Abstract
Recent studies have provided evidence for both intracellular and extracellular roles of the potent hepatotoxin microcystin (MC) in the bloom-forming cyanobacterium Microcystis. Here, we surveyed transcriptomes of the wild-type strain M. aeruginosa PCC 7806 and the microcystin-deficient ΔmcyB mutant under low light conditions with and without the addition of external MC of the LR variant (MC-LR). Transcriptomic data acquired by microarray and quantitative PCR revealed substantial differences in the relative expression of genes of the central intermediary metabolism, photosynthesis, and energy metabolism. In particular, the data provide evidence for a lower photosystem I (PSI)-to-photosystem II (PSII) ratio and a more pronounced carbon limitation in the microcystin-deficient mutant. Interestingly, only 6% of the transcriptional differences could be complemented by external microcystin-LR addition. This MC signaling effect was seen exclusively for genes of the secondary metabolism category. The orphan polyketide synthase gene cluster IPF38-51 was specifically downregulated in response to external MC-LR under low light. Our data suggest a hierarchical and light-dependent cross talk of secondary metabolites and support both an intracellular and an extracellular role of MC in Microcystis.
INTRODUCTION
Cyanobacteria of the genus Microcystis frequently occur as thick surface scums in freshwater lakes during summer (1). While these blooms are unpleasant, both visually and due to the presence of odors, a more serious problem is caused by their regular production of the potent hepatotoxin microcystin (MC) (2). Increasing eutrophication caused by urban and industrial developments, as well as global warming, is expected to amplify the severity of the problem (3). Microcystis cells are organized into colonies of diverse shapes (4) and possess constitutively present gas vesicles (5). Both features enable vertical migration in the water column, a phenomenon widely discussed as a competitive advantage over other phytoplankton species (6). Toxic genotypes commonly constitute only a subfraction within Microcystis communities present in blooms. The proportion of toxic and nontoxic genotypes, however, is dynamically changing, and the factors underlying these variations are not well understood (7).
In this respect, much research has been focused on deciphering the function and physiological role of microcystin in the MC-producing cyanobacteria (8). Microcystin is known to irreversibly inhibit eukaryotic protein phosphatases of types 1 and 2A (9). Indeed, some grazers and parasites have been shown to be sensitive to the toxin (10, 11). Nevertheless, a primary defense function of microcystin is increasingly questioned. One of the strongest arguments against this comes from an evolutionary analysis of the microcystin biosynthesis (mcy) genes that unambiguously showed that the corresponding genes already were present in the last common ancestor of all present cyanobacteria, long before the appearance of the first eukaryotic grazer (12). Numerous alternative roles have been discussed for microcystin in the literature, including both extra- and intracellular functions. Schatz et al., when studying a Microcystis sp. strain isolated from Lake Kinneret, Israel, suggested a role of microcystin as an infochemical and provided evidence for the autoinduction of Mcy proteins upon microcystin addition (13). Additionally, colony-size stimulating effects of extracellular microcystin for specific Microcystis strains were reported (14). Other studies have suggested intracellular functions for the toxin. The microcystin-deficient ΔmcyB mutant, for example, was shown to be more sensitive under high light stress and under inorganic carbon limitation (15–17). The facts that microcystin binds covalently to cysteines of proteins under stress conditions and that RubisCO is a predominant binding partner provide a mechanistic clue to understanding these observations (17). Differences in the carbon-nitrogen metabolism and redox maintenance also were discussed in a comparative proteomic study of toxic and nontoxic strains (18). Evidence for the binding of the global nitrogen regulator NtcA protein in the bidirectional mcy promoter region suggested regulatory links between the carbon-nitrogen metabolism and microcystin biosynthesis (18, 19). At present, it is not clear how much of the phenotypic and proteomic differences are governed on the transcriptional level, whether the intra- or the extracellular portion of microcystin has a larger impact on cellular physiology, and to what extent the intra- and extracellular roles are connected.
Here, we have analyzed differences in gene expression between the microcystin-producing wild-type (WT) strain Microcystis aeruginosa PCC 7806 and a microcystin-deficient mutant. Both were grown under low light conditions, with and without externally added microcystin of the LR variant form (MC-LR), which has l-leucine (L) and l-arginine (R) incorporated in the positions of variable residues within the microcystin structure. Microarray data reveal large transcriptomic differences between the wild type and mutants that were only fractionally complemented by extracellular microcystin-LR. Our study supports multiple, independent roles of microcystin for producing cyanobacteria.
MATERIALS AND METHODS
Experimental design and cultivation of cyanobacteria.
Axenic cultures of Microcystis aeruginosa wild-type strain PCC 7806 and its microcystin-deficient ΔmcyB mutant (MT) (20) were obtained from the Pasteur Culture Collection of Cyanobacteria (PCC). Both strains were precultivated in parallel in three biological replicates each, with enough time allowed for setting cells to similar physiological states. WT and MT replicates were cultured in BG-11 medium (21) at 16 μE m−2 s−1 and 25°C with identical starting cell densities (optical density at 750 nm [OD750] of 0.3) and were grown in parallel and maintained at exponential-phase ODs for at least 1 month before the actual experiment.
After precultivation, cultures were adapted to experimental light conditions (darkness [D], 0 μE m−2 s−1; low light [LL], 16 μE m−2 s−1; high light [HL], 70 μE m−2 s−1) at least 60 h before the addition of external MC-LR. For microarray experiments, at this point, all replicates were transferred to the multicultivator model MC 1000 by Photon Systems Instruments, Czech Republic, to allow for highly consistent cultivation, comprising illumination at 16 μE m−2 s−2 in identical vessels with equal external aeration. For reverse transcription-quantitative PCR (RT-qPCR) experiments, Microcystis replicates were kept in batch cultures in BG-11 at 25°C, with starting aeration and light adjustment to darkness, LL, or HL 60 h before MC treatment.
Aliquots of each replicate were treated with (i) the addition of MC-LR in BG-11 to a final concentration of 50 ng/ml or (ii) an equal volume of BG-11 in control aliquots. After 1 h of incubation, cultures were harvested and further processed for RNA use in microarrays or RT-qPCR. Cultures used in microarray experiments were cultivated and incubated under LL conditions only, while cultures intended for RT-qPCR were incubated under dark as well as LL and HL conditions. Principally, experimental settings for qPCR and microarray experiments were arranged similarly, with harvesting times at an OD750 of 0.5 to 0.7 for microarrays and 0.8 to 1.2 for qPCR.
RNA isolation and processing.
All harvesting steps were performed quickly, and samples were cooled to 4°C. Cell pellets obtained after centrifugation (10 min, 4,600 × g) were used for the isolation of total RNA with the phenol-based single-step method according to Chomczynski et al. (22), using either QIAzol (Qiagen) (microarray experiments) or TRIzol reagent (Life Technologies) (RT-qPCR) according to the manufacturers' protocols. Potential residual DNA was digested utilizing TURBO-DNase (Life Technologies), followed by RNA cleanup (RNeasy plant minikit; Qiagen) and verification of complete DNA digestion by PCR testing. DNase-treated RNA samples yielding no PCR products (mcyA-specific primers; for details, see Table S2 in the supplemental material) were considered DNA free and were used in downstream reactions.
Following an RNA integrity check with an RNA 6000 Nano chip on a 2100 Bioanalyzer (Agilent Technologies), 3 to 5 μg of DNA-free total RNA samples for microarrays served as templates in random primed reverse transcription reactions (Superscript II RT; Life Technologies) incorporating Cy3- and Cy5-labeled dUTPs into cDNA products of control and treatment samples, respectively. After subsequent cleanup (E.Z.N.A. MicroElute cleanup kit) and testing for Cy-dye incorporation efficiency (NanoDrop ND-1000; measurements were compared to those for control samples), Cy3- and Cy5-labeled cDNAs were hybridized to custom Microcystis sp. strain PCC 7806 microarrays (for array specifications, see “Microarray design” below) for 20 h at 65°C using a NimbleGen hybridization system 4 (NimbleGen/Roche) and scanned afterwards with an Agilent Technologies DNA microarray scanner G2565CA.
cDNA preparation for RT-qPCR followed a different protocol. After DNA digestion and test PCRs, total RNA quality was checked by RNA agarose gel electrophoresis. Subsequent reverse transcription using random primers and Maxima RT (Thermo Scientific) yielded cDNA directly used in RT-qPCR.
Microarray design.
Custom Microcystis sp. strain PCC 7806 oligonucleotide microarrays (with 8 arrays on one glass slide and 60,000 spots on each array) were used in this study. The 60,000 spots on each of the arrays covered 4,691 genes of the whole PCC 7806 genome (88.6%) (23), plus some of the intergenic regions, using 60-mer oligonucleotides in an average number of 5 probes per gene. The array design was based partly on that of Straub et al. (14), with modifications made at the Micro-Array Department, Amsterdam, Netherlands. Oligonucleotide probes for each gene were designed by the eArray protocol (Agilent Technologies). Each oligonucleotide was randomly printed in duplicate across one array by means of SurePrint technology (Agilent Technologies). For probe information, see Data Set S3 in the supplemental material.
Three biological replicates of each condition tested were separately hybridized as two-channel microarrays, with the fluorescent dyes Cy3 and Cy5 being labeled according to replicates representing a control condition sample on the one hand and its MC-LR-treated countersample on the other. A hybridization scheme was designed allowing for technical controls considering dye bias as well as effects resulting from manufacturing differences on different slides and arrays, respectively. Each test condition (3 replicates) was hybridized on each of the used slides, being labeled at least once with any of the used fluorescent dyes.
Data processing.
Statistical analysis of the microarray data was computed in the R environment (version R3.0.2 [24]), utilizing the freely available Bioconductor software (25), with the limma (26) package for normalization. Normalization employed within-array normalization (loess method, normexp background correction; offset, 50) and between-array normalization (aquantile method), followed by separation of the two color channels. Due to technical failure, one of the WT array triplicates, 1× WT(+MC-LR), was omitted from the statistical analysis. Reduced statistical power due to one missing replicate was carefully considered during the interpretation of the data and might have led to an overestimation of P values in a few cases. Accordingly, some genes just below the thresholds of differential expression were discussed. Based on the obtained intensity lists for red (Cy5) and green (Cy3) channels after normalization, hierarchical clustering (Euclidian), multidimensional scaling (27), and principal component analysis (PCA) (28) were performed. An analysis of variance (ANOVA) calculation provided probe-specific values of fold changes, log2-scaled absolute expression, and P values. In order to assess differential gene expression, P values for each of the 2 to 7 probes per gene had to be combined into a single value that could be evaluated as being clearly below or above the threshold of differential gene expression. This was achieved by applying Fisher's method (29), which considers each probe of a gene as a replicate and additionally takes into account that similarly low P values of all probes per gene are not likely to occur due to chance but reflect a more significant expression value than one value alone would show. Following this, lists of differentially expressed genes were extracted, setting thresholds to P ≤ 0.1 and log2-fold changes of ≥1 and ≤−1 (see Data Set S2 in the supplemental material).
For physiological interpretation of the array results, assignments of functional gene categories (see Table S1 in the supplemental material) to each gene were conducted using both a Microcystis sp. strain PCC 7806 annotation and Synechocystis sp. strain PCC 6803 annotation obtained from CyanoBase (Kazusa genome resources; http://genome.microbedb.jp/cyanobase/).
Single-gene transcript quantification.
RT-qPCR was designed to test for relative expression levels of five genes from biosynthesis clusters of secondary metabolites considered representative indicators for the metabolite itself (mcyA, microcystin; mcnB, cyanopeptolin; mcaE, microcyclamide; aerJ, aeruginosin; IPF47, polyketide synthase I/III [PKSI/PKSIII] metabolite). Primer design was arranged to generate PCR amplicons between 80 and 140 nucleotides without forming primer dimers, and primers were tested prior to RT-PCR (for primer details, see Table S2 in the supplemental material). RT-qPCR was conducted based on a SYBR green system by Bioline (SensiMix SYBR low-ROX kit) according to the manufacturer's instructions but downscaling reaction mixtures to a 10-μl final volume for measurements in 384-well plates, utilizing the real-time cycler LightCycler480 and attributed software by Roche. cDNA of each biological replicate was tested by qPCR in technical triplicates and one nontranscription control, setting cDNA template concentrations to 100 ng in each reaction. qPCR specifications were 95°C for 10 min; 45 cycles of 95°C for 15 s and 60°C for 60 s; melting curve at 95°C for 5 s, 65°C for 60 s, and 97°C for 1 s; and a final extension at 40°C.
After PCR cycling, data extraction and subsequent threshold cycle (CT) calculation, including baseline correction and determination of PCR efficiency, were performed by the LC480Conversion and LinRegPCR software, which is freely provided by the Heart Failure Research Center (HFRC) in Amsterdam, Netherlands.
Relative expression levels were determined after Pfaffl (30), considering corrections for actual PCR efficiencies for each gene and normalizing to rnpB (a constitutively expressed RNA component of RNase P) expression levels as an endogenous reference. Every expression level was quantified relative to the expression level found in WT control samples cultivated under LL conditions that had not been treated with external microcystin-LR.
Statistical testing included standard error calculations for replicate groups, error propagation, and testing for significant differences between MC-LR-treated and corresponding control samples using two-tailed t testing.
RESULTS AND DISCUSSION
Loss of MC can be complemented only partially via external addition of MC-LR.
Transcriptomes of wild-type PCC 7806 and the microcystin-deficient ΔmcyB mutant were compared under low light conditions (16 μmol photons m−2 s−1), where the two strains did not show any difference in growth according to optical density measurements but differed significantly in their chlorophyll a (Chl a)-to-phycocyanin (PC) ratios (31). Extracellular MC is negligible under these conditions, whereas under high light considerable amounts are released into the medium (32). To assess the impact of extracellular MC-LR on gene expression, 50 ng/ml of the peptide (corresponding to maximum amounts that were found in the medium of wild-type cultures under high light conditions [32]) were added to parallel biological replicates and compared to untreated control aliquots. Prior to the transcriptional analysis, genomes of wild-type PCC 7806 and the ΔmcyB mutant were compared using DNA/DNA hybridization. Even though a few differences could be observed (see Data Set S1 in the supplemental material), major secondary mutations or a reorganization of the genome by active transposons could be excluded (see Fig. S1). Analysis of transcriptomes rendered a complex data set. Considering the degree of natural variation between the different samples, thorough validation and testing by statistical methods were utilized for quality assessment and a first survey. When hierarchical complete linkage clustering and multidimensional scaling were applied, the transcriptome profiles of the wild type were well separated from those of the mutant independent of the MC-LR treatment (see Fig. S2). A very clear separation of wild-type and mutant data sets also was seen when principal component analysis (PCA) was used for dimensionality reduction (Fig. 1A). None of the analyzed component spaces, however, showed a segregation of transcriptome samples in response to external microcystin-LR treatment. This can be taken as a first indication that the impact of external MC-LR on Microcystis gene expression is low under the conditions tested and might affect only a small number of genes. A more detailed look at differentially expressed genes revealed 213 genes showing pronounced differences (log2-fold change of >1 and <−1) between wild-type and mutant control transcriptomes. Of those, 166 were downregulated and 47 were upregulated in the mutant compared to the wild type (Fig. 1C; also see Fig. S3 and Data Set S2). The MC-LR treatment resulted in significant changes for 12 genes in the wild type and mutant. All 12 genes (annotated as individual protein files IPF38 to IPF51) were downregulated in response to the addition of MC-LR in both the wild type and the mutant. However, the same genes were strongly upregulated in the untreated ΔmcyB mutant samples compared to the wild type. For this small fraction of genes, external MC-LR addition could complement the loss of MC in the mutant (Fig. 1C). Whereas this clearly indicates that the presence of MC-LR is transduced as a signal with an impact on gene expression, the MC-LR treatment does not result in global transcriptional reprogramming. Thus, the majority of transcriptional differences observed between the wild type and mutant in this study may relate to the intracellular role of MC rather than to its extracellular role. The total number of differentially expressed genes accounts for approximately 4.5% of the genome. In comparison to other transcriptional studies, where low nutrient (N and P) cultivation resulted in ∼1 to 14% (33) and 5 to 33% (34) differential gene expression, microcystin can be considered a major player in the adaptation of the cellular physiological state.
FIG 1.
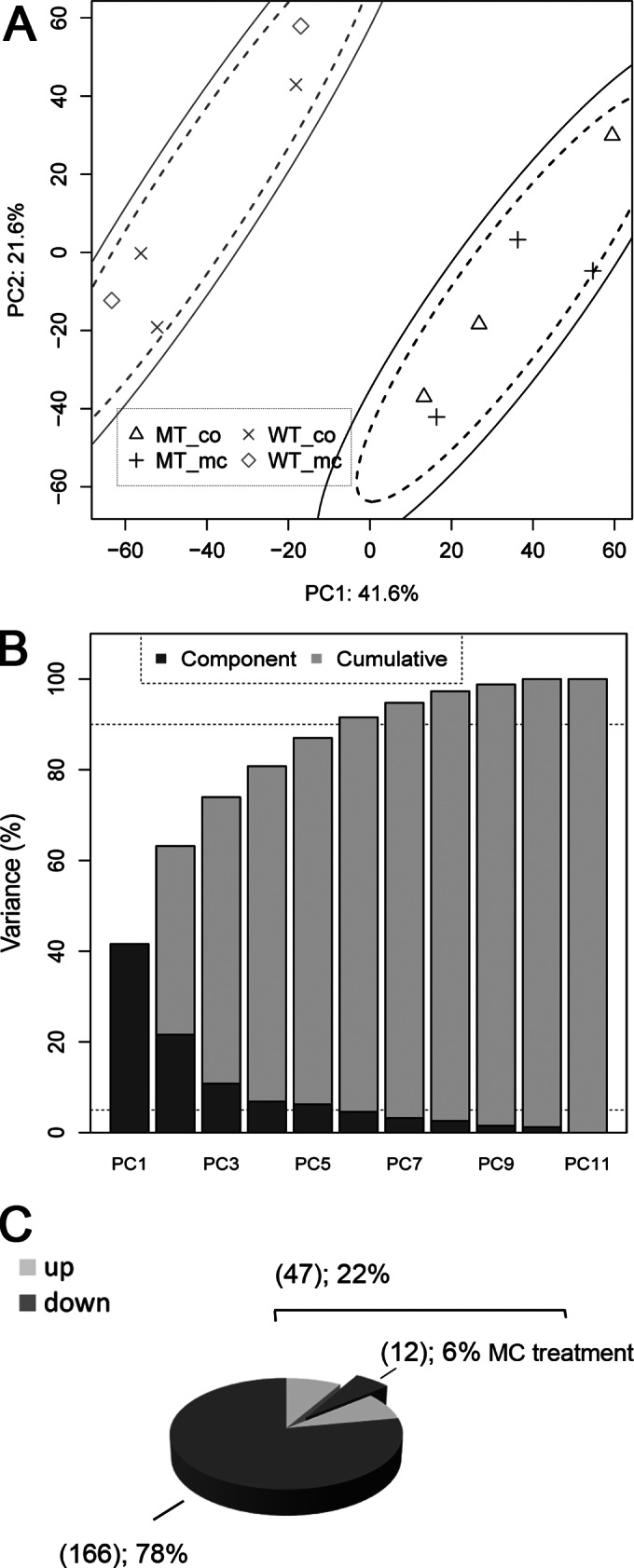
(A) PCA of log2-normalized gene expression data for M. aeruginosa WT and ΔmcyB mutant (MT) cells. As both the 85% (dashed line) and the 95% confidence (solid line) ellipses for PC1 (explaining 41.6% of the overall variance) and PC2 (21.6% variance) separate the genotypes, the cluster separation for the genotypes is highly significant. No separation was observed between controls (co) and MC-LR addition to the cultivation medium (mc). (B) As visible in the scree plot on the right by dashed lines for 5 and 90% variance, the first six principal components together carry more than 90% of the data set variance. (C) Number of genes significantly downregulated (log2-fold change of <−1) or upregulated (log2-fold change of >1) with P values of <0.1 in the ΔmcyB mutant cells compared to the wild type. For 6% of the differentially upregulated genes in the mutant, these effects could be reversed by external MC-LR addition, which provides complementation of the MC knockout in the mutant. No further genes were affected by the external MC-LR treatment under the tested conditions.
Functional assignment of differentially expressed genes.
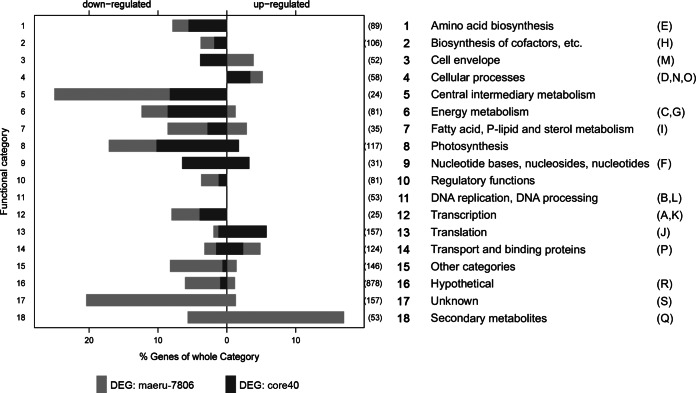
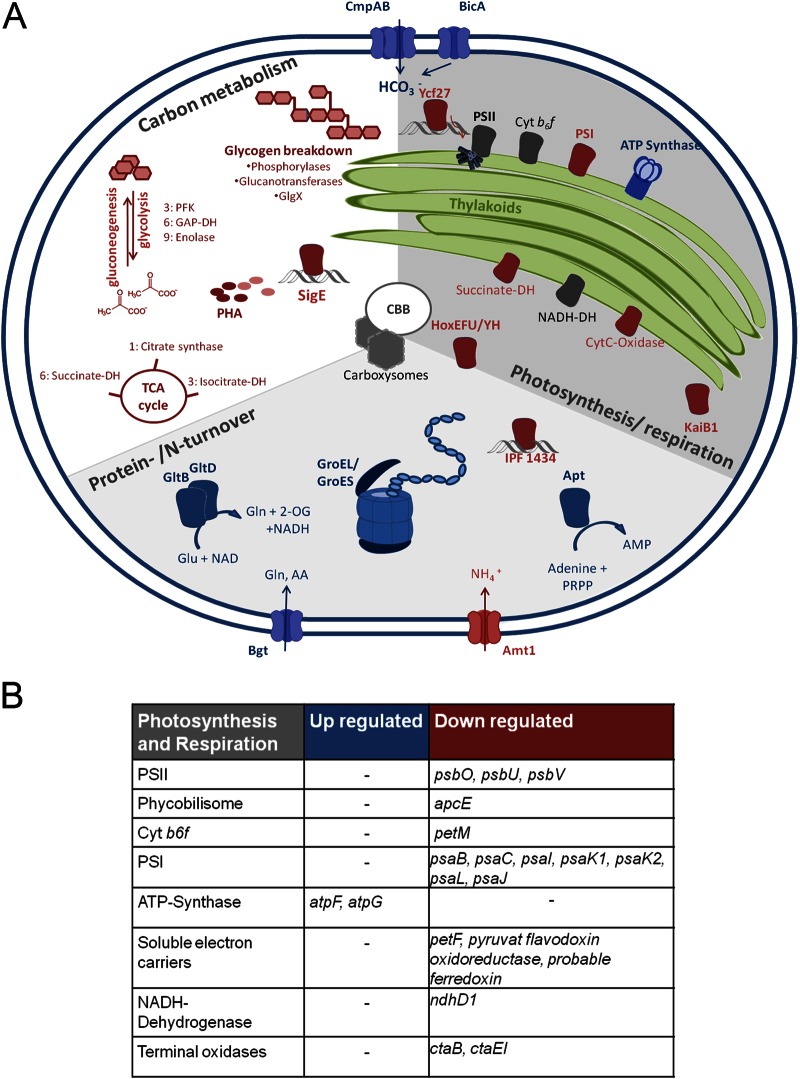
In order to extract more functional information, we took a closer look at the gene category assignments (according to Cyanobase and the Clusters of Orthologous Groups [COG] database) of differentially expressed candidates. Given the importance of secondary metabolites in Microcystis and the frequent misassignment to other functional categories, we additionally classified 52 genes into a secondary metabolism subcategory (category 18) (Fig. 2). Notably, more than 20% of genes of the intermediary metabolism category were transcribed at significantly lower levels in the mutant (Fig. 2). Downregulation in the MC-free mutant also was seen for 17% of the photosynthesis and respiration genes and 12% of the energy metabolism genes (Fig. 2). Considering that the two Microcystis strains did not show any difference in growth, this unidirectional advance of primary metabolism gene expression in the wild type was unexpected. The detailed analysis of the photosynthesis and respiration category revealed a number of genes encoding components of photosystem I (PSI) that were downregulated in the ΔmcyB mutant, although the psaA gene was missing from this list (Fig. 3; also see Fig. S3). psbW, -V, and -O (but not the core genes) were the only genes representing photosystem II (PSII) on the list of genes downregulated in the ΔmcyB mutant. This indicates different PSI-to-PSII ratios in the wild type and mutant with a lower PSI expression in the mutant than in the wild type, while PSII expression did not change significantly. Interestingly, an altered PSI-to-PSII ratio already was proposed by Hesse et al. (31), who have observed clear differences in the Chl a-to-PC ratios in the wild type and mutant under low light conditions (31). Thus, the transcriptome data are in good agreement with physiological parameters obtained from the same strains. The adjustment of PSI-to-PSII ratios is one of the adaptation mechanisms of cyanobacteria to changing light conditions (35). Central intermediate metabolism genes downregulated in the mutant include several candidates involved in glycogen degradation and polyhydroxyalkanoate biosynthesis, suggesting that the wild type and mutant follow different carbon storage routes (Fig. 3; also see Fig. S3). The mutant also shows a lower abundance of a number of genes encoding glycolysis and tricarboxylic acid (TCA) cycle enzymes, genes related to nitrogen metabolism and transport, and hox genes encoding bidirectional hydrogenase (Fig. 3; also see Fig. S3).
FIG 2.
Assignment of genes significantly downregulated or upregulated in the ΔmcyB mutant, compared to those the wild-type M. aeruginosa PCC 7806, to functional categories according to Cyanobase (http://genome.microbedb.jp/cyanobase/Synechocystis). Parentheses after the bars contain the total number of genes of that category. Equivalent COG category assignments (for a key, see Table S1 in the supplemental material) are shown in brackets on the right. DEG, differentially expressed genes; core40, conserved cyanobacterial genes that share orthologous genes in at least 15 further cyanobacterial genomes; maeru-7806, genes specific to PCC 7806 (27).
FIG 3.
Illustration of representative genes significantly up- or downregulated in the mutant. See Fig. S3 in the supplemental material for more details. Red components, downregulated in mutant; blue components, upregulated; gray, no difference. Cyt, cytochrome; DH, dehydrogenase; PRPP, phosphoribosyl pyrophosphate; AA, amino acid; 2-OG, 2-oxoglutarate; PHA, polyhydroxyalkanoate; CBB, Calvin-Benson-Bassham cycle.
The ΔmcyB mutant, on the other hand, shows higher transcript amounts for a number of genes assigned to translation, cellular processes, and transport. Remarkable candidates of the latter category are the bicarbonate transporter gene cmpA, the associated permease gene cmpB, and the bicarbonate transporter gene bicA (Fig. 3). There is increasing evidence for a differential adaptation of the wild type and ΔmcyB mutant to inorganic carbon limitation, which is also found in naturally toxic and nontoxic strains in the field (16, 36). Thus, the upregulation of cmpAB and bicA may be a direct consequence of a more pronounced carbon limitation in the mutant. Remarkably, the two chaperonin-encoding genes groES and groEL also were more strongly transcribed in the ΔmcyB mutant. This indicates a greater need for assistance with protein folding in the absence of microcystin. A protein-stabilizing effect of MC already was discussed in our previous proteomic study (17). The gene most strongly upregulated in the MC-free mutant (log-fold change of >4) was apt, encoding adenine phosphoribosyltransferase. The enzyme synthesizes AMP through a salvage pathway from adenine. The mutant further showed higher expression levels for two subunits of the ATP synthase, AtpG and AtpF. Subunits of the ATP synthase complex also were upregulated in the mutant in our recent proteomic study (17). The gene category showing the highest degree of upregulation in the mutant is the secondary metabolite category. This group includes the cluster IPF38-51, which also responded to external MC-LR. The cross talk between different secondary metabolites will be discussed in more detail below.
Besides the differentially transcribed genes in the microcystin-deficient ΔmcyB mutant, it is interesting which gene candidates do not show differential expression in our analysis. In particular, this includes genes encoding enzymes of the Calvin-Benson-Bassham (CBB) cycle that were revealed as major differences in a recent proteomic comparison of the two strains (17). This finding is in agreement with the hypothesis that the differential accumulation of RbcL and further CBB proteins is due to differences in protein stability resulting from MC binding rather than transcriptional variation.
Effect of MC on regulatory genes.
Although our study provides clear evidence for an intracellular role of MC, we cannot easily anticipate which mechanism underlies the transcriptional changes. One option is the interaction of MC with one or more global regulators, as previously proposed by Wilhelm and Boyer (37). Three transcriptional regulators were significantly downregulated in the mutant: IPF1434, encoding an HTH-type transcriptional regulator, IPF4842, encoding the alternative circadian clock protein KaiB1, and ycf27, encoding the light-dependent transcriptional regulator RpaB. IPF1434 is sporadically encoded in a number of Microcystis, Anabaena, and Cyanothece genomes; however, not all toxic Microcystis strains possess the gene. The role of alternative KaiB proteins is not yet known; however, the kaiB1 gene showed a circadian rhythm that was the opposite of that of the original kaiB gene in PCC 7806 (14). The gene is conserved in Microcystis and other cyanobacteria, and the corresponding protein could well be an interaction partner of MC. The essential response regulator RpaB has been linked to the regulation of excitation energy transfer from the phycobilisome to PSI (38). Interestingly, the gene product also has been connected to kaiBC gene expression and circadian oscillations (39). Whether or not the differential expression of rpaB and kaiB1 indicates major rhythmic differences in the wild type and mutant cannot be concluded from the present experiments, as the study was not designed for circadian analysis. Differentially expressed regulatory genes further include the sigE sigma factor (IPF5730), which was strongly downregulated in the mutant. The factor was previously connected with the expression of sugar catabolism genes in Synechocystis sp. strain PCC 6803. Studies have stated that SigE is responsible for the regulation of glycogen degradation in a light- and N-dependent manner (40, 41). SigE-controlled glycogen-degrading genes, like glgX, glgP, and polyhydroxyalkanoate (PHA) synthesis genes, as well as phosphofructokinase (pfkA), all were downregulated in our survey, pointing to a possible interrelation of MC and SigE. Taken together, the three above-mentioned candidates are known global regulators in cyanobacteria that could be responsible for the different transcriptional programs observed for the wild type and mutant in this study.
Impact of microcystin on secondary metabolite gene expression.
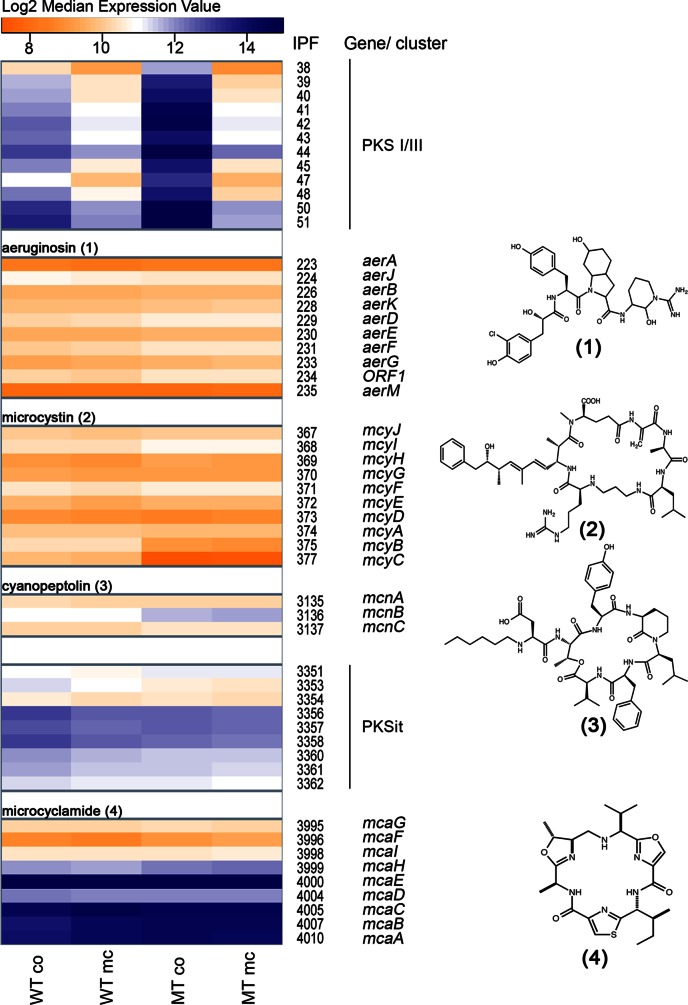
All genes directly responding to an MC-LR signal belong to the secondary metabolism category (Fig. 1C). Moreover, autoinduction of microcystin production as well as cross talk with other secondary metabolites was reported in an earlier study (13). This prompted us to have a closer look at the entire secondary metabolite category (Fig. 4). Of the six gene clusters analyzed, four are known to direct the biosynthesis of microcystin, cyanopeptolin, aeruginosin, and microcyclamide (Fig. 4). Two further clusters are supposed to be involved in the synthesis of so-far cryptic compounds. One cluster (IPF38-51) encodes a unique modular PKS complex comprised of a PKSIII chalcone type synthase (23). PKSIII members were only recently discovered in bacteria and typically are involved in the synthesis of aromatic scaffolds. The second cluster (IPF3351-3362) encodes an iterative type I PKS potentially involved in the synthesis of an enediyne type of compound (23). When the four data sets (wild type and mutant with and without MC-LR treatment) were compared, only the pksI-pksIII cluster exhibited pronounced transcriptional differences between the wild type and mutant. The gene cluster was upregulated and reached very high expression levels in the ΔmcyB mutant. The addition of MC-LR led to a strong downregulation of this cluster in both the wild type and mutant. The aeruginosin (IPF223-235), microcystin (IPF367-377), and cyanopeptolin (IPF3135-3137) gene clusters, on the other hand, did not show significant differences in the two genotypes or between treatments (Fig. 4). An exception was seen for mcyB and mcyC, two genes of the microcystin biosynthesis gene cluster. Whereas the rest of the cluster (mcyADEFGHIJ) does not show considerable differences in expression, mcyB and mcyC are transcribed at lower levels in the mutant. This phenomenon likely is due to the fact that the chloramphenicol resistance cassette is inserted into the mcyB gene and affects the expression of genes downstream, but not upstream, of mcyB. Furthermore, no significant difference between the wild type and mutant was seen for the enediyne type gene cluster (PKSit) and the microcyclamide gene cluster (IPF3995-4010) (Fig. 4). Microcyclamide belongs to the ribosomally produced and posttranslationally modified peptides (RiPPS) (42), and the corresponding gene cluster showed a much higher expression level than the nonribosomal peptide synthetase (NRPS) type gene clusters involved in the synthesis of microcystin, aeruginosin, and cyanopeptolin. The microcyclamide precursor gene mcaE (IPF4000) belonged to the overall strongest transcripts in the entire data set. This can be seen as a parallel to a recent metatranscriptomic study of Microcystis that has found another RiPP type secondary metabolite precursor transcript, encoding microviridin, as one of the most abundant transcripts (43).
FIG 4.
Heatmap showing expression levels of secondary metabolite genes in WT PCC 7806 and the ΔmcyB mutant (MT) with (mc) and without (co) MC-LR treatment. Expression levels are given as median log2 values as indicated in the color code bar at the top. The assignment of four of the six gene clusters to the biosynthesis of aeruginosin (1), microcystin (2), cyanopeptolin (3), and microcyclamide (4) is indicated on the right along with IPF numbers of genes and gene abbreviations. Two cryptic clusters encode polyketide synthase complexes presumably involved in the synthesis of an aromatic polyketide (PKSI/PKSIII) and an enediyne type polyketide (PKSit).
In conclusion, the response of the pksI-pksIII gene cluster to external MC-LR is highly specific. The gene cluster apparently is under the strict hierarchical control of MC. Notably, the number of transcriptional differences upon MC addition is far lower than the number of overall transcriptional differences in the ΔmcyB mutant. These facts suggest that under the conditions tested, microcystin-LR is not actively transported into the cells but is perceived and transduced by a receptor-mediated signaling cascade. Previous experiments showed hardly any MC import into cells, which further supports the idea of an MC-receptor signal cascade. The observed reception of an extracellular MC signal might be part of bacterial cell-cell communication, which is often organized as a hierarchical network. Examples for these cell-cell communication models were investigated in quorum-sensing (QS) systems, which are well known in Gram-negative bacteria. Quorum sensing usually is mediated by diffusible signals that accumulate under higher cell densities and then often controls the expression of other QS signals and more complex secondary metabolites (44). A recent study using Pseudomonas aeruginosa as a model has provided strong evidence that, by using only multiple and interconnected signals, the cells can infer both their social and physical environments (44). Although the secondary metabolites in Microcystis are not freely diffusible, our data provide increasing hints for an interrelation between MC and the cryptic PKSI/PKSIII metabolite that seemingly are part of a hierarchical cross talk network. This in turn implies that the metabolites reflect the social and/or physical status of individual cells or colonies. One of the key aspects of quorum sensing, however, is the autoinduction of genes involved in the biosynthesis of the signaling compound.
Comprehensive analysis of the transcriptional network between secondary metabolites in Microcystis.
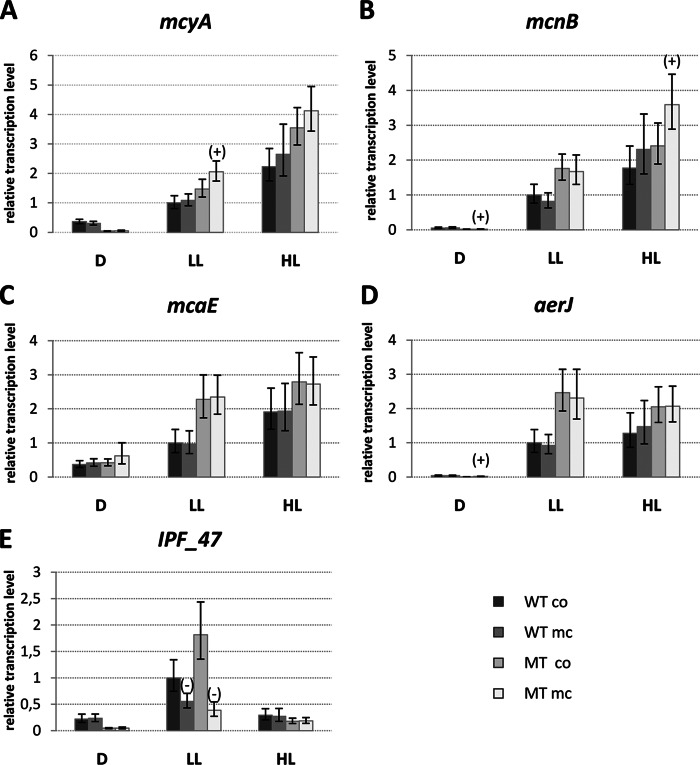
It was somewhat surprising that the MC-LR signal did not have an impact on the expression of MC biosynthesis genes itself, particularly as the autoregulation of MC already was proposed on the protein level in reference 13. Earlier studies of our group, however, already had shown that the MC autoinduction effect depends on the light conditions applied during cultivation (see Fig. S4 in the supplemental material). An autoinduction of mcy genes was observed primarily above a critical threshold of light (see Fig. S4). Therefore, we were curious as to whether MC-LR signal reception and response generally are dependent on light conditions, so we initiated a more comprehensive RT-PCR study for the secondary metabolite gene category. Wild-type and ΔmcyB mutant cultures were grown to late exponential phase (OD750, 1.0) and adjusted to three different light conditions for 2 days. At this time point, cultures were split and 50 ng/ml MC-LR (final concentration) was added to half of the cultures for 1 h, whereas the other replicates served as controls. Five genes were analyzed: mcyA, representing the MC biosynthesis gene cluster; IPF47, representing the cryptic pksI-pksIII gene cluster; mcaE, representing the microcyclamide gene cluster; aerJ, representing the aeruginosin gene cluster; and mcnB, representing the cyanopeptolin gene cluster (Fig. 5). The microcystin gene cluster showed the highest transcript accumulation under high light conditions, as previously reported (45) (Fig. 5A). The same trend was seen for the cyanopeptolin and microcyclamide biosynthesis genes (Fig. 5B and C). The aeruginosin biosynthesis gene aerJ showed similar transcript levels under low light and high light (Fig. 5D). In sharp contrast to the other four secondary metabolite clusters, the pksI-pksIII gene cluster showed the highest transcript accumulations under low light and a clear downregulation under high light in both the wild type and mutant (Fig. 5E). Thus, the cluster revealed a reciprocal transcriptional pattern compared to those of mcyA and mcnB with respect to light. Under most of the conditions tested, secondary metabolite gene clusters were upregulated in the ΔmcyB mutant compared to the wild type. Although MC-LR addition eventually led to an increase in mcyA transcription, the effect was negligible and significant only in the ΔmcyB mutant under low light conditions (Fig. 5A). A slight MC-LR induction effect also was observed for the cyanopeptolin biosynthesis gene mcnB under high light conditions in the ΔmcyB mutant (Fig. 5B). Similar to the microarray study (studying MC effects in mid-log-phase cultures), the strongest response to MC-LR addition (downregulation) was observed for the pksI-pksIII gene cluster under low light conditions (Fig. 5E). Taken together, the RT-PCR study indicates that the MC signaling effect strongly depends on the light conditions used for cultivation. The pksI-pksIII gene cluster was most sensitive to an external MC-LR signal, whereas the previously reported autoinduction effect on mcy gene expression was almost negligible in the present study and observed only above a critical threshold of light (see Fig. S4 in the supplemental material). Thus, MC signaling cannot be considered a bona fide quorum-sensing phenomenon; rather, it reflects a light-dependent hierarchical network between secondary metabolites of Microcystis in which microcystin and the cryptic PKSI/PKSIII metabolite are opposing players.
FIG 5.
RT-PCR showing relative expression levels of secondary metabolite biosynthesis genes mcyA (microcystin; A), mcnB (cyanopeptolin; B), mcaE (microcyclamide; C), aerJ (aeruginosin; D), and IPF47 (PKSI/PKSIII biosynthesis, presumably aromatic polyketide; E) in WT PCC 7806 and the ΔmcyB mutant (MT) without (co) and with (mc) MC-LR treatment under conditions of darkness (D; 0 μE m−2 s−1), low light (LL; 16 μE m−2 s−1), and high light (HL; 70 μE m−2 s−1). Expression rates were calculated as ratios to WT-LL-co levels. Statistical testing is indicated by error bars, representing standard errors, while the significance of MC-LR addition in relation to the control samples of the same genotype grown under the same light conditions and analyzed by two-tailed Student's t test is illustrated on respective bars as significantly upregulated (+) and significantly downregulated (−).
Even though our study can provide only a snapshot of the role of MC on global transcription in M. aeruginosa PCC 7806, there is clear evidence that the transcriptional differences observed are due primarily to the loss of the intracellular MC portion. Previous studies have provided hints that MC plays a global role in the redox-dependent protein stability of Microcystis (17, 46). Microcystin-binding proteins include phycobilisome antenna proteins that showed an altered accumulation in the wild type and ΔmcyB mutant (17, 31). Hence, the differential transcript accumulation of photosynthesis-related genes observed in this study might be a consequence of a different light perception of the two strains. Differences observed for the carbon uptake and carbon metabolism genes, on the other hand, may relate to the differential accumulation of Calvin-Benson-Bassham cycle enzymes upon MC binding (17). The secondary metabolism category stands out from the global transcriptional pattern. For one thing, it is the category in which most genes are upregulated upon the loss of MC. For another thing, for this category only, the loss of MC can be complemented by the external addition of MC. The distinct transcriptional profiles argue for a multifaceted role of MC and different mechanisms that underlie the variations observed in the transcriptomes of wild-type PCC 7806 and the mutant. It remains elusive as to whether MC signaling can cause a global transcriptional reprogramming under conditions not tested in this study. It also still is unclear what impact the MC signaling has in field colonies that are experiencing very different light conditions during vertical migration in the water column and due to shadowing within a bloom.
Supplementary Material
ACKNOWLEDGMENTS
We thank Martijs J. Jonker (Microarray Department, University of Amsterdam) for his sophisticated consulting on the design of a robust microarray hybridization scheme. Vladimir Karsikov helped greatly with consultation on statistical computation. Furthermore, we thank Wim A. Ensink (Microarray Department, University of Amsterdam) for his committed work and advice as well as technical conduction of the microarrays. We thank Sarah Ongley for copyediting and further input.
This work was supported by a project of the German Research Foundation (Di910/5-1) to E.D. Further support by the European Cooperation in Science and Technology COST Action (ES1105 CYANOCOST) was provided to finance a short-term scientific mission (to A.K.M.). We acknowledge use of DNA microarrays that were acquired with support from a Netherlands Organisation for Scientific Research (NWO) TOP grant (to J. Huisman).
We have no conflict of interest related to the manuscript.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02601-14.
REFERENCES
- 1.Paerl HW, Otten TG. 2013. Harmful cyanobacterial blooms: causes, consequences, and controls. Microb Ecol 65:995–1010. doi: 10.1007/s00248-012-0159-y. [DOI] [PubMed] [Google Scholar]
- 2.Dittmann E, Fewer DP, Neilan BA. 2013. Cyanobacterial toxins: biosynthetic routes and evolutionary roots. FEMS Microbiol Rev 37:23–43. doi: 10.1111/j.1574-6976.2012.12000.x. [DOI] [PubMed] [Google Scholar]
- 3.Paerl HW, Huisman J. 2009. Climate change: a catalyst for global expansion of harmful cyanobacterial blooms. Environ Microbiol Rep 1:27–37. doi: 10.1111/j.1758-2229.2008.00004.x. [DOI] [PubMed] [Google Scholar]
- 4.Via-Ordorika L, Fastner J, Kurmayer R, Hisbergues M, Dittmann E, Komarek J, Erhard M, Chorus I. 2004. Distribution of microcystin-producing and non-microcystin-producing Microcystis sp. in European freshwater bodies: detection of microcystins and microcystin genes in individual colonies. Syst Appl Microbiol 27:592–602. doi: 10.1078/0723202041748163. [DOI] [PubMed] [Google Scholar]
- 5.Thomas RH, Walsby AE. 1985. Buoyancy regulation in a strain of Microcystis. J Gen Microbiol 131:799–809. [Google Scholar]
- 6.Rabouille S, Thebault JM, Salencon MJ. 2003. Simulation of carbon reserve dynamics in Microcystis and its influence on vertical migration with Yoyo model. C R Biol 326:349–361. doi: 10.1016/S1631-0691(03)00123-9. [DOI] [PubMed] [Google Scholar]
- 7.Briand E, Escoffier N, Straub C, Sabart M, Quiblier C, Humbert JF. 2009. Spatiotemporal changes in the genetic diversity of a bloom-forming Microcystis aeruginosa (cyanobacteria) population. ISME J 3:419–429. doi: 10.1038/ismej.2008.121. [DOI] [PubMed] [Google Scholar]
- 8.Neilan BA, Pearson LA, Muenchhoff J, Moffitt MC, Dittmann E. 2013. Environmental conditions that influence toxin biosynthesis in cyanobacteria. Environ Microbiol 15:1239–1253. doi: 10.1111/j.1462-2920.2012.02729.x. [DOI] [PubMed] [Google Scholar]
- 9.Runnegar M, Berndt N, Kong SM, Lee EYC, Zhang LF. 1995. In vivo and in vitro binding of microcystin to protein phosphatase 1 and phosphatase 2A. Biochem Biophys Res Commun 216:162–169. doi: 10.1006/bbrc.1995.2605. [DOI] [PubMed] [Google Scholar]
- 10.Rohrlack T, Christiansen G, Kurmayer R. 2013. Putative antiparasite defensive system involving ribosomal and nonribosomal oligopeptides in cyanobacteria of the genus Planktothrix. Appl Environ Microbiol 79:2642–2647. doi: 10.1128/AEM.03499-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rohrlack T, Dittmann E, Henning M, Börner T, Kohl JG. 1999. Role of microcystins in poisoning and food ingestion inhibition of Daphnia galeata caused by the cyanobacterium Microcystis aeruginosa. Appl Environ Microbiol 65:737–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rantala A, Fewer DP, Hisbergues M, Rouhiainen L, Vaitomaa J, Börner T, Sivonen K. 2004. Phylogenetic evidence for the early evolution of microcystin synthesis. Proc Natl Acad Sci U S A 101:568–573. doi: 10.1073/pnas.0304489101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schatz D, Keren Y, Vardi A, Sukenik A, Carmeli S, Borner T, Dittmann E, Kaplan A. 2007. Towards clarification of the biological role of microcystins, a family of cyanobacterial toxins. Environ Microbiol 9:965–970. doi: 10.1111/j.1462-2920.2006.01218.x. [DOI] [PubMed] [Google Scholar]
- 14.Straub C, Quillardet P, Vergalli J, de Marsac NT, Humbert JF. 2011. A day in the life of Microcystis aeruginosa strain PCC 7806 as revealed by a transcriptomic analysis. PLoS One 6:e16208. doi: 10.1371/journal.pone.0016208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jähnichen S, Ihle T, Petzoldt T, Benndorf J. 2007. Impact of inorganic carbon availability on microcystin production by Microcystis aeruginosa PCC 7806. Appl Environ Microbiol 73:6994–7002. doi: 10.1128/AEM.01253-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van de Waal DB, Verspagen JM, Finke JF, Vournazou V, Immers AK, Kardinaal WE, Tonk L, Becker S, Van Donk E, Visser PM, Huisman J. 2011. Reversal in competitive dominance of a toxic versus non-toxic cyanobacterium in response to rising CO2. ISME J 5:1438–1450. doi: 10.1038/ismej.2011.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zilliges Y, Kehr JC, Meissner S, Ishida K, Mikkat S, Hagemann M, Kaplan A, Börner T, Dittmann E. 2011. The cyanobacterial hepatotoxin microcystin binds to proteins and increases the fitness of Microcystis under oxidative stress conditions. PLoS One 6:e17615. doi: 10.1371/journal.pone.0017615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alexova R, Haynes PA, Ferrari BC, Neilan BA. 2011. Comparative protein expression in different strains of the bloom-forming cyanobacterium Microcystis aeruginosa. Mol Cell Proteomics 10:M110.003749. doi: 10.1074/mcp.M110.003749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ginn HP, Pearson LA, Neilan BA. 2010. NtcA from Microcystis aeruginosa PCC 7806 is autoregulatory and binds to the microcystin promoter. Appl Environ Microbiol 76:4362–4368. doi: 10.1128/AEM.01862-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dittmann E, Neilan BA, Erhard M, von Döhren H, Börner T. 1997. Insertional mutagenesis of a peptide synthetase gene that is responsible for hepatotoxin production in the cyanobacterium Microcystis aeruginosa PCC 7806. Mol Microbiol 26:779–787. doi: 10.1046/j.1365-2958.1997.6131982.x. [DOI] [PubMed] [Google Scholar]
- 21.Rippka R, Deruelles J, Waterbury JB, Herdman M, Stanier RY. 1979. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol 111:1–61. doi: 10.1099/00221287-111-1-1. [DOI] [Google Scholar]
- 22.Chomczynski P, Downs TR, Frohman LA. 1988. Feedback regulation of growth hormone (GH)-releasing hormone gene expression by GH in rat hypothalamus. Mol Endocrinol 2:236–241. doi: 10.1210/mend-2-3-236. [DOI] [PubMed] [Google Scholar]
- 23.Frangeul L, Quillardet P, Castets AM, Humbert JF, Matthijs HC, Cortez D, Tolonen A, Zhang CC, Gribaldo S, Kehr JC, Zilliges Y, Ziemert N, Becker S, Talla E, Latifi A, Billault A, Lepelletier A, Dittmann E, Bouchier C, de Marsac NT. 2008. Highly plastic genome of Microcystis aeruginosa PCC 7806, a ubiquitous toxic freshwater cyanobacterium. BMC Genomics 9:274. doi: 10.1186/1471-2164-9-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.R Development Core Team. 2011. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 25.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smyth GK, Speed T. 2003. Normalization of cDNA microarray data. Methods 31:265–273. doi: 10.1016/S1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- 27.Cox TF, Cox MAA. 2001. Multidimensional scaling, 2nd ed. Chapman and Hall, London, United Kingdom. [Google Scholar]
- 28.Groth D, Hartmann S, Klie S, Selbig J. 2013. Principal components analysis. Methods Mol Biol 930:527–547. doi: 10.1007/978-1-62703-059-5_22. [DOI] [PubMed] [Google Scholar]
- 29.Mosteller F, Fisher RA. 1948. Questions and answers. Am Stat. 2:30–31. [Google Scholar]
- 30.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hesse K, Dittmann E, Börner T. 2001. Consequences of impaired microcystin production for light-dependent growth and pigmentation of Microcystis aeruginosa PCC 7806. FEMS Microbiol Ecol 37:39–43. doi: 10.1111/j.1574-6941.2001.tb00851.x. [DOI] [Google Scholar]
- 32.Wiedner C, Visser PM, Fastner J, Metcalf JS, Codd GA, Mur LR. 2003. Effects of light on the microcystin content of Microcystis strain PCC 7806. Appl Environ Microbiol 69:1475–1481. doi: 10.1128/AEM.69.3.1475-1481.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Steffen MM, Dearth SP, Dill BD, Li Z, Larsen KM, Campagna SR, Wilhelm SW. 2014. Nutrients drive transcriptional changes that maintain metabolic homeostasis but alter genome architecture in Microcystis. ISME J 8:2080–2092. doi: 10.1038/ismej.2014.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harke MJ, Gobler CJ. 2013. Global transcriptional responses of the toxic cyanobacterium, Microcystis aeruginosa, to nitrogen stress, phosphorus stress, and growth on organic matter. PLoS One 8:e69834. doi: 10.1371/journal.pone.0069834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muramatsu M, Sonoike K, Hihara Y. 2009. Mechanism of downregulation of photosystem I content under high-light conditions in the cyanobacterium Synechocystis sp. PCC 6803. Microbiology 155:989–996. doi: 10.1099/mic.0.024018-0. [DOI] [PubMed] [Google Scholar]
- 36.Sandrini G, Matthijs HC, Verspagen JM, Muyzer G, Huisman J. 2014. Genetic diversity of inorganic carbon uptake systems causes variation in CO2 response of the cyanobacterium Microcystis. ISME J 8:589–600. doi: 10.1038/ismej.2013.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilhelm SW, Boyer GL. 2011. Ecology: healthy competition. Nat Climate Change 1:300–301. doi: 10.1038/nclimate1202. [DOI] [Google Scholar]
- 38.Ashby MK, Houmard J, Mullineaux CW. 2002. The ycf27 genes from cyanobacteria and eukaryotic algae: distribution and implications for chloroplast evolution. FEMS Microbiol Lett 214:25–30. [DOI] [PubMed] [Google Scholar]
- 39.Hanaoka M, Takai N, Hosokawa N, Fujiwara M, Akimoto Y, Kobori N, Iwasaki H, Kondo T, Tanaka K. 2012. RpaB, another response regulator operating circadian clock-dependent transcriptional regulation in Synechococcus elongatus PCC 7942. J Biol Chem 287:26321–26327. doi: 10.1074/jbc.M111.338251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Osanai T, Kanesaki Y, Nakano T, Takahashi H, Asayama M, Shirai M, Kanehisa M, Suzuki I, Murata N, Tanaka K. 2005. Positive regulation of sugar catabolic pathways in the cyanobacterium Synechocystis sp. PCC 6803 by the group 2 sigma factor SigE. J Biol Chem 280:30653–30659. doi: 10.1074/jbc.M505043200. [DOI] [PubMed] [Google Scholar]
- 41.Osanai T, Oikawa A, Azuma M, Tanaka K, Saito K, Hirai MY, Ikeuchi M. 2011. Genetic engineering of group 2 sigma factor SigE widely activates expressions of sugar catabolic genes in Synechocystis species PCC 6803. J Biol Chem 286:30962–30971. doi: 10.1074/jbc.M111.231183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, Bulaj G, Camarero JA, Campopiano DJ, Challis GL, Clardy J, Cotter PD, Craik DJ, Dawson M, Dittmann E, Donadio S, Dorrestein PC, Entian KD, Fischbach MA, Garavelli JS, Goransson U, Gruber CW, Haft DH, Hemscheidt TK, Hertweck C, Hill C, Horswill AR, Jaspars M, Kelly WL, Klinman JP, Kuipers OP, Link AJ, Liu W, Marahiel MA, Mitchell DA, Moll GN, Moore BS, Muller R, Nair SK, Nes IF, Norris GE, Olivera BM, Onaka H, Patchett ML, Piel J, et al. . 2013. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat Prod Rep 30:108–160. doi: 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Penn K, Wang J, Fernando SC, Thompson JR. 2014. Secondary metabolite gene expression and interplay of bacterial functions in a tropical freshwater cyanobacterial bloom. ISME J 8:1866–1878. doi: 10.1038/ismej.2014.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cornforth DM, Popat R, McNally L, Gurney J, Scott-Phillips TC, Ivens A, Diggle SP, Brown SP. 2014. Combinatorial quorum sensing allows bacteria to resolve their social and physical environment. Proc Natl Acad Sci U S A 111:4280–4284. doi: 10.1073/pnas.1319175111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaebernick M, Neilan BA, Börner T, Dittmann E. 2000. Light and the transcriptional response of the microcystin biosynthesis gene cluster. Appl Environ Microbiol 66:3387–3392. doi: 10.1128/AEM.66.8.3387-3392.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meissner S, Steinhauser D, Dittmann E. 15 July 2014. Metabolomic analysis indicates a pivotal role of the hepatotoxin microcystin in high light adaptation of Microcystis. Environ Microbiol doi: 10.1111/1462-2920.12565. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.