Abstract
The alphaproteobacterium Hyphomonas neptunium proliferates by a unique budding mechanism in which daughter cells emerge from the end of a stalk-like extension emanating from the mother cell body. Studies of this species so far have been hampered by the lack of a genetic system and of molecular tools allowing the regulated expression of target genes. Based on microarray analyses, this work identifies two H. neptunium promoters that are activated specifically by copper and zinc. Functional analyses show that they have low basal activity and a high dynamic range, meeting the requirements for use as a multipurpose expression system. To facilitate their application, the two promoters were incorporated into a set of integrative plasmids, featuring a choice of two different selection markers and various fluorescent protein genes. These constructs enable the straightforward generation and heavy metal-inducible synthesis of fluorescent protein fusions in H. neptunium, thereby opening the door to an in-depth analysis of polar growth and development in this species.
INTRODUCTION
Bacteria are a phylogenetically diverse group of organisms whose study has provided important insights into the mechanisms that mediate the spatiotemporal organization of cells. However, most of our knowledge on bacterial cell biology so far has come from the analysis of only a few well-established model species, such as Escherichia coli, Bacillus subtilis, and Caulobacter crescentus, which typically exhibit a rod-like morphology and divide by symmetric or asymmetric binary fission.
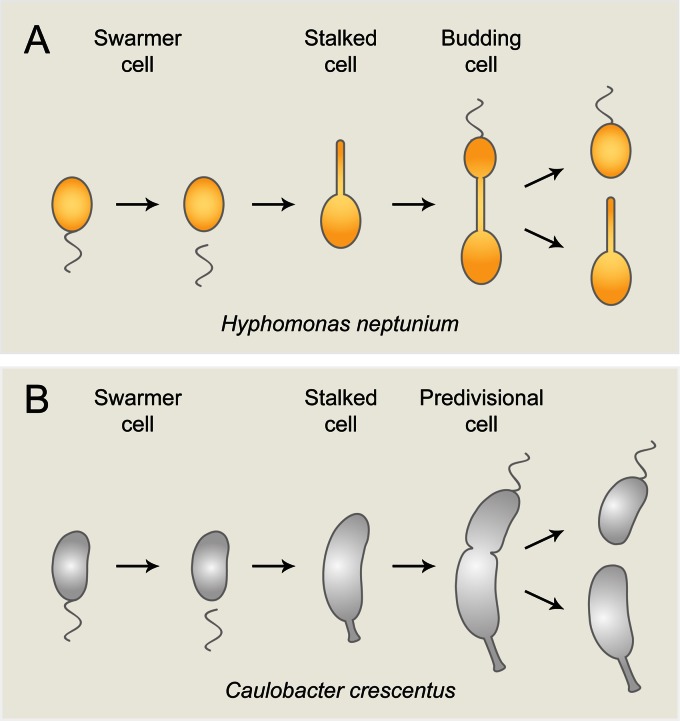
To further our understanding of subcellular organization in bacteria, we have started to investigate the marine alphaproteobacterium Hyphomonas neptunium (1), a representative of the stalked budding bacteria (2, 3). Similar to other members of this polyphyletic bacterial group, H. neptunium is characterized by a unique mode of reproduction that involves the formation of buds at the tip of a stalk emanating from the mother cell body (Fig. 1A). During the budding process, the nascent daughter cell is equipped with a single polar flagellum at the pole opposite the stalk. Cytokinesis then gives rise to a motile swarmer cell, which initially is unable to replicate, and an immotile stalked cell, which immediately enters a new round of budding and cell division (4). At a defined time in the cell cycle, the swarmer cell undergoes a differentiation process during which it sheds the flagellum and establishes a stalk at the opposite pole. Subsequently, a bud emerges from the tip of the stalk, setting the stage for the next division event.
FIG 1.
Life cycles of H. neptunium and C. crescentus. (A) Life cycle of H. neptunium. Cells proliferate by budding, using the stalk as a reproductive structure. (B) Life cycle of the closely related model bacterium C. crescentus. Cells divide by asymmetric binary fission. See the text for a detailed description.
H. neptunium was isolated from the harbor of Barcelona (Spain). Based on morphological criteria, it was originally described as Hyphomicrobium neptunium (1). Later, DNA-DNA hybridization experiments, 5S rRNA sequence analyses, and metabolic profiling revealed a close phylogenetic relationship to members of the genus Hyphomonas (5, 6). Interestingly, 16S rRNA-based phylogenetic studies identify H. neptunium as a member of the Rhodobacterales (7). However, 23S rRNA sequence analysis and concatenated protein alignments strongly suggest its re-classification as a member of the Caulobacterales (8). This notion is supported by a comparative genomics study which indicated a close phylogenetic relationship of H. neptunium with the stalked model bacterium C. crescentus (9). The life cycle of H. neptunium indeed bears resemblance to that of C. crescentus (Fig. 1B), and most of the key cell cycle regulators identified in C. crescentus are conserved in its relative (10). Nevertheless, there are significant differences between the two species with respect to their cellular organization and mode of proliferation. Importantly, the stalks are positioned at opposite poles, namely, the old pole in C. crescentus and the new pole in H. neptunium. Moreover, whereas the C. crescentus stalk is devoid of cytoplasm and physiologically separated from other regions of the cell by diffusion barriers (11, 12), its H. neptunium counterpart is an integral part of the cell connecting the mother and daughter cell compartments. The unique reproductive strategy of H. neptunium raises several important questions. For instance, it remains to be clarified how the conserved cell cycle regulatory circuitry of C. crescentus has adapted to the needs of the H. neptunium cell layout. Another open issue is how cell growth, budding, and asymmetric cell division are regulated in time and space and how these processes are coordinated with other cell cycle events, such as chromosome replication. Moreover, it is unclear how newly replicated DNA is moved through the stalk to be deposited in the daughter cell compartment. Finally, H. neptunium may represent an interesting model to study cell aging, as the number of offspring produced by one mother cell appears to be quite limited (2, 13).
The study of H. neptunium so far has been hampered by the lack of genetic methods and tools. To address this issue, we have set out to establish a reliable transformation protocol and suitable antibiotic selection markers. Moreover, we have identified two tightly controlled heavy metal-inducible promoters and incorporated them into a series of integrating plasmids designed for the straightforward construction and regulated synthesis of native and fluorescently tagged proteins in H. neptunium. This molecular toolbox opens the door to the genetic manipulation of stalked budding bacteria, paving the way for in-depth studies of this fascinating but poorly investigated bacterial group.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains used in this study are listed in Table 1. Hyphomonas neptunium LE670 and its derivatives were cultivated aerobically in Difco marine broth 2216 (MB) (BD Biosciences) at 28°C under vigorous shaking (210 rpm) in baffled flasks. Media were supplemented with kanamycin (100/200) or rifampin (1/2) when appropriate (μg/ml in liquid/solid medium). E. coli strains were grown at 37°C, and media were supplemented with antibiotics at the following concentrations (μg/ml in liquid/solid media): kanamycin (30/50) and rifampin (25/50). To cultivate E. coli WM3064, 300 μM 2,6-diaminopimelic acid (DAP) was added to the growth medium. E. coli TOP10 (Invitrogen) was used for cloning purposes.
TABLE 1.
Strains used in this study
| Strain | Genotype/description | Reference/source |
|---|---|---|
| H. neptunium | ||
| LE670 | Wild type (ATCC 15444) | Leifson (1) |
| EC85 | LE670 PCu::PCu-pleD (HNE_2284)-mCherry | This study |
| EC35 | LE670 PCu::PCu-pleD (HNE_2284)M1N-mCherry | This study |
| SE107 | LE670 PZn::PZn-pleD (HNE_2284)-egfp | This study |
| SE114 | LE670 PZn::PZn-pleD (HNE_2284)M1N-egfp | This study |
| E. coli | ||
| TOP10 | Cloning strain | Invitrogen |
| WM3064 | thrB1004 pro thi rpsL hsdS lacZΔM15 RP4-1360 Δ(araBAD)567 ΔdapA1341::[erm pir(wt)] | W. Metcalf (unpublished data) |
Transformation of H. neptunium.
Plasmids were transferred into H. neptunium by conjugation using E. coli strain WM3064 (dap mutant) as a donor. To this end, early-stationary-phase cultures of H. neptunium (2 ml) and E. coli WM3064 carrying the plasmid of interest (1 ml) were harvested by centrifugation. Each of the two pellets was washed with 1 ml MB medium and then resuspended in 100 μl medium containing 300 μM DAP. The two suspensions were mixed and spotted onto an MB-agar plate containing 300 μM DAP (without antibiotics). After overnight incubation at 28°C, the cells were scraped off the plate, washed twice in MB medium (without DAP), and finally resuspended in 1 ml MB medium. Dilutions of the suspension (undiluted, 1:10, and 1:100) were plated on selective MB-agar plates and incubated for 5 days at 28°C.
Construction of plasmids and strains.
The oligonucleotides used for plasmid construction are listed in Table 2. To generate pEC1, a fragment containing the upstream region of HNE_1486 was amplified by PCR using primers oEC1 and oEC2 and cut with the restriction endonucleases BlpI and NdeI. In parallel, HNE_2284 (pleD) was PCR amplified using primers oAJ18 and oOL16, followed by restriction of the product with NdeI and KpnI. The two fragments then were ligated into pXCHYC-2 (14) digested with BlpI and KpnI. For the construction of pEC41, HNE_2284 was PCR amplified using primers oSE69 and oOL16. The product was digested with AseI and KpnI and ligated with NdeI/KpnI-treated pEC1. In order to construct pCCFPN-2, pCCFPC-2, pCCFPN-3, pCCFPC-3, pCVENN-2, pCVENC-2, pCVENN-3, pCVENC-3, pCCHYN-2, pCCHYC-2, pCCHYN-3, and pCCHYC-3 (see Fig. 8), the upstream region of HNE_1486 was isolated from pEC1 by restriction with BlpI and NdeI and ligated with BlpI/NdeI-treated pXCFPN-2, pXCFPC-2, pXCFPN-3, pXCFPC-3, pXVENN-2, pXVENC-2, pXVENN-3, pXVENC-3, pXCHYN-2, pXCHYC-2, pXCHYN-3, and pXCHYC-3 (14), respectively. To generate pZGFPC-2 (see Fig. 8), the upstream region of HNE_2372 was PCR amplified using primers oSE1 and oSE5. The product was digested with HindIII and NdeI and ligated into the similarly treated pXGFPC-2 (14). For the construction of pSE10, HNE_2284 was PCR amplified using primers oAJ18 and oOL16. The PCR product was treated with NdeI and KpnI and ligated into pZGFPC-2 digested with the same enzymes. To create pSE46, HNE_2284 was PCR amplified using primers oSE69 and oOL16. The product was treated with AseI and KpnI and ligated into pZGFPC-2 digested with NdeI and KpnI. To generate pZCFPN-2, pZCFPC-2, pZGFPN-2, pZVENN-2, pZVENC-2, pZCHYN-2, and pZCHYC-2 (see Fig. 8), the upstream region of HNE_2372 was isolated from pSE4 by restriction with ClaI and NdeI and ligated with ClaI/NdeI-treated pXCFPN-2, pXCFPC-2, pXGFPN-2, pXVENN-2, pXVENC-2, pXCHYN-2, and pXCHYC-2 (14), respectively.
TABLE 2.
Oligonucleotides used in this study
| Name | Sequencea |
|---|---|
| oSE1 | atAAGCTTgagagaccgcgtcaatgaccagcg |
| oSE5 | aattaaCATATGtttttcctcggattgatccagaaccg |
| oSE69 | aattATTAATactgcccgcatcctcgtcgtggac |
| oEC1 | aaGCTTAGCctccggaaaagaatactggtacgtt |
| oEC2 | ttttCATATGgaagagttcctgtgaatgatcggtcc |
| oAJ18 | atatCATATGactgcccgcatcctcgtcgtggac |
| oOL16 | atatGGTACCggcggcgatgctctcgac |
Restriction sites are indicated by capital letters.
FIG 8.
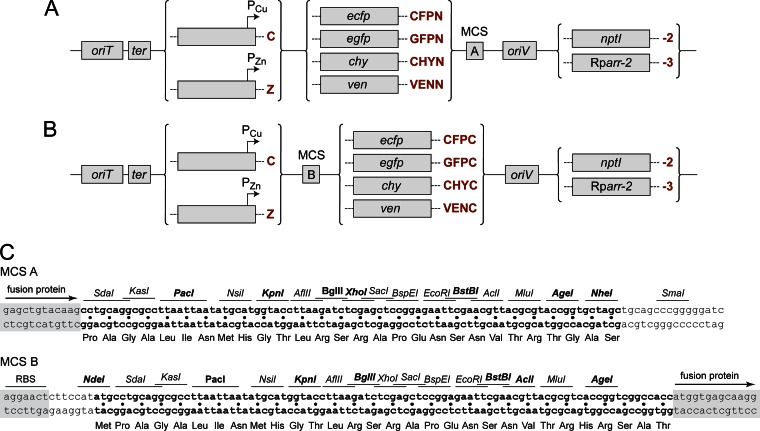
Integrative plasmids for copper- and zinc-inducible gene expression. (A and B) Schematic of integrative plasmids designed for the inducible expression of N-terminal (A) and C-terminal (B) fluorescent protein fusions under the control of the PCu or PZn promoter, respectively. Plasmid names are determined with the following format: each construct carries the prefix p, followed by an acronym describing its characteristic features and resistance gene, as given in boldface red print (e.g., pZCFPN-2). (C) Sequence and context of the multiple cloning sites. The locations of Shine-Dalgarno motifs (RBS; as present in PCu-containing constructs) and fusion tags (exemplified by mCherry) are indicated. Restriction sites that are unique in all plasmids carrying the respective MCS are highlighted in boldface, whereas those that are unique in only a subset of constructs are shown in normal print. The following gene names were used: RK2 origin of transfer (oriT), E. coli rrnBT1T2 transcriptional terminator (ter), pUC/pMB1 origin of replication (oriV), neomycin phosphotransferase I (KanR, nptI), rifampin ADP-ribosyltransferase (RifR, Rparr-2), enhanced green fluorescent protein (egfp), enhanced cyan fluorescent protein (ecfp), mCherry (chy), and Venus (ven).
To generate strains EC85, EC35, SE107, and SE114, H. neptunium LE670 was transformed with the integrative plasmids pEC1, pEC41, pSE10, or pSE46, respectively. The proper insertion of the plasmids at the HNE_1486 or HNE_2372 locus was verified by colony PCR.
Analysis of copper and zinc toxicity.
Stock solutions of CuSO4 (20 mM), ZnCl2, and ZnSO4 (1 M) were prepared in deionized water and filter sterilized. MB media (10 ml) containing increasing concentrations of the two salts were inoculated with a preculture of H. neptunium to an OD600 (optical density at 600 nm) of 0.05. After the incubation of the cultures at 28°C for 24 h, growth of the cells was assessed spectrophotometrically by determining the cell density at the OD600. To analyze the effect of heavy metals on the growth rate, a preculture of H. neptunium was diluted into 20 ml MB medium supplemented with CuSO4 and/or ZnSO4. Samples were taken every 2 h and analyzed for their cell density (OD600). Growth curves were generated by fitting the data to a suitable model (15) using the Solver function of Microsoft Excel 2007.
Extraction of total RNA from H. neptunium.
Total RNA was extracted with Roti-Aqua phenol-chloroform/isoamyl alcohol (Roth, Germany). To this end, cells were collected by centrifugation (10 min at 9,500 × g) and washed with 1 ml AE buffer (20 mM sodium acetate, 1 mM EDTA; adjusted to pH 5.5 with acetic acid). The cell sediment was resuspended in 600 μl ice-cold AE buffer and transferred to phase-lock tubes (5Prime, Germany). After the addition of 900 μl of hot (60°C) phenol-chloroform-isoamyl alcohol and 10 μl of 25% SDS (wt/vol), the samples were incubated at 60°C for 10 to 15 min, with the tubes being inverted gently every 2 to 3 min. After incubation on ice for 20 to 30 min, the samples were centrifuged (15 min at 16,000 × g and 4°C). The supernatant was transferred to a new phase-lock tube, mixed with 62.5 μl of a 2 M sodium acetate solution (pH 5.2), and centrifuged again for 15 min. The final aqueous layer then was transferred to a 2-ml Eppendorf tube. After the addition of 2.5 volumes of ice-cold ethanol (>96%, vol/vol), the samples were incubated overnight at −80°C. The next day, the samples were thawed on ice and centrifuged for 30 min at 16,000 × g and 4°C. The pellet was washed twice with ice-cold ethanol (70%, vol/vol) and dried at room temperature. RNA then was dissolved in 50 μl of diethyl pyrocarbonate (DEPC)-treated water (Roth, Germany) for at least 1 h on ice. Contaminating DNA was digested using the Turbo DNA-free kit (Ambion), and RNasin (Promega) was added to inhibit RNases. The quality and quantity of RNA was determined spectrophotometrically and by agarose gel electrophoresis.
Microarray analysis.
A culture of H. neptunium was diluted into fresh MB medium and grown overnight to an OD600 of 0.5. Subsequently, 2 ml of the culture was withdrawn (sample t0), and heavy-metal stock solution was added to obtain a final concentration of 0.5 mM CuSO4 and 0.5 mM ZnCl2, respectively. Twenty min and 60 min postinduction, further 2-ml samples were taken (samples t20 and t60). Immediately after withdrawal, cells were harvested by centrifugation (10 min at 9,500 × g). Total RNA then was isolated using the phenol-chloroform–isoamyl alcohol extraction protocol. Two independent samples were prepared for each time point (on different days) to provide biological replicates. All further steps, including chip design and synthesis, quality control, sample labeling, washing, and detection, as well as the primary data analysis, were performed by Febit (Heidelberg, Germany). The Geniom biochips used featured 15,000 60-mer oligonucleotide probes, including hybridization and blank controls. The probe design was based on the genome sequence of H. neptunium ATCC 15444 (9) deposited in the GenBank database (accession number CP000158) (16). For each gene sequence, up to four specific oligonucleotides were synthesized directly on the Geniom biochips. For 181 genes, it was not possible to design specific probes. After background correction, the median of the signals measured for each set of gene-specific probes was determined. These compressed background-subtracted raw data then were normalized using variance-stabilizing normalization (VSN) to account for technical variability between different arrays. The data obtained were analyzed further using Microsoft Excel 2007 and Microsoft Access 2003 to identify genes whose expression levels had significantly (more than 2-fold) changed at the 20-min (t20 compared to t0) and 60-min (t60 compared to t0) time points.
Determination of the MICs of antibiotics.
To determine the MICs of antibiotics, H. neptunium cells were diluted into MB medium containing various concentrations of antibiotics and incubated for 3 days. Subsequently, the cell densities were measured spectrophotometrically, and the minimal concentration sufficient to inhibit growth was determined.
Microscopy.
Cultures were grown to exponential phase in MB medium and induced with CuSO4 and ZnSO4, respectively, at the indicated concentrations. Cells were immobilized on pads made of 1% agarose and analyzed using a Zeiss Axio Observer.Z1 microscope (Zeiss, Germany) equipped with a Zeiss Plan-Apochromat 100×/1.46-numeric-aperture oil differential interference contrast (DIC) M27 objective, a Chroma ET-mCherry filter set, and a pco.edge sCMOS camera (PCO, Germany). Images were processed with Metamorph 7.7.5 (Universal Imaging Group, USA), Photoshop CS2, and Illustrator CS5 (Adobe Systems, USA).
Immunoblot analysis.
Cells were pelleted, resuspended in 2× SDS sample buffer (100 μl per 1 OD600 unit), and incubated for 15 min at 99°C. Proteins then were separated on 11% SDS-polyacrylamide gels and transferred to a polyvinylidene fluoride (PVDF) membrane (Immobilon, Merck Millipore, Germany). Immunodetection was performed with rabbit antisera directed against mCherry and green fluorescent protein (GFP) (Sigma, Germany) at dilutions of 1:10,000 using standard procedures. Goat anti-rabbit immunoglobulin G conjugated to horseradish peroxidase (PerkinElmer, USA) was used as the secondary antibody. Immunocomplexes were visualized using the Western Lightning plus-ECL chemiluminescence reagent (PerkinElmer, USA) and Amersham Hyperfilm ECL X-ray films (GE Healthcare). For quantitative analyses, signals were recorded with a ChemiDoc MP imaging system (Bio-Rad) and quantified using the Image Lab 5.0 software (Bio-Rad). Data were fitted to the Briggs-Haldane equation using the Solver function of Microsoft Excel 2010.
Microarray accession number.
The microarray data have been deposited in the Gene Expression Omnibus (GEO) database (17) under GEO accession number GSE62447.
RESULTS
Identification of heavy metal-inducible promoters.
H. neptunium is unable to utilize sugars as the sole carbon source (1), although its genome encodes all enzymes of the glycolytic pathway (9). Therefore, it may not contain sugar-regulated promoters, which are widely used for inducible gene expression in its relative, C. crescentus (18), and many other bacteria. Prompted by the successful application of heavy metal-inducible expression systems in other organisms (19–22), we investigated whether this strategy also would be applicable to H. neptunium. In doing so, we specifically focused on copper and zinc, two heavy metals with moderate toxicity that also serve as essential trace elements (23, 24).
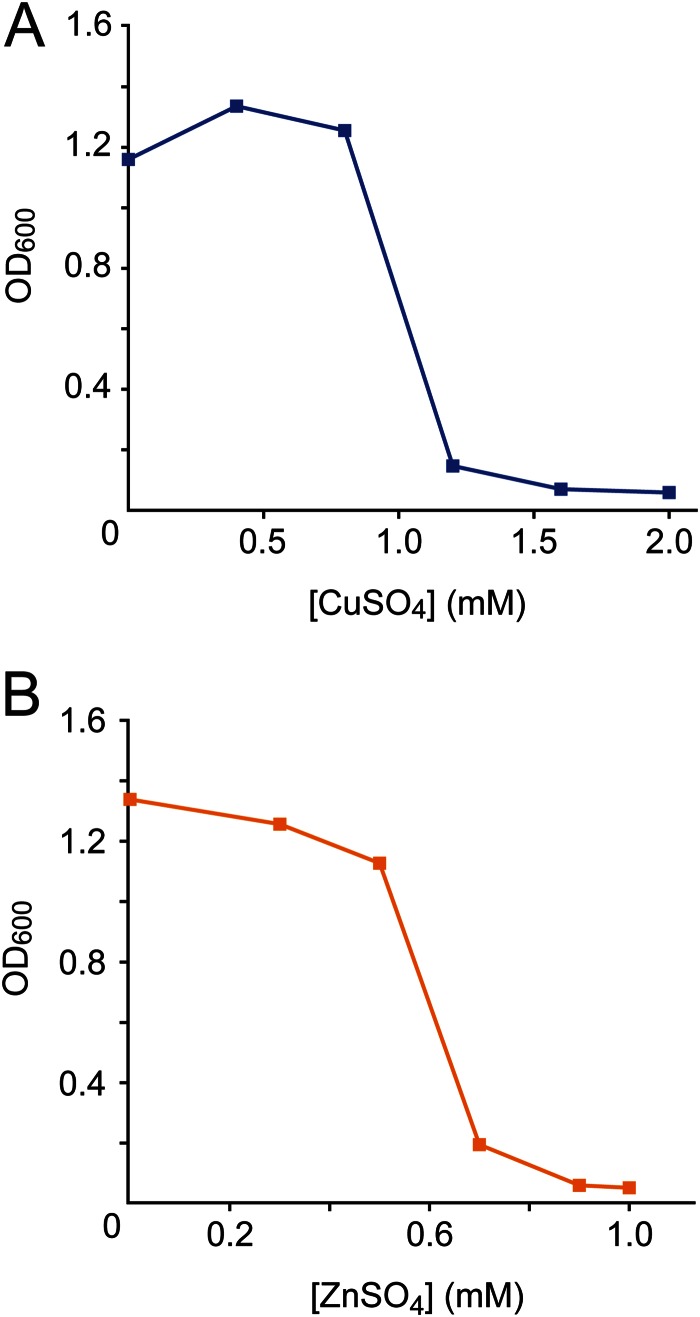
As a first step, the sensitivity of H. neptunium to copper and zinc was tested in a growth assay. The determination of the MICs showed that cells were able to grow in media containing up to 0.8 mM CuSO4 or 0.5 mM ZnSO4 (Fig. 2). Within the permissive concentration range, neither of the two metals had a significant effect on the steady-state growth rate. However, a noticeable lag was observed after shifting cells from standard to Zn2+-containing medium at metal concentrations higher than 0.3 mM (see Fig. S1 in the supplemental material). This lag was cell titer dependent and largely absent when the shift was performed at higher optical densities (OD600 of ≥0.25; data not shown). When added concomitantly, the two metals had an approximately additive effect (see Fig. S2). Of note, due to the lack of a suitable minimal medium, rich medium (marine broth) was used for all analyses in this work. As some of its components may form complexes with heavy-metal ions, the effective and nominal metal concentrations may differ to some extent.
FIG 2.
Effect of various concentrations of copper or zinc on H. neptunium growth. MB medium supplemented with various concentrations of CuSO4 (A) or ZnSO4 (B) was inoculated with a preculture of H. neptunium (initial OD600 of 0.05). Growth was assessed spectrophotometrically after 24 h of incubation.
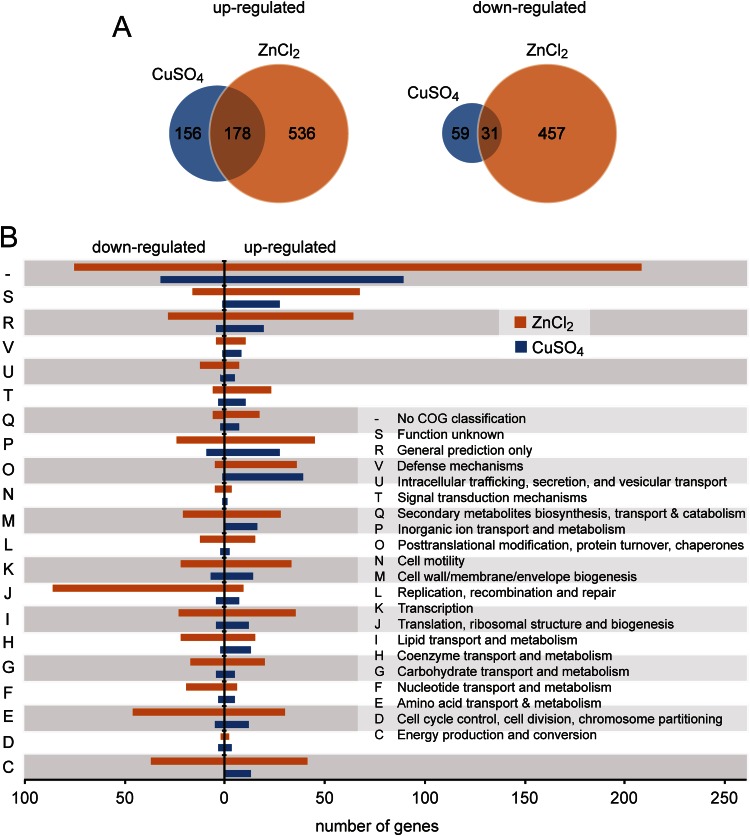
Having established the useful range of heavy-metal concentrations, we went on to determine the set of genes regulated in response to extracellular copper and zinc using microarray analysis. To this end, H. neptunium first was cultivated in the absence of heavy metals. CuSO4 or ZnCl2 (0.5 mM) then was added, and cells were withdrawn after 20-min and 60-min intervals. After extraction of RNA, samples were submitted to custom microarray analysis. In total, we were able to determine the expression levels of 3,387 out of the 3,568 genes annotated in the H. neptunium genome (9). The transcriptome data show that a considerable fraction of these genes were differentially expressed under conditions of heavy-metal stress (Table 3 and Fig. 3A). For both metals, there were clear differences in the expression patterns between the 20-min and 60-min time points (Fig. 3B; also see Fig. S3 in the supplemental material), suggesting that the immediate response and long-term adaptation to copper and zinc involve distinct but overlapping gene sets. The levels of many transcripts changed specifically in response to copper or zinc, but there was also a large group of genes that showed the same regulatory pattern in the presence of either heavy metal (Fig. 3A). The total number of deregulated genes was higher for cells treated with zinc. The strongest responses, however, were observed for copper-inducible genes, 10 of which were upregulated more than 100-fold by 20 min after induction (see Fig. S4A). Most of these highly regulated genes are located in two putative operons (HNE_1700-1708 and HNE_1483-1485) encoding known copper resistance determinants, such as copper-translocating P-type ATPases, multicopper oxidases, and periplasmic copper-binding proteins (25). In contrast, the majority of the 10 most highly zinc-regulated genes encode either uncharacterized proteins or subunits of the RND (resistance/nodulation/division) family efflux pumps (see Fig. S4B), which have been implicated previously in zinc resistance (26). Thus, H. neptunium appears to be equipped with specific defense systems that counteract the physiological stress exerted by elevated heavy-metal concentrations in the environment. To gain insight into the global effects of copper and zinc stress, we sorted all significantly (more than 2-fold) deregulated genes according to their COG (clusters of orthologous groups) classification (27). Many members of the two regulons lacked a COG category or are uncharacterized so far. The remaining genes covered a wide range of functions, indicating that heavy-metal stress leads to a global adjustment of cellular physiology.
TABLE 3.
Genes deregulated in response to heavy-metal stress
| Gene regulation state | Percentage of H. neptunium genes differentially expressed in response to: |
|||
|---|---|---|---|---|
| CuSO4 |
ZnCl2 |
|||
| 20 min | 60 min | 20 min | 60 min | |
| Upregulated | 9.9 | 10 | 21.1 | 18.4 |
| Downregulated | 2.7 | 4.1 | 14.4 | 11.6 |
FIG 3.
Genes significantly regulated upon heavy metal stress. (A) Venn diagram showing the number of genes whose expression is significantly regulated in cells treated for 20 min with ZnCl2 or CuSO4. (B) Functional categorization of the genes showing a significant change in expression levels after exposure of the cells for 20 min to ZnCl2 or CuSO4 based on their COG classification.
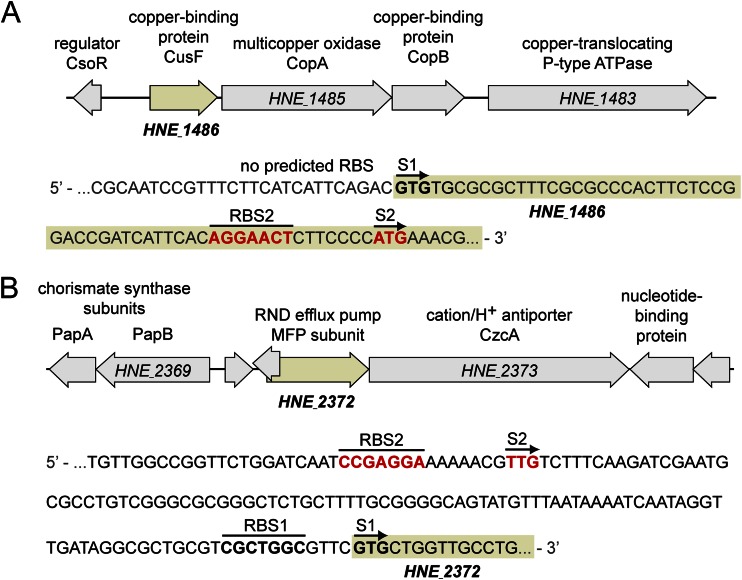
In order to identify promoters suitable for the establishment of a regulatable expression system in H. neptunium, the microarray data were scanned for genes that (i) showed low basal expression levels, (ii) were highly upregulated in response to heavy-metal stress, (iii) displayed high expression levels throughout the first hour of induction, and (iv) were specifically induced by either copper or zinc. Two candidate genes fulfilling these criteria (Table 4) then were chosen for further analysis. One of them, HNE_1486, was highly activated by copper and codes for a conserved protein with similarity to the periplasmic copper-binding protein CusF from E. coli (28, 29). It is located at the beginning of a multicistronic operon containing four conserved copper resistance determinants (Fig. 4A; also see Fig. S5 in the supplemental material). The second gene, HNE_2372, was activated by zinc and encodes the membrane fusion protein (MFP) subunit of an RND family efflux pump. Notably, HNE_2372 lies immediately upstream of a gene for a CzcA-like cation/H+ antiporter (Fig. 4B), which may represent the transporter subunit of the efflux system (26).
TABLE 4.
Transcription patterns of genes selected for promoter studies
| Locus tag | Annotation | Fold change in transcript levels in response to: |
|||
|---|---|---|---|---|---|
| CuSO4 |
ZnCl2 |
||||
| 20 min | 60 min | 20 min | 60 min | ||
| HNE_1486 | Periplasmic copper-binding protein CusF | 371 | 110 | 8.4 | 2.5 |
| HNE_2372 | RND family efflux pump (MFP subunit) | 3.6 | 2.3 | 68 | 78 |
FIG 4.
Chromosomal context of the heavy metal-inducible promoters analyzed in this study. (A) Chromosomal context of HNE_1486. The schematic on the top depicts the open reading frames surrounding the HNE_1486 gene. The sequence at the bottom shows the 5′ and upstream region of HNE_1486. The annotated start codon and its corresponding Shine-Dalgarno sequence (RBS) are shown in boldface. The experimentally confirmed start codon with its corresponding RBS is indicated in boldface red. (B) Chromosomal context of HNE_2372 (shown as described for panel A).
Functional characterization of the copper- and zinc-inducible promoters.
We next set out to adapt the regulatory elements that control the transcription and translation of HNE_1486 and HNE_2372 for the inducible expression of other genes of interest. As a reporter for these studies, we used a fluorescently (mCherry- or enhanced GFP-) tagged derivative of the predicted diguanylate cyclase HNE_2284, a homolog of the developmental regulator PleD from C. crescentus (30, 31), that localizes dynamically to the poles of the H. neptunium cell (O. Leicht and M. Thanbichler, unpublished data).
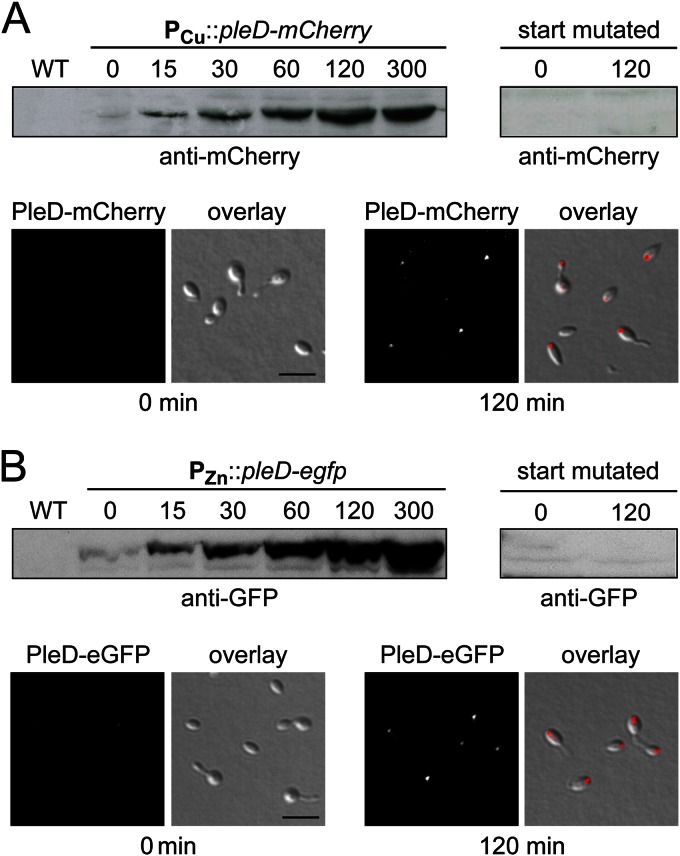
Given the problems associated with the automated prediction of open reading frames, we first reinvestigated the translational start sites of the two genes. To this end, different PCR fragments containing the chromosomal region (∼2 kb) upstream of (i) the annotated start codons or (ii) other potential start codons in the vicinity of the predicted start sites were fused to the pleD-mCherry or pleD-egfp reporter gene. Plasmids containing the fusion constructs then were integrated at the chromosomal HNE_1486 and HNE_2372 loci, respectively, by single homologous recombination. Analysis of the resulting strains by immunoblotting and fluorescence microscopy revealed that, in the case of HNE_1486, the synthesis of the reporter was observed only for a fragment extending to a potential translational start site located 54 bp downstream of the annotated start codon (Fig. 4A and 5A) (data not shown). In the case of the HNE_2372 fragments, in contrast, the inducible expression of the reporter fusion was observed when using either the annotated start site (data not shown) or an alternative start site located further (102 bp) upstream (Fig. 4B and 5B). Mutagenesis of the different start codons confirmed that the original annotation was wrong, and translation of the two genes indeed initiated at the newly determined positions (Fig. 5A and B, upper right).
FIG 5.
Analysis of the heavy metal-inducible promoters PCu and PZn. (A) Expression levels of pleD-mCherry under the control of PCu. Strain EC85 (PCu::PCu-pleD-mCherry) was grown to exponential phase in MB medium and induced with 300 μM CuSO4. At the indicated time points (min), samples were taken and subjected to immunoblot analysis with an anti-red fluorescent protein (RFP) antibody (upper left panel). In addition, cells withdrawn before (0 min) and 120 min after induction were analyzed by DIC and fluorescence microscopy (lower panel). Scale bar, 3 μm. As a control to verify the start codon, strain EC35 (PCu::PCu-pleDM1N-mCherry) was cultivated as described above. Samples were taken at the indicated time points after induction and probed with an anti-mCherry antibody (upper right panel). (B) Expression of pleD-egfp under the control of PZn. Strain SE107 (PZn::PZn-pleD-egfp) and the control strain SE114 (PZn::PZn-pleD-egfp) were grown to exponential phase in MB medium, induced with 500 μM ZnSO4, and analyzed as described for panel A, using an anti-GFP antibody. WT, wild type.
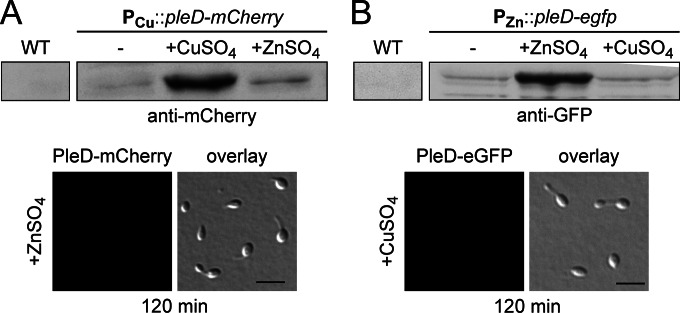
After having identified two chromosomal fragments that contained all regulatory elements required for copper- and zinc-regulated gene expression (Fig. 4), we further analyzed the characteristics of the two corresponding promoters (here named PCu and PZn, respectively). Consistent with the transcriptome data, immunoblot analysis confirmed that both PCu and PZn had only low basal activity in the absence of inducer (Fig. 5A and B, upper left). This tight control makes them suitable for use in protein depletion studies, although problems could arise with proteins of interest that are of very low abundance in the cell (data not shown). Under inducing conditions, by contrast, the reporter levels increased significantly with increasing heavy metal concentrations, with PCu showing a higher dynamic range than PZn (Fig. 6; also see Fig. S6 in the supplemental material). To use the two promoters independently of each other, it was necessary to verify that they responded only to their cognate inducer. The transcriptional data already indicated a relatively low degree of cross-activation (Table 4). To determine to what extent this effect translated into changes at the protein level, we compared the accumulation of the reporter fusions synthesized under the control of PCu or PZn under different inducing conditions (Fig. 7). This analysis showed that PCu mediated slightly higher protein levels in the presence of zinc than it did in the absence of inducer. A PZn-controlled reporter fusion, in contrast, did not show noticeable changes in its basal expression levels upon exposure of the cells to copper. Thus, the effects of regulatory cross talk are largely negligible, although they may be relevant for certain applications and vary depending on the metal concentrations used.
FIG 6.
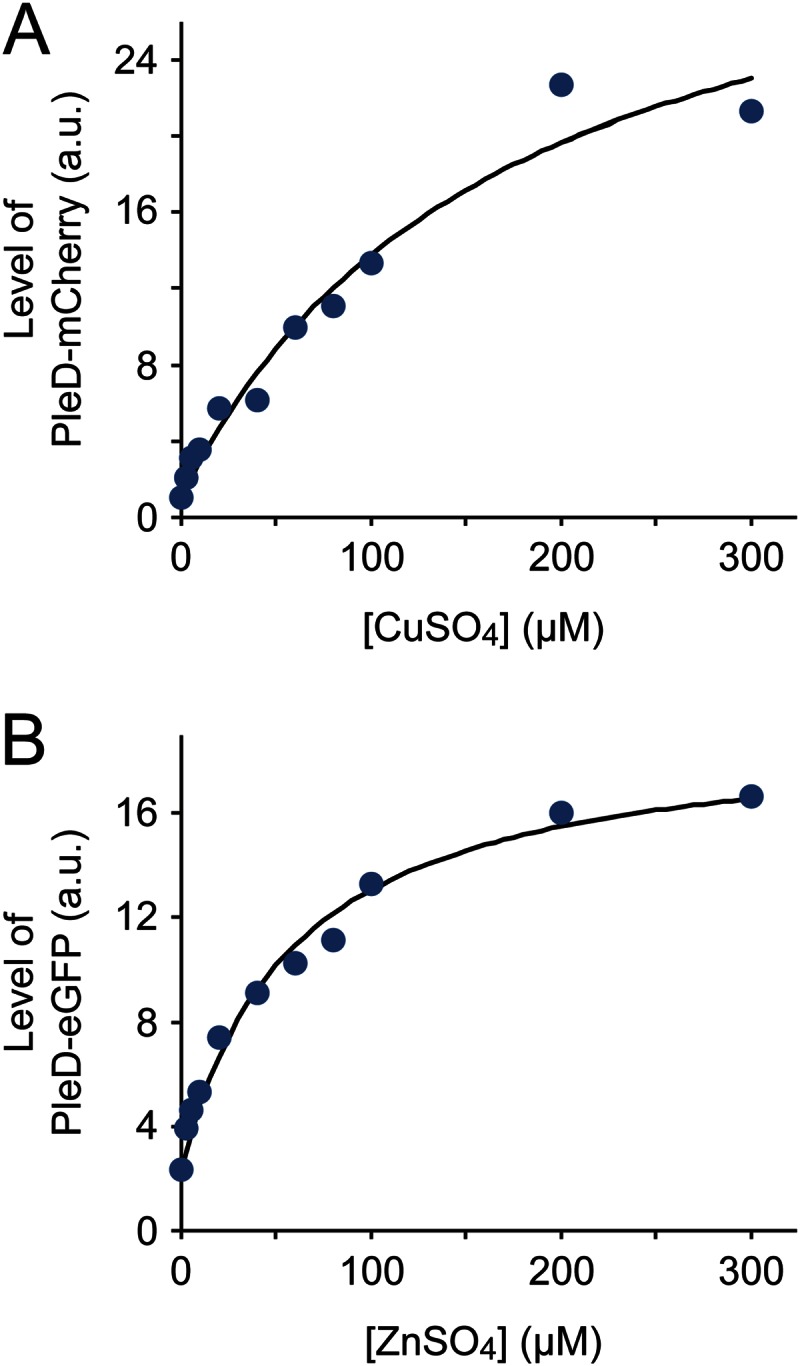
Effect of inducer concentrations on PCu and PZn activity. (A) Accumulation of PleD-mCherry expressed under the control of PCu at various copper concentrations. Exponentially growing cells of strain EC85 (PCu::PCu-pleD-mCherry) were induced for 1 h with different concentrations of CuSO4. Subsequently, cells were subjected to immunoblot analysis with an anti-mCherry antibody, and the PleD-mCherry signals were quantified. Shown are the normalized average values (n = 3) in arbitrary units (a.u.). (B) Accumulation of PleD-eGFP expressed under the control of PZn at various zinc concentrations. Cells of strain SE107 (PZn::PZn-pleD-egfp) were induced for 2 h with different concentrations of ZnSO4 and analyzed as described above (n = 2), using an anti-GFP antibody for immunodetection. Note that the signal intensities plotted in panels A and B are not directly comparable.
FIG 7.
Responsiveness of PCu and PZn to different heavy metals. (A) Analysis of PCu. Exponentially growing cells of strain EC85 (PCu::PCu-pleD-mCherry) were induced with 0.3 mM CuSO4 or 0.5 mM ZnSO4. Samples were taken before (0 min) and 120 min after the addition of the metal solutions and probed with an anti-mCherry antibody (upper panel). In addition, cells growing for 120 min in the presence of the indicated heavy metal were analyzed by DIC and fluorescence microscopy. Scale bars, 3 μm. (B) Analysis of PZn. Cells of strain SE107 (PZn::PZn-pleD-egfp) were analyzed as described for panel A, using an anti-GFP antibody for immunodetection.
Construction of a plasmid set for heavy metal-inducible gene expression in H. neptunium.
The results described above demonstrate that PCu and PZn are suitable for use as a multipurpose gene expression system in H. neptunium. Therefore, we aimed at constructing a comprehensive set of vectors that would facilitate their application in molecular and cell biological studies. To this end, it was first necessary to identify suitable antibiotic resistance markers that allowed the reliable selection of transformants. Growth analyses revealed that, under the given experimental conditions, a number of commonly used antibiotics had no or only a slight toxic effect on H. neptunium (data not shown). However, rifampin, chloramphenicol, and kanamycin effectively inhibited cell growth with MICs in the standard range (rifampin, 0.25 μg/ml; chloramphenicol, 5 μg/ml; kanamycin, 50 μg/ml). We were able to identify rifampin (Rparr-2) and kanamycin (nptI) resistance determinants that were functional in H. neptunium but have not yet succeeded in establishing a suitable chloramphenicol resistance cassette.
Building on existing vector backbones bearing (i) either the Rparr-2 or nptI gene, (ii) a narrow-host-range pUC replication origin for propagation in E. coli, and (iii) an R6K origin of transfer for conjugative transfer of the plasmid into H. neptunium (14), we then constructed a series of integrative plasmids containing the PCu and PZn promoters (Fig. 8). The promoter fragments comprised about 2 kb of the HNE_1486 or HNE_2372 upstream region, ending at the start codons defined in this study (Fig. 4). Their length allows the efficient integration of the constructs at the chromosomal HNE_1486 or HNE_2372 locus by single homologous recombination. To position the genes to be expressed at a proper distance from the respective Shine-Dalgarno sequences, an NdeI restriction site (5′-CATATG-3′) was engineered at the translational start sites. It is followed by an extensive multiple cloning site and one of several different fluorescent protein genes (ecfp, egfp, venus, and mCherry), arranged so as to allow the generation of translational fusions carrying either an N-terminal or C-terminal fluorescent tag. Of note, H. neptunium shows high autofluorescence at the excitation/emission wavelengths of GFP, which restricts the use of this tag to fusion proteins that accumulate to high levels in the cell. Upstream of the promoter fragments, all plasmids contain a strong Rho-independent transcription terminator to prevent transcriptional read-through from genes located in the flanking regions.
Whereas the PCu- and PZn-containing plasmids are designed for ectopic gene expression, it also may be desirable to generate fusion constructs that integrate at the native locus of a gene, obviating the need for an inducible promoter. For this purpose, and for general applications requiring only a basic plasmid backbone without promoters and fluorescent protein genes, the reader is referred to a previously described plasmid collection (14), which offers a wide choice of suitable Rparr-2- and nptI-bearing constructs. Interestingly, we found that cells grew very poorly after transformation with pBBR-based broad-host-range plasmids (data not shown), which so far has prevented the use of replicating plasmids in H. neptunium. This effect could be due to the inadequate expression of the resistance genes or, potentially, to inefficient segregation of the plasmids through the stalk structure.
DISCUSSION
Heavy metals are essential trace elements that serve as cofactors for a variety of different proteins. However, at elevated concentrations, they strongly impair cell viability. The mechanisms underlying heavy-metal toxicity are still under debate, and several different pathways have been proposed. For instance, copper and zinc can replace the cognate metal cofactor of metalloproteins, thereby compromising their activity (32, 33). Moreover, due to their thiophilicity, they are potent inhibitors of enzymes whose activity depends on active-site thiols and iron-sulfur clusters (34, 35). Finally, copper ions can mediate a Fenton-like reaction, producing reactive oxygen species that cause severe damage to proteins, lipids, and other molecules (36). Given these rather general effects, heavy-metal stress can affect a variety of different cellular functions. Consistently, and in line with results obtained with other species (37, 38), H. neptunium cells exposed to copper or zinc display significant changes in their transcriptome that affect diverse aspects of their physiology. Most prominently, however, they highly upregulate specific defense systems involved in metal binding and export, including P-type ATPases and RND efflux pumps.
In this work, two promoters that were highly activated in the presence of copper and zinc were characterized in detail and adopted for use as general-purpose expression systems in H. neptunium. They allow tightly regulated protein synthesis and reach maximum activity at metal concentrations that have no effect on cell growth (see Fig. S6 in the supplemental material). However, the toxicity of copper and zinc may vary depending on the type and pH value of the culture medium, as well as on the aeration state of the cells. Therefore, it is advisable to revalidate the induction conditions whenever the cultivation parameters are changed. This particularly applies to the use of zinc, which is less well tolerated by H. neptunium than copper. Caution also should be taken when both heavy metals are used concomitantly. In this case, the concentrations used for induction should be reduced to values well below the MICs to avoid adverse effects on cellular physiology.
The functionality of the tools developed in this work has been verified by various protein localization and depletion studies conducted in our laboratory (unpublished). With the possibility to genetically manipulate H. neptunium and tightly control the expression of target genes, the way is paved for in-depth studies of this unique bacterial species. It will be interesting to unravel the molecular mechanisms underlying its unusual way of proliferation and determine the selective advantages conferred by it.
Supplementary Material
ACKNOWLEDGMENTS
We thank Julia Rosum and Stephanie Wick for excellent technical assistance.
This work was supported by the German Research Foundation (DFG; SFB 987) and the Max Planck Society. E.C. acknowledges support from the DFG Research Training Group GRK 1216.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03104-14.
REFERENCES
- 1.Leifson E. 1964. Hyphomicrobium neptunium sp. n. Antonie Van Leeuwenhoek 30:249–256. doi: 10.1007/BF02046730. [DOI] [PubMed] [Google Scholar]
- 2.Moore RL. 1981. The biology of Hyphomicrobium and other prosthecate, budding bacteria. Annu Rev Microbiol 35:567–594. doi: 10.1146/annurev.mi.35.100181.003031. [DOI] [PubMed] [Google Scholar]
- 3.Hirsch P. 1974. Budding bacteria. Annu Rev Microbiol 28:391–444. doi: 10.1146/annurev.mi.28.100174.002135. [DOI] [PubMed] [Google Scholar]
- 4.Wali TM, Hudson GR, Danald DA, Weiner RM. 1980. Timing of swarmer cell cycle morphogenesis and macromolecular synthesis by Hyphomicrobium neptunium in synchronous culture. J Bacteriol 144:406–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moore RL, Weiner RM, Gebers R. 1984. Genus Hyphomonas Pongratz 1957 nom. rev. emend., Hyphomonas polymorpha Pongratz 1957 nom. rev. emend., and Hyphomonas neptunium (Leifson 1964) comb. nov. emend. (Hyphomicrobium neptunium). Int J Syst Bacteriol 34:71–73. doi: 10.1099/00207713-34-1-71. [DOI] [Google Scholar]
- 6.Nikitin DI, Vishnewetskaya OY, Chumakov KM, Zlatkin IV. 1990. Evolutionary relationship of some stalked and budding bacteria (genera Caulobacter, “Hyphobacter”, Hyphomonas and Hyphomicrobium) as studied by the new integral taxonomical method. Arch Microbiol 153:123–128. doi: 10.1007/BF00247808. [DOI] [PubMed] [Google Scholar]
- 7.Garrity GM, Bell JA, Lilburn T. 2005. Family I. Rhodobacteraceae fam. nov., p 161 In Brenner DJ, Krieg NR, Staley JT, Garrity GM (ed), Bergey's manual of systematic bacteriology, 2nd ed, vol 2 Springer, New York, NY. [Google Scholar]
- 8.Badger JH, Eisen JA, Ward NL. 2005. Genomic analysis of Hyphomonas neptunium contradicts 16S rRNA gene-based phylogenetic analysis: implications for the taxonomy of the orders “Rhodobacterales” and Caulobacterales. Int J Syst Evol Microbiol 55:1021–1026. doi: 10.1099/ijs.0.63510-0. [DOI] [PubMed] [Google Scholar]
- 9.Badger JH, Hoover TR, Brun YV, Weiner RM, Laub MT, Alexandre G, Mrazek J, Ren Q, Paulsen IT, Nelson KE, Khouri HM, Radune D, Sosa J, Dodson RJ, Sullivan SA, Rosovitz MJ, Madupu R, Brinkac LM, Durkin AS, Daugherty SC, Kothari SP, Giglio MG, Zhou L, Haft DH, Selengut JD, Davidsen TM, Yang Q, Zafar N, Ward NL. 2006. Comparative genomic evidence for a close relationship between the dimorphic prosthecate bacteria Hyphomonas neptunium and Caulobacter crescentus. J Bacteriol 188:6841–6850. doi: 10.1128/JB.00111-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brilli M, Fondi M, Fani R, Mengoni A, Ferri L, Bazzicalupo M, Biondi EG. 2010. The diversity and evolution of cell cycle regulation in alpha-proteobacteria: a comparative genomic analysis. BMC Syst Biol 4:52. doi: 10.1186/1752-0509-4-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ireland MM, Karty JA, Quardokus EM, Reilly JP, Brun YV. 2002. Proteomic analysis of the Caulobacter crescentus stalk indicates competence for nutrient uptake. Mol Microbiol 45:1029–1041. doi: 10.1046/j.1365-2958.2002.03071.x. [DOI] [PubMed] [Google Scholar]
- 12.Schlimpert S, Klein EA, Briegel A, Hughes V, Kahnt J, Bolte K, Maier UG, Brun YV, Jensen GJ, Gitai Z, Thanbichler M. 2012. General protein diffusion barriers create compartments within bacterial cells. Cell 151:1270–1282. doi: 10.1016/j.cell.2012.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kysela DT, Brown PJ, Huang KC, Brun YV. 2013. Biological consequences and advantages of asymmetric bacterial growth. Annu Rev Microbiol 67:417–435. doi: 10.1146/annurev-micro-092412-155622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thanbichler M, Iniesta AA, Shapiro L. 2007. A comprehensive set of plasmids for vanillate- and xylose-inducible gene expression in Caulobacter crescentus. Nucleic Acids Res 35:e137. doi: 10.1093/nar/gkm818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang L. 2011. A new mechanistic growth model for simultaneous determination of lag phase duration and exponential growth rate and a new Bělehdrádek-type model for evaluating the effect of temperature on growth rate. Food Microbiol 28:770–776. doi: 10.1016/j.fm.2010.05.019. [DOI] [PubMed] [Google Scholar]
- 16.Benson DA, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. 2014. GenBank. Nucleic Acids Res 42:D32–D37. doi: 10.1093/nar/gkt1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, Yefanov A, Lee H, Zhang N, Robertson CL, Serova N, Davis S, Soboleva A. 2013. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res 41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meisenzahl AC, Shapiro L, Jenal U. 1997. Isolation and characterization of a xylose-dependent promoter from Caulobacter crescentus. J Bacteriol 179:592–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gomez-Santos N, Treuner-Lange A, Moraleda-Munoz A, Garcia-Bravo E, Garcia-Hernandez R, Martinez-Cayuela M, Perez J, Sogaard-Andersen L, Munoz-Dorado J. 2012. Comprehensive set of integrative plasmid vectors for copper-inducible gene expression in Myxococcus xanthus. Appl Environ Microbiol 78:2515–2521. doi: 10.1128/AEM.07502-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Llull D, Poquet I. 2004. New expression system tightly controlled by zinc availability in Lactococcus lactis. Appl Environ Microbiol 70:5398–5406. doi: 10.1128/AEM.70.9.5398-5406.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gebhart D, Bahrami AK, Sil A. 2006. Identification of a copper-inducible promoter for use in ectopic expression in the fungal pathogen Histoplasma capsulatum. Eukaryot Cell 5:935–944. doi: 10.1128/EC.00028-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mett VL, Lochhead LP, Reynolds PH. 1993. Copper-controllable gene expression system for whole plants. Proc Natl Acad Sci U S A 90:4567–4571. doi: 10.1073/pnas.90.10.4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nies DH. 1999. Microbial heavy-metal resistance. Appl Microbiol Biotechnol 51:730–750. doi: 10.1007/s002530051457. [DOI] [PubMed] [Google Scholar]
- 24.Duxbury T. 1981. Toxicity of heavy metals to soil bacteria. FEMS Microbiol Lett 11:217–220. doi: 10.1111/j.1574-6968.1981.tb06967.x. [DOI] [Google Scholar]
- 25.Bondarczuk K, Piotrowska-Seget Z. 2013. Molecular basis of active copper resistance mechanisms in Gram-negative bacteria. Cell Biol Toxicol 29:397–405. doi: 10.1007/s10565-013-9262-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choudhury R, Srivastava S. 2001. Zinc resistance mechanisms in bacteria. Curr Sci India 81:768–775. [Google Scholar]
- 27.Tatusov RL, Koonin EV, Lipman DJ. 1997. A genomic perspective on protein families. Science 278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 28.Delmar JA, Su CC, Yu EW. 2014. Bacterial multidrug efflux transporters. Annu Rev Biophys 43:93–117. doi: 10.1146/annurev-biophys-051013-022855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loftin IR, Franke S, Roberts SA, Weichsel A, Heroux A, Montfort WR, Rensing C, McEvoy MM. 2005. A novel copper-binding fold for the periplasmic copper resistance protein CusF. Biochemistry 44:10533–10540. doi: 10.1021/bi050827b. [DOI] [PubMed] [Google Scholar]
- 30.Paul R, Weiser S, Amiot NC, Chan C, Schirmer T, Giese B, Jenal U. 2004. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev 18:715–727. doi: 10.1101/gad.289504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hecht GB, Newton A. 1995. Identification of a novel response regulator required for the swarmer-to-stalked-cell transition in Caulobacter crescentus. J Bacteriol 177:6223–6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tottey S, Waldron KJ, Firbank SJ, Reale B, Bessant C, Sato K, Cheek TR, Gray J, Banfield MJ, Dennison C, Robinson NJ. 2008. Protein-folding location can regulate manganese-binding versus copper- or zinc-binding. Nature 455:1138–1142. doi: 10.1038/nature07340. [DOI] [PubMed] [Google Scholar]
- 33.McDevitt CA, Ogunniyi AD, Valkov E, Lawrence MC, Kobe B, McEwan AG, Paton JC. 2011. A molecular mechanism for bacterial susceptibility to zinc. PLoS Pathog 7:e1002357. doi: 10.1371/journal.ppat.1002357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Macomber L, Imlay JA. 2009. The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc Natl Acad Sci U S A 106:8344–8349. doi: 10.1073/pnas.0812808106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu FF, Imlay JA. 2012. Silver(I), mercury(II), cadmium(II), and zinc(II) target exposed enzymic iron-sulfur clusters when they toxify Escherichia coli. Appl Environ Microbiol 78:3614–3621. doi: 10.1128/AEM.07368-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dupont CL, Grass G, Rensing C. 2011. Copper toxicity and the origin of bacterial resistance—new insights and applications. Metallomics 3:1109–1118. doi: 10.1039/c1mt00107h. [DOI] [PubMed] [Google Scholar]
- 37.Baker J, Sitthisak S, Sengupta M, Johnson M, Jayaswal RK, Morrissey JA. 2010. Copper stress induces a global stress response in Staphylococcus aureus and represses sae and agr expression and biofilm formation. Appl Environ Microbiol 76:150–160. doi: 10.1128/AEM.02268-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamamoto K, Ishihama A. 2005. Transcriptional response of Escherichia coli to external copper. Mol Microbiol 56:215–227. doi: 10.1111/j.1365-2958.2005.04532.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.