Abstract
We compared the results of high-hydrostatic-pressure (HHP) inactivation of murine norovirus type 1 (MNV-1) and Tulane virus (TV) obtained by a porcine gastric mucin binding assay followed by quantitative reverse transcription-PCR (referred to here as the PGM-MB/PCR assay) and a plaque assay and evaluated HHP inactivation of a human norovirus (HuNoV) genogroup I genotype 1 (GI.1) strain and a HuNoV GII.4 strain by using the PGM-MB/PCR assay. Viruses were treated at different pressure levels for 2 min at 4 or 21°C in culture medium of neutral pH and in culture medium of pH 4 at 21°C. The log reductions of infectious MNV-1 and TV particles caused by HHP were assessed using the PGM-MB/PCR and plaque assays, while the log reductions of HuNoVs were assessed by the PGM-MB/PCR assay only. For TV and MNV-1, the two pressure inactivation curves obtained using the plaque and PGM-MB/PCR assays were almost identical at ≤2-log-reduction levels regardless of the treatment temperature and pH. Further increasing the pressure over the 2-log-reduction level resulted in higher log reductions of TV and MNV-1, as assessed by the plaque assay, but did not increase the log reductions, as assessed by the PGM-MB/PCR assay. HHP treatments could achieve maximum reductions of ∼3 and 3.5 log units for GI.1 and GII.4, respectively, as assessed by the PGM-MB/PCR assay. On the basis of these results, it can reasonably be concluded that the PGM-MB/PCR assay would very likely be able to estimate HHP inactivation of HuNoV at ≤2-log-reduction levels. It would also likely conservatively quantify HHP inactivation of the GI.1 strain at 2- to 3-log-reduction levels and the GII.4 strain at 2- to 3.5-log-reduction levels.
INTRODUCTION
Human norovirus (HuNoV) is the leading cause of foodborne illnesses in the United States (1). It has frequently been associated with a variety of ready-to-eat foods, including berries, salsa, and guacamole, and raw shellfish, such as oysters (2–4). Currently, the lack of suitable cell culture systems and practical small-animal models has been hindering research for developing and/or identifying effective processing methods to inactivate HuNoV (1, 5). Therefore, evaluation of the efficacy of processing treatments must rely on HuNoV surrogates. The accuracy of methods that use these surrogates is questionable since direct comparison of methods that use surrogates with methods that use HuNoV is not possible. Molecular biology methods, mostly quantitative reverse transcription-PCR (qRT-PCR), can be used for HuNoV quantification. However, these methods can detect only the presence of HuNoV RNA and cannot distinguish between infectious and noninfectious virus particles.
HuNoVs are reported to bind to histo-blood group antigens (HBGAs) in the human intestinal tract, and HBGAs are considered the attachment factors necessary for infection (6, 7). To initiate infection, HuNoVs need to be able to bind to HBGAs to enter the host cells. Porcine gastric mucin (PGM) contains HBGAs and has been shown to be able to bind to genogroup I (GI) and genogroup II (GII) HuNoV strains (8–10). In addition, the partially purified PGM powder used in this study contained 0.5 to 1.5% sialic acid (Sigma-Aldrich), which was reported to be the receptor for murine norovirus (MNV) (11). Utilizing the ability of PGM to bind to HuNoV, Tian et al. (9, 12, 13) and Pan et al. (14) used PGM-conjugated magnetic beads (PGM-MBs) to extract and concentrate HuNoV from food matrices, such as fresh produce, salad, and sewage. After binding, only bound virus particles can be quantified using qRT-PCR. Dancho et al. (15) first used this PGM-MB binding method followed by qRT-PCR assay (referred to here as the PGM-MB/PCR assay) to discriminate between infectious and noninfectious HuNoV treated by HHP, thermal, and UV treatments, based on the assumption that only the infectious virus could bind to PGM. Later, the same group (16) used the same approach to assess the efficacy of chlorine, chlorine dioxide, peroxyacetic acid, hydrogen peroxide, and trisodium phosphate for the inactivation of HuNoV. However, the validity of their assumption remains to be tested. It is possible that partially damaged and inactivated HuNoV can still bind to PGM and can subsequently be detected by qRT-PCR. Currently, it is impossible to verify the assumption since no tissue culture-based method for determination of HuNoV infectivity is available to generate data that can be compared with those obtained using the PGM-MB/PCR assay. Since a plaque assay is available for the two most commonly used HuNoV surrogates, MNV and Tulane virus (TV), inactivation results for these two viruses could be obtained using both the plaque assay and the PGM-MB/PCR assay and compared. If the results of the two assays were comparable, such findings would provide support for using the PGM-MB/PCR assay as a means to quantify infectious HuNoV.
HHP has been the most successfully and most widely used nonthermal processing technology in the food industry over the last 2 decades because treated foods retain their raw characteristics after processing and because it minimizes the loss of the nutritional value of the foods. It is commercially used to process various kinds of foods mainly to increase their shelf life and enhance food safety by inactivating pathogenic bacteria, such as Listeria monocytogenes, Salmonella, and Escherichia coli O157:H7 (17, 18). Sometimes it is commercially used as a processing aid; for example, it has been used to facilitate oyster shucking. Commercially HHP-treated foods include those that have been involved in HuNoV outbreaks, such as oysters, salsa, and guacamole (4, 19). Unfortunately, the HHP parameters designed for the processing of those foods do not take HuNoV into consideration, and it is not clear whether those HHP processing parameters would be able to eliminate HuNoV. It has been well documented that HHP is able to inactivate HuNoV and its surrogates, MNV, TV, and feline calicivirus (20–23). For example, our previous study showed that an HHP treatment of 350 MPa for 2 min at 21°C resulted in a 3.8-log reduction of TV at neutral pH and a 400-MPa treatment under the same conditions resulted in a 4.5-log reduction of MNV-1 (20). Using human volunteers for direct assessment of HuNoV inactivation, Leon et al. (24) were able to demonstrate that an HHP treatment of 600 MPa at 6°C for 5 min was able to inactivate an HuNoV genogroup I genotype 1 (GI.1) strain inoculated into oysters and protect human subjects from infection.
The objectives of this study were to (i) compare the data on the HHP inactivation of MNV-1 and TV obtained using both the plaque assay and the PGM-MB/PCR assay under two different HHP treatment temperatures and two different culture medium pH values and (ii) determine whether the HuNoV GI.1 strain and the GII.4 strain behaved similarly to the two surrogates under HHP using the PGM-MB/PCR assay.
MATERIALS AND METHODS
Virus and cell lines.
Detailed information about the viruses (HuNoV GI.1 and GII.4, MNV-1, and TV) and cell lines (RAW 264.7 cells for MNV-1 and LLC-MK2 cells for TV) used in this study was described in our previous studies (20, 21). For the two HuNoV strains, fecal suspensions were centrifuged at 4,000 × g for 20 min, filtered through a 0.22-μm-pore-size filter, aliquoted, and stored at −80°C until use. High-glucose Dulbecco's modified Eagle medium (DMEM; Life Technologies, Carlsbad, CA) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Life Technologies) was used to culture RAW 264.7 cells, while M199 medium (Corning, Manassas, VA) supplemented with 10% heat-inactivated FBS, penicillin G (100 U/ml), and streptomycin (100 μg/ml) was used to culture LLC-MK2 cells. Both RAW 264.7 and LLC-MK2 cells were cultured at 37°C under a 5% CO2 atmosphere. To grow the MNV-1 stock, confluent RAW 264.1 cells were infected with MNV-1 at a multiplicity of infection (MOI) of 1. After 1 h of incubation at 37°C under a 5% CO2 atmosphere, 25 ml of DMEM supplemented with 2% FBS was added. MNV-1 was harvested 2 days after inoculation by the use of three freeze-thaw cycles and centrifugation. Virus was stored at −80°C until use. The same procedures were followed to grow TV, except that cells were infected with TV at an MOI of 0.1 and 25 ml M199 medium supplemented with 10% FBS was used after 1 h of incubation.
Viral plaque assay.
MNV-1 and TV were quantified by plaque assay following the procedures published previously (20). Briefly, RAW 264.7 cells were seeded into six-well plates (Becton, Dickinson and Company, Franklin Lakes, NJ) at a density of 2 × 106 cells per well. After 24 h of incubation, cell monolayers were infected with 400 μl of a 10-fold dilution series of MNV-1 and the plates were incubated for 1 h at 37°C in a 5% CO2 atmosphere with gentle agitation every 10 to 15 min. Cells were overlaid with 2.5 ml of Eagle minimum essential medium (MEM) containing 0.5% agarose, 5% FBS, 0.12% sodium bicarbonate, penicillin G (100 U/ml), streptomycin (100 μg/ml), amphotericin B (0.25 μg/ml), 25 mM HEPES (pH 7.7), and 2 mM l-glutamine (Life Technologies). The plates were then incubated at 37°C in a 5% CO2 atmosphere for 2 days and fixed in 3.7% formaldehyde (37% formaldehyde [Fisher Scientific, Pittsburgh, PA] diluted 10-fold in phosphate-buffered saline [PBS] solution [pH 7.4]), and the plaques were visualized by staining with 0.05% (wt/vol) crystal violet in 10% ethanol. For the TV plaque assay, LLC-MK2 cells were seeded in six-well plates at a density of 4 × 105 cells per well. Cells were overlaid with 0.5 volume of M199 medium, 10% fetal bovine serum, penicillin G (100 U/ml), streptomycin (100 μg/ml), amphotericin B (0.25 μg/ml), and 0.5% agarose. The plates were then incubated at 37°C in a 5% CO2 atmosphere for 4 days before the cells were fixed with formaldehyde.
Preparation of PGM-MBs.
PGM-MBs were prepared as described previously (21). Briefly, 1 ml MagnaBind carboxyl-derivatized beads (Thermo Scientific, Waltham, MA) was washed three times with 1 ml PBS (pH 7.2), and a bead attractor (EMD Millipore) was used to separate the beads after each wash. One milliliter of 10 mg/ml type III mucin from porcine stomach (bound sialic acid, 0.5 to 1.5%; Sigma, St. Louis, MO) and 0.1 ml of 10 mg/ml 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC), both in conjugation buffer (0.1 M MES [2-(N-morpholino)ethanesulfonic acid], 0.9% NaCl, pH 4.7), were added to the beads. The mixture was rotated for 30 min on a Labquake shaker rotisserie (Thermo Scientific), washed three times with 1 ml PBS (pH 7.2), suspended in 1 ml PBS (pH 7.2) containing 0.05% sodium azide, and stored at 4°C.
PGM binding assay and RNA preparation.
The procedure described previously was used for the PGM binding assay (21). One hundred microliters of PGM-MBs was added to each sample, and each sample was incubated at room temperature for 15 min on a Labquake shaker rotisserie. The PGM-MBs were then separated by the magnetic bead attractor and washed three times with 1 ml PBS (pH 7.2). After that, the beads were resuspended in 140 μl of molecular biology-grade H2O and put on ice before the RNA was extracted with a QIAamp viral RNA minikit (Qiagen USA, Valencia, CA, USA) as described previously (21).
qRT-PCR quantification.
The primers and TaqMan probes and their concentrations used for qRT-PCR for HuNoV GI.1 and GII.4 were those described previously, with minor modifications (21). The concentration of the probe for the HuNoV GI.1 strain was 250 nM instead of 100 nM. The primers and probe for MNV-1 were adapted from those described in a previous publication (25), and the primers and probe for TV were designed by Xi Jiang at Cincinnati Children's Hospital Medical Center through personal communication. Primer FW-ORF1/ORF2 (CACGCCACCGATCTGTTCTG; 200 nM), primer RV-ORF1/ORF2 (GCGCTGCGCCATCACTC; 200 nM), and probe MGB-ORF1/ORF2 (FAM-CGCTTTGGAACAATG-MGBNFQ [where FAM is 6-carboxyfluorescein]; 200 nM), based on the sequence with GenBank accession number DQ285629, were used for MNV-1, while primer p1888-F (TCGCGCAGCGCACTTA; 900 nM), primer p1889-R (CAAGAATCCAGAACAACCAATATCA; 400 nM), and probe TVRdRp-P (FAM-CACCTTCTTGTGGGCCA-MGBNFQ; 175 nM), based on the sequence with GenBank accession number EU391643, were used for TV. PCR conditions and other PCR reagents were those described in a previous publication (21). Viral RNA extracted directly from virus using a QIAamp viral RNA minikit was serially diluted and applied as the standard for qRT-PCR.
HHP treatment of viruses.
The culture media for RAW 264.7 and LLC-MK2 cells were adjusted to pH 4 using concentrated HCl (∼12.1 M). The original culture media with neutral pH (∼pH 7.0 to 7.5) and culture media adjusted to pH 4 were sterilized by being filtered through 0.22-μm-pore-size filters (EMD Millipore, Billerica, MA). One volume of TV (∼5 × 105 to 5 × 106 PFU/ml) or MNV-1 (∼107 to 108 PFU/ml) stock was mixed with 9 volumes of the corresponding cell culture medium adjusted to pH 4 as well as the original medium. HuNoV GII.4 and GI.1 stocks were diluted 20 and 10 times, respectively, using the original MNV-1 culture medium or the MNV-1 culture medium adjusted to pH 4. Each of the four virus solutions (150 μl for HuNoVs and 1.5 ml for TV and MNV-1) was double bagged and double sealed in a sterile polyethylene stomacher pouch (Seward, Port Saint Lucie, FL). MNV-1, TV, HuNoV GI.1, and HuNoV GII.4 samples were pressurized at 150 to 550, 50 to 400, 250 to 575, and 100 to 450 MPa, respectively, using an Avure PT-1 pressure unit (Avure Technologies, Kent, WA) with temperature control and with water as a hydrostatic medium. Pressure treatments were conducted at 4 and 21°C for samples in the original cell culture media and 21°C for samples in the culture media adjusted to pH 4. The treatment time was 2 min for all the pressure treatments. The pressure come-up rate was approximately 22 MPa/s, and the pressure release time was <4 s. The pressurization times reported here did not include the pressure come-up time or the release time. Control or untreated samples were prepared in the same way as the pressure-treated samples. Negative controls containing all the reagents but not the viruses were also prepared. After the pressure treatments, 100 μl of each sample was mixed with 800 μl PBS and the mixture was incubated with 40 μg of RNase A (Life Technologies) at 37°C for 30 min. Viruses were then assayed using the PGM-MB/PCR assay as described above. TV and MNV-1 samples were also used directly for plaque assay after pressure treatments.
Statistical analysis.
Each experiment was replicated 3 times. Log reductions were the differences between the untreated samples and the treated samples. One-way analysis of variance (for comparison of more than 2 groups) following Tukey's post hoc comparisons or an independent-samples t test (for comparison of 2 groups) using SPSS software, version 22 (IBM SPSS Inc., Chicago, IL), was performed for statistical analysis. A P value of <0.05 was considered statistically significant.
RESULTS
Comparisons of results of plaque assay and PGM binding assay.
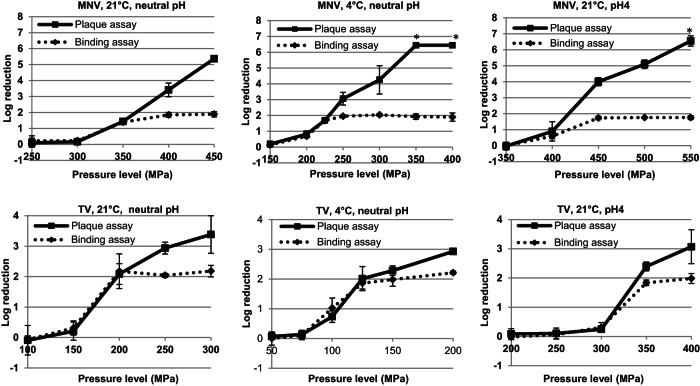
TV and MNV-1 in culture media with an approximately neutral pH were pressure treated at 21°C. The pressure inactivation curves of both viruses were obtained using the plaque and PGM-MB/PCR assays (Fig. 1). For TV, the two pressure inactivation curves obtained using the plaque and PGM-MB/PCR assays were almost identical at ≤2-log-reduction levels. Increasing the pressure to over 200 MPa resulted in higher log reductions for the plaque assay but not for the PGM-MB/PCR assay. The maximum reduction achieved using the PGM-MB/PCR assay was ∼2 log units. Similar pressure inactivation patterns were also observed for MNV-1. We further compared the pressure inactivation results between the plaque assay and the PGM-MB/PCR assay using a lower pressure treatment temperature, 4°C (Fig. 1). Patterns similar to those described above were again observed. The two pressure inactivation curves obtained using the plaque and PGM-MB/PCR assays were almost identical at ≤2-log-reduction levels, but the curves differed above the 2-log-reduction level. We then determined whether changing the medium pH to 4 would affect this pattern (Fig. 1). For TV, patterns similar to those described above were observed. However, the two pressure inactivation curves for MNV-1 obtained using the plaque and PGM-MB/PCR assays were identical only at ≤1-log-reduction levels but differed above the 1-log-reduction level. This is possibly because the 2-log-reduction level was reached at a pressure between 400 and 450 MPa, a range not included in the study.
FIG 1.
Pressure inactivation of TV and MNV-1 assessed by plaque assay and the PGM-MB/PCR assay. Data are the means from 3 replicates. Error bars represent 1 standard deviation. *, the titers were below the detection limit for some or all of the replicates.
Figure 1 also shows that a lower pressure treatment temperature significantly enhanced the pressure inactivation of TV and MNV-1. For example, plaque assay results showed that a 2.3-log reduction was achieved when TV was pressurized at 150 MPa and 4°C, but only a 0.2-log reduction was achieved at the same pressure level at 21°C. Pressurization of MNV-1 at 300 MPa and 4°C resulted in a 4.3-log reduction by the plaque assay, but only a 0.2-log reduction was noticed with the treatment at the same pressure level at 21°C. The results also showed that both TV and MNV-1 were more resistant to pressure at pH 4 than neutral pH. For example, plaque assay results showed that a treatment of 200 MPa for 2 min at 21°C resulted in a 2.1-log reduction of TV at neutral pH, but only a 0.1-log reduction was observed for the same treatment at pH 4. A treatment with 400 MPa for 2 min at 21°C resulted in a 3.4-log reduction of MNV-1 at neutral pH, but only a 0.9-log reduction was observed for the same treatment at pH 4. The results also indicated that MNV-1 was more resistant to pressure than TV. For example, a treatment of 200 MPa for 2 min at 21°C resulted in a 2.1-log reduction of TV. However, treatment at a higher pressure, 250 MPa, for 2 min at 21°C caused only a 0.1-log reduction of MNV-1.
HHP inactivation of HuNoVs.
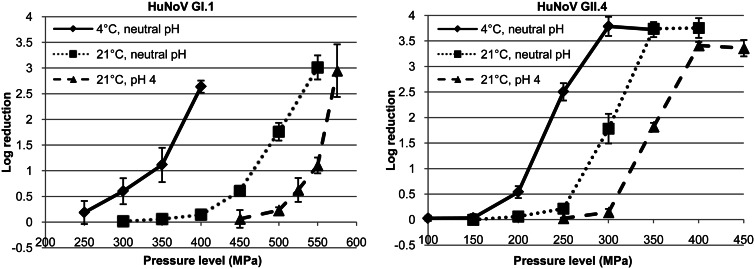
The pressure inactivation curves for GII.4 obtained using the PGM-MB/PCR assay showed patterns similar to those for the curves for TV and MNV-1 (Fig. 2). There was no virus inactivation at low pressure levels, and rapid inactivation was found after the pressure was increased above the critical pressure level. The maximum reduction achieved was ∼3.5 log units and was about 1.5 log units higher than that for TV and MNV-1. Increasing the pressure did not result in greater reductions, as assessed by the PGM-MB/PCR assay. Changing the treatment temperature and pH of the suspending medium affected the pressure sensitivity only of GII.4 and did not affect the shapes of the pressure inactivation curves. The pressure inactivation curves for GI.1 also showed patterns similar to those of the curves for GII.4, except that no maximum plateau was observed. However, unlike the GII.4 strain, which had a higher titer, so that the plateau was achieved when it was still above the detection limit, due to the low initial titer of the GI.1 stock, a ∼3-log reduction was the maximum that could be achieved. It is not known whether greater log reductions could be achieved by increasing the pressure levels.
FIG 2.
Pressure inactivation of HuNoV GI.1 and GII.4 strains assessed by PGM-MB/PCR assay. Data are the means from 3 replicates. Error bars represent 1 standard deviation.
Consistent with the results for TV and MNV-1 and our previously published results (21), both the GI.1 and the GII.4 strains were more sensitive to pressure at 4°C than at 21°C and at neutral pH than at pH 4. For example, results showed that a 2.6-log reduction was achieved when the GI.1 strain was pressurized at 400 MPa and 4°C, but only a 0.1-log reduction was achieved at the same pressure level at 21°C. Pressurization of the GII.4 strain at 300 MPa and 4°C resulted in a 3.8-log reduction, but only a 1.8-log reduction was noticed for the treatment at the same pressure level at 21°C. In addition, at pH 4, treatment at the same pressure level resulted in only a 0.1-log reduction for the GII.4 strain. Similarly, treatment at 500 MPa for 2 min at 21°C resulted in a 1.8-log reduction of the GI.1 strain at neutral pH, but only a 0.2-log reduction was achieved at pH 4 with the same treatment conditions. These results also show that the GI.1 strain was more resistant to pressure than the GII.4 strain that we tested. Considering the results obtained for MNV-1 and TV (Fig. 1), it is likely that the order of pressure resistance for the virus strains that we used in this study was GI.1 > MNV-1 > GII.4 > TV.
DISCUSSION
Since qRT-PCR quantifies only the total amount of RNA in a virus sample and is not able to distinguish between infectious and noninfectious viruses, PGM was used in this study to bind and collect potentially infectious HuNoV and exclude inactivated virions with capsids that had been rendered unable to bind to PGM by HHP. To test whether the PGM-MB/PCR assay would be able to provide accurate inactivation results, we used this assay to test the pressure inactivation of MNV-1 and TV, which are currently considered the most appropriate HuNoV surrogates, and compared the inactivation results with those obtained using the plaque assay. MNV belongs to the genus Norovirus and has biochemical features, a genome size, and a gene organization similar to those of HuNoV (1, 26, 27). However, MNV is still different from HuNoV in its pathogenesis. MNV infection in mice does not cause the same symptoms that HuNoV does in humans (1, 26, 27). Although MNV uses sialic acid as a functional receptor while HuNoV uses HBGAs as attachment factors (7), our results demonstrate that MNV-1 was still able to bind to PGM. Since the PGM used in this study contained 0.5 to 1.5% sialic acid, it is possible that MNV-1 specifically bound to the sialic acid contained in PGM. However, it is also possible that the binding of MNV-1 to PGM was unspecific. Nevertheless, the results are in agreement with those reported by Hirneisen and Kniel (28), who used a monoclonal anti-MNV primary antibody (IgG) to show that the level of MNV-1 attachment to PGM was significantly greater than its level of attachment to sialic acid, the receptor for MNV-1. The reason for this is not clear but is possibly due to different binding mechanisms (28). TV, a monkey calicivirus isolated at the Tulane National Primate Research Center, has a genomic sequence that closely resembles that of HuNoV, and more importantly, it recognizes the type A and B HBGAs, similar to HuNoV (29). Even though at this point it is not clear that the binding to PGM was specific for TV, as well as MNV, both TV and MNV-1 behaved similarly, with the results obtained by the PGM-MB/PCR assay agreeing well with those obtained by the plaque assay at approximately ≤2-log-reduction levels. It is likely that HHP treatment probably did not preferentially destroy specific binding sites on the virus capsids but more likely damaged the whole virus capsid. Due to the uncertainty of the specificity of PGM binding to different viruses, no conclusion regarding the details of the damage to virus particles caused by HHP can be drawn from this study.
It should be noted that for the PGM-MB/PCR assay to accurately quantify the RNA copies of infectious HuNoV, the following criteria must be met: (i) 100% of the infectious virus must bind to PGM-MB, (ii) 100% of the inactivated virus may not bind to PGM-MB, and (iii) no loss of infectious virus may be achieved during the whole assay process, from PGM-MB binding to final qRT-PCR quantification. Any deviations from those criteria could affect the accuracy of the results. Apparently, some inactivated TV and MNV-1 virions with damaged capsids could still bind to PGM-MBs, since there were differences between the log reductions of the RNA copy numbers assessed using the PGM-MB/PCR assay and the log reductions of the titers assessed using the plaque assay. It was surprising to observe that the log reduction of RNA copy numbers for TV and MNV-1 reached a maximum plateau of ∼2 log units. Further increasing the pressure over the 2-log-reduction level could inactivate more virus particles but did not further decrease their ability to bind to PGM. It seems that the capsids of a fraction of inactivated TV and MNV-1 were still able to bind to PGM, regardless of the treatment conditions used in the study. Since many factors are involved in the process of viral replication, which includes the initial step of attachment to host cells, and details on the damage incurred by the virions are unclear, it is difficult to speculate why only a ∼2-log reduction of TV and MNV-1 was achieved by HHP when the reductions were evaluated using the PGM-MB/PCR binding assay. Interestingly, greater than 2 log unit reductions of the RNA copy numbers could be achieved for HuNoV GI.1 and GII.4. HHP mainly targets the capsids and not the genomes of the viruses (22, 30, 31). A previous study has shown that HHP disrupts the structure of the viral capsid of MNV-1 and not the primary or secondary structure of the VP1 protein, which plays a role in viral attachment (22). A few studies have demonstrated that the pressurized capsid protein is still antigenic (22, 31). Interestingly, our results did not totally agree with some results of Lou et al. (22), who showed that no RNA was detected after HHP and RNase treatment. On the other hand, our results agree with those reported in a recent study by Cromeans et al. (32). In that study, it was found that the RNA copy number of TV was reduced by 1 log unit with treatment with 200 MPa of pressure, with no further reduction of the RNA copy number occurring up to a pressure of 800 MPa, while the RNA copy numbers of semipurified preparations of HuNoV GI.5 and GII.13 were reduced by 2 log units by treatment with 300 MPa of pressure and by 1 log unit by treatment with 400 MPa of pressure, respectively, with no further reductions of the RNA copy numbers occurring at a pressure of up to 800 MPa (32). Their results suggest that some of the capsids of the viruses, which were probably damaged to some extent, were still able to protect the RNA inside the capsids after pressure treatment. On the basis of our PCR results with the tested viruses, particularly for MNV-1 and TV, a portion of the damaged capsids was able to not only protect the RNA inside the capsids but also bind to PGM after the pressure treatments. Even though damage to the capsid binding sites can certainly lead to viral inactivation, for some virions, it is also possible that some of their other parts but not the binding sites were damaged, which consequently caused them to lose infectivity. In addition, it is still not clear if the binding to PGM of any of the viruses tested in this study was specific or unspecific. Therefore, some of the noninfectious viruses could still bind to PGM and be further quantified by the PGM-MB/PCR assay.
In addition to PGM, other binding agents in combination with qRT-PCR have also been evaluated for their ability to distinguish between infectious and inactivated viruses. Li et al. (33) used RAW 264.7 cells and ganglioside GD1a (the binding receptor of MNV-1) to bind MNV-1 and further quantified the bound virus by qRT-PCR to evaluate the inactivation of MNV-1 by heat and hydrogen peroxide. No correlation between the results of the PCR assay and those of the plaque assay was found. A recent study used an in situ capture qRT-PCR to determine the inactivation of TV and found that this method reflected inactivation of the virus by chlorine or ethanol treatment but not UV irradiation (34). The UV irradiation used in that study did not reduce the ability of TV to bind to its receptor, and subsequent PCR was still able to amplify the target fragment, even though the genome was damaged by UV irradiation and the virus was not able to replicate, as indicated by the results of a cell culture-based assay (34). Taken together, the effectiveness of using a binding method followed by PCR to distinguish between infectious and inactivated viruses might depend on the binding agents and processing methods. Therefore, before a cell culture system is available for HuNoV, it is important to carefully evaluate molecular biology-based methods when they are used to quantify the effectiveness of different inactivation methods.
The results from this study show that for the tested viruses, all of them were more sensitive to pressure at a refrigeration temperature than room temperature and at neutral pH than pH 4, in agreement with the findings of our previous studies (20, 21). Since the results of the plaque assay for both MNV-1 and TV matched those of the PGM-MB/PCR assay, the PGM-MB/PCR assay could provide some basic information on the HHP inactivation of HuNoVs. Although it is understandable that HuNoVs are acid tolerant because they are enteric viruses and need to be able to survive in the acidic environment of the stomach to infect the intestine, there is still no knowledge of why the viruses are more resistant to pressure at a lower pH from the biophysical point of view of the virus. Due to the influence of temperature and pH, the pressure treatment temperature and the formulations of different food products need to be taken into consideration when designing HHP treatments.
The human challenge trial is the only study available in the literature which roughly defined the pressure inactivation profile of HuNoV GI.1 (24). In that study, human subjects were challenged with pressure-treated and untreated oysters inoculated with HuNoV GI.1. When human volunteers were challenged with untreated oysters, 7 out of 15, or 47%, of the subjects became infected with HuNoV (24). Conversely, none of the 10 subjects challenged with oysters treated with 600 MPa of pressure at 6°C for 5 min became infected (24). Since the study was an endpoint study in which subjects were either infected or not infected with HuNoV, the extent to which HuNoV was inactivated was not clearly defined (20). Recently, it was found in our laboratory (35) that the same pressure treatment condition (600 MPa at 6°C for 5 min) could achieve >4-log reductions in the number of RNA copies of the same GI.1 strain inoculated into oysters, as assessed using the PGM-MB/PCR assay, which roughly agreed with the results from the human challenge study. Taking together the results from the study by Ye et al. (35) and the results from this study, it can be concluded that the PGM-MB/PCR assay would very likely be able to estimate the level of HHP inactivation of HuNoV at ≤2-log-reduction levels. It would also likely be able to conservatively quantify the level of HHP inactivation of the GI.1 strain at 2- to 3-log-reduction levels and the GII.4 strain at 2- to 3.5-log-reduction levels. Therefore, the PGM-MB/PCR assay is an important tool for studies of HHP inactivation of HuNoV until a cell culture system is developed. The results from this study also suggest that MNV-1 is probably a good surrogate for some GII.4 strains for use in HHP inactivation studies, since it was more pressure resistant than the GII.4 strain that we tested, but is not a good surrogate for the GI.1 strain, since it was more pressure sensitive than the GI.1 strain. In addition, MNV-1 behaved similarly to the GII.4 and the GI.1 strains under different HHP treatment conditions (conditions that differed by treatment temperature and pH). It should be noted that the PGM-MB/PCR assay may not be suitable for the evaluation of other processing methods. Therefore, the use of surrogates, such as MNV-1, has its merits in certain types of inactivation studies.
ACKNOWLEDGMENT
This project was supported by the Agriculture and Food Research Initiative Competitive Grants Program of the USDA National Institute of Food and Agriculture, NIFA award no. 2011-68003-30005.
REFERENCES
- 1.Li J, Predmore A, Divers E, Lou F. 2012. New interventions against human norovirus: progress, opportunities, and challenges. Annu Rev Food Sci Technol 3:331–352. doi: 10.1146/annurev-food-022811-101234. [DOI] [PubMed] [Google Scholar]
- 2.Falkenhorst G, Krusell L, Lisby M, Madsen SB, Bottiger B, Molbak K. 2005. Imported frozen raspberries cause a series of norovirus outbreaks in Denmark, 2005. Euro Surveill 10(9):pii=2795 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=2795. [DOI] [PubMed] [Google Scholar]
- 3.Suffredini E, Lanni L, Arcangeli G, Pepe T, Mazzette R, Ciccaglioni G, Croci L. 2014. Qualitative and quantitative assessment of viral contamination in bivalve molluscs harvested in Italy. Int J Food Microbiol 184:21–26. doi: 10.1016/j.ijfoodmicro.2014.02.026. [DOI] [PubMed] [Google Scholar]
- 4.Kendall ME, Mody RK, Mahon BE, Doyle MP, Herman KM, Tauxe RV. 2013. Emergence of salsa and guacamole as frequent vehicles of foodborne disease outbreaks in the United States, 1973–2008. Foodborne Pathog Dis 10:316–322. doi: 10.1089/fpd.2012.1328. [DOI] [PubMed] [Google Scholar]
- 5.Duizer E, Schwab KJ, Neill FH, Atmar RL, Koopmans MP, Estes MK. 2004. Laboratory efforts to cultivate noroviruses. J Gen Virol 85:79–87. doi: 10.1099/vir.0.19478-0. [DOI] [PubMed] [Google Scholar]
- 6.Marionneau S, Ruvoen N, Le Moullac-Vaidye B, Clement M, Cailleau-Thomas A, Ruiz-Palacois G, Huang P, Jiang X, Le Pendu J. 2002. Norwalk virus binds to histo-blood group antigens present on gastroduodenal epithelial cells of secretor individuals. Gastroenterology 122:1967–1977. doi: 10.1053/gast.2002.33661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ruvoen-Clouet N, Belliot G, Le Pendu J. 2013. Noroviruses and histo-blood groups: the impact of common host genetic polymorphisms on virus transmission and evolution. Rev Med Virol 23:355–366. doi: 10.1002/rmv.1757. [DOI] [PubMed] [Google Scholar]
- 8.Tian P, Jiang X, Zhong W, Jensen HM, Brandl M, Bates AH, Engelbrektson AL, Mandrell R. 2007. Binding of recombinant norovirus like particle to histo-blood group antigen on cells in the lumen of pig duodenum. Res Vet Sci 83:410–418. doi: 10.1016/j.rvsc.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 9.Tian P, Engelbrektson A, Mandrell R. 2008. Two-log increase in sensitivity for detection of norovirus in complex samples by concentration with porcine gastric mucin conjugated to magnetic beads. Appl Environ Microbiol 74:4271–4276. doi: 10.1128/AEM.00539-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tian P, Yang D, Jiang X, Zhong W, Cannon JL, Burkhardt W III, Woods JW, Hartman G, Lindesmith L, Baric RS, Mandrell R. 2010. Specificity and kinetics of norovirus binding to magnetic bead-conjugated histo-blood group antigens. J Appl Microbiol 109:1753–1762. doi: 10.1111/j.1365-2672.2010.04812.x. [DOI] [PubMed] [Google Scholar]
- 11.Taube S, Perry JW, Yetming K, Patel SP, Auble H, Shu L, Nawar HF, Lee CH, Connell TD, Shayman JA, Wobus CE. 2009. Ganglioside-linked terminal sialic acid moieties on murine macrophages function as attachment receptors for murine noroviruses. J Virol 83:4092–4101. doi: 10.1128/JVI.02245-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tian P, Yang D, Pan L, Mandrell R. 2012. Application of a receptor-binding capture quantitative reverse transcription-PCR assay to concentrate human norovirus from sewage and to study the distribution and stability of the virus. Appl Environ Microbiol 78:429–436. doi: 10.1128/AEM.06875-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tian P, Yang D, Mandrell R. 2011. A simple method to recover norovirus from fresh produce with large sample size by using histo-blood group antigen-conjugated to magnetic beads in a recirculating affinity magnetic separation system (RCAMS). Int J Food Microbiol 147:223–227. doi: 10.1016/j.ijfoodmicro.2011.04.013. [DOI] [PubMed] [Google Scholar]
- 14.Pan L, Zhang Q, Li X, Tian P. 2012. Detection of human norovirus in cherry tomatoes, blueberries and vegetable salad by using a receptor-binding capture and magnetic sequestration (RBCMS) method. Food Microbiol 30:420–426. doi: 10.1016/j.fm.2011.12.026. [DOI] [PubMed] [Google Scholar]
- 15.Dancho BA, Chen H, Kingsley DH. 2012. Discrimination between infectious and non-infectious human norovirus using porcine gastric mucin. Int J Food Microbiol 155:222–226. doi: 10.1016/j.ijfoodmicro.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 16.Kingsley DH, Vincent EM, Meade GK, Watson CL, Fan X. 2014. Inactivation of human norovirus using chemical sanitizers. Int J Food Microbiol 171:94–99. doi: 10.1016/j.ijfoodmicro.2013.11.018. [DOI] [PubMed] [Google Scholar]
- 17.Huang Y, Ye M, Chen H. 2013. Inactivation of Escherichia coli O157:H7 and Salmonella spp. in strawberry puree by high hydrostatic pressure with/without subsequent frozen storage. Int J Food Microbiol 160:337–343. doi: 10.1016/j.ijfoodmicro.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 18.Jofre A, Garriga M, Aymerich T. 2008. Inhibition of Salmonella sp., Listeria monocytogenes and Staphylococcus aureus in cooked ham by combining antimicrobials, high hydrostatic pressure and refrigeration. Meat Sci 78:53–59. doi: 10.1016/j.meatsci.2007.06.015. [DOI] [PubMed] [Google Scholar]
- 19.Iritani N, Kaida A, Abe N, Kubo H, Sekiguchi JI, Yamamoto SP, Goto K, Tanaka T, Noda M. 2014. Detection and genetic characterization of human enteric viruses in oyster-associated gastroenteritis outbreaks between 2001 and 2012 in Osaka City, Japan. J Med Virol 86:2019–2025. doi: 10.1002/jmv.23883. [DOI] [PubMed] [Google Scholar]
- 20.Li X, Ye M, Neetoo H, Golovan S, Chen H. 2013. Pressure inactivation of Tulane virus, a candidate surrogate for human norovirus and its potential application in food industry. Int J Food Microbiol 162:37–42. doi: 10.1016/j.ijfoodmicro.2012.12.016. [DOI] [PubMed] [Google Scholar]
- 21.Li X, Chen H, Kingsley DH. 2013. The influence of temperature, pH, and water immersion on the high hydrostatic pressure inactivation of GI.1 and GII.4 human noroviruses. Int J Food Microbiol 167:138–143. doi: 10.1016/j.ijfoodmicro.2013.08.020. [DOI] [PubMed] [Google Scholar]
- 22.Lou F, Neetoo H, Chen H, Li J. 2011. Inactivation of a human norovirus surrogate by high-pressure processing: effectiveness, mechanism, and potential application in the fresh produce industry. Appl Environ Microbiol 77:1862–1871. doi: 10.1128/AEM.01918-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen H, Hoover DG, Kingsley DH. 2005. Temperature and treatment time influence high hydrostatic pressure inactivation of feline calicivirus, a norovirus surrogate. J Food Prot 68:2389–2394. [DOI] [PubMed] [Google Scholar]
- 24.Leon JS, Kingsley DH, Montes JS, Richards GP, Lyon GM, Abdulhafid GM, Seitz SR, Fernandez ML, Teunis PF, Flick GJ, Moe CL. 2011. Randomized, double-blinded clinical trial for human norovirus inactivation in oysters by high hydrostatic pressure processing. Appl Environ Microbiol 77:5476–5482. doi: 10.1128/AEM.02801-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stals A, Baert L, Botteldoorn N, Werbrouck H, Herman L, Uyttendaele M, Van Coillie E. 2009. Multiplex real-time RT-PCR for simultaneous detection of GI/GII noroviruses and murine norovirus 1. J Virol Methods 161:247–253. doi: 10.1016/j.jviromet.2009.06.019. [DOI] [PubMed] [Google Scholar]
- 26.Karst SM, Wobus CE, Lay M, Davidson J, Virgin HW IV. 2003. STAT1-dependent innate immunity to a Norwalk-like virus. Science 299:1575–1578. doi: 10.1126/science.1077905. [DOI] [PubMed] [Google Scholar]
- 27.Wobus CE, Thackray LB, Virgin HW IV. 2006. Murine norovirus: a model system to study norovirus biology and pathogenesis. J Virol 80:5104–5112. doi: 10.1128/JVI.02346-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirneisen KA, Kniel KE. 2012. Comparison of ELISA attachment and infectivity assays for murine norovirus. J Virol Methods 186:14–20. doi: 10.1016/j.jviromet.2012.07.018. [DOI] [PubMed] [Google Scholar]
- 29.Farkas T, Cross RW, Hargitt E III, Lerche NW, Morrow AL, Sestak K. 2010. Genetic diversity and histo-blood group antigen interactions of rhesus enteric caliciviruses. J Virol 84:8617–8625. doi: 10.1128/JVI.00630-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lou F, Neetoo H, Li J, Chen H, Li J. 2011. Lack of correlation between virus barosensitivity and the presence of a viral envelope during inactivation of human rotavirus, vesicular stomatitis virus, and avian metapneumovirus by high-pressure processing. Appl Environ Microbiol 77:8538–8547. doi: 10.1128/AEM.06711-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang Q, Li D, Xu J, Wang J, Zhao Y, Li Z, Xue C. 2010. Mechanism of inactivation of murine norovirus-1 by high pressure processing. Int J Food Microbiol 137:186–189. doi: 10.1016/j.ijfoodmicro.2009.10.033. [DOI] [PubMed] [Google Scholar]
- 32.Cromeans T, Park GW, Costantini V, Lee D, Wang Q, Farkas T, Lee A, Vinje J. 2014. Comprehensive comparison of cultivable norovirus surrogates in response to different inactivation and disinfection treatments. Appl Environ Microbiol 80:5743–5751. doi: 10.1128/AEM.01532-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li D, Baert L, Van Coillie E, Uyttendaele M. 2011. Critical studies on binding-based RT-PCR detection of infectious noroviruses. J Virol Methods 177:153–159. doi: 10.1016/j.jviromet.2011.07.013. [DOI] [PubMed] [Google Scholar]
- 34.Wang D, Xu S, Yang D, Young GM, Tian P. 2014. New in situ capture quantitative (real-time) reverse transcription-PCR method as an alternative approach for determining inactivation of Tulane virus. Appl Environ Microbiol 80:2120–2124. doi: 10.1128/AEM.04036-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ye M, Li X, Kingsley DH, Jiang X, Chen H. 2014. Inactivation of human norovirus in contaminated oysters and clams by high hydrostatic pressure. Appl Environ Microbiol 80:2248–2253. doi: 10.1128/AEM.04260-13. [DOI] [PMC free article] [PubMed] [Google Scholar]