1 ΔndvB strain accumulated 10 mM over the course of the fermentation. The combined cellobiose and cellobionate production from 20 g/liter of cellulose was 10.5 g/liter for the F5 Δace-1 Δcre-1 ΔndvB strain.
FIG 3.
Cellobiose (A) and cellobionate (B) production by the F5 Δace-1 Δcre-1 and F5 Δace-1 Δcre-1 ΔndvB strains grown on 20 g/liter Avicel. The values shown are the means for biological triplicates. Error bars are the standard deviations.
In a separate experiment conducted identically to the above-described experiment, flasks were harvested at 167 h into the fermentation and analyzed for cell mass production. Based on the cell mass percentage in the insoluble fraction, the amounts of utilized and remaining Avicel were calculated, including the amount of Avicel directed toward cellobiose and cellobionate. The results are shown in Table 3. The F5 Δace-1 Δcre-1 strain hydrolyzed approximately 76% of the Avicel. However, only 18% of the hydrolyzed Avicel remained as the desired products, cellobiose and cellobionate, at the end of the fermentation. In comparison, the F5 Δace-1 Δcre-1 ΔndvB strain hydrolyzed only 62% of the Avicel, but 75% of this Avicel was directed toward cellobiose and cellobionate. Substantially less biomass was produced with the F5 Δace-1 Δcre-1 ΔndvB strain than with the parent strain.
TABLE 3.
Percentages of Avicel hydrolyzed and directed toward fermentable products for the F5 Δace-1 Δcre-1 and F5 Δace-1 Δcre-1 ΔndvB strains grown on 20 g/liter Avicela
| Strain | Amt (g) of Avicel |
Cellulose conversion (%) | Mycelium produced (g) | Yield (%) from consumed Avicel |
||
|---|---|---|---|---|---|---|
| Starting | Residual | Cellobiose and cellobionate (mol/mol)b | Mycelium mass (g/g) | |||
| F5 Δace-1 Δcre-1 | 1.00 | 0.24 ± 0.02 | 76 ± 2 | 0.12 ± 0.003 | 18 ± 2 | 15.7 ± 0.6 |
| F5 Δace-1 Δcre-1 ΔndvB | 1.00 | 0.38 ± 0.002 | 62 ± 0.2 | 0.02 ± 0.001 | 75 ± 2 | 3.3 ± 0.2 |
Errors are calculated based upon standard deviations and error propagation theory.
The molecular mass of Avicel was assumed to be 324 g/mol.
DISCUSSION
Previous studies have shown that the cre-1/creA genes encode a carbon catabolite repressor which regulates plant cell wall-degrading enzyme production in many filamentous fungi, such as T. reesei, Aspergillus niger, and N. crassa (16–18). CRE1/CreA regulate the production of enzymes associated with alternative carbon source utilization, including plant cell wall-degrading enzymes, in a double-lock manner (33–36). For example, CRE1 in T. reesei is known to bind to the promoters of the structure genes such as cbh1 and xyn1, which encode a key cellulase and a key hemicellulase (37, 38). It also represses the expression of a regulatory gene, xyl1, which encodes the main activator protein for cellulase and hemicellulase enzyme production in T. reesei (39, 40). The deletion of cre-1 in T. reesei led to enhanced cellulase and hemicellulase production (41, 42). The cre-1 gene in N. crassa also represses the production of the plant cell wall-degrading enzymes (16). More than 100 genes in the Δcre-1 mutant showed more than a 2-fold increase over levels in the wild-type strain under Avicel growth conditions. Among them are 16 cellulase genes and 7 predicted hemicellulase genes (16). Δcre-1 mutants of N. crassa grew faster on Avicel than the wild type (16).
In addition to cre-1, ace-1 is another important repressor for cellulase and hemicellulase gene expression (23). It was reported that it repressed cellulase gene expression (including that of cbh1 and xyn1) in T. reesei grown on cellulose (23, 43). The expression of ace-1 is also subject to the global CCR regulated by cre-1 in T. reesei (18). An orthologue of ace-1 was identified in N. crassa (25). However, its function was not yet characterized. In another study, it was found that mutations in cellulase genes, such as cbh1, could alter the effect of mutations of other regulatory genes in the zinc finger family on cellulase activity (25). Therefore, it is interesting to characterize the effect of cre-1 and/or ace-1 on cellulase expression with our mutant strain that contains deletions of six of seven bgl genes. Our data suggest that the F5 Δace-1 single deletion strain has no improvement of CDH and exoglucanases expression compared to the parent F5 strain. In contrast, the F5 Δcre-1 and F5 Δace-1 Δcre-1 double-knockout strains show an increase in exoglucanase and CDH expression compared to the parent strain. While CDH and exoglucanase expression increased for the F5 Δcre-1 and F5 Δace-1 Δcre-1 deletion strains, this did not lead to an increase in cellobiose concentration under the fermentation conditions tested.
Although a 50% increase in CDH activity was detected for the F5 Δace-1 Δcre-1 strain, cellobionate production was still not detected. This led to the assumption that N. crassa must have the ability to metabolize cellobionate. Recent literature suggested that the ndvB gene encodes a cellobionate phosphorylase enzyme, which converts cellobionate to α-d-glucose 1-phosphate and d-gluconic acid in an energy-efficient manner, both of which can be metabolized by N. crassa (26). We tested the effect of the ndvB gene deletion on cellobionate consumption, and our data suggest that deletion of ndvB can eliminate cellobionate consumption completely. Knocking out this gene resulted in a strain (F5 Δace-1 Δcre-1 ΔndvB) which generated 10 mM cellobionate and 20 mM cellobiose over the course of a 167-hour fermentation on 20 g/liter of Avicel. In contrast, the F5 Δace-1 Δcre-1 parent strain produced no detectable cellobionate and about 18 mM cellobiose at 96 h, and the cellobiose declined to about 13 mM by the end of the fermentation (180 h). While the cellulose conversion achieved was higher for the F5 Δace-1 Δcre-1 strain (76%) than for the F5 Δace-1 Δcre-1 ΔndvB strain (62%), a smaller fraction of fermentable sugars was produced. With the F5 Δace-1 Δcre-1 strain, only 18% of the utilized Avicel was converted to the desired products (cellobiose and cellobionate), compared to 75% for the F5 Δace-1 Δcre-1 ΔndvB strain, where only a small fraction of the Avicel is consumed for cell growth and maintenance. Additionally, we showed that the ndvB gene allows N. crassa to consume cellobiose in addition to cellobionate. The deletion of the ndvB gene effectively prevented the cellobiose concentration from declining after 96 h into the fermentation.
Over the course of the fermentation of the F5 Δace-1 Δcre-1 ΔndvB strain, cellobiose production reached a plateau while cellobionate production continuously increased, which indicates that the conversion of cellobiose to cellobionate was the limiting step. Cellobiose is a known inhibitor of cellulases, although there is much debate as to the mechanism of inhibition due to the complexity of the cellulase mixtures and hydrolysis of cellulose (44). Previous studies have shown that cellobiose is more inhibitory to CBHs than to EGs and BGLs and that the presence of CDH can alleviate the competitive inhibition of cellobiose on CBHs by oxidizing cellobiose to cellobionate (45, 46). Should the rate of cellobiose conversion to cellobionate be increased by means such as improving CDH production in the F5 Δace-1 Δcre-1 ΔndvB strain, more cellobiose could be directed to cellobionate, which can possibly relieve the inhibition of cellobiose on cellulase. Therefore, higher cellulase activities could be exploited for increased rates of hydrolysis of cellulosic substrates, a higher cellulose conversion, a shorter processing time, and a possible higher product yield. Improving the conversion of cellobiose to cellobionate will be one of the focuses of our future work.
In conclusion, knocking out the cre-1 gene in an N. crassa mutant with six of seven bgl genes deleted increased expression of exoglucanases and CDH, while no effect was observed for endoglucanase expression. Furthermore, knocking out the ndvB gene (strain F5 Δace-1 Δcre-1 ΔndvB) prevented metabolism of cellobiose and cellobionate and resulted in a significant improvement of cellulose conversion toward fermentable sugar and sugar-like products. While N. crassa was studied here as a model organism, the results obtained here could be extrapolated to industrially relevant organisms such as T. reesei to improve cellulose conversion toward fermentable products.
Supplementary Material
ACKNOWLEDGMENTS
This project was supported by Agriculture and Food Research Initiative Competitive Grant no. 2011-67009-20060 from the USDA National Institute of Food and Agriculture and by an EPA Star fellowship and Cota-Robles fellowship awarded to Amanda Hildebrand.
We thank Eric Walters for reading the paper.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02885-14.
REFERENCES
- 1.Lynd LR, Cushman J, Nichols R, Wyman C. 1991. Fuel ethanol from cellulosic biomass. Science 251:1318–1323. doi: 10.1126/science.251.4999.1318. [DOI] [PubMed] [Google Scholar]
- 2.Humbird D, Davis R, Tao L, Kinchin C, Hsu D, Aden A. 2011. Process design and economics for biochemical conversion of lignocellulosic biomass to ethanol. NREL/TP-5100-47764. NREL, Washington, DC. [Google Scholar]
- 3.Wooley R, Ruth M, Sheehan J, Ibsen K, Majdeski H, Galvez A. 1999. Lignocellulosic biomass to ethanol process design and economics utilizing co-current dilute acid prehydrolysis and enzymatic hydrolysis: current and futuristic scenarios. NREL, Washington, DC. [Google Scholar]
- 4.Himmel ME, Ding S-Y, Johnson DK, Adney WS, Nimlos MR, Brady JW, Foust TD. 2007. Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science 315:804–807. doi: 10.1126/science.1137016. [DOI] [PubMed] [Google Scholar]
- 5.Ingram LO, Aldrich HC, Borges ACC, Causey TB, Martinez A, Morales F, Saleh A, Underwood SA, Yomano LP, York SW, Zaldivar J, Zhou S. 1999. Enteric bacterial catalysts for fuel ethanol production. Biotechnol Prog 15:855–866. doi: 10.1021/bp9901062. [DOI] [PubMed] [Google Scholar]
- 6.Lynd L, Wyman C, Gerngross T. 1999. Biocommodity engineering. Biotechnol Prog 15:777–793. doi: 10.1021/bp990109e. [DOI] [PubMed] [Google Scholar]
- 7.Mosier N, Wyman C, Dale B, Elander R, Lee YY, Holtzapple M, Ladisch M. 2005. Features of promising technologies for pretreatment of lignocellulosic biomass. Bioresour Technol 96:673–686. doi: 10.1016/j.biortech.2004.06.025. [DOI] [PubMed] [Google Scholar]
- 8.Wu W, Hildebrand A, Kasuga T, Xiong X, Fan Z. 2013. Direct cellobiose production from cellulose using sextuple beta-glucosidase gene deletion Neurospora crassa mutants. Enzyme Microb Technol 52:184–189. doi: 10.1016/j.enzmictec.2012.12.010. [DOI] [PubMed] [Google Scholar]
- 9.Davis RH, Perkins DD. 2002. Neurospora: a model of model microbes. Nat Rev Genet 3:397–403. doi: 10.1038/nrg797. [DOI] [PubMed] [Google Scholar]
- 10.Dunlap JC, Borkovich KA, Henn MR, Turner GE, Sachs MS, Glass NL, McCluskey K, Plamann M, Galagan JE, Birren BW, Weiss RL, Townsend JP, Loros JJ, Nelson MA, Lambreghts R, Colot HV, Park G, Collopy P, Ringelberg C, Crew C, Litvinkova L, DeCaprio D, Hood HM, Curilla S, Shi M, Crawford M, Koerhsen M, Montgomery P, Larson L, Pearson M, Kasuga T, Tian C, Baştürkmen M, Altamirano L, Xu J. 2007. Enabling a community to dissect an organism: overview of the Neurospora functional genomics project. Adv Genet 57:49–96. doi: 10.1016/S0065-2660(06)57002-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galagan JE, Calvo SE, Borkovich KA, Selker EU, Read ND, Jaffe D, Fitzhugh W, Ma L, Smirnov S, Purcell S, Rehman B, Elkins T, Engels R, Wang S, Nielsen CB, Butler J, Endrizzi M, Qui D, Ianakiev P, Bell-pedersen D, Nelson MA, Werner-Washburne M, Selitrennikoff CP, Kinsey JA, Braun EL, Zelter A, Schulte U. 2003. The genome sequence of the filamentous fungus Neurospora crassa. Nature 422:859–868. doi: 10.1038/nature01554. [DOI] [PubMed] [Google Scholar]
- 12.Zhang R, Xu C, Shen Q, Kasuga T, Wu W, Szewczyk E, Ma D, Fan Z. 2013. Characterization of two cellobiose dehydrogenases and comparison of their contributions to total activity in Neurospora crassa. Int Biodeterior Biodegradation 82:24–32. doi: 10.1016/j.ibiod.2013.03.017. [DOI] [Google Scholar]
- 13.Beeson WT, Phillips CM, Cate JH, Marletta MA. 2012. Oxidative cleavage of cellulose by fungal copper-dependent polysaccharide monooxygenases. J Am Chem Soc 134:890–892. doi: 10.1021/ja210657t. [DOI] [PubMed] [Google Scholar]
- 14.Zhang J, Viikari L. 2012. Xylo-oligosaccharides are competitive inhibitors of cellobiohydrolase I from Thermoascus aurantiacus. Bioresour Technol 117:286–291. doi: 10.1016/j.biortech.2012.04.072. [DOI] [PubMed] [Google Scholar]
- 15.Fan Z, Wu W, Hildebrand A, Kasuga T, Zhang R, Xiong X. 2012. Novel biochemical route for fuels and chemicals production from cellulosic biomass. PLoS One 7:8. doi: 10.1371/journal.pone.0031693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun J, Glass NL. 2011. Identification of the CRE-1 cellulolytic regulon in Neurospora crassa. PLoS One 6:e25654. doi: 10.1371/journal.pone.0025654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stricker AR, Mach RL, de Graaff LH. 2008. Regulation of transcription of cellulases- and hemicellulases-encoding genes in Aspergillus niger and Hypocrea jecorina (Trichoderma reesei). Appl Microbiol Biotechnol 78:211–220. doi: 10.1007/s00253-007-1322-0. [DOI] [PubMed] [Google Scholar]
- 18.Portnoy T, Margeot A, Seidl-Seiboth V, Le Crom S, Ben Chaabane F, Linke R, Seiboth B, Kubicek CP. 2011. Differential regulation of the cellulase transcription factors XYR1, ACE2, and ACE1 in Trichoderma reesei strains producing high and low levels of cellulase. Eukaryot Cell 10:262–271. doi: 10.1128/EC.00208-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dowzer CEA, Kelly JM. 1991. Analysis of the creA gene, a regulator of carbon catabolite repression in Aspergillus nidulans PsP3sI. Mol Cell Biol 11:5701–5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ilmen M, Thrane C, Penttil M. 1996. The glucose repressor gene crel of Trichoderma: isolation and expression of a full-length and a truncated mutant form. Mol Gen Genet 251:451–460. doi: 10.1007/BF02172374. [DOI] [PubMed] [Google Scholar]
- 21.Ebbole DJ. 1998. Carbon catabolite repression of gene expression and conidiation in Neurospora crassa. Fungal Genet Biol 25:15–21. doi: 10.1006/fgbi.1998.1088. [DOI] [PubMed] [Google Scholar]
- 22.Ruijter G, Vanhanen S, Gielkens M, van de Vondervoort P, Visser J. 1997. Isolation of Aspergillus niger creA mutants and effects of the mutations on expression of arabinases and l-arabinose catabolic enzymes. Microbiology 143:2991–2998. doi: 10.1099/00221287-143-9-2991. [DOI] [PubMed] [Google Scholar]
- 23.Aro N, Ilmen M, Saloheimo A, Penttila M. 2003. ACEI of Trichoderma reesei is a repressor of cellulase and xylanase expression Appl Environ Microbiol 69:56–65. doi: 10.1128/AEM.69.1.56-65.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mach-Aigner AR, Pucher ME, Steiger MG, Bauer GE, Preis SJ, Mach RL. 2008. Transcriptional regulation of xyr1, encoding the main regulator of the xylanolytic and cellulolytic enzyme system in Hypocrea jecorina. Appl Environ Microbiol 74:6554–6562. doi: 10.1128/AEM.01143-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmoll M, Tian C, Sun J, Tisch D, Glass NL. 2012. Unravelling the molecular basis for light modulated cellulase gene expression—the role of photoreceptors in Neurospora crassa. BMC Genomics 13:127. doi: 10.1186/1471-2164-13-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nihira T, Saito Y, Nishimoto M, Kitaoka M, Igarashi K, Ohtsubo K, Nakai H. 2013. Discovery of cellobionic acid phosphorylase in cellulolytic bacteria and fungi. FEBS Lett 587:3556–3561. doi: 10.1016/j.febslet.2013.09.014. [DOI] [PubMed] [Google Scholar]
- 27.Szewczyk E, Kasuga T, Fan Z. 2013. Efficient sequential repetitive gene deletions in Neurospora crassa employing a self-excising β-recombinase/six cassette. J Microbiol Methods 92:236–243. doi: 10.1016/j.mimet.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 28.Szewczyk E, Kasuga T, Fan Z. 2014. A new variant of self-excising β-recombinase/six cassette for repetitive gene deletion and homokaryon purification in Neurospora crassa. J Microbiol Methods 100:17–23. doi: 10.1016/j.mimet.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 29.Vogel H. 1956. A convenient growth medium for Neurospora (Medium N). Microb Genet Bull 13:42–43. [Google Scholar]
- 30.Baminger U, Nidetzky B, Kulbe KD, Haltrich D. 1999. A simple assay for measuring cellobiose dehydrogenase activity in the presence of laccase 35:253–259. [DOI] [PubMed] [Google Scholar]
- 31.King BC, Donnelly MK, Bergstrom GC, Walker LP, Gibson DM. 2009. An optimized microplate assay system for quantitative evaluation of plant cell wall-degrading enzyme activity of fungal culture extracts. Biotechnol Bioeng 102:1033–1044. doi: 10.1002/bit.22151. [DOI] [PubMed] [Google Scholar]
- 32.Gessner MO, Bauchrowitz MA, Escautier M, Renouvelables R, Marvig J. 1991. Extraction and quantification of ergosterol as a measure of fungal biomass in leaf litter. Microb Ecol 285–291. [DOI] [PubMed] [Google Scholar]
- 33.Orejas M, Maccabe AP, Pe A, Ramo D. 1999. Carbon catabolite repression of the Aspergillus nidulans xlnA gene. Mol Microbiol 31:177–184. doi: 10.1046/j.1365-2958.1999.01157.x. [DOI] [PubMed] [Google Scholar]
- 34.Tamayo EN, Villanueva A, Hasper Aa, de Graaff LH, Ramón D, Orejas M. 2008. CreA mediates repression of the regulatory gene xlnR which controls the production of xylanolytic enzymes in Aspergillus nidulans. Fungal Genet Biol 45:984–993. doi: 10.1016/j.fgb.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 35.Mathieu M, Felenbok B. 1994. The Aspergillus nidulans CREA protein mediates glucose repression of the ethanol regulon at various levels through competition with the ALCR-specific transactivator. EMBO J 13:4022–4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kulmburg P, Mathieu M, Dowzer C, Kelly J. 1993. Specific binding sites in the alcRb and aIcA promoters of the ethanol regulon for the CREA repressor mediating carbon catabolite repression in Aspergillus nidulans. Mol Microbiol 7:847–857. doi: 10.1111/j.1365-2958.1993.tb01175.x. [DOI] [PubMed] [Google Scholar]
- 37.Takashima S, Iikura H, Nakamura A, Masaki H, Uozumi T. 1996. Analysis of Cre1 binding sites in the Trichoderma reesei cbh1 upstream region. FEMS Microbiol Lett 145:361–366. doi: 10.1111/j.1574-6968.1996.tb08601.x. [DOI] [PubMed] [Google Scholar]
- 38.Strauss J, Mach RL, Zeilinger S, Hartler G, Stoffler G, Wolschek M, Kubicek CP. 1995. Cre1, the carbon catabolite repressor protein from trichoderma reesei. FEBS Lett 376:103–107. doi: 10.1016/0014-5793(95)01255-5. [DOI] [PubMed] [Google Scholar]
- 39.Holstege FCP, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95:717–728. doi: 10.1016/S0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 40.Becker PB, Hörz W. 2002. ATP-dependent nucleosome remodeling. Annu Rev Biochem 71:247–273. doi: 10.1146/annurev.biochem.71.110601.135400. [DOI] [PubMed] [Google Scholar]
- 41.Bailey C, Arst HN. 1975. Carbon catabolite repression in Aspergillus nidulans. Eur J Biochem 51:573–577. doi: 10.1111/j.1432-1033.1975.tb03958.x. [DOI] [PubMed] [Google Scholar]
- 42.Thomas-Chollier M, Defrance M, Medina-Rivera A, Sand O, Herrmann C, Thieffry D, van Helden J. 2011. RSAT 2011: regulatory sequence analysis tools. Nucleic Acids Res 39:W86–W91. doi: 10.1093/nar/gkr377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mach RL, Schindler M, Kubicek CP. 1994. Transformation of Trichoderma reesei based on hygromycin B resistance using homologous expression signals. Curr Genet 25:567–570. doi: 10.1007/BF00351679. [DOI] [PubMed] [Google Scholar]
- 44.Holtzapple M, Cognata M, Hendrickson C. 1990. Inhibition of Trichoderma reesei cellulase by sugars and solvents. Biotechnol Bioeng 36:275–287. doi: 10.1002/bit.260360310. [DOI] [PubMed] [Google Scholar]
- 45.Teugjas H, Väljamäe P. 2013. Product inhibition of cellulases studied with 14C-labeled cellulose substrates. Biotechnol Biofuels 6:104. doi: 10.1186/1754-6834-6-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Igarashi K, Samejima M, Eriksson KE. 1998. Cellobiose dehydrogenase enhances Phanerochaete chrysosporium cellobiohydrolase I activity by relieving product inhibition. Eur J Biochem 253:101–106. doi: 10.1046/j.1432-1327.1998.2530101.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.