Abstract
Nitrification has an immense impact on nitrogen cycling in natural ecosystems and in wastewater treatment plants. Mathematical models function as tools to capture the complexity of these biological systems, but kinetic parameters especially of nitrite-oxidizing bacteria (NOB) are lacking because of a limited number of pure cultures until recently. In this study, we compared the nitrite oxidation kinetics of six pure cultures and one enrichment culture representing three genera of NOB (Nitrobacter, Nitrospira, Nitrotoga). With half-saturation constants (Km) between 9 and 27 μM nitrite, Nitrospira bacteria are adapted to live under significant substrate limitation. Nitrobacter showed a wide range of lower substrate affinities, with Km values between 49 and 544 μM nitrite. However, the advantage of Nitrobacter emerged under excess nitrite supply, sustaining high maximum specific activities (Vmax) of 64 to 164 μmol nitrite/mg protein/h, contrary to the lower activities of Nitrospira of 18 to 48 μmol nitrite/mg protein/h. The Vmax (26 μmol nitrite/mg protein/h) and Km (58 μM nitrite) of “Candidatus Nitrotoga arctica” measured at a low temperature of 17°C suggest that Nitrotoga can advantageously compete with other NOB, especially in cold habitats. The kinetic parameters determined represent improved basis values for nitrifying models and will support predictions of community structure and nitrification rates in natural and engineered ecosystems.
INTRODUCTION
Aerobic nitrite oxidation is the second microbially mediated part of nitrification, a key process in the global nitrogen cycle, catalyzed by autotrophic, slow-growing nitrite-oxidizing bacteria (NOB). Under nitrifying conditions, the growth of NOB is directly linked to the nitrite production rate and the kinetics of nitrite oxidation (1). Nitrification occurs in almost every aquatic and terrestrial ecosystem, in natural as well as in artificial environments like wastewater treatment plants (WWTPs). The intermediate nitrite hardly accumulates, but local or temporary peaks might appear especially when conditions suddenly change or adverse conditions like alkaline pH values impair NOB activity (2). In WWTPs, disturbances can cause nitrite peaks after destabilization of the NOB guild (3). In soils, nitrite concentrations can vary from ∼0.01 to ∼100 μg of nitrogen g of soil−1, with the highest values measured in fertilized samples (4). Therefore, the concentration of nitrite as the substrate of NOB varies and is regarded as one major factor providing niche differentiation (5). In the ecological context, field studies are important to characterize nitrifying communities with specific activities and affinities for nitrite, but for a better understanding of wastewater treatment processes, mathematical models are required (6). Such models include ecophysiological kinetic data, which are known primarily for ammonia-oxidizing bacteria (AOB) (7). They provide the substrate nitrite for the second major step of nitrification. However, the development of two-step nitrification models also requires kinetic parameters of NOB (8, 9).
Because of their earlier discovery and the ease with which pure cultures can be obtained, studies of the nitrite oxidation kinetics of NOB were previously performed mainly with Nitrobacter bacteria, initially regarded as a model NOB in WWTPs (10). The first cultures of Nitrospira, later identified as key NOB in activated sludge (11, 12), were obtained from 1986 on (13, 14). In the beginning, estimations of the oxidation kinetics of Nitrospira populations were done by microsensor measurements of biofilm samples (15, 16). Differences in growth kinetics between NOB can be described in terms of K and r strategists (16, 17). Nitrobacter bacteria (r strategists) have relatively high maximum nitrite oxidation activity (or growth rate) and low substrate affinities, while Nitrospira bacteria (K strategists) are characterized by low maximum activity and high substrate affinities. These contrary strategies were confirmed by biotechnological approaches in reactor systems (18–20) but not with pure cultures until now.
Besides WWTPs, members of the genus Nitrospira represent the most dominant nitrite oxidizers in many natural and engineered ecosystems in terms of diversity (12, 21–23). An open question is the relevance of the new candidate genus Nitrotoga (24) for the treatment of wastewaters. These NOB were detected primarily in soils (24) but also occur in cave biofilm (25), freshwater (26), and activated sludge (27–29).
Here we present a broad comparison of directly measured oxidation kinetics of nonmarine NOB by microsensor measurements. In addition to Nitrobacter, the study includes pure cultures of lineage I and II Nitrospira from freshwater and activated sludge and the first analysis of a highly enriched Nitrotoga culture from permafrost soil. Hence, we investigated representatives of three different phyla, namely, Alpha- and Betaproteobacteria and Nitrospirae (14, 24, 30). Not included are the marine NOB genera Nitrococcus and Nitrospina (31) within the phyla Gammaproteobacteria and “Nitrospinae” (75), respectively, and the novel strain Nitrolancea hollandica within the phylum Chloroflexi (32).
The lab-scale kinetic parameters provided show the nitrite-dependent niche differentiation of NOB and might be translated to full-scale mathematical models (like an activated-sludge model [33]) to improve the efficiency of biological nitrification in WWTPs and reactor systems.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Seven different NOB species of three genera were investigated in this study. Nitrobacter hamburgensis X14 and Nitrobacter winogradskyi strain “Engel” were both isolated from soil (34, 35), and Nitrobacter vulgaris Ab1 was from sewage (36). The isolates of “Candidatus Nitrospira defluvii” A17 (lineage I) and Nitrospira strain BS10 (lineage II) were derived from activated sludge (37; B. Nowka, S. Off, H. Daims, and E. Spieck, unpublished data), and Nitrospira moscoviensis M-1 (lineage II) originated from a heating water system (14). “Candidatus Nitrotoga arctica” 6680 was highly enriched from permafrost soil (24). Although no pure culture was investigated here, Nitrotoga was the only type of cells visible by light microscopy when grown in mineral salts medium with nitrite.
All Nitrobacter and Nitrospira bacteria were grown as batch cultures in 500 ml mineral medium with the following composition: 1 liter of demineralized water, different concentrations of NaNO2 as the sole energy source, 0.007 g of CaCO3, 0.5 g of NaCl, 0.05 g of MgSO4·7H2O, 0.15 g of KH2PO4, and the following trace elements: 33.8 μg of MnSO4·H2O, 49.4 μg of H3BO3, 43.1 μg of ZnSO4·7H2O, 37.1 μg of (NH4)6Mo7O24, 973.0 μg of FeSO4·7H2O, and 25.0 μg of CuSO4·5H2O. For the “Ca. Nitrotoga arctica” enrichment, a modified trace element composition was used: 100.0 μg of MnCl2·4H2O, 30.0 μg of H3BO3, 144.0 μg of ZnSO4·7H2O, 36.0 μg of Na2MoO4·2H2O, 2.1 mg of FeSO4·7H2O, 2.0 μg of CuCl2·2H2O, 190.0 μg of CoCl2·6H2O, and 24.0 μg of NiCl2·6H2O. Before demineralized water was added, these supplements were dissolved in 12.5 ml of HCl (25%) (38). The pH was adjusted to 8.4 to 8.6 and changed to 7.4 to 7.6 2 days after autoclaving. The cultures were started with inocula of 1% (vol/vol) and incubated in the dark at temperatures of 17°C (“Ca. Nitrotoga arctica”), 28°C (all Nitrobacter strains, “Ca. Nitrospira defluvii,” and strain BS10), and 37°C (N. moscoviensis). After the first detection of nitrite consumption, the Nitrospira and Nitrobacter cultures were stirred moderately (100 to 300 rpm). The “Ca. Nitrotoga arctica” enrichment was incubated without agitation.
Chemical analyses.
Nitrite and nitrate concentrations were determined by high-performance liquid chromatography (HPLC) via ion pair chromatography with a LiChrospher RP-18 column (5 μm, 4 by 125 mm; Merck KGaA, Darmstadt, Germany) (39) and UV detection in an automated system (LaChrom Elite HPLC system; VWR International GmbH, Darmstadt, Germany). Cell protein concentrations were measured by the bicinchoninic acid method (40) after cell lysis in 0.15 M NaOH and incubation at 90°C for 30 min.
Disruption of “Ca. Nitrospira defluvii” aggregates and microcolonies.
Since “Ca. Nitrospira defluvii” forms extensive cell aggregates and microcolonies under the growth conditions used, we developed a method for disruption to enable single-cell counting. In the first step, 1 ml of cell culture was shaken with a Mikro-Dismembrator S (Sartorius Stedim Biotech GmbH, Göttingen, Germany) for 40 min at a frequency of 1,500 oscillations/min in a 3-ml polytetrafluoroethylene shaking flask with a chromium steel grinding ball (diameter, 5 mm). Afterwards, the cell culture was diluted in 10 ml of distilled H2O (dH2O) and sonicated for 10 min at a frequency of 24 kHz and an amplitude of 0.5 (UP200S Ultrasonic Processor; Hielscher Ultrasonics GmbH, Teltow, Germany). This procedure removed the bulk of the extracellular polymeric substances and resulted in single cells suitable for cell counting.
Cell counting.
Aliquots of cell cultures were diluted in 10 to 15 ml of dH2O. Cells were collected by filtration with a Milli Pure Filter System (Sartorius GmbH, Göttingen, Germany) with membrane filters with a 0.2-μm pore size (Merck Millipore, Billerica, MA). For cell counting, the filters were dried and pieces of the filters were subsequently stained with 0.01 mg ml−1 DAPI (4′,6-diamidino-2-phenylindole) for 3 min immersed in dH2O and 70% ethanol for a few seconds. Cells were counted on dried filters by fluorescence microscopy (Axio Imager.M2; Carl Zeiss, Göttingen, Germany) and by use of a counting ocular (area of a square: 0.0004 mm2 at ×1,000 magnification). Cell counts were calculated by the following equation: cellsoverall = [filter area (380 mm2)/square area (0.0004 mm2)] · (cell count per square). The cell count per square was generated from the average cell number of 30 squares.
Activity measurements.
Activity measurements were performed with early-stationary-phase cells in nitrite-limited mineral medium by the following procedure. Cultures (500 ml) of “Ca. Nitrospira defluvii” and the three Nitrobacter strains were started with a nitrite concentration of 9 mM, and N. moscoviensis was started with a nitrite concentration of 5.7 mM. To avoid inhibition of cell growth, cultures of Nitrospira strain BS10 and “Ca. Nitrotoga arctica” were started with low nitrite concentrations of 0.3 and 0.7 mM, respectively, and replenished with these amounts when nitrite was consumed. Nitrite consumption and nitrate production were monitored frequently by HPLC. Between 12 and 48 h after complete nitrite consumption (early stationary phase), 50-ml aliquots of the cultures were transferred to 100-ml flasks, stirred, and incubated in thermostat-regulated rooms at corresponding temperatures until the start of the measurements (“Ca. Nitrotoga arctica” at 17°C; all Nitrobacter strains, “Ca. Nitrospira defluvii,” and strain BS10 at 28°C; N. moscoviensis at 37°C). Nitrite-dependent oxygen consumption was measured in a microrespiration system (Unisense AS, Denmark). This system consisted of a one-channel oxygen sensor amplifier (OXY-Meter), a Clark-type oxygen microsensor (OX-MR; polarized for at least 48 h before use), a stirring system with glass-coated magnets, 2-ml glass chambers with glass stoppers, a rack for eight chambers, and the data acquisition software MicOx 3.0. The response time (90%) of the oxygen microsensor was <15 s, and the oxygen uptake of the microsensor was below 1 nM day−1 (41). All measurements were carried out in a recirculated water bath in thermostat-regulated rooms with stirring at 200 rpm. Subsamples of the early-stationary-phase cells were transferred to the 2-ml glass chambers, sealed with glass stoppers, and immersed in the water bath. The microrespiration sensor was inserted through a capillary hole inside the glass stopper. The measurements started with an initial equilibration for 15 to 30 min until the signal from the sensor was stable; nitrite from stock solutions was then added through a second capillary hole with a syringe.
Calculation of oxidation kinetics and other properties.
Nitrite oxidation kinetics were estimated from multiple oxygen consumption rates at various defined nitrite concentrations. Measurements of every nitrite-oxidizing strain were performed with at least three independently grown cultures. The amounts of consumed nitrite were calculated from oxygen consumption according to the ratio of nitrite oxidation to oxygen consumption of 1:0.5. By using the data analysis software SigmaPlot 12.0 (Systat Software GmbH, Erkrath, Germany), oxygen uptake rates were plotted according to the total nitrite concentration and kinetic characteristics were obtained by fitting a Michaelis-Menten kinetic with the following equation to the data: V = (Vmax · [S])/(Km + [S]). Here, V is activity, Vmax is the maximum specific activity (μmol/mg protein/h), Km is the half-saturation constant for nitrite oxidation (μM), and [S] is the nitrite concentration (μM). Although the experiments were not performed with purified enzymes, we prefer to use the terms Km and Vmax, since we determined kinetics in short-term activity assays within a few hours, where growth can be neglected.
Nitrite consumption and cell growth were determined at the nitrite concentrations shown at the corresponding temperatures (described above) in 150- to 500-ml Erlenmeyer flasks containing mineral medium inoculated with 1% (vol/vol) early-stationary-phase cells. Subsamples were withdrawn for determination of nitrite and nitrate concentrations (1 ml), cell counts (1 to 10 ml), and protein concentrations (50 ml) as described above. Values of generation time (g in hours) and cell activity (k in fmol NO2− cell−1 h−1) were calculated from the exponential growth phase as described previously (42). Protein growth yield (Yp in mg protein mmol−1 nitrite) and cell growth yield (Yc in log cell counts mmol−1 nitrite) were calculated on the basis of cells or protein produced per millimole of nitrite oxidized.
RESULTS
Growth of NOB in batch cultures.
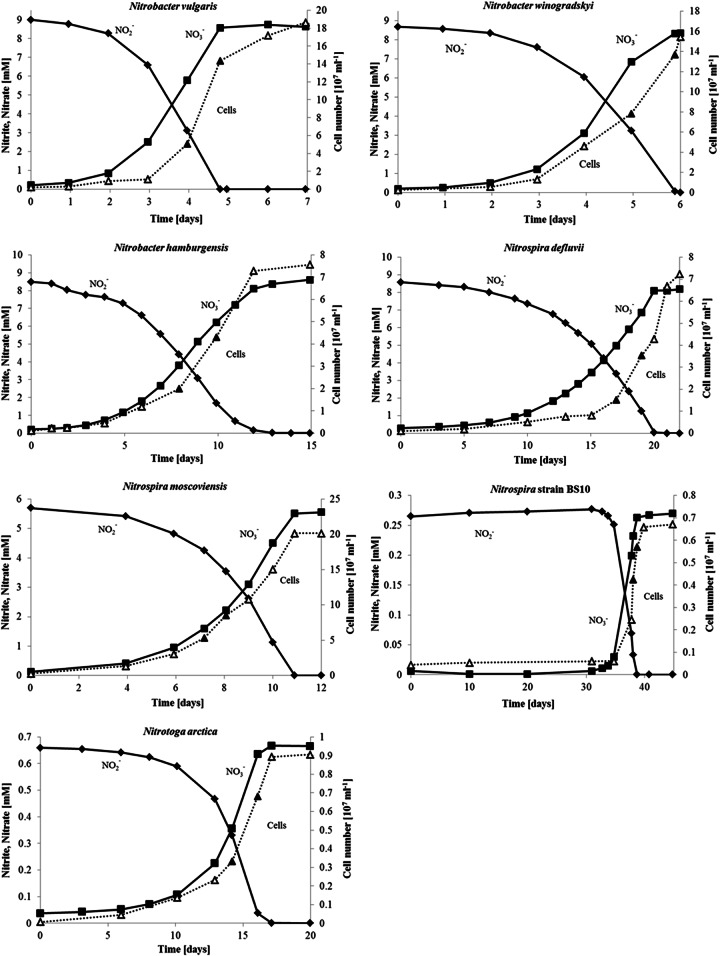
Prior to the respirometry measurements, the cells were grown in batch cultures in nitrite-limited mineral medium. Figure 1 summarizes the exemplary cell growth, nitrite consumption, and equivalent nitrate formation of such NOB batch cultures. Since the growth of Nitrospira strain BS10 and “Ca. Nitrotoga arctica” was inhibited by high nitrite concentrations, the substrate concentrations were reduced to 0.3 and 0.7 mM nitrite, respectively. In contrast, the Nitrobacter cultures and “Ca. Nitrospira defluvii” grew well with 9 mM nitrite and N. moscoviensis grew well with 5.7 mM nitrite. For Nitrobacter, minimum generation times (g) of exponentially growing cells (Table 1) between 13 h (N. vulgaris) and 43 h (N. hamburgensis) were obtained. The generation times of the Nitrospira cultures ranged from 32 h (N. moscoviensis) to 37 h (“Ca. Nitrospira defluvii,” strain BS10), and the slowest growth was that of “Ca. Nitrotoga arctica” (g = 44 h). Cell activities (k) were calculated during the exponential growth phase by simultaneous determination of cell counts and nitrite oxidation rates (Table 1). The results are, by tendency, consistent with the Vmax values measured via oxygen consumption (see below). N. vulgaris showed the highest cell activity (13.1 fmol of NO2− cell−1 h−1), and N. moscoviensis showed the lowest (0.6 fmol of NO2− cell−1 h−1). Nitrite-dependent growth yields of produced protein (Yp) and cell concentrations (Yc) are listed in Table 1. In general, Nitrospira cultures (0.122 to 0.213 mg of protein mmol−1 nitrite) revealed higher yields of nitrite conversion for protein synthesis than Nitrobacter (0.083 to 0.108 mg of protein mmol−1 nitrite) and Nitrotoga (0.102 mg of protein mmol−1 nitrite) cultures, but all of the species produced comparable cell amounts per unit of nitrite (Table 1).
FIG 1.
Growth of NOB in nitrite-limited mineral medium as batch cultures. Nitrobacter vulgaris, Nitrobacter hamburgensis, Nitrobacter winogradskyi, “Ca. Nitrospira defluvii,” Nitrospira moscoviensis, and Nitrospira strain BS10 were pure cultures, and “Ca. Nitrotoga arctica” was an enrichment culture. The initial provided nitrite was set to noninhibiting concentrations. After complete depletion of nitrite, the cultures entered the stationary phase. The time points at which generation times were calculated are marked by filled triangles.
TABLE 1.
Growth parameters of selected NOB cultures
| Organism (temp [°C]) | Generation time (g)j | Max cell sp act (k)k | Growth yield |
|
|---|---|---|---|---|
| Protein (Yp)l | Cells (Yc)m | |||
| Nitrobacter vulgarisa (28) | 13 (12)d | 13.1 | 0.099 | 10.32 |
| Nitrobacter hamburgensisa (28) | 43 (40, 63–84)e | 2.9 (3.7–7.4, 1.0–3.3)c,f | 0.108 | 9.95 |
| Nitrobacter winogradskyia (28) | 26 (8–14, 41–87)g | 2.5 (1.9–3.7, 12)h | 0.083 | 10.26 |
| “Ca. Nitrospira defluvii”a (28) | 37 | 2.8 | 0.122 | 9.93 |
| Nitrospira moscoviensisa (37) | 32 (12)i | 0.6 | 0.213 (0.120)i | 10.55 |
| Nitrospira strain BS10a (28) | 37 | 2.1 | 0.142 | 10.4 |
| “Ca. Nitrotoga arctica”b (17) | 44 | 2 | 0.102 | 10.14 |
Oxidation kinetics.
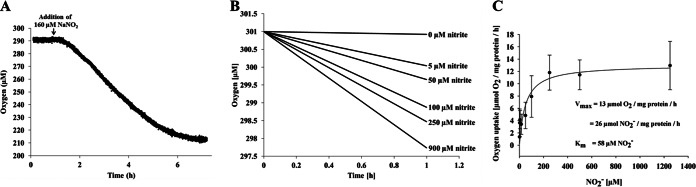
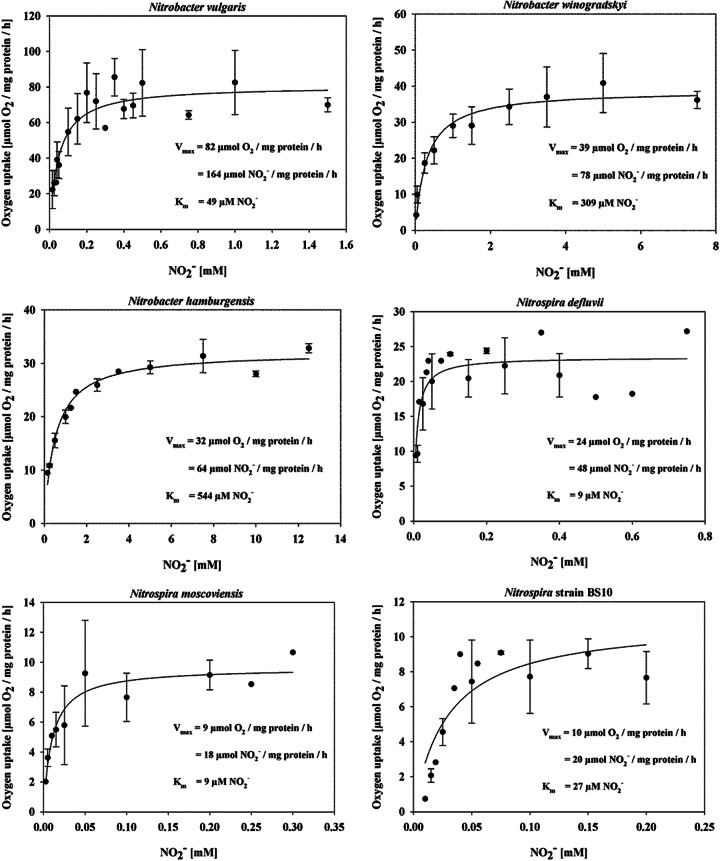
Key parameters of nitrite oxidation kinetics, maximum specific activities (Vmax), and half-saturation constants (Km), were measured by microsensor-based oxygen consumption of the Nitrotoga enrichment (Fig. 2) and the six NOB isolates (Fig. 3; Table 2). Nitrite-depleted, early-stationary-phase cells of all of the cultures consumed very low concentrations of oxygen and started O2 uptake rapidly, within a few minutes after nitrite addition (Fig. 2A). The oxygen consumption rates were dependent on the nitrite concentrations provided (Fig. 2B) and followed Michaelis-Menten kinetics (Fig. 2C). The highest maximum oxidation activities obtained were those of members of the genus Nitrobacter. N. vulgaris from activated sludge showed by far the highest activity (164 ± 9 μmol of NO2− mg of protein−1 h−1), followed by soil-derived N. winogradskyi and N. hamburgensis (78 ± 5 and 64 ± 1 μmol of NO2− mg of protein−1 h−1, respectively). All of the Nitrospira strains studied reached lower maximum activities, though lineage I “Ca. Nitrospira defluvii” (48 ± 2 μmol of NO2− mg of protein−1 h−1) enriched (37) and later isolated from activated sludge (Nowka et al., unpublished data) reached nitrite consumption rates similar to those of Nitrobacter strains from soil. Both of the lineage II Nitrospira isolates investigated, N. moscoviensis from heating water and strain BS10 from activated sludge, showed less than half of this maximum activity. “Ca. Nitrotoga arctica” (26 ± 3 μmol of NO2− mg of protein−1 h−1) oxidized nitrite with a slightly higher maximum activity than both lineage II Nitrospira isolates.
FIG 2.
Nitrite oxidation kinetics of “Ca. Nitrotoga arctica.” (A) Nitrite-dependent oxygen uptake of early-stationary-phase cells at 17°C. After equilibration of the cell aliquot and a stable oxygen concentration signal for ∼1 h, the experiment was started by the addition of 160 μM NaNO2. (B) Nitrite-dependent oxygen uptake of early-stationary-phase cells at 17°C at the nitrite concentrations shown. (C) Michaelis-Menten plot of oxygen uptake according to the nitrite concentration. The experiments were performed with early-stationary-phase cells at 17°C. Average values and standard deviations were calculated from 3-fold measurements. The kinetic parameters were calculated by fitting a Michaelis-Menten equation to the data.
FIG 3.
Nitrite oxidation kinetics of NOB. Michaelis-Menten plots of oxygen uptake at various nitrite concentrations are shown. Experiments were performed with early-stationary-phase cultures at defined incubation temperatures of 28°C (Nitrobacter vulgaris, Nitrobacter hamburgensis, Nitrobacter winogradskyi, “Ca. Nitrospira defluvii,” Nitrospira strain BS10) and 37°C (Nitrospira moscoviensis). Average values and standard deviations were calculated from 3-fold measurements. The kinetic parameters were calculated by fitting a Michaelis-Menten equation to the data. See experimental details and calculations in Materials and Methods.
TABLE 2.
Kinetic constants of selected NOB cultures
| Organism (temp [°C]) | Mean max sp actf ± SD | Mean saturation constant for activityg ± SD |
|---|---|---|
| Nitrobacter vulgarisa (28) | 164 ± 9 | 49 ± 11 |
| Nitrobacter hamburgensisa (28) | 64 ± 1 | 544 ± 55 (540–1,370, 706–1,240)c,d |
| Nitrobacter winogradskyia (28) | 78 ± 5 | 309 ± 92 (36–260, 170–1,380)e |
| “Ca. Nitrospira defluvii”a (28) | 48 ± 2 | 9 ± 3 |
| Nitrospira moscoviensisa (37) | 18 ± 1 | 9 ± 3 |
| Nitrospira strain BS10a (28) | 20 ± 2 | 27 ± 11 |
| “Ca. Nitrotoga arctica”b (17) | 26 ± 3 | 58 ± 28 |
A comparison of the Km values also revealed a clear differentiation of nitrite affinities between the NOB investigated. “Ca. Nitrospira defluvii” and N. moscoviensis had by far the greatest affinity for nitrite of the seven NOB investigated (both 9 ± 3 μM NO2−), whereas strain BS10 featured a Km value three times as high (27 ± 11 μM NO2−). Of the three Nitrobacter strains investigated, N. vulgaris showed the best adaption to low nitrite concentrations (49 ± 11 μM NO2−), though its affinity was still 1.8 to 5.4 times lower than that of the Nitrospira species. The lowest affinities measured were those of N. hamburgensis and N. winogradskyi (544 ± 55 and 309 ± 92 μM NO2−, respectively). The nitrite affinity of “Ca. Nitrotoga arctica” (58 ± 28 μM NO2−) was comparable to that of N. vulgaris, in the midrange of the NOB studied.
We compared the Km values we determined with those of other studies (Fig. 4), which demonstrated that the affinities of NOB for nitrite differ over 2 orders of magnitude. The highest affinities found were those of N. moscoviensis retrieved from heating water and “Ca. Nitrospira defluvii” obtained from activated sludge, whereas the soil-derived NOB had relatively high Km values, with “Ca. Nitrotoga arctica” being most competitive representative. By tendency, the affinities of NOB originating from activated sludge and freshwater were higher than those of NOB from soil.
FIG 4.
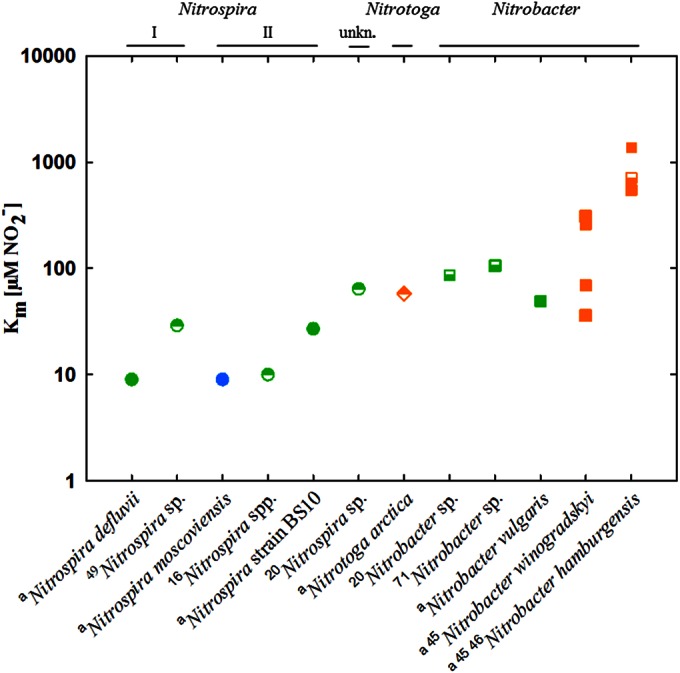
Nitrite affinities of NOB. Km values of members of the genera Nitrospira (circles), Nitrotoga (diamond), and Nitrobacter (squares) are shown (pure cultures, filled symbols; enrichment cultures, half-filled symbols). Cultures were derived from activated sludge (green), freshwater (blue), and soil (orange). Phylogenetic affiliations, including Nitrospira lineages I and II, are displayed at the top (unkn., unknown). The values shown were obtained from this study (indicated by a superscript letter a) and references 16, 20, 45, 46, 49, and 71 (indicated by superscript numbers).
DISCUSSION
Knowledge of the community and activity of nitrifying bacteria is essential to improve operation strategies and design biotechnological processes in WWTPs (6). Ammonia oxidation, as the initial step of nitrification, is catalyzed by AOB and ammonia-oxidizing archaea, which revealed significantly different affinities for ammonia (43). Nitrification kinetics measured by respirometry often lack information about nitrite oxidation (9), and the range of nitrite affinities of NOB besides Nitrobacter is still a black box. Our goal was to generate kinetic data for most of the known NOB species of the genera Nitrobacter, Nitrospira, and Nitrotoga to close this gap. Although scientific interest in activated-sludge modeling may be declining (44), the behavior of single organisms in activated-sludge systems might enhance the understanding of such processes.
Reported key growth parameters of pure NOB cultures like the maximum specific activity (Vmax) and the half-saturation constant for nitrite (Km) are known mainly for Nitrobacter (1, 45, 46). The current data on Nitrospira were determined with enrichments but not with pure cultures (16, 20). Since the oxidation kinetics of cells are affected by different growth conditions, values can fluctuate greatly (45, 46) and shift through different growth phases with changing substrate concentrations (47). Therefore, the aim of this study was to determine the nitrite oxidation kinetics of NOB under standardized laboratory conditions, adjusting only strain-specific temperatures and substrate preferences. We are aware that these batch culture experiments rely on artificial growth conditions with the disadvantage of changing growth conditions (e.g., substrate concentration, cell density) in the system over time (48). Since the natural conditions of the NOB studied most are probably more complex and diverse, the results obtained should be seen as one vital component for the determination of NOB kinetics.
Nitrite oxidation kinetics.
Previous studies demonstrated that the availability of nitrite is an important factor for the competition of NOB, suggesting different nitrite oxidation kinetics for members of the genera Nitrobacter and Nitrospira (16, 18–20). Moreover, coexisting Nitrospira populations revealed distinct preferences for nitrite (49). These findings indicate that substrate availability may account for the distribution of the different NOB in natural and artificial ecosystems. The comparison of Vmax and Km values for nonmarine NOB strains, in addition to results from the literature, strongly supports this assumption (Table 2 and Fig. 4), revealing specific adaptations for Nitrospira, Nitrobacter, Nitrotoga, and their sublineages, and a distinct clustering of populations obtained from different ecosystems.
Nitrospira.
Members of the genus Nitrospira showed by far the highest affinities of the NOB investigated here. With a Km value of 9 ± 3 μM nitrite, “Ca. Nitrospira defluvii” and N. moscoviensis had the lowest half-saturation constant determined for Nitrospira so far. Interestingly, these species belong to phylogenetically separate Nitrospira lineages I and II, which are widely distributed in natural habitats and were reported to be the main nitrite oxidizers in man-made ecosystems like WWTPs (12, 37, 50). This dominance can be explained by their advantage of oxidizing nitrite with high affinity and an energetically favorable periplasmic nitrite oxidoreductase (NXR) (51). It is interesting that their Km values often resemble in situ nitrite concentrations. In the WWTP, where “Ca. Nitrospira defluvii” and strain BS10 originated, nitrite concentrations of 7 to 8 μM in activated-sludge samples (52) and 21 to 571 μM in sewage influents (Hamburg Wasser, personal communication, 2014) were measured. Nitrospira bacteria have commonly been detected in oligotrophic ecosystems such as subsurface fluids (53) and freshwaters with nitrite at nanomolar concentrations and below the detection limit, respectively (54, 55). These findings and the oxidation kinetics measured in this study support the suggested classification of Nitrospira bacteria as K strategists (16). Intriguingly, Nitrospira bacteria are not restricted to environments with low nitrite concentrations but can also be found at millimolar concentrations reported in subsurface fluids (56) or sequencing batch reactors (12). If such elevated nitrite levels are only transient, “Ca. Nitrospira defluvii” competes with the low-affinity r strategists of the genus Nitrobacter. However, continuously high nitrite concentrations were shown to select for Nitrobacter (19, 57).
Comparing the three Nitrospira species studied, lineage II N. moscoviensis and strain BS10 revealed considerably lower maximum activities than lineage I “Ca. Nitrospira defluvii,” which supports the hypothesis of Maixner et al. (49) that lineage I Nitrospira bacteria outcompete lineage II Nitrospira bacteria at high nitrite concentrations. Interestingly, the Vmax of “Ca. Nitrospira defluvii” was much higher than that of N. moscoviensis, although the experimental temperature was 9°C lower. This finding suggests a high efficiency or a higher expression of the NXR inside “Ca. Nitrospira defluvii.”
In contrast to the maximum activities, the nitrite affinities determined allow no differentiation between the Nitrospira strains from different phylogenetic lineages tested. The indicated advantage of “Ca. Nitrospira defluvii” over strain BS10 at low as well as high nitrite concentrations raises the question of whether there are other selective factors promoting the growth of strain BS10 in wastewater treatment, such as pH, affinity for oxygen, availability of organics, and temperature.
Nitrobacter.
While Nitrobacter species showed the highest maximum activities (Vmax) of NOB, their affinities for the substrate nitrite are generally low. One reason besides the kinetics of the NXR might be low affinities of the transporters that are needed to shuttle nitrite across the cytoplasmic membrane. There are narK homologs identified in the genomes of N. hamburgensis and N. winogradskyi that could function as nitrite/nitrate transporters (58, 59), but their kinetics await experimental clarification. However, the Km values for nitrite indicate a great diversity of Nitrobacter ecotypes, which is consistent with a large genetic heterogeneity within Nitrobacter communities (60, 61). In this study, N. vulgaris derived from activated sludge revealed the highest nitrite affinity and maximum activity of the species of this genus investigated. The low nitrite affinities of soil-derived isolates of both N. winogradskyi and N. hamburgensis indicate that environmental factors other than nitrite concentration may account for Nitrobacter distribution. It was recently demonstrated that Nitrospira dominated in nutrient-limited environments such as surface soil (62), whereas Nitrobacter preferred a high supply of organic matter like that present in the rhizosphere or after fertilization (63, 64). Indeed, the potential of growing mixotrophically and chemoorganotrophically (65, 66) and even anaerobically via nitrate reduction (67) gives Nitrobacter a versatile metabolism, which may compensate for its low affinity for nitrite.
Nitrotoga.
The nitrite oxidation kinetics of “Ca. Nitrotoga arctica” bacteria from permafrost soil revealed that they are rather K strategists like Nitrospira. Both NOB are characterized by a wide periplasmic space, which contains the NXR in Nitrospira (51, 68) and in Nitrotoga (E. Spieck, unpublished data). The maximum specific activity was slightly higher than that of N. moscoviensis and strain BS10 but lower than the activity of other NOB, especially the soil-derived Nitrobacter strains. The Km value of Nitrotoga connects the affinity ranges of Nitrospira and Nitrobacter (Fig. 4) and indicates better adaption of “Ca. Nitrotoga arctica” than of the soil-inhabiting Nitrobacter strains to low nitrite concentrations. However, it should be considered that the measurements of Nitrotoga kinetics were performed with an enrichment culture and the presence of heterotrophic bacteria (despite a very low number) could have supported the activity and growth of “Ca. Nitrotoga arctica.” Therefore, final conclusions have to await the isolation of these NOB and the availability of pure cultures to confirm the results. The preferred incubation temperature of 17°C, in contrast to the higher preferred incubation temperatures of the other cultures (28 to 37°C), shows why Nitrotoga is apparently widely distributed in cold-affected environments (24, 26), in particular, in pristine soils (69) or freshwater systems (25). Nevertheless, Nitrotoga is not restricted to nutrient-limiting habitats, since it is also abundant in sewage (27, 29). Nitrotoga overgrew Nitrospira and Nitrobacter during long-term cultivation at 5 and 10°C, respectively (27, 70), but extended competition experiments with Nitrotoga and the other genera of NOB are missing so far and will be a subject for the future.
ACKNOWLEDGMENTS
This research was funded by the Vienna Science and Technology Fund, WWTF (project LS09-40), and the German Research Foundation, DFG (project SP 667/10-1).
We thank Hamburg Wasser for support.
REFERENCES
- 1.Prosser JI. 1989. Autotrophic nitrification in bacteria. Adv Microb Physiol 30:125–181. [DOI] [PubMed] [Google Scholar]
- 2.Van Cleemput O, Samater AH. 1996. Nitrite in soils: accumulation and role in the formation of gaseous N compounds. Fertilizer Res 45:81–89. [Google Scholar]
- 3.Graham DW, Knapp CW, van Vleck ES, Bloor K, Lane TB, Graham CE. 2007. Experimental demonstration of chaotic instability in biological nitrification. ISME J 1:385–393. doi: 10.1038/ismej.2007.45. [DOI] [PubMed] [Google Scholar]
- 4.Su H, Cheng Y, Oswald R, Behrendt T, Trebs I, Meixner FX, Andreae MO, Cheng P, Zhang Y, Pöschl U. 2011. Soil nitrite as a source of atmospheric HONO and OH radicals. Science 333:1616–1618. doi: 10.1126/science.1207687. [DOI] [PubMed] [Google Scholar]
- 5.Both GJ, Laanbroek HJ. 1991. The effect of the incubation period on the result of MPN enumerations of nitrite-oxidizing bacteria: theoretical considerations. FEMS Microbiol Lett 85:335–344. doi: 10.1111/j.1574-6968.1991.tb04760.x. [DOI] [Google Scholar]
- 6.Okabe S, Aoi Y, Satoh H, Suwa Y. 2011. Nitrification in wastewater treatment, p 405–433. In Ward BB, Arp DJ, Klotz MG (ed), Nitrification. ASM Press, Washington, DC. [Google Scholar]
- 7.Koops HP, Pommerening-Röser A. 2001. Distribution and ecophysiology of the nitrifying bacteria emphasizing cultured species. FEMS Microbiol Ecol 37:1–9. doi: 10.1111/j.1574-6941.2001.tb00847.x. [DOI] [Google Scholar]
- 8.Chandran K, Smets BF. 2005. Optimizing experimental design to estimate ammonia and nitrite oxidation biokinetic parameters from batch respirograms. Water Res 39:4969–4978. doi: 10.1016/j.watres.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 9.Bouskill NJ, Tang J, Riley WJ, Brodie EL. 2012. Trait-based representation of biological nitrification: model development, testing, and predicted community composition. Front Microbiol 3:364. doi: 10.3389/fmicb.2012.00364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henze M, Harremoes P, LaCour J, Jansen J, Arvin E (ed). 1997. Wastewater treatment. Biological and chemical processes, 2nd ed. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 11.Juretschko S, Timmermann G, Schmid M, Schleifer KH, Pommerening-Röser A, Koops HP, Wagner M. 1998. Combined molecular and conventional analyses of nitrifying bacterium diversity in activated sludge: Nitrosococcus mobilis and Nitrospira-like bacteria as dominant populations. Appl Environ Microbiol 64:3042–3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daims H, Nielsen JL, Nielsen PH, Schleifer KH, Wagner M. 2001. In situ characterization of Nitrospira-like nitrite-oxidizing bacteria active in wastewater treatment plants. Appl Environ Microbiol 67:5273–5284. doi: 10.1128/AEM.67.11.5273-5284.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watson SW, Bock E, Valois FW, Waterbury JB, Schlosser U. 1986. Nitrospira marina gen. nov. sp. nov.: a chemolithotrophic nitrite-oxidizing bacterium. Arch Microbiol 144:1–7. doi: 10.1007/BF00454947. [DOI] [Google Scholar]
- 14.Ehrich S, Behrens D, Lebedeva E, Ludwig W, Bock E. 1995. A new obligately chemolithoautotrophic, nitrite-oxidizing bacterium, Nitrospira moscoviensis sp. nov. and its phylogenetic relationship. Arch Microbiol 164:16–23. [DOI] [PubMed] [Google Scholar]
- 15.Okabe S, Satoh H, Watanabe Y. 1999. In situ analysis of nitrifying biofilms as determined by in situ hybridization and the use of microelectrodes. Appl Environ Microbiol 65:3182–3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schramm A, de Beer D, van den Heuvel JC, Ottengraf S, Amann R. 1999. Microscale distribution of populations and activities of Nitrosospira and Nitrospira spp. along a macroscale gradient in a nitrifying bioreactor: quantification by in situ hybridization and the use of microsensors. Appl Environ Microbiol 65:3690–3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andrews JH, Harris RF. 1986. r- and K-selection and microbial ecology. Adv Microb Ecol 9:99–147. doi: 10.1007/978-1-4757-0611-6_3. [DOI] [Google Scholar]
- 18.Kim DJ, Kim SH. 2006. Effect of nitrite concentration on the distribution and competition of nitrite-oxidizing bacteria in nitratation reactor systems and their kinetic characteristics. Water Res 40:887–894. doi: 10.1016/j.watres.2005.12.023. [DOI] [PubMed] [Google Scholar]
- 19.Nogueira R, Melo LF. 2006. Competition between Nitrospira spp. and Nitrobacter spp. in nitrite-oxidizing bioreactors. Biotechnol Bioeng 95:169–175. doi: 10.1002/bit.21004. [DOI] [PubMed] [Google Scholar]
- 20.Blackburne R, Vadivelu VM, Yuan Z, Keller J. 2007. Kinetic characterisation of an enriched Nitrospira culture with comparison to Nitrobacter. Water Res 41:3033–3042. doi: 10.1016/j.watres.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 21.Hovanec TA, Taylor LT, Blakis A, DeLong EF. 1998. Nitrospira-like bacteria associated with nitrite oxidation in freshwater aquaria. Appl Environ Microbiol 64:258–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freitag TE, Chang L, Clegg CD, Prosser JI. 2005. Influence of inorganic nitrogen management regime on the diversity of nitrite-oxidizing bacteria in agricultural grassland soils. Appl Environ Microbiol 71:8323–8334. doi: 10.1128/AEM.71.12.8323-8334.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lebedeva EV, Alawi M, Fiencke C, Namsaraev B, Bock E, Spieck E. 2005. Moderately thermophilic nitrifying bacteria from a hot spring of the Baikal rift zone. FEMS Microbiol Ecol 54:297–306. doi: 10.1016/j.femsec.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 24.Alawi M, Lipski A, Sanders T, Pfeiffer EM, Spieck E. 2007. Cultivation of a novel cold-adapted nitrite oxidizing betaproteobacterium from the Siberian Arctic. ISME J 1:256–264. doi: 10.1038/ismej.2007.34. [DOI] [PubMed] [Google Scholar]
- 25.Chen Y, Wu L, Boden R, Hillebrand A, Kumaresan D, Moussard H, Baciu M, Lu Y, Murrell CJ. 2009. Life without light: microbial diversity and evidence of sulfur- and ammonium-based chemolithotrophy in Movile Cave. ISME J 3:1093–1104. doi: 10.1038/ismej.2009.57. [DOI] [PubMed] [Google Scholar]
- 26.White CP, Debry RW, Lytle DA. 2012. Microbial survey of a full-scale, biologically active filter for treatment of drinking water. Appl Environ Microbiol 78:6390–6394. doi: 10.1128/AEM.00308-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alawi M, Off S, Kaya M, Spieck E. 2009. Temperature influences the population structure of nitrite-oxidizing bacteria in activated sludge. Environ Microbiol Rep 1:184–190. doi: 10.1111/j.1758-2229.2009.00029.x. [DOI] [PubMed] [Google Scholar]
- 28.Benedek T, Tancsics A, Szilagyi N, Toth I, Farkas M, Szoboszlay S, Krifaton C, Hartman M, Kriszt B. 2014. Analysis of biofilm bacterial communities responsible for carbon removal through a reactor cascade treating wastewater. World J Microbiol Biotechnol 30:977–987. doi: 10.1007/s11274-013-1516-9. [DOI] [PubMed] [Google Scholar]
- 29.Lücker S, Schwarz J, Gruber-Dorninger C, Spieck E, Wagner M, Daims H. 2 September 2014. Nitrotoga-like bacteria are previously unrecognized key nitrite oxidizers in full-scale wastewater treatment plants. ISME J doi: 10.1038/ismej.2014.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winogradsky S. 1890. Recherches sur les organismes de la nitrification. Ann Inst Pasteur 4:213–231. [Google Scholar]
- 31.Watson SW, Waterbury JB. 1971. Characteristics of two marine nitrite oxidizing bacteria, Nitrospina gracilis nov. gen. nov. sp. and Nitrococcus mobilis nov. gen. nov. sp. Arch Mikrobiol 77:203–230. [Google Scholar]
- 32.Sorokin DY, Lucker S, Vejmelkova D, Kostrikina NA, Kleerebezem R, Rijpstra WI, Damste JS, Le Paslier D, Muyzer G, Wagner M, van Loosdrecht MC, Daims H. 2012. Nitrification expanded: discovery, physiology and genomics of a nitrite-oxidizing bacterium from the phylum Chloroflexi. ISME J 6:2245–2256. doi: 10.1038/ismej.2012.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gujer W, Henze M, Mino T, van Loosdrecht M. 1999. Activated sludge model no. 3. Water Sci Technol 39:183–193. doi: 10.1016/S0273-1223(98)00785-9. [DOI] [Google Scholar]
- 34.Sundermeyer H, Bock E. 1981. Energy metabolism of autotrophically and heterotrophically grown cells of Nitrobacter winogradskyi. Arch Microbiol 130:250–254. doi: 10.1007/BF00459529. [DOI] [Google Scholar]
- 35.Bock E, Sundermeyer-Klinger H, Stackebrandt E. 1983. New facultative lithoautotrophic nitrite-oxidizing bacteria. Arch Microbiol 136:281–284. doi: 10.1007/BF00425217. [DOI] [Google Scholar]
- 36.Bock E, Koops HP, Möller UC, Rudert M. 1990. A new facultatively nitrite oxidizing bacterium, Nitrobacter vulgaris sp. nov. Arch Microbiol 153:105–110. doi: 10.1007/BF00247805. [DOI] [Google Scholar]
- 37.Spieck E, Hartwig C, McCormack I, Maixner F, Wagner M, Lipski A, Daims H. 2006. Selective enrichment and molecular characterization of a previously uncultured Nitrospira-like bacterium from activated sludge. Environ Microbiol 8:405–415. doi: 10.1111/j.1462-2920.2005.00905.x. [DOI] [PubMed] [Google Scholar]
- 38.Widdel F, Bak F. 1992. Gram-negative mesophilic sulfate-reducing bacteria, p 3352–3378. In Balows A, Tr̈uper HG, Dworkin M, Harder W, Schleifer K-H (ed), The prokaryotes, 2nd ed. Springer, New York, NY. [Google Scholar]
- 39.Meincke M, Bock E, Kastrau D, Kroneck PMH. 1992. Nitrite oxidoreductase from Nitrobacter hamburgensis: redox centers and their catalytic role. Arch Microbiol 158:127–131. doi: 10.1007/BF00245215. [DOI] [Google Scholar]
- 40.Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. 1985. Measurement of protein using bicinchoninic acid. Anal Biochem 150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 41.Gundersen JK, Ramsing NB, Glud RN. 1998. Predicting the signal of O2 microsensors from physical dimensions, temperature, salinity, and O2 concentration. Limnol Oceanogr 43:1932–1937. [Google Scholar]
- 42.Belser LW, Schmidt EL. 1980. Growth and oxidation kinetics of three genera of ammonia oxidizing nitrifiers. FEMS Microbiol Lett 7:213–216. doi: 10.1111/j.1574-6941.1980.tb01628.x. [DOI] [Google Scholar]
- 43.Martens-Habbena W, Berube PM, Urakawa H, de la Torre JR, Stahl DA. 2009. Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature 461:976–979. doi: 10.1038/nature08465. [DOI] [PubMed] [Google Scholar]
- 44.Gujer W. 2006. Activated sludge modelling: past, present and future. Water Sci Technol 53:111–119. doi: 10.2166/wst.2006.082. [DOI] [PubMed] [Google Scholar]
- 45.Both GJ, Gerards S, Laanbroek HJ. 1992. Kinetics of nitrite oxidation in two Nitrobacter species grown in nitrite-limited chemostats. Arch Microbiol 157:436–441. doi: 10.1007/BF00249101. [DOI] [Google Scholar]
- 46.Laanbroek HJ, Bodelier PLE, Gerards S. 1994. Oxygen consumption kinetics of Nitrosomonas europaea and Nitrobacter hamburgensis grown in mixed continuous cultures at different oxygen concentrations. Arch Microbiol 161:156–162. doi: 10.1007/BF00276477. [DOI] [Google Scholar]
- 47.Martens-Habbena W, Stahl DA. 2011. Nitrogen metabolism and kinetics of ammonia-oxidizing archaea. Methods Enzymol 496:465–487. doi: 10.1016/B978-0-12-386489-5.00019-1. [DOI] [PubMed] [Google Scholar]
- 48.Kovárová-Kovar K, Egli T. 1998. Growth kinetics of suspended microbial cells: from single-substrate-controlled growth to mixed-substrate kinetics. Microbiol Mol Biol Rev 62:646–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maixner F, Noguera DR, Anneser B, Stoecker K, Wegl G, Wagner M, Daims H. 2006. Nitrite concentration influences the population structure of Nitrospira-like bacteria. Environ Microbiol 8:1487–1495. doi: 10.1111/j.1462-2920.2006.01033.x. [DOI] [PubMed] [Google Scholar]
- 50.Pester M, Maixner F, Berry D, Rattei T, Koch H, Lücker S, Nowka B, Richter A, Spieck E, Lebedeva E, Loy A, Wagner M, Daims H. 2014. NxrB encoding the beta subunit of nitrite oxidoreductase as functional and phylogenetic marker for nitrite-oxidizing Nitrospira. Environ Microbiol 16:3055–3071. doi: 10.1111/1462-2920.12300. [DOI] [PubMed] [Google Scholar]
- 51.Lücker S, Wagner M, Maixner F, Pelletier E, Koch H, Vacherie B, Rattei T, Sinninghe Damsté J, Spieck E, Le Paslier D, Daims H. 2010. A Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. Proc Natl Acad Sci U S A 107:13479–13484. doi: 10.1073/pnas.1003860107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kruse M, Zumbrägel S, Bakker E, Spieck E, Eggers T, Lipski A. 2013. The nitrite-oxidizing community in activated sludge from a municipal wastewater treatment plant determined by fatty acid methyl ester-stable isotope probing. Syst Appl Microbiol 36:517–524. doi: 10.1016/j.syapm.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 53.Swanner ED, Templeton AS. 2011. Potential for nitrogen fixation and nitrification in the granite-hosted subsurface at Henderson Mine, CO. Front Microbiol 2:254. doi: 10.3389/fmicb.2011.00254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martiny AC, Albrechtsen HJ, Arvin E, Molin S. 2005. Identification of bacteria in biofilm and bulk water samples from a nonchlorinated model drinking water distribution system: detection of a large nitrite-oxidizing population associated with Nitrospira spp. Appl Environ Microbiol 71:8611–8617. doi: 10.1128/AEM.71.12.8611-8617.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mußmann M, Ribot M, von Schiller D, Merbt SN, Augspurger C, Karwautz C, Winkel M, Battin TJ, Marti E, Daims H. 2013. Colonization of freshwater biofilms by nitrifying bacteria from activated sludge. FEMS Microbiol Ecol 85:104–115. doi: 10.1111/1574-6941.12103. [DOI] [PubMed] [Google Scholar]
- 56.Gihring TM, Moser DP, Lin L-H, Davidson M, Onstott TC, Morgan L, Milleson M, Kieft TL, Trimarco E, Balkwill DL, Dollhopf ME. 2006. The distribution of microbial taxa in the subsurface water of the Kalahari Shield, South Africa. Geomicrobiol J 23:415–430. doi: 10.1080/01490450600875696. [DOI] [Google Scholar]
- 57.Bartosch S, Wolgast I, Spieck E, Bock E. 1999. Identification of nitrite-oxidizing bacteria with monoclonal antibodies recognizing the nitrite oxidoreductase. Appl Environ Microbiol 65:4126–4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Starkenburg SR, Chain PS, Sayavedra-Soto LA, Hauser L, Land ML, Larimer FW, Malfatti SA, Klotz MG, Bottomley PJ, Arp DJ, Hickey WJ. 2006. Genome sequence of the chemolithoautotrophic nitrite-oxidizing bacterium Nitrobacter winogradskyi Nb-255. Appl Environ Microbiol 72:2050–2063. doi: 10.1128/AEM.72.3.2050-2063.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Starkenburg SR, Larimer FW, Stein LY, Klotz MG, Chain PS, Sayavedra-Soto LA, Poret-Peterson AT, Gentry ME, Arp DJ, Ward B, Bottomley PJ. 2008. Complete genome sequence of Nitrobacter hamburgensis X14 and comparative genomic analysis of species within the genus Nitrobacter. Appl Environ Microbiol 74:2852–2863. doi: 10.1128/AEM.02311-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Grundmann GL, Normand P. 2000. Microscale diversity of the genus Nitrobacter in soil on the basis of analysis of genes encoding rRNA. Appl Environ Microbiol 66:4543–4546. doi: 10.1128/AEM.66.10.4543-4546.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vanparys B, Spieck E, Heylen K, Wittebolle L, Geets J, Boon N, De Vos P. 2007. The phylogeny of the genus Nitrobacter based on comparative rep-PCR, 16S rRNA and nitrite oxidoreductase gene sequence analysis. Syst Appl Microbiol 30:297–308. doi: 10.1016/j.syapm.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 62.Ke X, Angel R, Lu Y, Conrad R. 2013. Niche differentiation of ammonia oxidizers and nitrite oxidizers in rice paddy soil. Environ Microbiol 15:2275–2292. doi: 10.1111/1462-2920.12098. [DOI] [PubMed] [Google Scholar]
- 63.Attard E, Poly F, Commeaux C, Laurent F, Terada A, Smets BF, Recous S, Roux XL. 2010. Shifts between Nitrospira- and Nitrobacter-like nitrite oxidizers underlie the response of soil potential nitrite oxidation to changes in tillage practices. Environ Microbiol 12:315–326. doi: 10.1111/j.1462-2920.2009.02070.x. [DOI] [PubMed] [Google Scholar]
- 64.Wertz S, Leigh AK, Grayston SJ. 2012. Effects of long-term fertilization of forest soils on potential nitrification and on the abundance and community structure of ammonia oxidizers and nitrite oxidizers. FEMS Microbiol Ecol 79:142–154. doi: 10.1111/j.1574-6941.2011.01204.x. [DOI] [PubMed] [Google Scholar]
- 65.Smith AJ, Hoare DS. 1968. Acetate assimilation by Nitrobacter agilis in relation to its “obligate autotrophy.” J Bacteriol 95:844–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Steinmüller W, Bock E. 1976. Growth of Nitrobacter in the presence of organic matter. I. Mixotrophic growth. Arch Microbiol 108:299–304. [DOI] [PubMed] [Google Scholar]
- 67.Freitag A, Rudert M, Bock E. 1987. Growth of Nitrobacter by dissimilatoric nitrate reduction. FEMS Microbiol Lett 48:105–109. doi: 10.1111/j.1574-6968.1987.tb02524.x. [DOI] [Google Scholar]
- 68.Spieck E, Ehrich S, Aamand J, Bock E. 1998. Isolation and immunocytochemical location of the nitrite-oxidizing system in Nitrospira moscoviensis. Arch Microbiol 169:225–230. doi: 10.1007/s002030050565. [DOI] [PubMed] [Google Scholar]
- 69.Schmidt SK, Nemergut DR, Miller AE, Freeman KR, King AJ, Seimon A. 2009. Microbial activity and diversity during extreme freeze-thaw cycles in periglacial soils, 5400 m elevation, Cordillera Vilcanota, Peru. Extremophiles 13:807–816. doi: 10.1007/s00792-009-0268-9. [DOI] [PubMed] [Google Scholar]
- 70.Karkman A, Mattila K, Tamminen M, Virta M. 2011. Cold temperature decreases bacterial species richness in nitrogen-removing bioreactors treating inorganic mine waters. Biotechnol Bioeng 108:2876–2883. doi: 10.1002/bit.23267. [DOI] [PubMed] [Google Scholar]
- 71.Vadivelu VM, Yuan Z, Fux C, Keller J. 2006. Stoichiometric and kinetic characterisation of Nitrobacter in mixed culture by decoupling the growth and energy generation processes. Biotechnol Bioeng 94:1176–1188. doi: 10.1002/bit.20956. [DOI] [PubMed] [Google Scholar]
- 72.Watson WS, Bock E, Harms H, Koops HP, Hooper AB. 1989. Nitrifying bacteria, p 1808–1834. In Holt JG, Staley JT, Bryant MP, Pfennig N (ed), Bergey's manual of systematic bacteriology. Williams & Wilkins, Baltimore, MD. [Google Scholar]
- 73.Belser LW. 1979. Population ecology of nitrifying bacteria. Annu Rev Microbiol 33:309–333. doi: 10.1146/annurev.mi.33.100179.001521. [DOI] [PubMed] [Google Scholar]
- 74.Hunik JH, Meijer HJG, Tramper J. 1993. Kinetics of Nitrobacter agilis at extreme substrate, product and salt concentrations. Appl Microbiol Biotechnol 40:442–448. [Google Scholar]
- 75.Spieck E, Keuter S, Wenzel T, Bock E, Ludwig W. 2014. Characterization of a new marine nitrite oxidizing bacterium, Nitrospina watsonii sp. nov., a member of the newly proposed phylum “Nitrospinae.” Syst Appl Microbiol 37:170–176. [DOI] [PubMed] [Google Scholar]