Abstract
Propionyl coenzyme A (propionyl-CoA) is an important intermediate during the biosynthesis and catabolism of intracellular carbon storage of poly(3-hydroxybutyrate-co-3-hydroxyvalerate) (PHBV) in haloarchaea. However, the haloarchaeal propionyl-CoA carboxylase (PCC) and its physiological significance remain unclear. In this study, we identified a PCC that catalyzed propionyl-CoA carboxylation with an acetyl-CoA carboxylation side activity in Haloferax mediterranei. Gene knockout/complementation demonstrated that the PCC enzyme consisted of a fusion protein of a biotin carboxylase and a biotin-carboxyl carrier protein (PccA [HFX_2490]), a carboxyltransferase component (PccB [HFX_2478]), and an essential small subunit (PccX [HFX_2479]). Knockout of pccBX led to an inability to utilize propionate and a higher intracellular propionyl-CoA level, indicating that the PCC enzyme is indispensable for propionyl-CoA utilization. Interestingly, H. mediterranei DBX (pccBX-deleted strain) displayed multiple phenotypic changes, including retarded cell growth, decreased glucose consumption, impaired PHBV biosynthesis, and wrinkled cells. A propionyl-CoA concentration equivalent to the concentration that accumulated in DBX cells was demonstrated to inhibit succinyl-CoA synthetase of the tricarboxylic acid cycle in vitro. Genome-wide microarray analysis showed that many genes for glycolysis, pyruvate oxidation, PHBV accumulation, electron transport, and stress responses were affected in DBX. This study not only identified the haloarchaeal PCC for the metabolism of propionyl-CoA, an important intermediate in haloarchaea, but also demonstrated that impaired propionyl-CoA metabolism affected global metabolism in H. mediterranei.
INTRODUCTION
Haloarchaea represent a distinct group of Archaea that typically inhabit hypersaline environments, in which nutrient supplies could vary considerably over time. Therefore, most of these extremophiles have developed the adaptation mechanism of depositing poly(3-hydroxybutyrate-co-3-hydroxyvalerate) (PHBV) intracellularly to store carbon and energy when carbon sources are oversupplied and utilizing PHBV in the absence of exogenous carbon sources (1, 2). During the process of PHBV biosynthesis and utilization, propionyl coenzyme A (propionyl-CoA) is an important intermediate metabolite. In addition, propionyl-CoA is also an essential intermediate of the methylaspartate cycle, a pathway for acetate assimilation in haloarchaea (3). In Bacteria and Eukarya, propionyl-CoA metabolism has been extensively studied, as excess propionyl-CoA inside the cell causes toxic effects (4, 5). For example, deficiencies in propionyl-CoA utilization affect polyketide synthesis, cell growth, and morphology of conidia of Aspergillus fumigatus (5) or lead to the serious disease propionic acidemia in humans (4). However, the enzymes for propionyl-CoA metabolism as well as the physiological roles of propionyl-CoA metabolism remain unclear for haloarchaea.
Propionyl-CoA carboxylase (PCC) is the key enzyme for propionyl-CoA metabolism by catalyzing the carboxylation of propionyl-CoA to methylmalonyl-CoA. It is widely distributed in the three domains of life. PCC is typically composed of three functional components: the biotin carboxylase (BC), the biotin-carboxyl carrier protein (BCCP), and the carboxyltransferase (CT) (6). The crystal structure of a bacterial PCC holoenzyme has been reported, and a similar structure for human PCC has been obtained by using cryo-electron microscopy reconstruction (7). Recently, an acyl-CoA carboxylase with almost equal acetyl-CoA carboxylase (ACC) and PCC activities was characterized for the thermophilic archaeon Metallosphaera sedula (8). The ACC/PCC enzyme is responsible for two important carboxylation steps in the autotrophic carbon fixation cycle of the 3-hydroxypropionate/4-hydroxybutyrate cycle (9). Whether a similar ACC/PCC enzyme occurred in haloarchaea remained to be determined.
Haloferax mediterranei is a metabolically versatile haloarchaeon and has been used as a model for studies of haloarchaeal metabolism, and especially PHBV biosynthesis, for decades (10). This strain can accumulate a large amount of PHBV with a high 3-hydroxyvalerate (3HV) content (∼10 mol%) from many unrelated carbon sources (11). In the absence of an exogenous carbon source, PHBV is degraded for nutritional purposes, and a large quantity of propionyl-CoA was able to be produced. Bioinformatic analysis revealed that potential genes encoding propionyl-CoA carboxylase, methylmalonyl-CoA epimerase, and methylmalonyl-CoA mutase of the methylmalonyl-CoA pathway for propionyl-CoA utilization are present in the genome of H. mediterranei (10). Besides, knockout of the potential gene for methylmalonyl-CoA mutase in this strain led to the loss of the capability to grow on sodium propionate (our unpublished data). These results suggested that the methylmalonyl-CoA pathway, in which PCC is the carboxylating enzyme, is the only pathway for propionyl-CoA utilization in H. mediterranei. It is noteworthy that during PHBV biosynthesis, the 3-hydroxypropionate pathway, one of the four propionyl-CoA-supplying pathways, assimilates acetyl-CoA and CO2 into propionyl-CoA for the synthesis of the 3HV monomer (12). Acetyl-CoA carboxylation is a key step in this pathway; that is, two important carboxylation reactions, acetyl-CoA carboxylation and propionyl-CoA carboxylation, occurred in H. mediterranei for propionyl-CoA synthesis and utilization, respectively.
Therefore, in this study, we used H. mediterranei as a model strain to identify the haloarchaeal acyl-CoA carboxylase for propionyl-CoA metabolism and to investigate its importance in the global metabolic network.
MATERIALS AND METHODS
Strains and culture conditions.
The strains used in this study are listed in Table 1. Escherichia coli JM109 was cultured in lysogeny broth (LB) (13) at 37°C. When needed, 100 μg ml−1 of ampicillin was added to the medium. As for H. mediterranei strains, cells reached the late exponential phase after 36 h of growth in nutrient-rich AS-168 medium (5 g Casamino Acids, 5 g yeast extract, 1 g sodium glutamate, 3 g trisodium citrate, 2 g KCl, 20 g MgSO4 · 7H2O, 200 g NaCl, 5 mg FeSO4 · 7H2O, and 0.036 mg MnCl2 · 4H2O [pH 7.0] per liter) (14) at 37°C and were used as the seed culture. The seed culture was then inoculated into MG medium [1 g yeast extract, 1 g sodium glutamate, 20 g MgSO4 · 7H2O, 2 g KCl, 37.5 mg KH2PO4, 200 g NaCl, 5 mg FeSO4 · 7H2O, 0.036 mg MnCl2 · 4H2O, 15 g piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES), and 10 g glucose [pH 7.0] per liter) (15) with a 5% (vol/vol) inoculum size for additional cultivation to the exponential growth phase or stationary growth phase. For H. mediterranei DF50 (a uracil auxotroph mutant of H. mediterranei ATCC 33500) and DF50-based knockout mutants, uracil was supplied in the medium at a final concentration of 50 μg ml−1 (16). For the strains carrying the integration plasmid pHFX, the expression plasmid pWL502, or their derivatives, modified AS-168 medium, in which 5 g liter−1 of yeast extract was removed, and modified MG (mMG) medium, with 2 g liter−1 NH4Cl instead of 1 g liter−1 yeast extract, were used (16, 17). For examination of the ability of the strains to metabolize propionate, 7.5 ml of the seed culture was harvested, washed with a sterile 20% NaCl solution, and inoculated into 150 ml basal medium (18) with 2 g liter−1 of sodium propionate as the sole carbon source.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Reference |
|---|---|---|
| Strains | ||
| E. coli JM109 | recA1 supE44 endA1 hsdR17 gyrA96 relA1 thi | 13 |
| H. mediterranei DF50 | pyrF-deleted mutant of H. mediterranei ATCC 33500 | 16 |
| H. mediterranei DBX | pccBX-deleted mutant of H. mediterranei DF50 | This study |
| H. mediterranei ΔyccB | yccB-deleted mutant of H. mediterranei DF50 | This study |
| H. mediterranei DA | pccA-disrupted heterozygous single-crossover mutant (pccA+::ΔpccA) of H. mediterranei DF50 | This study |
| Plasmids | ||
| pHFX | 4.0 kb; integration vector containing pyrF and its native promoter | 16 |
| pHFXDpccBX | 5.1 kb; pHFX-derived integration vector for knockout of pccBX | This study |
| pHFXDyccB | 5.3 kb; pHFX-derived integration vector for knockout of yccB | This study |
| pHFXpccA | 4.6 kb; pHFX-derived integration vector for disruption of pccA | This study |
| pWL502 | 7.9 kb; expression vector containing pyrF and its native promoter | 17 |
| pWLpccB | 9.7 kb; pWL502-derived expression vector for expression of PccB | This study |
| pWLpccX | 8.4 kb; pWL502-derived expression vector for expression of PccX | This study |
| pWLpccBX | 10.0 kb; pWL502-derived expression vector for expression of PccBX | This study |
RT-PCR.
H. mediterranei DF50 cells were grown in MG medium until mid-exponential phase and harvested for total RNA isolation. Total RNA was isolated by using TRIzol reagent (Invitrogen-Life Technologies, USA). Reverse transcription (RT)-PCR was performed as described previously (14), except that the primers used for the RT reaction were random hexamer primers (Thermo Scientific); the primers for PCR are listed in Table 2.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′–3′)a | Use(s) |
|---|---|---|
| RTBX-F | TTCGCTAACCCGTACACG | RT-PCR of pccBX |
| RTBX-R | TGGAGATGTGCGCCGACT | |
| DBX-F1 | TAGCGGAAGCTTATCCCCGTCGCGTTCCTC | Knockout of pccBX and mutant verification |
| DBX-R1 | CGACGTACCGCTTACCAGCCGTGAAAGTAGCGTCCC | |
| DBX-F2 | GGGACGCTACTTTCACGGCTGGTAAGCGGTACGTCG | |
| DBX-R2 | GCGATAGGTACCAGTCACCATCGGCAGTTC | |
| DyccB-F1 | ATAGGATCCTGGGTAACGGCTCGCTGT | Knockout of yccB and mutant verification |
| DyccB-R1 | CTCACCCGACTCCCGACAGCCAGTAACTCAGTCAAG | |
| DyccB-F2 | CTTGACTGAGTTACTGGCTGTCGGGAGTCGGGTGAG | |
| DyccB-R2 | ATAGGTACCTAGCCGGGGTTGTCTCAC | |
| DA-F | TGCTGGATCCCGACCGAGCCCGCAGACT | Disruption of pccA |
| DA-R | GCGATAGGTACCGGGCTTCCGCGTTGATGC | |
| pyrF | GAGTACGGACTGGCAGAC | Mutant verification of pccA+::ΔpccA |
| DA-F | TGCTGGATCCCGACCGAGCCCGCAGACT | |
| AF | GATGGAGCGACTGGGTGA | |
| AR | CGTCGGTGAGCATGAGGC | |
| X-F1 | ATACCATGGTTTCCGGTCCGTTTTCTC | Expression of PccX |
| X-R1 | GAGCAGGTCCTCCGTCATGCCAGATCACCTTCTCTC | |
| X-F2 | GAGAGAAGGTGATCTGGCATGACGGAGGACCTGCTC | |
| X-R2 | ATAGGATCCACCGCTTACCAGTCAGAA | |
| B-F | ATACCATGGCGCTCTGTGTTTCCGGTC | Expression of PccB |
| B-R | ATAGGATCCGCAGGTCCTCCGTCATAG | |
| BX-F | TGCTGGATCCATTTGACAGCTAGTTGAT | Expression of PccBX |
| BX-R | TGCTGGATCCTCAGAACCGGTCAGTACG |
Sequences of the restriction sites are shown in boldface type.
Mutant construction and verification.
The plasmids and primers used for mutant construction and verification are listed in Tables 1 and 2, respectively. Mutant construction was performed as previously described (12). As for the knockout mutants, only those mutants with a complete loss of the target gene were selected by PCR verification. Gene complementation was performed by transformation of the H. mediterranei DF50-derived mutant with the corresponding pWL502-derived plasmid (17). The polyethylene glycol-mediated method was used for the transformation of H. mediterranei strains (19).
Enzyme assay.
H. mediterranei cells were grown in MG or mMG medium until mid-exponential phase and harvested by centrifugation (5,000 × g for 20 min at 4°C). Cell pellets (0.5 g [wet weight]) were suspended in 0.6 ml of 100 mM Tris-HCl buffer (pH 7.8) containing 2 M KCl and trace amounts of DNase and dithiothreitol (DTT). The cell suspension was then transferred into a 1.5-ml Eppendorf tube containing 1.1-g glass beads with a diameter of 0.10 to 0.25 mm. After prechilling, the Eppendorf tube was placed onto the vortex shaker of the Retsch Mixer Mill MM 200 system. The “bead-beating” process was performed at an agitation speed of 30 s−1 for 10 min. The lysate was centrifuged for 10 min (10,000 × g at 4°C) to obtain the supernatant for the enzyme assay. ACC and PCC activities were detected radiochemically by determining acetyl-CoA- or propionyl-CoA-dependent fixation of 14CO2 as described previously by Khomyakova et al. (3), except that the reaction mixture also contained 15 mM unlabeled NaHCO3. Succinyl-CoA synthetase activity was measured colorimetrically at 412 nm by recording the release of CoA from succinyl-CoA at 37°C, with 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB) as a CoA-detecting agent (20). The reaction mixture contained 100 mM Tris-HCl (pH 7.8), 2 M KCl, 5 mM MgCl2, 0.5 mM GDP, 5 mM KH2PO4, 1 mM DTNB, 0.14 mM succinyl-CoA, and cell extract at a final concentration of 1.5 mg protein ml−1. When inhibition of the succinyl-CoA synthetase with propionyl-CoA was examined, propionyl-CoA was added to the reaction mixture to a final concentration of 0.2 or 0.4 mM. During the enzyme assay, the reaction mixture without DTNB, succinyl-CoA, and cell extract was first incubated at 37°C for 2 min and set as a blank control. DTNB was then added to the reaction mixture. The cell extract was added until the optical density at 412 nm (OD412) became stable. When the OD412 became stable again, succinyl-CoA was then added to start the enzyme reaction. The reaction mixture without the addition of cell extract was set as the negative control. The protein concentration in the crude extracts was determined by using the Bradford method (21). All enzyme activities were expressed as nmol min−1 mg−1 protein.
Scanning electron microscopy and transmission electron microscopy analyses.
Both exponential- and stationary-phase H. mediterranei cells in MG or mMG medium were collected for scanning electron microscopy (SEM) analysis, and stationary-phase cells were collected for transmission electron microscopy (TEM) analysis. The SEM sample was prepared according to standard procedures (22), with some modifications. Briefly, high-salt SP buffer was used for fixing and rinsing cells, as described previously by Cai et al. (17). After that, the cells were dehydrated through a graded series of absolute ethanol and dried with CO2 at critical point in a critical-point drier (catalog number CPD030; Bal-Tec). The samples were then subjected to gold plating (catalog number SCD005; Bal-Tec) and observed with an FEI Quanta 200 SEM instrument at a 10-kV accelerating voltage. TEM analysis was performed according to a procedure described previously by Cai et al. (17).
Measurement of cell growth by the diphenylamine colorimetric method and determination of the residual glucose concentration in the culture.
The growth of H. mediterranei strains was monitored by the diphenylamine colorimetric method as described previously by Zhao et al. (23), with minor modifications. Harvested cells (∼20 mg [wet weight]) were washed twice with a 20% NaCl solution. Before the addition of diphenylamine reagent to the cell pellet, 100 μl of a 20% NaCl solution was used to resuspend the haloarchaeal cells.
The residual glucose concentration in the culture was determined enzymatically by using a biosensor analyzer (SBA-40C; Institute of Biology, Shandong Academy of Sciences, Shandong, China) after centrifugation of the culture to remove cells from the medium.
PHBV accumulation and 13C nuclear magnetic resonance analysis.
H. mediterranei strains cultured in MG or mMG medium until stationary phase were subjected to PHBV accumulation analysis. The PHBV content in the cells and its monomer composition were determined by gas chromatography (GC) analysis as previously described (14). For 13C nuclear magnetic resonance (NMR) analysis of PHBV, H. mediterranei strains were cultured by using a two-stage cultivation strategy (12). During the second stage, 10 g liter−1 of unlabeled glucose and 2 g liter−1 of NaH13CO3 (99%; Cambridge Isotope Laboratories) were supplied as carbon sources. At the end of fermentation, intracellular PHBV was isolated (24). The NMR spectra were recorded by using an Agilent DD2 500-MHz NMR spectrometer and processed by using MestRec software, as previously described (12).
High-performance liquid chromatography–electrospray ionization–tandem mass spectrometry detection of propionyl-CoA.
H. mediterranei cells (both wild-type and mutant strains) cultured in MG medium were used for high-performance liquid chromatography–electrospray ionization–tandem mass spectrometry (HPLC-ESI-MS/MS) detection of intracellular propionyl-CoA. Samples were prepared from both mid-exponential- and stationary-phase cells, as described previously by Peyraud et al. (25). HPLC-ESI-MS/MS analysis was performed as previously described (12). Based on the standard curve of propionyl-CoA (Sigma-Aldrich, USA) generated by HPLC-ESI-MS/MS, the intracellular content of propionyl-CoA in H. mediterranei cells was determined as nmol per OD595 DNA content. In order to obtain the intracellular propionyl-CoA concentration (mM), it was necessary to know the free intracellular water content in relation to the OD595 DNA content. For H. mediterranei cells, cells with a DNA content of 1 OD595 were ∼15 mg (dry weight) (our unpublished data), in which the free intracellular water content was estimated to be 7.5 μl (20, 26). We used this value for the calculation of the intracellular propionyl-CoA concentration; that is, the metabolite content of 1 nmol per OD595 DNA content equaled a concentration of ∼0.13 mM.
Microarray analysis of chemostat cultures of H. mediterranei strains.
H. mediterranei strains were cultured in custom-built chemostats with 120-ml working volumes. The cultures were fed with MG medium, in which glucose was the growth-limiting nutrient. The dilution rate, which at steady state equals the specific growth rate, was 0.05 h−1. The growth conditions in the chemostats were kept constant (temperature of 37°C at pH 7.0). Every 8 h, a 2-ml sample was taken and used to measure cell growth and residual glucose levels in the culture, as described above. Total RNA of H. mediterranei strains was isolated from chemostat cultures by using TRIzol reagent (Invitrogen-Life Technologies, USA). The oligonucleotide microarrays were designed by CapitalBio and manufactured by Agilent Technologies based on the H. mediterranei genome sequence. Three biological replicates were analyzed by using SAM (Significance Analysis of Microarray, version 2.23b) software (12). Genes exhibiting an expression change of >2.0-fold and a q value of <0.05 were considered to be significantly altered.
Protein sequence analysis.
Sequence homology was analyzed by using the BLAST service (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Domain annotation was carried out by using the NCBI conserved domain database (CDD) (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). Prediction of protein secondary structure was performed with the NPS@ Web server (41). Phylogenetic tree analysis was performed with MEGA4 software by using the neighbor-joining method. The topology of the phylogenetic tree was evaluated by bootstrap analysis based on 1,000 replicates.
RESULTS
Bioinformatic analysis of potential genes encoding the acyl-CoA carboxylase.
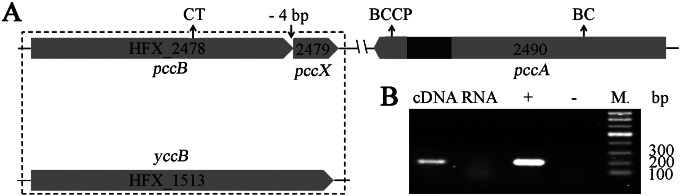
The BCCP, BC, and CT components of ACC/PCC from M. sedula are encoded by Msed_0147, Msed_0148, and Msed_1375, respectively (9). Based on their amino acid sequences, in silico analysis of the H. mediterranei genome was carried out and revealed the presence of genes potentially encoding the ACC/PCC components. In contrast to ACC/PCC from M. sedula, in H. mediterranei, BC and BCCP were fused into a single protein of 65.5 kDa encoded by pccA (HFX_2490). The N-terminal region (residues 1 to 441) and the C-terminal region (residues 535 to 601) served as the BC component and the BCCP component, respectively (Fig. 1A). Additionally, there were two genes potentially encoding the CT component. The pccB (HFX_2478) gene encoded a protein of 56.9 kDa, exhibiting 54% identity to the CT component from M. sedula, and yccB (HFX_1513) encoded a protein of 63.6 kDa, exhibiting 34% identity (Fig. 1A). Interestingly, pccX (HFX_2479), encoding a putative small protein of 9.2 kDa, was located immediately downstream of pccB and overlapped pccB by 4 bp (Fig. 1A). RT-PCR revealed that pccX was cotranscribed with pccB (Fig. 1B). Considering that pccB is a candidate gene encoding the CT component of the acyl-CoA carboxylase, pccX might also encode a component of the acyl-CoA carboxylase.
FIG 1.
Genetic organization of the potential acyl-CoA carboxylase genes in H. mediterranei. (A) Schematic diagram of the gene organization of potential acyl-CoA carboxylase genes. (B) RT-PCR determination of the cotranscription pattern of the pccBX gene cluster. For the “cDNA” lane, the cDNA reverse transcribed from the RNA of DF50 was the template; for the “RNA” lane, the RNA extracted from DF50 was the template; for the “+” lane, the genome was the template (positive control); the “−” lane represents the negative control; and the “M.” lane represents the marker.
Determination of PCC-encoding genes by enzyme assays.
Since we have identified potential genes encoding the acyl-CoA carboxylase, ACC and PCC activities were then assayed in crude extracts of H. mediterranei strain DF50 (a uracil auxotroph strain of H. mediterranei ATCC 33500). The results demonstrated that the PCC activity (0.63 ± 0.07 nmol min−1 mg−1 protein) was much higher than the ACC activity (0.07 ± 0.02 nmol min−1 mg−1 protein) in the crude extract. To determine the genes encoding the enzyme with PCC activity, we attempted to obtain in-frame markerless knockout mutants of pccBX, yccB, and pccA, respectively, based on the H. mediterranei DF50 strain. However, complete deletion of pccA failed, indicating its indispensability for cell growth. As pccA is the single-copy gene that encodes BC and BCCP, it may be shared by other biotin-dependent carboxylases in H. mediterranei, such as pyruvate carboxylase and a variety of acyl-CoA carboxylases (6). Alternatively, we tried to disrupt pccA by single-crossover recombination, resulting in a heterozygous mutant strain with both wild-type and disrupted gene copies. The phenomenon of heterozygosity is due to the multiple copies of haloarchaeal genomes (12, 27). Enzymatic assays of the crude extract prepared from exponential-phase cells showed that the PCC activity of mutant strain DBX (pccBX-deleted mutant) (Table 1) or DA (heterozygous pccA+::ΔpccA mutant) (Table 1) was undetectable. In contrast, deletion of yccB showed PCC activity of 0.42 ± 0.04 nmol min−1 mg−1 protein. As for the complemented DBX strains, the PCC activity of the complemented strains DBX(pccB) and DBX(pccX) was undetectable, while the PCC activity of DBX(pccBX) was 0.37 ± 0.15 nmol min−1 mg−1 protein. From these results, we deduced that PccB, PccX, and PccA were indispensable for PCC activity in H. mediterranei, while YccB did not play a significant role for PCC activity, and the role of YccB remained to be further determined. Therefore, the PCC enzyme in H. mediterranei consisted of the PccB, PccX, and PccA subunits. It is noteworthy that the PCC activity of the DBX(pccBX) strain was lower than that of the DF50 strain. The phenomenon of partial complementation of mutant phenotypes is widely found in different organisms (28). In the complemented DBX(pccBX) strain, the pccBX genes under the control of their native promoter were carried on a plasmid instead of being integrated into their original sites in the genome. The phenomenon of partial complementation might be caused by the differences in the gene context compared to the DF50 strain, as an inappropriate ratio or level of the PCC subunits in the complemented strain would affect PCC activity.
13C NMR analysis of the acetyl-CoA carboxylation activity by the PCC enzyme.
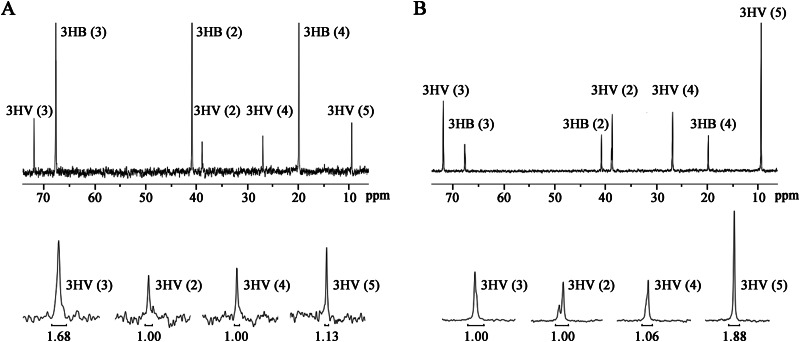
Regarding the low ACC activity in the crude extract in vitro, we performed 13C labeling experiments to determine if the PCC enzyme catalyzed acetyl-CoA carboxylation in vivo. It has been reported that the carbon atom of H13CO3− is incorporated as the C-1 atom of propionyl-CoA through the 3-hydroxypropionate pathway (path IV) (see Fig. S1 in the supplemental material), in which acetyl-CoA carboxylation is a key step, and then as the C-3 atom of the 3HV monomer of PHBV (12). Accordingly, PHBV samples extracted from DF50 and DBX cells grown with 10 g liter−1 glucose and 2 g liter−1 NaH13CO3 were subjected to 13C NMR spectroscopy analysis. Interestingly, compared with the molar ratio of the carbon atom set [3HV(2):3HV(3):3HV(4):3HV(5)] of PHBV of 1.00:1.68:1.00:1.13 for the DF50 strain (Fig. 2A), the ratio of that for the DBX strain was 1.00:1.00:1.06:1.88 (Fig. 2B). This result clearly revealed that H13CO3− was not assimilated into the C-3 position of the 3HV monomer in the DBX strain, indicating that the 3-hydroxypropionate pathway was blocked. Therefore, the PCC enzyme was also responsible for the carboxylation of acetyl-CoA in H. mediterranei.
FIG 2.
13C NMR spectroscopy analysis of PHBV extracted from H. mediterranei strains DF50 (A) and DBX (B) grown on glucose and NaH13CO3. Each peak corresponds to a certain carbon atom in 3HB or 3HV. A portion of the same spectrum is magnified for better visualization. The peak area of 3HV(2) is set as the standard of 1.0, and the relative areas of other peaks are compared with it.
As for the enrichment of 13C in 3HV C-5, which is derived from propionyl-CoA C-3, in the DBX strain (Fig. 2A and B), it might be attributed to H13CO3− incorporation into PHBV by pyruvate/phosphoenolpyruvate carboxylase (29). H13CO3− is first incorporated into oxaloacetate C-4. Oxaloacetate C-4 could enter into propionyl-CoA C-3 via the aspartate/2-oxobutyrate pathway (path II) (see Fig. S1 in the supplemental material). In the DBX strain, the 3-hydroxypropionate pathway (path IV) was blocked. The aspartate/2-oxobutyrate pathway (path II) might be upregulated, leading to the enrichment of 13C in 3HV C-5 (see Fig. S1 in the supplemental material). Taken together, results from ACC/PCC assays of crude extracts and 13C NMR showed that the PCC enzyme consisting of PccA, PccB, and PccX catalyzed the carboxylation of propionyl-CoA and also catalyzed the acetyl-CoA carboxylation side reaction.
Involvement of the PCC enzyme in propionate utilization.
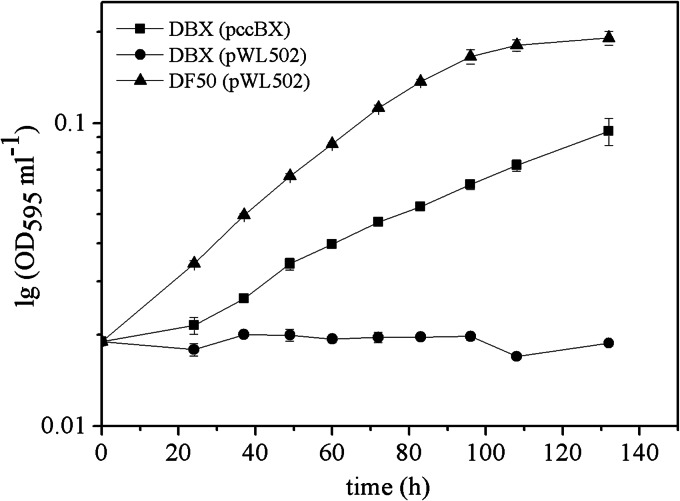
To investigate the role of the PCC enzyme in the utilization of propionate, we cultured DF50, DBX, and the complemented DBX strain with the pccBX genes under the control of their native promoter on basal medium with sodium propionate as the sole carbon source. As H. mediterranei can accumulate a large amount of PHBV in the form of bright and refractive intracellular granules, the optical density at 600 nm does not accurately reflect cell growth (30). In this study, the diphenylamine colorimetric method (shown as an OD595 curve on a semilog scale) was used to quantify DNA for the measurement of cell growth (23). The OD595 curves indicated that DF50 could grow on sodium propionate as the sole carbon source, while DBX could not (Fig. 3). The complemented DBX(pccBX) strain could also grow on sodium propionate, although it exhibited a lower growth rate than that of DF50 (Fig. 3). These results verified that the PCC enzyme was responsible for propionate utilization, and no other propionate utilization pathway occurred in H. mediterranei. The phenomenon of partial complementation of mutant phenotypes was consistent with that observed for the enzyme assay of PCC activity.
FIG 3.
Growth curves of H. mediterranei DF50(pWL502), DBX(pWL502), and the complemented strain DBX(pccBX), using sodium propionate as the sole carbon source. Cell growth was quantified by the diphenylamine colorimetric method for quantification of the DNA content and is shown as the OD595 ml−1 culture on a semilog scale.
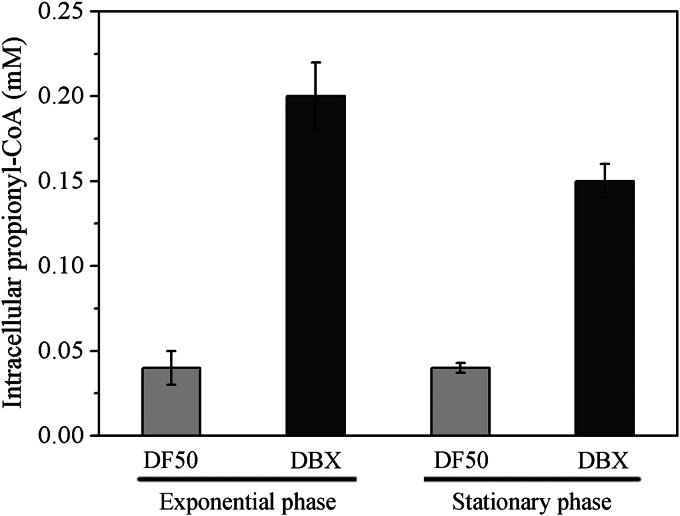
Excessive intracellular propionyl-CoA in DBX cells.
As the PCC enzyme in H. mediterranei was responsible for propionate utilization, deficiency of the PCC enzyme might lead to an excessive propionyl-CoA level in vivo. Thus, we investigated whether propionyl-CoA accumulated in DBX cells during growth on MG medium. CoA-ester pools extracted from mid-exponential- and stationary-phase DF50 and DBX cells were analyzed by HPLC-ESI-MS/MS. Remarkably, the intracellular propionyl-CoA concentration was almost constant for mid-exponential- and stationary-phase DF50 (0.04 mM) and DBX (0.15 to 0.20 mM) cells (Fig. 4). However, the propionyl-CoA level in DBX cells was 3 to 4 times higher than that in DF50 cells in the same growth phase (Fig. 4). Therefore, it was clear that the knockout of the pccBX genes led to a higher level of propionyl-CoA in vivo. It is noteworthy that the effects of an excessive propionyl-CoA level on global metabolism have been commonly identified in bacteria and eukarya (4, 5). Hence, we investigated the effects of the pccBX knockout on overall cell metabolism in H. mediterranei.
FIG 4.
Intracellular level of propionyl-CoA (in mM) in H. mediterranei DF50 and DBX cells at both exponential and stationary phases.
Knockout of pccBX affects cell growth and glucose utilization.
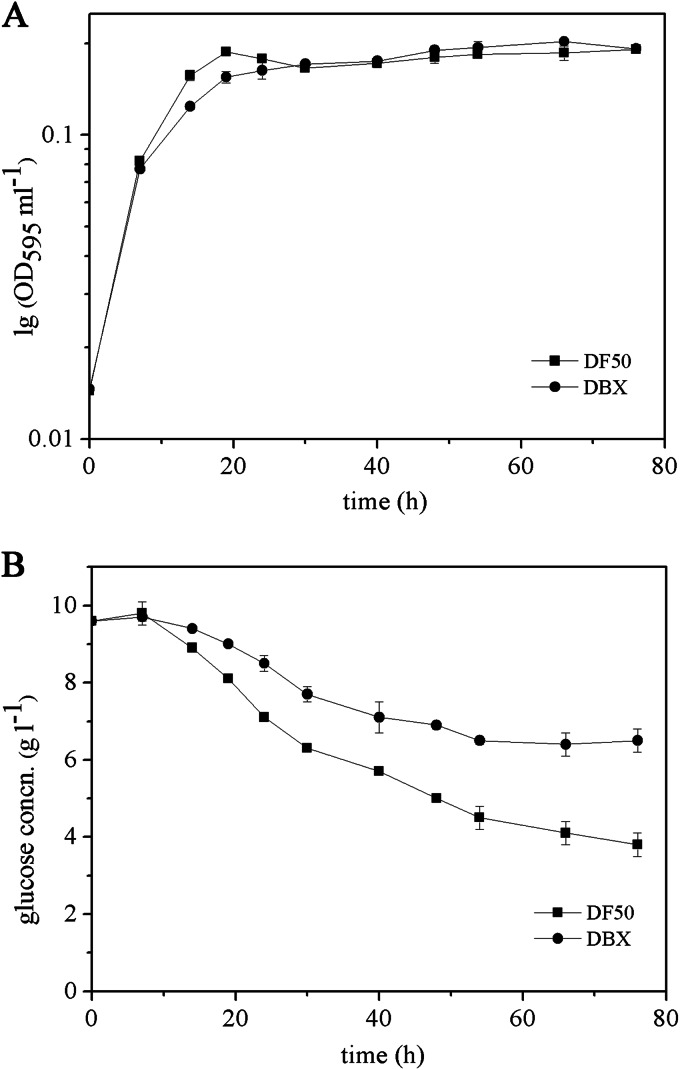
We examined the effects of the pccBX knockout on cell growth in MG medium with glucose as a major carbon source. The OD595 curves indicated that the DBX strain exhibited a lower growth rate in the mid-exponential and late exponential phases than the DF50 strain, but the DF50 and DBX strains grew to approximately equivalent levels overall (Fig. 5A). In addition, we monitored the consumption of glucose in MG medium by the DF50 and DBX strains. The residual glucose concentration in the medium of the DBX strain was always higher than that of the DF50 strain (Fig. 5B), indicating that the DBX strain consumed less glucose than the DF50 strain during cultivation. These results suggested that the knockout of pccBX somewhat affected cell growth in MG medium and glucose utilization, in contrast to the inability to utilize propionate by the DBX strain.
FIG 5.
Measurement of the growth of H. mediterranei strains DF50 and DBX in MG medium. (A) Time course of the DNA content of the DF50 and DBX strains, which was quantified by the diphenylamine colorimetric method and is shown as the OD595 ml−1 culture on a semilog scale. (B) Time course of the residual glucose concentrations in cultures of the DF50 and DBX strains.
Knockout of pccBX affects PHBV biosynthesis.
As PHBV biosynthesis involves two important metabolites of acetyl-CoA and propionyl-CoA from primary metabolism (15), and the PCC enzyme was involved in propionyl-CoA metabolism, we determined the PHBV accumulation capability of the DBX strain. In nitrogen-limiting MG medium, knockout of pccBX resulted in an ∼86% decrease in PHBV accumulation and a significant increase in 3HV content (Table 3). This result suggested that the PHBV accumulation capability was significantly affected in the DBX strain.
TABLE 3.
PHBV accumulation in H. mediterranei strainsa
| Medium | Strain | Mean CDWb (g liter−1) ± SD | Mean PHBV content (wt%) ± SD | Mean 3HV fraction (mol%) ± SD | Mean PHBV concn (g liter−1) ± SD |
|---|---|---|---|---|---|
| MG | DF50 | 9.0 ± 0.2 | 23.3 ± 0.9 | 8.0 ± 0.2 | 2.1 ± 0.1 |
| DBX | 7.0 ± 0.3 | 4.0 ± 0.1 | 33.3 ± 1.9 | 0.3 ± 0.0 | |
| mMG | DF50(pWL502) | 3.4 ± 0.1 | 51.6 ± 1.2 | 9.4 ± 0.4 | 1.8 ± 0.1 |
| DBX(pWL502) | 3.2 ± 1.3 | 6.0 ± 1.6 | 59.6 ± 4.1 | 0.2 ± 0.0 | |
| DBX(pccB) | 2.5 ± 0.1 | 7.4 ± 0.1 | 16.2 ± 0.4 | 0.2 ± 0.0 | |
| DBX(pccX) | 2.9 ± 0.2 | 5.8 ± 0.7 | 64.6 ± 2.0 | 0.2 ± 0.0 | |
| DBX(pccBX) | 2.5 ± 0.0 | 30.0 ± 1.1 | 5.7 ± 0.3 | 0.7 ± 0.0 |
All data are expressed as means ± standard deviations from three independent experiments.
CDW, cell dry weight.
Different DBX complementation strains were also subjected to PHBV accumulation analysis to further examine the subunit composition of the PCC enzyme. As these complementation strains carried pWL502-derived plasmids containing the pyrF gene as a selection marker (17), mMG medium was used, in which yeast extract was replaced with NH4Cl. Similarly to that in MG medium, the amount of PHBV accumulated by the DBX strain (with empty plasmid pWL502) was only 11% of that accumulated by the DF50 strain (with empty plasmid pWL502) (Table 3). Complementation of the DBX strain with pccB or pccX alone did not have any significant effect on the PHBV concentration, while complementation with the pccBX genes partially restored the PHBV concentration (Table 3). These complementation results were consistent with those of the enzyme assay of PCC activity and further confirmed the subunit components of PccB, PccX, and PccA for the PCC enzyme.
Knockout of pccBX leads to morphological abnormalities.
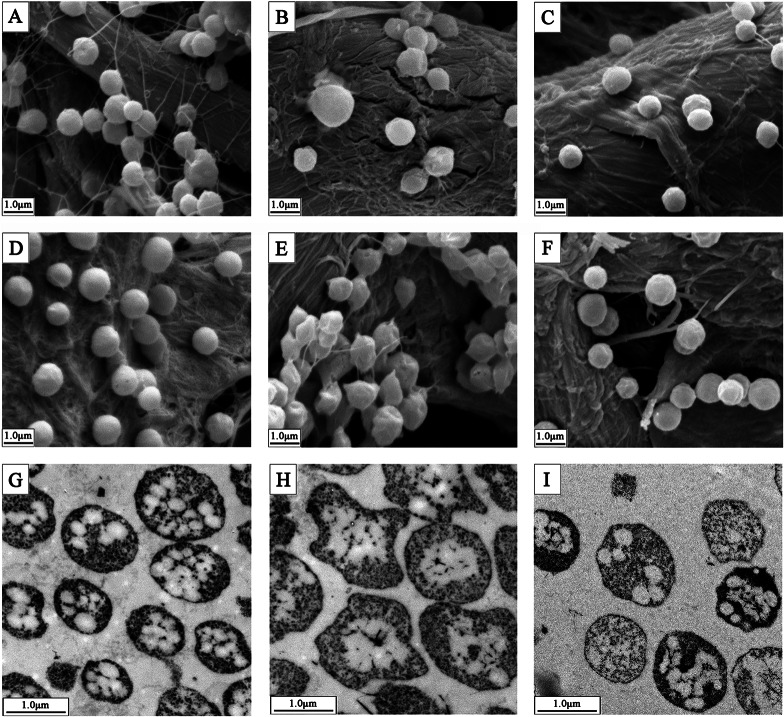
It captured our attention that a conversion from a smooth to a rough colony morphology occurred after we knocked out the pccBX genes. Further examination by SEM revealed that the DF50 strain displayed a regular morphology (Fig. 6A), while the DBX strain exhibited a wrinkled morphology (Fig. 6B). Notably, the morphological abnormalities of the wrinkled morphology of the DBX strain were more pronounced in stationary phase (Fig. 6D and E) than in exponential phase (Fig. 6A and B). Complementation of the DBX strain with the pccBX genes almost restored the regular morphology of the DF50 strain in both exponential (Fig. 6C) and stationary (Fig. 6F) phases. These results indicated that the morphological changes of the DBX strain were indeed due to the deletion of the pccBX genes. Cells of strains DF50, DBX, and DBX(pccBX) in stationary phase were also examined by TEM. Consistently, the DF50 strain displayed a regular morphology (Fig. 6G), deletion of pccBX resulted in a wrinkled cell morphology (Fig. 6H), and complementation with the pccBX genes almost restored the cell morphology (Fig. 6I). Interestingly, many PHBV granules were observed in DF50 cells (Fig. 6G), but few were observed in DBX cells (Fig. 6H), and complementation with the pccBX genes partially restored the number of PHBV granules (Fig. 6I). The phenotype of intracellular PHBV granules was consistent with the PHBV concentration described above. These results suggested that the knockout of pccBX had a significant impact on cell morphology and the number of PHBV granules.
FIG 6.
SEM and TEM images of H. mediterranei DF50, DBX, and complemented strain DBX(pccBX). (A) SEM image of DF50 in exponential phase. (B) SEM image of DBX in exponential phase. (C) SEM image of DBX(pccBX) in exponential phase. (D) SEM image of DF50 in stationary phase. (E) SEM image of DBX in stationary phase. (F) SEM image of DBX(pccBX) in stationary phase. (G) TEM image of DF50 in stationary phase. (H) TEM image of DBX in stationary phase. (I) TEM image of DBX(pccBX) in stationary phase.
Inhibition of succinyl-CoA synthetase by propionyl-CoA in vitro.
We have demonstrated that knockout of pccBX led to an intracellular accumulation of excessive propionyl-CoA and multiple phenotype changes in H. mediterranei, which raised the question of whether there was a link between an altered propionyl-CoA level and phenotype changes. It has been reported that excessive propionyl-CoA can inhibit bacterial and mammalian metabolism by inhibiting key enzymes in primary metabolism (31, 32). We then investigated the inhibitory effect of propionyl-CoA on the in vitro activity of succinyl-CoA synthetase, a key enzyme in the tricarboxylic acid (TCA) cycle. Crude extracts of DF50 cells were used for enzyme assays. We found that in the presence of 0.2 mM propionyl-CoA, which equaled the intracellular propionyl-CoA concentration of exponential-phase DBX cells, the activity of succinyl-CoA synthetase was inhibited by ∼20%, and in the presence of 0.4 mM propionyl-CoA, the activity was inhibited by ∼55%. These results provided insight into the possible mechanism of the effect of the pccBX knockout on cell metabolism in H. mediterranei. It appeared that the deletion of key genes encoding the PCC enzyme would result in a significant accumulation of propionyl-CoA in vivo, which would then inhibit CoA-dependent enzymes such as succinyl-CoA synthetase (5, 20, 32). This could partially inhibit the TCA cycle, an important process of primary metabolism.
Microarray analysis of the impact of the pccBX knockout on cell metabolism.
To further investigate the impact of the pccBX knockout on overall cell metabolism at the transcriptomic level, we performed microarray analysis. The total RNA used for microarray analysis was isolated from chemostat cultures of the DF50 and DBX strains, in which the cells were grown at a fixed specific growth rate. Thus, any difference between the gene expression patterns of the two strains would be caused by the deletion of the pccBX genes and not secondary consequences of the difference in growth rates (33). In general, the changes in the transcript levels of the DBX strain compared with the DF50 strain were consistent with the phenotype changes described above. The significantly changed genes in these metabolic pathways are listed in Table 4. For example, the expressions of the genes encoding the key enzymes for glycolysis (glyceraldehyde-3-phosphate dehydrogenase [HFX_0447]) and pyruvate oxidation (pyruvate:ferredoxin oxidoreductase [HFX_1370 and HFX_1371]) were downregulated in the DBX strain (Table 4), consistent with retarded cell growth and decreased glucose consumption. Additionally, regarding the affected PHBV accumulation in the DBX strain, the expression of the gene cluster for PHBV biosynthesis (enoyl-CoA hydratase [HFX_5217], polyhydroxyalkanoate [PHA] synthesis regulator [HFX_5218], and PHA synthase [HFX_5220 and HFX_5221]) was also largely downregulated (Table 4). Interestingly, the expressions of a considerable number of genes encoding proteins involved in the electron transport chain (HFX_0290, HFX_0941 to HFX_0943, HFX_1755, HFX_1927, HFX_2125, HFX_5092, HFX_5106, HFX_5107, HFX_6166, and HFX_6295), reactive oxygen species scavengers (alkylhydroperoxidase-like protein [HFX_1388], NADPH2:quinone reductase [HFX_2665], and superoxide dismutase [HFX_6014]), and stress response proteins (HFX_1094, HFX_1208, HFX_1928, HFX_6007, HFX_6020, and HFX_6431) were significantly upregulated in the DBX strain (Table 4). These results indicated that the redox state of the DBX cells might be disturbed, leading to oxidative stress. Oxidative stress might cause some damage to the components of DBX cells, as the expressions of the key genes involved in membrane biogenesis (hydroxymethylglutaryl-CoA synthase [HFX_2424]) (34) and DNA repair (A/G-specific adenine glycosylase [HFX_2887]) (35) were largely upregulated (Table 4). Hence, the wrinkled cell morphology of the DBX strain might be caused by oxidative damage to the cell membrane. Additionally, the expression levels of some genes encoding transcriptional regulators (HFX_1248, HFX_1377, HFX_2426, HFX_5154, HFX_6042, and HFX_6204) were also altered (Table 4). It is likely that these transcriptional regulators were activated or inhibited to regulate the expression of certain pathways for restoring balance in the metabolite pool. Therefore, knockout of the pccBX genes seemed to have disturbed metabolic homeostasis, thereby leading to the up- or downregulation of particular sets of genes to cope with the imbalance.
TABLE 4.
Significantly up- and downregulated genes for specific pathways in strain DBX
| Classification | Locus tag | Gene | Mean fold change ± SDa |
|---|---|---|---|
| Glycolysis | HFX_0447 | gapA | −4.7 ± 0.8 |
| Pyruvate oxidation | HFX_1370 | porB | −2.2 ± 0.6 |
| HFX_1371 | porA | −2.3 ± 0.4 | |
| PHBV biosynthesis | HFX_5217 | maoC | −5.3 ± 0.7 |
| HFX_5218 | phaR | −11.1 ± 3.4 | |
| HFX_5220 | phaE | −5.5 ± 0.1 | |
| HFX_5221 | phaC | −3.9 ± 0.4 | |
| Electron transport chain | HFX_0290 | etfA | 2.1 ± 0.3 |
| HFX_0428 | cydB | −2.2 ± 0.5 | |
| HFX_0941 | cbaE | 2.3 ± 0.6 | |
| HFX_0942 | hcpB | 3.4 ± 1.4 | |
| HFX_0943 | cbaB | 2.2 ± 0.9 | |
| HFX_1755 | glbN | 2.9 ± 0.8 | |
| HFX_1927 | fixA | 2.6 ± 1.5 | |
| HFX_2125 | nolA | 2.9 ± 1.2 | |
| HFX_5092 | pcy | 2.5 ± 1.4 | |
| HFX_5106 | narC | 6.9 ± 2.6 | |
| HFX_5107 | narB | 6.2 ± 3.2 | |
| HFX_6166 | hcpA | 6.5 ± 2.2 | |
| HFX_6295 | fnr | 2.5 ± 0.9 | |
| Reactive oxygen species scavengers | HFX_1388 | aphD | 15.3 ± 2.4 |
| HFX_2665 | yfmJ1 | 2.1 ± 0.1 | |
| HFX_6014 | sodA | 3.8 ± 0.7 | |
| Stress response | HFX_1094 | usp26 | 2.1 ± 0.5 |
| HFX_1208 | usp | 2.9 ± 1.6 | |
| HFX_1928 | uspA | 3.5 ± 1.9 | |
| HFX_2556 | uspA | −2.2 ± 0.7 | |
| HFX_5134 | usp22 | −4.2 ± 0.9 | |
| HFX_6007 | uspA | 2.3 ± 0.2 | |
| HFX_6020 | usp6 | 2.1 ± 0.3 | |
| HFX_6431 | usp31 | 2.2 ± 1.0 | |
| Transcriptional regulators | HFX_1248 | prrC | −2.1 ± 0.2 |
| HFX_1377 | cdcp | 2.9 ± 1.3 | |
| HFX_2426 | tcrg | 2.0 ± 0.4 | |
| HFX_5154 | dbp | −3.3 ± 0.2 | |
| HFX_6042 | iclR | 2.5 ± 0.4 | |
| HFX_6204 | copG | −2.1 ± 0.2 | |
| Membrane biogenesis | HFX_2424 | mvaB | 5.3 ± 0.5 |
| DNA repair | HFX_2887 | mutY | 7.2 ± 1.8 |
Data represent the means ± standard deviations from three independent biological replicates.
DISCUSSION
In this study, we identified the PCC enzyme in H. mediterranei, which was demonstrated to be indispensable for the assimilation of propionyl-CoA, an essential intermediate for haloarchaea. The PCC enzyme catalyzed the carboxylation of propionyl-CoA with an acetyl-CoA carboxylation side activity in H. mediterranei. This is reasonable because the 3-hydroxypropionate pathway involving acetyl-CoA carboxylation is just one of the four propionyl-CoA-supplying pathways, while the methylmalonyl-CoA pathway involving propionyl-CoA carboxylation is the only pathway for propionyl-CoA utilization in H. mediterranei. It is noteworthy that the subunit composition of the PCC enzyme in H. mediterranei is different from that of the ACC/PCC reported for the archaeon M. sedula but similar to that of acyl-CoA carboxylases from actinomycetes (8, 36). In M. sedula, BC and BCCP components exist as separate subunits, while in H. mediterranei, they are fused into a single protein. Additionally, the PCC enzyme in H. mediterranei possesses a supplementary small subunit, PccX. Thus far, this type of small subunit has been found only in acyl-CoA carboxylases from actinomycetes (36). The auxiliary ε subunit from actinomycetes characteristic of the helix structure might be responsible for the interaction of the BC and CT components (36, 37). Prediction of the secondary structure revealed that the PccX subunit in H. mediterranei was also rich in helix structures; thus, it might also be involved in the subunit interaction. However, the primary structure of PccX from H. mediterranei was not comparable to that of its counterparts in actinomycetes, and its secondary structure was different from that in actinomycetes (see Table S1 in the supplemental material), indicating that PccX from H. mediterranei possessed some unique features that are distinct from those of the ε subunit in actinomycetes. In addition, presence-absence analyses revealed that a similar subunit composition of PccA, PccB, and PccX was widespread in haloarchaea. However, the explanation for why the subunit composition of haloarchaeal acyl-CoA carboxylases is similar to that of actinomycetes remains unclear. Coincidently, phylogenetic tree analysis of the CT components of acyl-CoA carboxylases from archaea and bacteria revealed that the CTs from haloarchaea were more related to their bacterial counterparts than to those from thermophilic archaea (see Fig. S2 and Table S2 in the supplemental material), suggesting that lateral gene transfer might have occurred between haloarchaeal and bacterial acyl-CoA carboxylase genes.
Interestingly, knockout of the pccBX genes led to a higher level of propionyl-CoA in vivo. This seems reasonable, as the propionyl-CoA utilization pathway involving propionyl-CoA carboxylation was blocked. Additionally, although the 3-hydroxypropionate pathway involving the PCC enzyme for propionyl-CoA supply was blocked in the DBX strain, there are three other propionyl-CoA-supplying pathways (12). In contrast to the increased propionyl-CoA content in vivo, knockout of the pccBX genes resulted in a decreased acetyl-CoA supply, as reflected by the affected PHBV accumulation and the increased 3HV molar fraction in the DBX strain. This is understandable, as the activity of the CoA-dependent enzymes involved in glycolysis and pyruvate oxidation could be inhibited by the excessive intracellular propionyl-CoA concentration (5, 20), thereby leading to a decreased acetyl-CoA supply. Microarray analysis of the pccBX knockout also revealed that the expression of key genes involved in glycolysis and pyruvate oxidation was inhibited. Therefore, we concluded that the knockout of the pccBX genes affected the overall cellular metabolic network that was far beyond propionyl-CoA generation and consumption.
Accompanied by an altered CoA-ester pool, multiple phenotype changes, including retarded cell growth, decreased glucose utilization, lower-level PHBV production, and abnormal morphology, were observed for the DBX strain. The inhibitory effect of excessive propionyl-CoA has been reported for some organisms. For example, in Aspergillus nidulans, an excessive level of propionyl-CoA in the cell can inhibit cell growth and polyketide synthesis (20, 38). In Rhodopseudomonas sphaeroides, excessive propionyl-CoA in vivo leads to a retardation of cell growth (32). It has been generally demonstrated that the inhibitory effect of propionyl-CoA on cell metabolism is due to the inhibition of CoA-dependent enzymes such as pyruvate dehydrogenase and succinyl-CoA synthetase, which are important enzymes in primary metabolism (5, 20, 32). Similarly, the activity of succinyl-CoA synthetase in crude extracts of H. mediterranei strain DF50 was also inhibited by excessive propionyl-CoA in vitro. Therefore, the phenotype changes of the DBX strain were at least partially caused by the direct or indirect inhibitory effect of excessive propionyl-CoA on cellular metabolism. In addition to this kind of inhibitory effect, we cannot exclude the possibility of an intracellular depletion of free coenzyme A-SH (CoASH) caused by increased propionyl-CoA concentrations (20). Decreased CoASH concentrations could also result in a strong disturbance of cell metabolism, as reported previously for E. coli (39, 40). To understand the global metabolic changes caused by the knockout of the pccBX genes, we investigated the gene expression profile changes of the DBX strain at the omics level. Microarray analysis revealed that the expressions of many genes encoding key enzymes of certain metabolic pathways, proteins involved in cell stress, and transcriptional regulators were significantly affected in the DBX strain. Therefore, we deduced that the knockout of the pccBX genes might cause cell stress and transduce signals that affect the global metabolic network and also the gene regulatory network to restore metabolic homeostasis in the DBX strain. However, details of the mechanisms need to be further investigated.
In conclusion, we identified the PCC enzyme in H. mediterranei, which consists of the PccA, PccB, and PccX subunits and represents a novel type of acyl-CoA carboxylase in archaea. The PCC enzyme was demonstrated to be indispensable for the assimilation of propionyl-CoA. Additionally, we have demonstrated that impaired propionyl-CoA metabolism affected the global metabolic network in H. mediterranei. As a large quantity of propionyl-CoA is involved in the biosynthesis and catabolism of PHBV in haloarchaea, and propionyl-CoA is also an essential intermediate of acetate assimilation via the methylaspartate cycle in haloarchaea (3), the PCC enzyme is not only important for haloarchaea to maintain the balance in propionyl-CoA pool but also indispensable for haloarchaea to assimilate propionyl-CoA, e.g., from PHBV catabolism in the absence of an exogenous carbon source and assimilation of acetyl-CoA to enter central carbon metabolism.
Supplementary Material
ACKNOWLEDGMENTS
The work described in this paper was supported by the National Natural Science Foundation of China (grant no. 31330001, 31370096, and 31000023).
We thank Ivan Berg (Fakultät Biologie, Albert Ludwigs Universität Freiburg) for helpful discussion, Farshad Borjian (Fakultät Biologie, Albert Ludwigs Universität Freiburg) for help with enzyme assays, and Guomin Ai (Institute of Microbiology, Chinese Academy of Sciences) for help with HPLC-ESI-MS/MS analysis.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03167-14.
REFERENCES
- 1.Han J, Hou J, Liu H, Cai S, Feng B, Zhou J, Xiang H. 2010. Wide distribution among halophilic archaea of a novel polyhydroxyalkanoate synthase subtype with homology to bacterial type III synthases. Appl Environ Microbiol 76:7811–7819. doi: 10.1128/AEM.01117-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jendrossek D, Handrick R. 2002. Microbial degradation of polyhydroxyalkanoates. Annu Rev Microbiol 56:403–432. doi: 10.1146/annurev.micro.56.012302.160838. [DOI] [PubMed] [Google Scholar]
- 3.Khomyakova M, Bükmez Ö, Thomas LK, Erb TJ, Berg IA. 2011. A methylaspartate cycle in haloarchaea. Science 331:334–337. doi: 10.1126/science.1196544. [DOI] [PubMed] [Google Scholar]
- 4.Deodato F, Boenzi S, Santorelli FM, Dionisi-Vici C. 2006. Methylmalonic and propionic aciduria. Am J Med Genet C Semin Med Genet 142C:104–112. doi: 10.1002/ajmg.c.30090. [DOI] [PubMed] [Google Scholar]
- 5.Maerker C, Rohde M, Brakhage AA, Brock M. 2005. Methylcitrate synthase from Aspergillus fumigatus. Propionyl-CoA affects polyketide synthesis, growth and morphology of conidia. FEBS J 272:3615–3630. doi: 10.1111/j.1742-4658.2005.04784.x. [DOI] [PubMed] [Google Scholar]
- 6.Tong L. 2013. Structure and function of biotin-dependent carboxylases. Cell Mol Life Sci 70:863–891. doi: 10.1007/s00018-012-1096-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang CS, Sadre-Bazzaz K, Shen Y, Deng B, Zhou ZH, Tong L. 2010. Crystal structure of the α6β6 holoenzyme of propionyl-coenzyme A carboxylase. Nature 466:1001–1005. doi: 10.1038/nature09302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hügler M, Krieger RS, Jahn M, Fuchs G. 2003. Characterization of acetyl-CoA/propionyl-CoA carboxylase in Metallosphaera sedula. Carboxylating enzyme in the 3-hydroxypropionate cycle for autotrophic carbon fixation. Eur J Biochem 270:736–744. doi: 10.1046/j.1432-1033.2003.03434.x. [DOI] [PubMed] [Google Scholar]
- 9.Berg IA, Kockelkorn D, Buckel W, Fuchs G. 2007. A 3-hydroxypropionate/4-hydroxybutyrate autotrophic carbon dioxide assimilation pathway in Archaea. Science 318:1782–1786. doi: 10.1126/science.1149976. [DOI] [PubMed] [Google Scholar]
- 10.Han J, Zhang F, Hou J, Liu X, Li M, Liu H, Cai L, Zhang B, Chen Y, Zhou J, Hu S, Xiang H. 2012. Complete genome sequence of the metabolically versatile halophilic archaeon Haloferax mediterranei, a poly(3-hydroxybutyrate-co-3-hydroxyvalerate) producer. J Bacteriol 194:4463–4464. doi: 10.1128/JB.00880-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu Q, Han J, Zhou L, Zhou J, Xiang H. 2008. Genetic and biochemical characterization of the poly(3-hydroxybutyrate-co-3-hydroxyvalerate) synthase in Haloferax mediterranei. J Bacteriol 190:4173–4180. doi: 10.1128/JB.00134-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han J, Hou J, Zhang F, Ai G, Li M, Cai S, Liu H, Wang L, Wang Z, Zhang S, Cai L, Zhao D, Zhou J, Xiang H. 2013. Multiple propionyl coenzyme A-supplying pathways for production of the bioplastic poly(3-hydroxybutyrate-co-3-hydroxyvalerate) in Haloferax mediterranei. Appl Environ Microbiol 79:2922–2931. doi: 10.1128/AEM.03915-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 14.Han J, Lu Q, Zhou L, Zhou J, Xiang H. 2007. Molecular characterization of the phaECHm genes, required for biosynthesis of poly(3-hydroxybutyrate) in the extremely halophilic archaeon Haloarcula marismortui. Appl Environ Microbiol 73:6058–6065. doi: 10.1128/AEM.00953-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hou J, Feng B, Han J, Liu H, Zhao D, Zhou J, Xiang H. 2013. Haloarchaeal-type β-ketothiolases involved in poly(3-hydroxybutyrate-co-3-hydroxyvalerate) synthesis in Haloferax mediterranei. Appl Environ Microbiol 79:5104–5111. doi: 10.1128/AEM.01370-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu H, Han J, Liu X, Zhou J, Xiang H. 2011. Development of pyrF-based gene knockout systems for genome-wide manipulation of the archaea Haloferax mediterranei and Haloarcula hispanica. J Genet Genomics 38:261–269. doi: 10.1016/j.jgg.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 17.Cai S, Cai L, Liu H, Liu X, Han J, Zhou J, Xiang H. 2012. Identification of the haloarchaeal phasin (PhaP) that functions in polyhydroxyalkanoate accumulation and granule formation in Haloferax mediterranei. Appl Environ Microbiol 78:1946–1952. doi: 10.1128/AEM.07114-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hou J, Han J, Cai L, Zhou J, Lü Y, Jin C, Liu J, Xiang H. 2014. Characterization of genes for chitin catabolism in Haloferax mediterranei. Appl Microbiol Biotechnol 98:1185–1194. doi: 10.1007/s00253-013-4969-8. [DOI] [PubMed] [Google Scholar]
- 19.Cline SW, Lam WL, Charlebois RL, Schalkwyk LC, Doolittle WF. 1989. Transformation methods for halophilic archaebacteria. Can J Microbiol 35:148–152. doi: 10.1139/m89-022. [DOI] [PubMed] [Google Scholar]
- 20.Brock M, Buckel W. 2004. On the mechanism of action of the antifungal agent propionate. Propionyl-CoA inhibits glucose metabolism in Aspergillus nidulans. Eur J Biochem 271:3227–3241. doi: 10.1111/j.1432-1033.2004.04255.x. [DOI] [PubMed] [Google Scholar]
- 21.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 22.Muller F, Brissac T, Le Bris N, Felbeck H, Gros O. 2010. First description of giant Archaea (Thaumarchaeota) associated with putative bacterial ectosymbionts in a sulfidic marine habitat. Environ Microbiol 12:2371–2383. doi: 10.1111/j.1462-2920.2010.02309.x. [DOI] [PubMed] [Google Scholar]
- 23.Zhao Y, Xiang S, Dai X, Yang K. 2013. A simplified diphenylamine colorimetric method for growth quantification. Appl Microbiol Biotechnol 97:5069–5077. doi: 10.1007/s00253-013-4893-y. [DOI] [PubMed] [Google Scholar]
- 24.Han J, Li M, Hou J, Wu L, Zhou J, Xiang H. 2010. Comparison of four phaC genes from Haloferax mediterranei and their function in different PHBV copolymer biosyntheses in Haloarcula hispanica. Saline Systems 6:9. doi: 10.1186/1746-1448-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peyraud R, Kiefer P, Christen P, Massou S, Portais JC, Vorholt JA. 2009. Demonstration of the ethylmalonyl-CoA pathway by using 13C metabolomics. Proc Natl Acad Sci U S A 106:4846–4851. doi: 10.1073/pnas.0810932106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Christian JH, Waltho JA. 1962. Solute concentrations within cells of halophilic and non-halophilic bacteria. Biochim Biophys Acta 65:506–508. doi: 10.1016/0006-3002(62)90453-5. [DOI] [PubMed] [Google Scholar]
- 27.Lange C, Zerulla K, Breuert S, Soppa J. 2011. Gene conversion results in the equalization of genome copies in the polyploid haloarchaeon Haloferax volcanii. Mol Microbiol 80:666–677. doi: 10.1111/j.1365-2958.2011.07600.x. [DOI] [PubMed] [Google Scholar]
- 28.Wei J, Tian Y, Niu G, Tan H. 2014. GouR, a TetR family transcriptional regulator, coordinates the biosynthesis and export of gougerotin in Streptomyces graminearus. Appl Environ Microbiol 80:714–722. doi: 10.1128/AEM.03003-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Falb M, Müller K, Köenigsmaier L, Oberwinkler T, Horn P, von Gronau S, Gonzalez O, Pfeiffer F, Bornberg-Bauer E, Oesterhelt D. 2008. Metabolism of halophilic archaea. Extremophiles 12:177–196. doi: 10.1007/s00792-008-0138-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pieper-Fürst U, Madkour MH, Mayer F, Steinbüchel A. 1995. Identification of the region of a 14-kilodalton protein of Rhodococcus ruber that is responsible for the binding of this phasin to polyhydroxyalkanoic acid granules. J Bacteriol 177:2513–2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brass EP. 1992. Interaction of carnitine and propionate with pyruvate oxidation by hepatocytes from clofibrate-treated rats: importance of coenzyme A availability. J Nutr 122:234–240. [DOI] [PubMed] [Google Scholar]
- 32.Maruyama K, Kitamura H. 1985. Mechanisms of growth inhibition by propionate and restoration of the growth by sodium bicarbonate or acetate in Rhodopseudomonas sphaeroides S. J Biochem 98:819–824. [DOI] [PubMed] [Google Scholar]
- 33.Hayes A, Zhang N, Wu J, Butler PR, Hauser NC, Hoheisel JD, Lim FL, Sharrocks AD, Oliver SG. 2002. Hybridization array technology coupled with chemostat culture: tools to interrogate gene expression in Saccharomyces cerevisiae. Methods 26:281–290. doi: 10.1016/S1046-2023(02)00032-4. [DOI] [PubMed] [Google Scholar]
- 34.VanNice JC, Skaff DA, Wyckoff GJ, Miziorko HM. 2013. Expression in Haloferax volcanii of 3-hydroxy-3-methylglutaryl coenzyme A synthase facilitates isolation and characterization of the active form of a key enzyme required for polyisoprenoid cell membrane biosynthesis in halophilic archaea. J Bacteriol 195:3854–3862. doi: 10.1128/JB.00485-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Denver DR, Swenson SL, Lynch M. 2003. An evolutionary analysis of the helix-hairpin-helix superfamily of DNA repair glycosylases. Mol Biol Evol 20:1603–1611. doi: 10.1093/molbev/msg177. [DOI] [PubMed] [Google Scholar]
- 36.Gago G, Diacovich L, Arabolaza A, Tsai SC, Gramajo H. 2011. Fatty acid biosynthesis in actinomycetes. FEMS Microbiol Rev 35:475–497. doi: 10.1111/j.1574-6976.2010.00259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lombard J, Moreira D. 2011. Early evolution of the biotin-dependent carboxylase family. BMC Evol Biol 11:232. doi: 10.1186/1471-2148-11-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang YQ, Brock M, Keller NP. 2004. Connection of propionyl-CoA metabolism to polyketide biosynthesis in Aspergillus nidulans. Genetics 168:785–794. doi: 10.1534/genetics.104.027540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jackowski S, Rock CO. 1986. Consequences of reduced intracellular coenzyme A content in Escherichia coli. J Bacteriol 166:866–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vallari DS, Jackowski S. 1988. Biosynthesis and degradation both contribute to the regulation of coenzyme A content in Escherichia coli. J Bacteriol 170:3961–3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Combet C, Blanchet C, Geourjon C, Deleage G. 2000. NPS@: network protein sequence analysis. Trends Biochem Sci 25:147–150. doi: 10.1016/S0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.