Abstract
Human-associated bacteria dominate the built environment (BE). Following decontamination of floors, toilet seats, and soap dispensers in four public restrooms, in situ bacterial communities were characterized hourly, daily, and weekly to determine their successional ecology. The viability of cultivable bacteria, following the removal of dispersal agents (humans), was also assessed hourly. A late-successional community developed within 5 to 8 h on restroom floors and showed remarkable stability over weeks to months. Despite late-successional dominance by skin- and outdoor-associated bacteria, the most ubiquitous organisms were predominantly gut-associated taxa, which persisted following exclusion of humans. Staphylococcus represented the majority of the cultivable community, even after several hours of human exclusion. Methicillin-resistant Staphylococcus aureus (MRSA)-associated virulence genes were found on floors but were not present in assembled Staphylococcus pan-genomes. Viral abundances, which were predominantly enterophages, human papilloma virus, and herpesviruses, were significantly correlated with bacterial abundances and showed an unexpectedly low virus-to-bacterium ratio in surface-associated samples, suggesting that bacterial hosts are mostly dormant on BE surfaces.
INTRODUCTION
The analysis of microbial diversity of indoor environments, collectively termed the built environment (BE), is important because of its potential impact on human health. It is estimated that humans in industrialized countries spend as much as 90% of their lives indoors (1, 2). Indeed, for billions of humans, the “great indoors” comprises the new human ecosystem. BEs contain an enormous variety of potential microhabitats for microorganisms and are continually colonized by human and outdoor-associated microbiota (3–5). Understanding the ecological dynamics of the microbiota in BEs may help us develop strategies to define and promote an indoor microbiome that minimizes disease risk (2).
While it has long been known that viable bacteria can be cultured from virtually any surface in an indoor ecosystem, we know relatively little about the true diversity and viability of the indoor microbiome. In the past, studies of microbial diversity relied mainly on culture-based techniques (3, 6). However, the application of culture-independent sequencing techniques to the study of BE microbiology has already greatly expanded our understanding of the origin and diversity of BE microbes (2).
Comparisons of sequence data collected in one location to other existing data sets generated by the same approaches allow inference of the likely environmental origins of BE communities (e.g., human skin, soil, etc.) (7). Additionally, the impact of season and geographic location on bacterial community composition (5, 8) has revealed an extraordinary variability in BE-associated microbial diversity. However, with a few exceptions, most studies have involved single-time-point samplings of surfaces. While this allows for a characterization of microbial diversity and comparative analysis between surfaces, replicated time series studies need to be undertaken in order to understand the formation, stability, and dynamics of BE communities (9). In addition, most BE work has focused on bacterial communities, and there are few studies looking at viral community diversity (2). These are significant gaps that need to be filled in order to understand the distribution and behavior of the microbes that inhabit our BEs.
Restrooms are a shared public space with clear disease transmission potential (4). However, the potential for disease transmission from a surface fomite relies on the accumulation and continued viability of pathogenic taxa. A prior amplicon-sequencing study investigating the biogeography of restroom surfaces established putative colonization sources, gender-specific microbial signatures, and surface-specific community structure for restroom microbial communities (4). This spatial study revealed the dominance of human-associated microbes on restroom surfaces but did not examine questions of community assembly dynamics, temporal stability, or viability. In addition, this study focused on bacterial diversity and did not investigate patterns in viral abundance and diversity. Using a combination of 16S rRNA amplicon sequencing, shotgun metagenomics, culturing, culture-independent bacterial and viral abundance estimates, and building science measurements, we addressed the following questions. What are the successional dynamics of BE microbial communities? How stable are these communities over different timescales? Do we see reproducible assembly of the same microbial community? How and when do different source environments contribute to BE microbial communities? How long do microbes remain viable on BE surfaces, and do we see persistently viable human pathogens? What are the structure and diversity of the surface-associated viral communities and how does viral abundance relate to bacterial abundance? Finally, what effects do environmental factors (e.g., temperature, humidity, and occupancy) have on the diversity and abundance of microbiota? To answer these questions, we characterized the microbial community structure, function, and abundance on floors, toilet seats, and soap dispensers over time in four restrooms (high use and low use, male and female) following decontamination of each surface. Initially, surfaces were analyzed hourly, and then, they were analyzed daily for up to 8 weeks. To determine the influence of humans as a dispersal source, surfaces were sterilized again, and following 4 h of use, humans were excluded from the restrooms, and longitudinal changes in community structure and viability of floor-associated bacterial communities were determined.
MATERIALS AND METHODS
Sample collection.
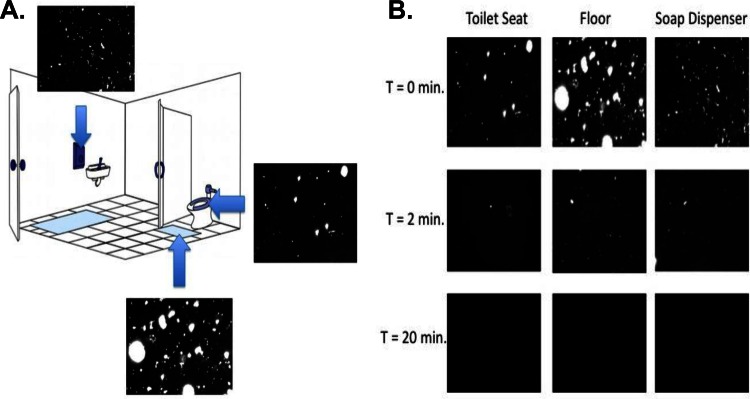
A visual summary of the experimental design, sampling locations, and decontamination results are included in Fig. 1 and Fig. S1 in the supplemental material. For detailed information about each study, see below.
FIG 1.
(A) Samples were collected from three surfaces in both female and male restrooms at San Diego State University. The surfaces analyzed were the toilet seat, the floor in front of the toilet, and the soap dispenser pump. Epifluorescence microscopy confirmed that bacteria and virus-like particles (VLPs) are present on all three surfaces. (Restroom drawing modified from Flores et al. [4]). (B) Epifluorescence microscopy images show selected restroom surfaces that are DNA and RNA free after 20 min of treatment with 10% bleach; T is the length of time the surface was soaked in bleach.
Eight-week study.
Samples were collected from three surfaces (the toilet seat, the floor in front of the toilet, and the soap dispenser pump) in two public restrooms on the third floor of the North Life Sciences building at San Diego State University (SDSU). Samples were collected once a week for an 8-week period between 22 November 2011 and 31 January 2012. For all studies presented in this article, the floor samples were collected from an area beginning directly below the edge of the toilet bowl and extending outwards away from the toilet. Eight hours prior to sample collection, each surface was decontaminated using a 10% bleach solution that was allowed to sit on the surface for approximately 20 min. After the 20-min bleach treatment, the surfaces were rinsed with sterile water (DNA- and RNA-free molecular biology grade water). The treated surfaces were shown to be DNA and RNA free by epifluorescence microscopy using SYBR Gold (Invitrogen) staining. Approximately 8 h after decontamination, sterile rayon-tipped swabs (MacroPur Swab P) that had been moistened with sterile 1× phosphate-buffered saline (PBS) were used to obtain surface samples from the targeted restroom surfaces. Areas of 50.8 cm × 50.8 cm were swabbed for each sample. The entire surface was passed over once with the swab, which took approximately 45 s. After surface swabbing, the tips of the swabs were broken off into 1.5-ml microtubes containing 500 μl of sterile 1× PBS solution. The swab tips were then immediately vortexed for 10 s. One hundred microliters of solution was removed and fixed in 100 μl of 4% paraformaldehyde for later analysis. The remaining samples were then stored at −20°C until further processing.
Eight-hour study.
Samples were collected every hour over an 8-h period from the floor in front of the toilet in two restrooms for females and two restrooms for males in the North Life Sciences building at SDSU. For sampling purposes, the floor in front of each toilet was partitioned into eight equally sized rectangles to ensure that we did not resample the same surface more than once in the 8-h period. This experiment was conducted on two different days (30 November 2012 and 5 December 2012) and on two different floors (first and third). The female and male restrooms on the third floor are open to everyone and are used frequently throughout the day. The restrooms on the first floor are locked and reserved for faculty and staff use only and are used much less frequently. An hour before sample collection began, the floor in front of each toilet was soaked in 10% bleach for 20 min as described above. Once an hour for eight time points, a sterile rayon-tipped swab that was moistened with sterile 1× PBS was used to swab one of the eight randomly selected rectangles (25.4 cm × 25.4 cm) on the floor. The entire floor surface was passed over once with the swab, which took approximately 30 s. The soap pump handles and the surfaces of the toilet seats were swabbed, making sure the entire surface was passed over once. Sampling and storage for molecular work and microscopy were performed as described previously. The time series samples were completed in the four restrooms on the two sampling days.
Month-long study.
Samples were collected from three restroom surfaces (the toilet seat, the floor in front of the toilet, and the soap dispenser pump) in the same four restrooms used in the 8-h study every other day beginning 30 January 2013 and ending 27 February 2013. All sampled surfaces were treated with bleach as described above for the morning of 30 January 2013. This was the only time during the course of this experiment that the restroom surfaces were treated with bleach. The first samples were collected in the afternoon of 30 January 2013. Subsequent samples were collected every other afternoon until 27 February 2013, yielding 15 sampling time points. Floor areas (50.8 cm × 50.8 cm) were swabbed for each sample. The entire surface was passed over once with the swab, which took approximately 45 s. The soap pump handles and the surfaces of the toilet seats were swabbed, making sure the entire surface was passed over once. Sampling and storage for molecular work and microscopy were performed as described previously.
Human-free study.
In high-use public restrooms for males and females at San Diego State University, the floors (in front of the toilet seat in two stalls) were decontaminated as described above. The restroom floor was cleaned at 8:00 a.m., and the restrooms were opened for use as needed for 4 h. At 12:00, the restrooms were locked for the remainder of the day to allow for sample collection without restroom use. For sampling purposes, the floor in front of each toilet was partitioned into five equally sized rectangles (25.4 cm × 25.4 cm) to ensure that we did not resample the same surface more than once in the 8-h period. The entire surface was passed over once with the swab, which took approximately 30 s. Using aseptic technique, a double-tipped CultureSwab (BD, Franklin Lakes, NJ) was dipped in 0.02-μm-filtered 1× PBS and used to collect floor surface samples at 12:00, 14:00, 16:00, 18:00, and 20:00 h. One of the two swabs was broken off into microtubes filled with 500 μl of 1× PBS to be used for 16S rRNA gene sequencing and microscopy (as described above). The other swab was broken off into 600 μl of 0.02-μm-filtered 1× PBS to be used later for culturing. After the two microtubes were vortexed, 100 μl was removed from each 500-μl tube and added to 100 μl of 4% paraformaldehyde and later analyzed by microscopy. From each 600-μl tube intended for culturing, 80 μl was added to 5 ml of tryptic soy broth (TSB) (BD, Franklin Lakes, NJ) in 15-ml-capacity Falcon tubes for culturing at 4 conditions, to mimic room temperature and human body temperature: aerobic 25°C and 37°C and anaerobic 25°C and 37°C. For the anaerobic conditions, AnaeroGen (Oxoid, Lenexa, KS) packets were used to remove oxygen inside an airtight jar containing anaerobic samples, and oxygen indicator strips were used to verify the absence of oxygen for the entire culture period. For all samples, including negative and positive controls, the lids were cracked slightly to ensure air exchange as typical for culturing. Aerobic samples were incubated for 2 days, and anaerobic samples were incubated for 4 days. After the incubation period, the samples were stored in a −20°C freezer until they were sent to Argonne National Laboratory for DNA extraction, PCR, and sequencing.
Microscopy.
Epifluorescence microscopy was used to ensure that all samples collected contained microbes and virus-like particles (VLPs) and to estimate the abundance of both bacteria and VLPs in each sample. We used methods developed by Caporaso et al. in the Rohwer laboratory (10). Briefly, 100 μl of each of the paraformaldehyde-fixed samples collected from restrooms was suspended in 5 ml of sterile water and then filtered onto 0.02-μm Whatman Anodisc filter membranes. The filters were then stained with 1× SYBR Gold for 10 min in the dark before the filters were rinsed and mounted onto slides. The slides were visualized using the Leica microscope at the Electron Microscope Facility at San Diego State University. Image-Pro Plus software was used to record digital images of the slides and generate estimates of both bacterial and viral abundance.
Bacterial DNA extraction.
Bacterial DNA was extracted directly from swab tips and residual 1× PBS collection buffer using the PowerSoil DNA isolation kit (Mo Bio Laboratories) following the protocol of Flores et al. (4). The 8-h study and the 1-month study used the same protocol, but DNA was extracted using the PowerSoil-htp 96-well Soil DNA isolation kit (Mo Bio Laboratories). Extracted DNA was sent to Argonne National Laboratory for sequencing.
DNA amplicon and metagenome sequencing.
Amplicon sequencing was performed using primers designed to be massively multiplexed and cover the V4-V5 hypervariable region of the 16S rRNA gene using the standard methods outlined by the Earth Microbiome Project (EMP) (http://www.earthmicrobiome.org/emp-standard-protocols/16S/) (11). Samples were sequenced on the Illumina MiSeq platform, at the Argonne National Laboratory core sequencing facility (11). Shotgun metagenomic sequencing for single-read annotation was performed on 7 surface swab samples (F1FH811302012, F1MH211302012, F3FH412052012, F3FH711302012, F3MH611302012, F3MH811302012, and F3MH412052012). Metagenomic libraries were prepared using 1 ng of genomic DNA and the Nextera XT protocol according to the manufacturer's instructions (Illumina). Metagenomes were run through the MG-RAST annotation pipeline (12). Shotgun metagenomes were sequenced for an additional eight samples (S1Post25A12pmD3, S1Post25A4pmD3, S1Post37A12pmD1, S1Post37A8pmD1, S1Post37A8pmD3, S1Post37An12pmD1, S1Post37An4pmD1, and S1Post37An6pmD1) from cultured swabs for genome assemblies. Libraries were prepared as described above.
Sequence analysis.
QIIME (v. 1.7.0; QIIME stands for quantitative insights into microbial ecology; www.qiime.org) was used to filter reads and cluster operational taxonomic units (OTUs) as described previously (11, 13). Approximately 18% of the ∼17 million raw amplicon reads were removed during quality filtering, leaving ∼14 million reads for downstream analysis. Briefly, the open reference OTU picking script (pick_open_reference_otus.py) (14) was employed, where sequences were first clustered with the Greengenes (May 2013) reference database (15); OTUs that did not cluster with known taxa (at 97% identity) in the database were then clustered de novo. Singleton sequences were removed prior to downstream analyses. Representative sequences for each OTU were aligned using PyNast, with a minimum alignment overlap of 75 bp (16). Alignments were used to build a phylogenetic tree (FastTree [17]). We computed alpha-diversity metrics among substrates using the alpha_diversity.py script in QIIME (Shannon entropy, species richness, and phylogenetic diversity), at the same sequence depth for all samples (3,700 sequences per sample). The beta_diversity_through_plots.py script was used to compute beta-diversity distances between samples (weighted UniFrac) and to construct principal coordinate (PCoA) plots and account for both the phylogenetic composition (18) and relative abundance of taxa. Beta-diversity comparisons were done using ANOSIM (compare_categories.py; QIIME). We tested whether the abundances of particular OTUs differed significantly between surfaces or sampling times using analysis of variance (ANOVA; adjusted by Bonferroni's correction) with the otu_category_significance.py script. ANOVAs and linear regressions were run using the R software package to compare Shannon diversity to the metadata (19). QIIME was used to calculate the core microbial communities for different surfaces, times, and dates. Taxonomic distributions across sample categories were calculated (from phylum to genus levels) using the summarize_taxa_through_plots.py script in QIIME. The two-dimensional (2D) histogram of PCoA space was generated using Matplotlib (20).
Quality trimming and de novo metagenome assembly.
Raw shotgun sequence reads (paired end; average insert size, 180 bp) were quality trimmed using the high-throughput sequence analysis toolkit Nesoni (http://www.vicbioinformatics.com/software.nesoni.shtml) with the following parameter settings: adaptor, clip; match, 10; max errors, 1; clip, ambiguous yes; quality, 10; and length, 70. One of the samples (S1Post25A12pmD3) failed to sequence well, and 94.6% of sequences were lost during quality trimming. The remaining 7 assembly samples were high quality and lost only 5.6% of their raw reads, on average, during quality filtering. Quality trimmed data from 8 human free samples were assembled into scaffolds using Velvet (version 1.2.10) (21) set at the following parameter values, k = 51; exp_cov, 55; cov_cutoff, 5; ins_length, 180; ins_length_sd, 20; min_contig_lgth, 250; and scaffolding, yes. Taxonomic status was assigned to the metagenomes using MetaPhlAn (22).
Metagenome-based recovery of genomes.
Scaffolds (minimum length, 300 bp) from human free metagenome assemblies were clustered into bins using tetranucleotide frequency (TNF) usage anomaly Z-statistics, read depth, and percent G+C profiles. Briefly, a tetranucleotide frequency-based matrix was constructed for each assembly (scaffolds; minimum length, 3 kb) using custom R script. TNF values were arcsine square root transformed before clustering was performed via hierarchical agglomerative clustering with squared Euclidean distance and ward criterion in pv-clust package (23). A correlation-based (minimum pairwise R2 value = 0.9) subgraph was constructed for each cluster. Each subgraph was manually checked for read depth and percent G+C profiles and outliers (standard of the mean [SEM] ± 1) were excluded from further analysis. Taxonomic status was assigned to the reconstructed bins using BLAST2LCA program (https://github.com/emepyc/Blast2lca), and phylogenetically divergent contigs were also removed. The completeness of the reconstructed draft genomes (n = 6) was estimated based on the single-copy marker gene profiles (24). Paired-end read information was used to iteratively increase the length of draft genomes using the PRICE assembler (25). Metagenome raw data, assemblies, and reconstructed population genomes were compared against mecA (broad-spectrum beta-lactam resistance) and staphylococcal chromosome cassette mec (SCCmec) (mobile genetic element that carries the mecA gene) reference gene sequences (26) using BLASTX. Automated genome annotations of reconstructed Staphylococcus population genomes (see Table S1 in the supplemental material) were performed using RAST (27) and KAAS (28) servers. A whole-genome-based maximum likelihood phylogenetic tree was constructed for the reconstructed Staphylococcus genomes (Table S1) and reference genomes using PhyoPhlAn (29).
Accession number and data availability.
Raw amplicon data are available in the SRA database under accession number SRP049338. 16S and genome assembly raw data, along with sample metadata, can be accessed on FigShare (http://dx.doi.org/10.6084/m9.figshare.899218), and MG-RAST-annotated samples can be accessed on the MG-RAST webserver under project number 8313 (http://metagenomics.anl.gov/linkin.cgi?project=8313).
RESULTS AND DISCUSSION
Approximately 14 million high-quality 16S rRNA V4 amplicons representing 77,990 distinct operational taxonomic units (OTUs) were generated from 602 samples, along with bacterial and viral abundance counts. A total of 4.5 million metagenomic reads were sequenced from seven floor samples for single-read annotation, and 34 million reads were generated from eight cultured enrichments for genome reassembly.
Longitudinal analysis surface-associated microbial succession.
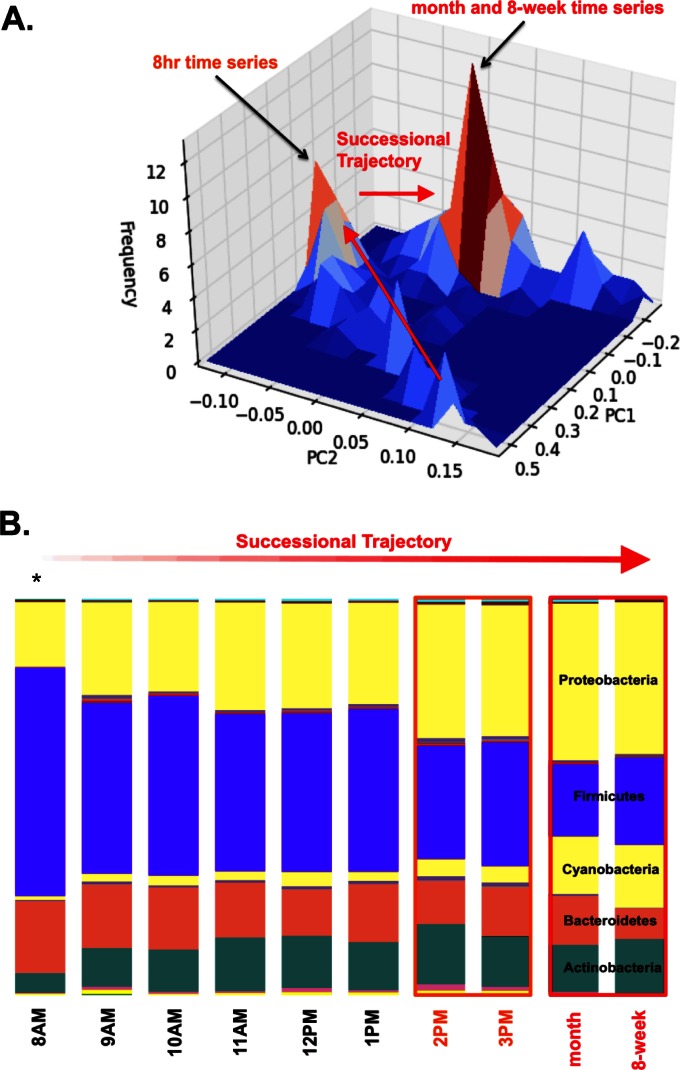
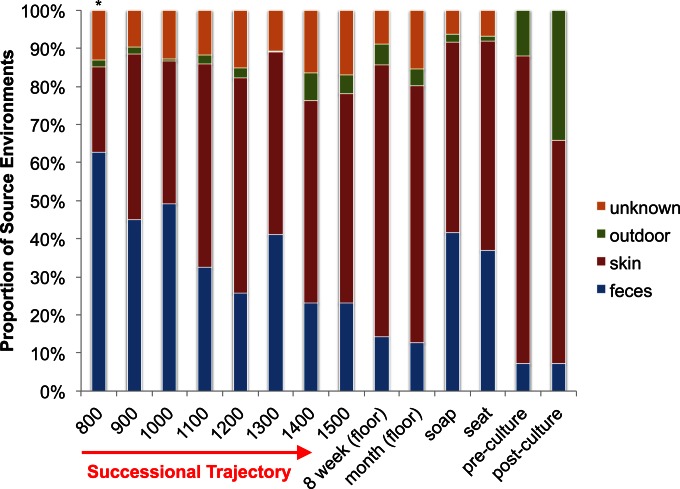
The communities associated with each surface (floor, soap dispenser, and toilet seats) converged upon a confined region within PCoA space within 5 h following decontamination, and the resulting late-successional surface community structure did not differ significantly across 8 weeks of continued sampling (ANOSIM, P > 0.8; Fig. 2A and 3). Floor communities showed a rapid reduction in the relative abundances of Firmicutes and Bacteroidetes, concomitant with a relative increase in the abundances of Proteobacteria, Cyanobacteria (78.3% of which are annotated as “Chloroplast,” which are likely to be derived from dietary plant biomass [30] or from plant material tracked in from outdoors) and Actinobacteria over the course of a day (Fig. 2B). Succession was highly reproducible for floor-associated communities, showing equivalent trajectories in four different restrooms over two separate days (Fig. 3A to C), and beta-diversity distance was significantly correlated with time (mantel r = 0.226, P < 0.0001). Floor-associated communities quickly develop toward a meta-stable region within PCoA space (the smaller of the two peaks in Fig. 2A; Fig. 3C). However, over a slightly longer time frame, a more stable optimum is defined by a further reduction in Firmicutes and expansion in Proteobacteria and Cyanobacteria (Fig. 2). A Bayesian classifier known as SourceTracker (7), trained on the Earth Microbiome Project (EMP) database (includes both human-associated and environmental samples), showed that the early successional community is dominated by taxa associated with feces that are likely aerosolized by toilet flushing (Fig. 4) and are largely displaced by skin- and outdoor-associated taxa within 8 h (Fig. 2B and 4). The stable communities present on toilet seats and soap dispensers were comprised of ∼45% feces- and ∼45% skin-associated taxa (Fig. 4).
FIG 2.
(A) A 2D histogram of floor samples (including 8-h, 8-week, and month-long experiments) in principal coordinate space (weighted UniFrac). Peaks denote the areas within principal coordinate space where samples are found most frequently (regions of stability). The smaller peak corresponds to the later time points in the 8-h study. The larger peak shows the stable community state that remains relatively fixed in the 9-week and month-long samplings (corresponding to the community structures highlighted in red in panel B). We observed only early successional community composition in the 8-h time series. Over longer timescales, the community was consistently found in the late-successional state. (B) Composition of the microbial community along its successional trajectory. The asterisk above the 8 a.m. time point denotes the sample taken directly following rigorous decontamination of the floor surface with bleach. The orange and red boxes surrounding time points refer to the average community states characteristic of the two peaks (labeled with the corresponding colors [orange and red]) seen in panel A.
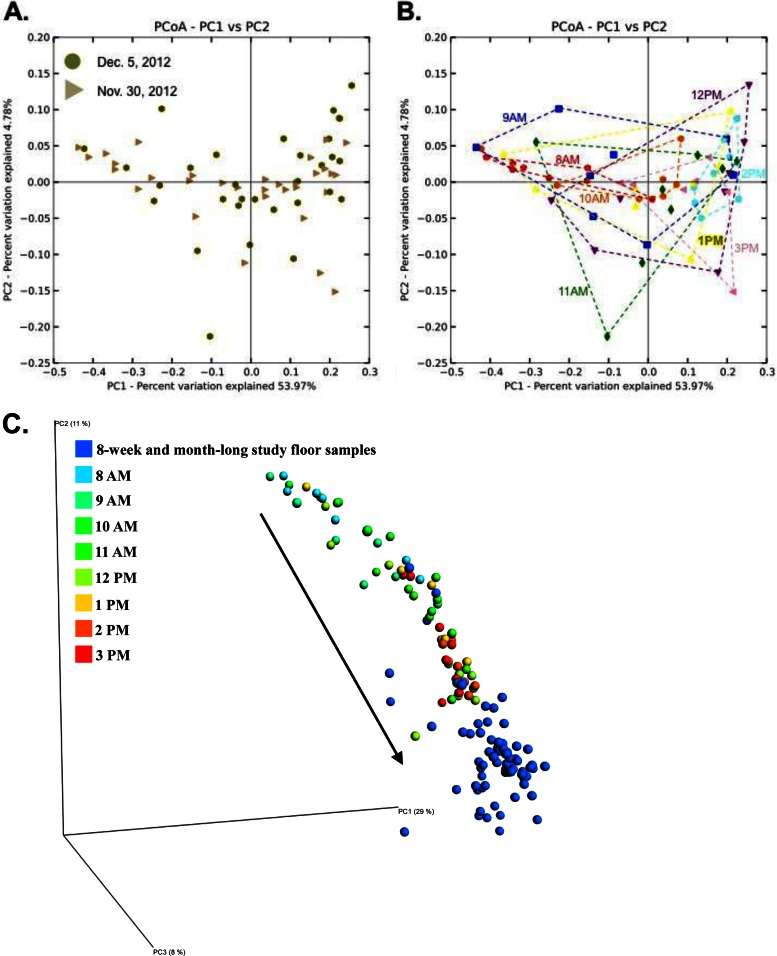
FIG 3.
Principal coordinate (PCoA) plots of floor microbial communities over different timescales. (A) Replicate 8-h time series experiments cluster on top of one another. (B) Same plot as in panel A, but with samples colored by sample collection time point, and time point replicates encapsulated by convex hulls. (C) Samples from the 8-h experiment (rainbow colors) show a larger spread than samples taken from longer-term studies (8-week and 1-month with daily sampling; dark blue), showing that succession is rapid and occurs within 5 to 8 h. The black arrow in panel C shows the successional trajectory.
FIG 4.
Relative proportions of OTUs derived from particular source environments (as determined using SourceTracker). The first eight samples represent the averages of replicate samples taken at each hour for the 8-h study. The 8-week and month bars represent the averages of all replicates for floor samples only. The soap and seat bars show the average of replicates for those surfaces across the 8-week and month-long studies. The pre- and postculture bars represent the averages from the human exclusion study (floor samples prior to culturing and after culturing). The asterisk above the 8 a.m. bar indicates that the floor was decontaminated prior to taking this sample. The source environment database was constructed using Earth Microbiome Project (EMP) data (closed reference OTUs; Greengenes release from May 2013). The outdoor category includes database samples from many outdoor environments: freshwater, freshwater microbial mat, freshwater sediment, bird nest, hot spring water, hot spring microbial mat, ice, marine biofilm, marine water, marine sediment, hypersaline water, sand, sandstone, and soil.
Environmental factors influencing surface-associated microbial community structure.
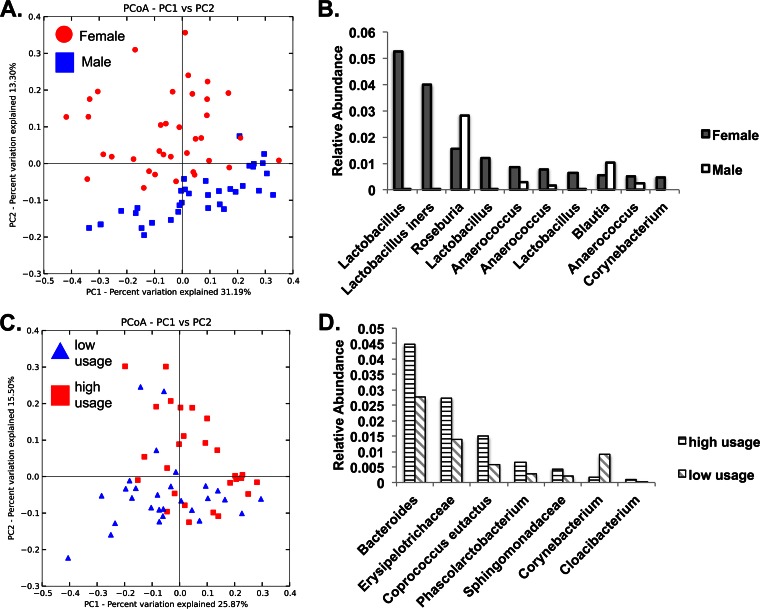
Microbial communities clustered significantly based on sample surface (ANOSIM R = 0.7428, P < 0.001). Only toilet seat samples clustered based on the sex of the population using the restroom (ANOSIM R = 0.199, P = 0.001; Fig. 5A), with Lactobacillus and Anaerococcus dominating toilet seats in the restrooms for females (31–33), and the gut-associated Roseburia and Blautia being more abundant on toilet seats in the restrooms for males (ANOVA, false discovery rate [FDR]-corrected P < 0.05; Fig. 5B). High-use and low-use restrooms had significantly different toilet seat and soap dispenser microbial communities (ANOSIM R = 0.142 and P = 0.001 and ANOSIM R = 0.091 and P = 0.001, respectively; Fig. 5C), but floor-associated communities were not significantly different. Fecal-associated OTUs were found at a greater relative abundance on high-use toilet seats (e.g., Bacteroidetes and Coprococcus), while skin-associated OTUs were more prevalent on low-usage seats (e.g., Corynebacterium; Fig. 5D).
FIG 5.
(A) PCoA (weighted UniFrac) shows a clear separation of toilet seat-associated microbial communities based on gender. (B) Top 10 most abundant OTUs that show significant differences in abundance between the toilet seats in male and female restrooms (most resolved taxonomic annotations are shown on the x axis). (C) Toilet seat samples cluster separately based on restroom usage frequency (high versus low). (D) Seven OTUs that exhibit significantly different abundances between high- and low-usage frequencies (most resolved taxonomic annotations are listed along the x axis).
Changes in community structure after human exclusion.
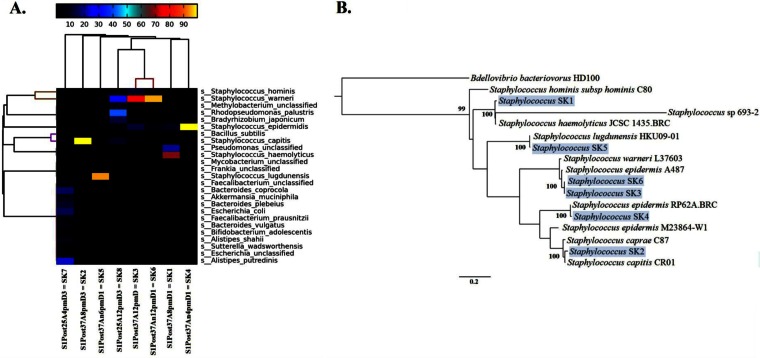
In the 8 h following the exclusion of humans, there was no significant reduction in the prevalence of fecal microbiota (Pearson's R = −0.1093; P = 0.45). This persistence of fecal microbiota, as in the late-successional floor community, is potentially due to dormant or endospore-forming taxa (e.g., Firmicutes; Fig. 2B). We suggest that active fecal microbiota would experience a significant shock when transiting between the warm, moist, anaerobic conditions of the host to the colder, drier, aerobic floor. Thus, we propose that fecal taxa able to enter into a dormant phase can persist for longer periods of time on restroom surfaces, although further work needs to be done to quantify dormancy in these BE systems. The floor community structure, following human exclusion, resembled the late-successional floor communities, while the viable organisms that grew in culture from these same samples resembled a subset of the communities associated with soap dispensers and toilet seats (weighted Unifrac; see Fig. S2 in the supplemental material). These viable communities were not significantly different across time or between culture conditions (i.e., anaerobic or aerobic, at 25°C or 37°C; weighted UniFrac; ADONIS, P > 0.8). Despite the lack of statistical difference in beta-diversity between culture conditions, we did see notable differences in the source environments. Skin- and outdoor-associated taxa comprised 68 to 98% of the cultured communities, and fecal taxa represented 0 to 15% of the cultured communities. On average, both pre- and postculture communities had a larger percentage of outdoor-associated taxa than other samples from the prior experiments, which suggests that, over time, human-associated taxa are displaced in the absence of dispersal (Fig. 4). Looking specifically at different culture conditions, ambient temperature (25°C), under both aerobic and anaerobic culture conditions, showed equal proportions of environment- and skin-associated taxa in the cultures (∼1:1). However, at human body temperature (37°C), skin-associated microbiota were dominant (∼70% of the community), regardless of oxygen potential. Several Staphylococcus taxa were positively correlated with incubation temperature (Bonferroni-corrected Pearson's P < 0.05), and several OTUs shared 100% nucleotide similarity across the 16S rRNA V4 region with Staphylococcus aureus, which is the most common cause of skin and soft tissue infections (34). The presence of Staphylococcus isolates was verified by metagenome assembly of several pan-genomes from cultured samples (Fig. 6; see Table S1 in the supplemental material), which shows that opportunistic pathogens remain viable on surfaces for many hours following human exclusion. The assembled pan-genomes made up large fractions of the total number of shotgun reads from most of the sequenced culture metagenomes (Table S2). Table S3 shows the distribution of annotated genes across SEED subsystem functional categories for each pan-genome.
FIG 6.
Species-level metagenomic diversity from metagenomic assemblies of postculture swab samples from the human free study. (A) Species-level (SK1-SK8) relative abundance patterns across eight metagenomes, with hierarchical clustering of both rows and columns (average linkage clusters, using Bray-Curtis distance), reporting only the 25 most abundant species annotations (according to the 90th percentile of the abundances). The heatmap key shows percent relative abundance. (B) Rooted tree showing the phylogenetic positions of six Staphylococcus population genomes (shown on a light blue background), along with reference strains and Bdellovibrio bacteriovorus HD100 as the outgroup. The tree was constructed using PhyloPhlAn (29) with concatenated amino acid sequences from ∼400 conserved proteins. Values assigned to internal nodes within the phylogeny represent bootstrap support (bootstrap values of <0.50 are not reported). Bar, 0.2 changes per amino acid position.
Composition of the surface-associated core microbial community.
Of the taxa that were ubiquitous across samples, most were associated with phyla that are known to dominate the human gut (except for Corynebacterium). While skin-associated OTUs dominated overall, they were not represented in this core community, because they were likely derived from a more diverse seed bank, as the human skin microbiome is more variable through time than the gut microbiome is (35). Concordantly, Shannon diversity rises significantly from morning to afternoon (in the 8-h study), which supports the idea that late-successional taxa are derived from more-diverse sources (R2 = 0.282, P < 0.0001; see Fig. S3A in the supplemental material).
Viral/bacterial ratios and the composition of the viral metagenome.
Bacterial and viral abundances, as determined by epifluorescence microscopy, were significantly positively correlated with each other (R = 0.764, P < 0.0001; see Fig. S3B in the supplemental material) and negatively correlated with temperature (R2 = 0.048 and R2 = 0.153, respectively; P < 0.03). Bacterial abundance was positively, albeit weakly, correlated with humidity (R2 = 0.0412, P < 0.02). Viral abundance was significantly lower in high-usage restrooms (ANOVA, P < 0.002), but no significant relationship existed between either viral or bacterial abundance and gender. Absolute viral and bacterial abundance show no discernible trend across time, as the bacterial community reaches an abundance plateau (∼6.2 × 103 cells cm−2, as determined by microscopy) within 1 h after decontamination, suggesting limited growth and a high rate of dispersal. The viral/bacterial (v/b) ratio was ∼1:1, which is around 10 to 20 times lower than considered typical for environmental samples (36). We speculate that this lowered ratio suggests microbial dormancy (e.g., sporulation, persister cells, or lowered metabolic rates), as phage lytic cycles cannot occur in dormant cells. Even if lytic cycles could occur, the low bacterial density observed on these surfaces may further limit the spread of phage (37).
In the human exclusion experiment, v/b ratios tended to be lower (0.2 to 0.3). The ratios differed across culture conditions as well (0.204 ± 0.253 [mean ± standard deviation {SD}] for aerobic at 25°C, 0.040 ± 0.006 for anaerobic at 25°C, 0.211 ± 0.131 for aerobic at 37°C, and 0.338 ± 0.151 for anaerobic at 37°C). Most of the phages detected on restroom surfaces were enterophages (within Microviridae; see Fig. S4 in the supplemental material), and because there were few gut microbes that persisted on surfaces or that grew in culture it follows that phage abundance would remain low in the absence of their hosts. The fact that the 25°C, anaerobic condition consistently yielded the lowest v/b ratio may indicate that most phages/hosts are unlikely to have encountered this condition in their original environmental context (e.g., the temperature of the anaerobic gut environment is 37°C, and non-host-associated environments that are likely to be dispersal sources for restrooms, like soils, tend to be aerobic). The most abundant viral group within Microviridae was enterophage ϕX174, indicating that most of the viral sequences detected on restroom surfaces were derived from the gut. Human papillomaviruses (within the Papillomaviridae family) and human herpesviruses (within Herpesvirales) were also detected in high abundance, which are associated with skin and epithelium (38, 39).
Metagenome data show the prevalence of bacterial virulence factors.
In addition to standard cellular functions such as carbohydrate and protein metabolism (see Fig. S5A in the supplemental material), virulence-associated functions were present, including fluoroquinolone resistance and heavy metal efflux and multidrug efflux pumps (Fig. S5B). Staphylococcus methicillin resistance pathways were the sixth most abundant category (representing 3% of “Bacterial” sequences; Fig. S5C). S. aureus is increasing in prevalence outside the hospital environment (34), and these results suggest that it might be a common constituent of public restroom surfaces. Culturing work showed a high prevalence of staphylococcal species, but the assembled pan-genomes of cultured organisms did not contain methicillin resistance genes.
Conclusions.
Compared to host-associated environments, restroom surfaces are dry, barren, and resource poor. As such, these surfaces probably do not support considerable microbial growth, as evidenced by low cell densities. Continual dispersal, dormancy, and cell death appear to be the dominant forces shaping community structure through time, with minor contributions from cell growth and competition. The prevalence of skin-associated, rather than feces-associated taxa, in the late-successional community suggests that organisms are selected for their ability to persist in a dry, aerobic environment, which is a very different environment from the gut. Human-associated microbiota, including Staphylococcus strains, can remain viable on BE surfaces for many hours after their dispersal agents are removed. This suggests that common BE surfaces may be significant fomites for viable human pathogens.
Supplementary Material
ACKNOWLEDGMENTS
S.M.G. was supported by an EPA STAR Graduate Fellowship and the National Institutes of Health Training Grant 5T-32EB-009412. We acknowledge funding from the Alfred P. Sloan Foundation's Microbiology of the Built Environment Program. This work was completed in part with resources provided by the University of Chicago Research Computing Center.
We thank Sarah Owens and Jarrad Hampton-Marcell at Argonne National Laboratory for help with DNA extraction, amplification, and sequencing. Thanks to Reto Trappitsch for help with Matplotlib and Maureen L. Coleman for helpful discussions. We also thank Julia Bell for help with study organization and planning.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03117-14.
REFERENCES
- 1.Custovic A, Taggart S, Woodcock A. 1994. House dust mite and cat allergen in different indoor environments. Clin Exp Allergy 24:1164. doi: 10.1111/j.1365-2222.1994.tb03323.x. [DOI] [PubMed] [Google Scholar]
- 2.Kelley ST, Gilbert JA. 2013. Studying the microbiology of the indoor environment. Genome Biol 14:202. doi: 10.1186/gb-2013-14-2-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kembel SW, Jones E, Kline J, Northcutt D, Stenson J, Womack AM, Bohannan BJ, Brown G, Green JL. 2012. Architectural design influences the diversity and structure of the built environment microbiome. ISME J 6:1469–1479. doi: 10.1038/ismej.2011.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flores GE, Bates ST, Knights D, Lauber CL, Stombaugh J, Knight R, Fierer N. 2011. Microbial biogeography of public restroom surfaces. PLoS One 6:e28132. doi: 10.1371/journal.pone.0028132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rintala H, Pitkäranta M, Toivola M, Paulin L, Nevalainen A. 2008. Diversity and seasonal dynamics of bacterial community in indoor environment. BMC Microbiol 8:56. doi: 10.1186/1471-2180-8-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Angenent LT, Kelley ST, Amand AS, Pace NR, Hernandez MT. 2005. Molecular identification of potential pathogens in water and air of a hospital therapy pool. Proc Natl Acad Sci U S A 102:4860–4865. doi: 10.1073/pnas.0501235102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knights D, Kuczynski J, Charlson ES, Zaneveld J, Mozer MC, Collman RG, Bushman FD, Knight R, Kelley ST. 2011. Bayesian community-wide culture-independent microbial source tracking. Nat Methods 8:761–763. doi: 10.1038/nmeth.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hewitt KM, Gerba CP, Maxwell SL, Kelley ST. 2012. Office space bacterial abundance and diversity in three metropolitan areas. PLoS One 7:e37849. doi: 10.1371/journal.pone.0037849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knight R, Jansson J, Field D, Fierer N, Desai N, Fuhrman JA, Hugenholtz P, van der Lelie D, Meyer F, Stevens R. 2012. Unlocking the potential of metagenomics through replicated experimental design. Nat Biotechnol 30:513–520. doi: 10.1038/nbt.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sano E, Carlson S, Wegley L, Rohwer F. 2004. Movement of viruses between biomes. Appl Environ Microbiol 70:5842–5846. doi: 10.1128/AEM.70.10.5842-5846.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glass EM, Meyer F. 2011. The metagenomics RAST server: a public resource for the automatic phylogenetic and functional analysis of metagenomes, p 325–331. In de Bruijn FJ. (ed), Handbook of molecular microbial ecology I: metagenomics and complementary approaches. John Wiley & Sons, Inc, Hoboken, NJ. [Google Scholar]
- 13.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rideout JR, He Y, Navas-Molina JA, Walters WA, Ursell LK, Gibbons SM, Chase J, McDonald D, Gonzalez A, Robbins-Pianka A. 2014. Subsampled open-reference clustering creates consistent, comprehensive OTU definitions and scales to billions of sequences. PeerJ 2:e545. doi: 10.7717/peerj.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. 2012. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. 2010. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lozupone C, Knight R. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.R Core Development Team. 2008. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 20.Hunter JD. 2007. Matplotlib: a 2D graphics environment. Comput Sci Eng 9:90–95. doi: 10.1109/MCSE.2007.55. [DOI] [Google Scholar]
- 21.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haft DH, Tovchigrechko A. 2012. High-speed microbial community profiling. Nat Methods 9:793–794. doi: 10.1038/nmeth.2080. [DOI] [PubMed] [Google Scholar]
- 23.Suzuki R, Shimodaira H. 2006. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22:1540–1542. doi: 10.1093/bioinformatics/btl117. [DOI] [PubMed] [Google Scholar]
- 24.Wu M, Scott AJ. 2012. Phylogenomic analysis of bacterial and archaeal sequences with AMPHORA2. Bioinformatics 28:1033–1034. doi: 10.1093/bioinformatics/bts079. [DOI] [PubMed] [Google Scholar]
- 25.Ruby JG, Bellare P, DeRisi JL. 2013. PRICE: software for the targeted assembly of components of (meta) genomic sequence data. G3 3:865–880. doi: 10.1534/g3.113.005967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements (IWG-SCC). 2009. Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob Agents Chemother 53:4961–4967. doi: 10.1128/AAC.00579-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. 2007. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res 35:W182–W185. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Segata N, Börnigen D, Morgan XC, Huttenhower C. 2013. PhyloPhlAn is a new method for improved phylogenetic and taxonomic placement of microbes. Nat Commun 4:2304. doi: 10.1038/ncomms3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kelley ST, Dobler S. 2011. Comparative analysis of microbial diversity in Longitarsus flea beetles (Coleoptera: Chrysomelidae). Genetica 139:541–550. doi: 10.1007/s10709-010-9498-0. [DOI] [PubMed] [Google Scholar]
- 31.Vásquez A, Jakobsson T, Ahrné S, Forsum U, Molin G. 2002. Vaginal Lactobacillus flora of healthy Swedish women. J Clin Microbiol 40:2746–2749. doi: 10.1128/JCM.40.8.2746-2749.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma B, Forney LJ, Ravel J. 2012. Vaginal microbiome: rethinking health and disease. Annu Rev Microbiol 66:371–389. doi: 10.1146/annurev-micro-092611-150157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SS, McCulle SL, Karlebach S, Gorle R, Russell J, Tacket CO. 2011. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci U S A 108:4680–4687. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.LeBlanc DM, Reece EM, Horton JB, Janis JE. 2007. Increasing incidence of methicillin-resistant Staphylococcus aureus in hand infections: a 3-year county hospital experience. Plast Reconstr Surg 119:935–940. doi: 10.1097/01.prs.0000252270.84195.41. [DOI] [PubMed] [Google Scholar]
- 35.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. 2009. Bacterial community variation in human body habitats across space and time. Science 326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weinbauer MG. 2004. Ecology of prokaryotic viruses. FEMS Microbiol Rev 28:127–181. doi: 10.1016/j.femsre.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 37.Lewis K. 2010. Persister cells. Annu Rev Microbiol 64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 38.Davison AJ, Eberle R, Ehlers B, Hayward GS, McGeoch DJ, Minson AC, Pellett PE, Roizman B, Studdert MJ, Thiry E. 2009. The order Herpesvirales. Arch Virol 154:171–177. doi: 10.1007/s00705-008-0278-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walboomers JM, Jacobs MV, Manos MM, Bosch FX, Kummer JA, Shah KV, Snijders PJ, Peto J, Meijer CJ, Munoz N. 1999. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol 189:12–19. doi:. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.