Abstract
Brucella is an expanding genus of major zoonotic pathogens, including at least 10 genetically very close species occupying a wide range of niches from soil to wildlife, livestock, and humans. Recently, we have shown that in the new species Brucella microti, the glutamate decarboxylase (Gad)-dependent system (GAD system) contributes to survival at a pH of 2.5 and also to infection in mice by the oral route. In order to study the functionality of the GAD system in the genus Brucella, 47 isolates, representative of all known species and strains of this genus, and 16 strains of the closest neighbor genus, Ochrobactrum, were studied using microbiological, biochemical, and genetic approaches. In agreement with the genome sequences, the GAD system of classical species was not functional, unlike that of most strains of Brucella ceti, Brucella pinnipedialis, and newly described species (B. microti, Brucella inopinata BO1, B. inopinata-like BO2, and Brucella sp. isolated from bullfrogs). In the presence of glutamate, these species were more acid resistant in vitro than classical terrestrial brucellae. Expression in trans of the gad locus from representative Brucella species in the Escherichia coli MG1655 mutant strain lacking the GAD system restored the acid-resistant phenotype. The highly conserved GAD system of the newly described or atypical Brucella species may play an important role in their adaptation to acidic external and host environments. Furthermore, the GAD phenotype was shown to be a useful diagnostic tool to distinguish these latter Brucella strains from Ochrobactrum and from classical terrestrial pathogenic Brucella species, which are GAD negative.
INTRODUCTION
Brucellae are the etiologic agents of brucellosis, the most widespread bacterial zoonosis, infecting livestock and humans (human incidence, 500,000/year). The disease is endemic in the Mediterranean, in Near East and Middle East countries, and in Latin America (1, 2).
These pathogens are classified as different species on the basis of specific phenotypic traits and their natural hosts (3): Brucella melitensis (isolated from goats and sheep), Brucella abortus (cattle and bison), Brucella suis (pigs and wild boar), and Brucella canis (dogs). Transmitted to humans via the mucosal, cutaneous, respiratory or, most frequently, oral route, these species may induce undulant fever (Malta fever) and a wide range of clinical manifestations, including encephalitis and endocarditis. With a few exceptions, Brucella species isolated from marine mammals (Brucella pinnipedialis and Brucella ceti) have not been described as human pathogens. Two other species are nonpathogenic for humans: Brucella ovis (sheep) and Brucella neotomae (desert woodrat) (3). Being known for 20 years or longer, these eight species are considered “classical”. More recently, three “new” species/strains were identified: B. microti from common vole, red fox, and also soil (4–7) and Brucella inopinata BO1 and B. inopinata-like BO2 from patients with a breast implant infection (8, 9) and a chronic destructive pneumonia (10), respectively. Potentially novel Brucella spp., described as “atypical” because they do not match the phenotypical criteria defined for the classical species (like B. melitensis), were isolated from baboons (11), Australian rodents (12), and African bullfrogs (13). Thus, it has become increasingly clear that the genus Brucella comprises species which are able to occupy a vast range of niches from soil to wildlife animals, from domesticated livestock to humans.
Orally acquired bacteria, such as Escherichia coli, Shigella flexneri, and Listeria monocytogenes, possess the ability to withstand acidic environments, such as those found in fermented food, gastric juice, distal gut lumen, and the macrophagic phagosome. To counteract the deleterious effects of acid stress, the above-mentioned bacteria activate specific systems, among which the glutamate decarboxylase (Gad)-dependent system (GAD system) was shown to be by far the most effective (14–18). Upon exposure to an extremely low pH (mimicking that of the gastric compartment, ≤2.5), the GAD system becomes activated: a molecule of glutamate (Glu) is taken up via the inner membrane antiporter GadC and decarboxylated into γ-aminobutyrate (GABA) by the cytosolic enzyme Gad, with concomitant intracellular consumption of a proton; GadC promotes the export of GABA in exchange for an incoming Glu molecule (19), coupled to an electrogenic antiport (20). E. coli possesses two cytoplasmic isoforms of glutamate decarboxylase (GadA and GadB), and the gene coding for GadC is cotranscribed with gadB (15).
A comparative genome analysis of B. microti and several classical Brucella species revealed the occurrence of two sequences homologous to those of E. coli gadB and gadC, which contain in all cases except for B. microti either stop codons or frameshift mutations, generating truncated/potentially nonfunctional proteins (21). B. microti grows faster and is more resistant to pH 4.5 in vitro than classical Brucella strains, such as B. suis 1330 (22). The B. microti GAD system is fully active, i.e., it confers resistance to extreme acid stress (pH 2.5), and also contributes to infection transmitted through ingestion in mice (23).
In order to elucidate whether the occurrence of an operative GAD system correlates with a better adaptation to acidic environments, a collection of 47 strains, including all currently known species of the genus Brucella, was thoroughly investigated for GAD and GABA export activities. Furthermore, the survival of a subgroup of 17 strains among those that were GAD positive and negative was analyzed for acid resistance (or survival) at pH 2.5 in the presence of Glu. Finally, the role of the gadBC locus of these strains in glutamate-dependent acid resistance (AR) was corroborated by functional complementation of an acid-sensitive gadABC mutant of E. coli.
MATERIALS AND METHODS
Bacterial strains and media.
Sixty-nine bacterial strains were analyzed for the presence of GAD activity and for GABA export: 47 belong to Brucella, 16 to Ochrobactrum, and 6 to E. coli (see Table S1 in the supplemental material). Brucella and Ochrobactrum strains were grown in tryptic soy (TS) broth (Becton-Dickinson) and E. coli in Luria-Bertani (LB) broth (Difco) at 37°C, supplemented when appropriate with chloramphenicol and kanamycin at 25 and 50 μg/ml, respectively. B. pinnipedialis and B. ovis were cultured in a 5%-CO2 atmosphere and in TS medium with 10% fetal calf serum.
Construction and complementation of E. coli gad triple mutant strains.
A triple ΔgadA ΔgadBC mutant of the E. coli K-12 strain MG1655 was used for heterologous expression of the gadBC operon of Brucella species representative of the GAD-positive and GAD-negative species. This E. coli mutant strain was generated by using the one-step inactivation procedure of Datsenko and Wanner (24). In this specific case, two consecutive gene-specific inactivation steps using plasmid pKD13 as the template for amplification of the kanamycin cassette (Kanr) were performed. Briefly, a gadA mutant was constructed by replacement of its open reading frame (ORF) with Kanr and, after excision of this cassette, the entirety of gadBC was replaced with Kanr. Thus, to accomplish the mutation in E. coli, a linear DNA substrate was constructed by PCR amplification of the Kanr cassette-encoding gene using bipartite primers. Usually, these primers consist of 60 to 70 nucleotides (nt) to generate the gene replacement, with 40 to 50 nt of homology to the target region where the cassette has to be inserted, followed by 20 nt (highlighted in italics in sequences below) to prime the drug-resistant cassette in pKD13. The primers used in this study were the following: ΔgadA_for, 5′-TTCGAAATGGACCAGAAGCTGTTAACGGATTTCCGCTCATGTAGGCTGGAGCTGCTTC-3′; ΔgadA_rev, 5′-TCAGGTGTGTTTAAAGCTGTTCTGCTGGGCAATACCCTGATTCCGGGGATCCGTCGACC; ΔgadB_for, GATTTAAGGTCGGAACTACTCGATTCACGTTTTGGTGCGTGTAGGCTGGAGCTGCTTC-3′; ΔgadC_rev, 5′- TTAGTGTTTCTTGTCATTCATCACAATATAGTGTGGTGAAATTCCGGGGATCCGTCGACC-3′. Homologous and heterologous complementations of the triple mutant were obtained by transformation with the vector pBBR1MCS carrying the complete gadBC operon of E. coli or Brucella strains, each including its native promoter, as reported (23). A 3.6-kb DNA fragment of each Brucella species containing the gadB and gadC genes was amplified by PCR using the long-extend and high-fidelity Platinum Taq polymerase (Invitrogen) and the gadBC-op_PstI-For (GCCCTGCAGCCGAGCTTATTGCGCTAATATC) and gadBC-op_XbaI-Rev (GCCTCTAGAATCGCATCTGATGAGCTTGAC) primers (Sigma). In the above primers, the sequence in bold has no homology with the target genes and is included to increase stability of the 5′ end, which contains the restriction sites (underlined) for PstI and XbaI, respectively. The resulting clones were selected on the basis of their double resistance to Kan and Cm, the latter provided by pBBR1MCS.
Glutamate-dependent acid resistance assay.
Assays with Brucella spp. and E. coli wild-type and mutant strains were carried out in minimal media as previously described (15, 23), namely, modified Gerhardt's (GMM without glutamate) for Brucella spp. and Vogel Bonner's medium with 0.4% glucose (EG) for E. coli; both media were brought to pH 2.5 with HCl and used in the presence/absence of 1.5 mM Glu.
Qualitative and quantitative GAD assays and GABA measurements.
The qualitative GAD assay (25) was performed as described for E. coli and B. microti. For quantitative assay of GAD activity in E. coli strains, the cells from a 50-ml overnight culture grown in LB broth at pH 5, containing 0.4% glucose (LBG), were harvested by centrifugation and resuspended in 1 ml of a solution containing 1 mM pyridoxal 5′-phosphate, 1 mM dithiothreitol, and protease inhibitors (cOmplete Mini, EDTA free; Roche). Bacteria were lysed by sonication and then centrifuged at 12,000 rpm for 15 min at 4°C to remove cells debris. The cleared cell lysates were immediately assayed for protein content (26) and GAD activity (27). The concentration of GABA exported by E. coli and by Brucella in acidified medium was measured using a commercial biochemical GABase assay (Sigma), as described previously (15, 23).
RESULTS
An in silico analysis reveals potentially functional GadB and GadC proteins in several Brucella species.
Comparing the putative protein sequences of GadB and GadC (available from NCBI, Broad Institute, and PATRIC databases), several Brucella species, in addition to B. microti, were expected to have an active GAD system. These include B. ceti, B. inopinata BO1, B. inopinata-like BO2, and B. pinnipedialis. On the contrary, the analysis of the sequences homologous to those of the gadB and gadC genes in other classical Brucella species (B. abortus, B. canis, B. melitensis, B. neotomae, B. ovis, and B. suis) showed evidence of point or frameshift mutations. In gadB, most of the mutations fall within the nucleotide region coding for the large cofactor- and substrate-binding domain (19, 28), spanning from amino acid (aa) residue 58 to 346, referring to the numbering of E. coli GadB (Fig. 1 and Table 1; see also Table S1 and Fig. S1 in the supplemental material). The gadC sequences are much more conserved, and any change in the sequence affects the overall length of the polypeptide chain, which will result is shorter forms truncated in the N-terminal region, i.e., missing the first 42 to 117 aa in the sequence. These shorter polypeptides will likely be unable to localize in the membrane and perform an antiport (Fig. 1 and Table 1; see also Table S1 and Fig. S2), even in the presence of an active GadB.
FIG 1.
Schematic representation of the gadB (dark-gray arrow) and gadC (light-gray arrow) loci in the Brucella species and strains for which genome sequences (complete or partial) are available. The arrow length is proportional to the gene length, whereas the distance between the two genes is arbitrary, though always referring to two adjacent loci. The corresponding locus tags are shown above each arrow. Dashed interrupted arrows indicate the occurrence of frameshift mutations or stop codons. Squared arrows mark genes shorter than the canonical length for GadB (464 aa) and GadC (510 to 513 aa).
TABLE 1.
Representative Brucella strains for which the GAD phenotype (Rice test) was assayed and exported GABA was measureda
| Species and strain | Natural host | Protein length (aa) |
GAD Rice testb | Exported GABA (mM) | |
|---|---|---|---|---|---|
| GadB | GadC | ||||
| B. abortus | |||||
| ATCC 23448 bv. 1 (544) | Cattle | 155/286 | 510 | − | 0.00 ± 0.00 |
| ATCC 27565 bv. 1 (S19) | Cattle | 155/274 | 510 | − | 0.00 ± 0.00 |
| B. canis | |||||
| ATCC 23365 bv.1 | Dog | 319/136 | 426 | − | 0.00 ± 0.00 |
| B. ceti | |||||
| B1/94 bv.1 | Porpoise | 464 | 513 | + | 0.25 ± 0.00 |
| M490/95/1 | Common seal | 464 | 513 | + | 0.53 ± 0.01 |
| M644/93/1 | Common dolphin | 464 | 513 | + | 1.83 ± 0.37 |
| B202R | Minke whale | ND | ND | + | 0.26 ± 0.08 |
| B. inopinata | |||||
| BO1 | Human | 464 | 509 | + | 0.12 ± 0.04 |
| B. inopinata-like | |||||
| BO2 | Human | 464 | 471c | + | 0.02 ± 0.02 |
| B. melitensis | |||||
| ATCC 23456 bv. 1 (16 M) | Goat | 167/304 | 510 | − | 0.00 ± 0.00 |
| B. microti | |||||
| CCM4915 | Common vole | 464 | 510 | + | 0.20 ± 0.04 |
| CCM4916 | Common vole | ND | ND | + | 0.65 ± 0.09 |
| B. neotomae | |||||
| 5K33 | Desert woodrat | 263/201 | 510 | − | 0.00 ± 0.00 |
| B. ovis | |||||
| ATCC 25840 bv. 1 (63/290) | Sheep | 455 | 291/206 | +w | 0.00 ± 0.00 |
| B. pinnipedialis | |||||
| M163/99/10 | Hooded seal | 464 | 513 | + | 0.33 ± 0.06 |
| B2/94 bv. 1 | Common seal | 464 | 510 | +w | 0.04 ± 0.04 |
| Brucella spp. | |||||
| Br1 | African bullfrog | ND | ND | +s | 0.17 ± 0.04 |
| Br2 | African bullfrog | ND | ND | + | 1.18 ± 0.47 |
| Br3 | African bullfrog | ND | ND | + | 0.25 ± 0.18 |
| Br4 | African bullfrog | ND | ND | + | 1.04 ± 0.25 |
| B. suis | |||||
| ATCC 23444 bv. 1 (1330) | Swine | 319/136 | 426 | − | 0.00 ± 0.00 |
| ATCC 23445 bv. 2 (Thomsen) | Swine | 454 | 336d | + | 0.00 ± 0.00 |
The natural host of each strain and the lengths of their putative GadB and GadC proteins are provided. For GadB, the numbers before and after the character “/” indicate the respective sizes of the two consecutive fragments annotated as decarboxylases in the presence of a stop codon. ND, not determined (sequence unavailable).
The qualitative GAD test is based on the color change of the Rice reagent. Following the increase in pH that accompanies the decarboxylation of glutamic acid, the pH indicator bromocresol green turns from yellow (negative) to blue (positive). −, negative (yellow); +, positive (blue); +w, weakly positive (green); +s, slowly positive (blue color develops slowly during incubation of >8 h).
Shorter form, truncated at N terminus, with probably decreased ability to export GABA.
Shorter form, truncated at N terminus, probably unable to export GABA.
GAD activity and GABA export in Brucella strains correlate with the integrity of the gadBC locus.
To determine which Brucella species are GAD positive, a qualitative GAD test (Rice assay) was employed for a collection of 47 strains belonging to all 10 recognized species of the genus and also including yet-unclassified atypical strains (Table 1; see also Table S1 in the supplemental material for the full list). In this rapid test, initially described for E. coli (25), the proton-consuming GAD activity is indirectly demonstrated by a change in color (i.e., from yellow to green and blue) of an unbuffered assay solution (at pH 3.4) containing bromocresol green, a pH indicator, as pH increases (GAD Rice test in Table 1; see also Table S1). The test revealed that most of the strains of classical brucellae were GAD negative (yellow), even after 24 h of incubation of the bacteria in the GAD reagent. These included B. abortus (10/10), B. canis (1/1), B. melitensis (3/3), B. neotomae (1/1), B. ovis (1/2), and B. suis (4/4, one strain of each biovar, 1, 3, 4, and 5). Only the two reference strains, B. ovis ATTC 25840 bv. 1 (63/290) and B. suis ATCC 23445 bv. 2 (Thomsen), were GAD positive, with green and blue colors, respectively (Table 1; see also Table S1). In contrast, within 4 h, several strains of the newly described Brucella species were found to be GAD positive (blue) (Table 1; see also Table S1), i.e., B. microti (12/12 strains), B. ceti (4/4), B. inopinata BO1, B. inopinata-like BO2, and 3 out of 4 Brucella sp. strains isolated from bullfrogs. After a prolonged incubation period (≥8 h), the initially negatively tested strain isolated from bullfrogs (Brucella sp. Br1 in Table 1; see also Table S1) turned blue in the Rice test. The results of the Rice test were in agreement with the putative sequences of the GadB proteins, as far as those were available (Table 1, column 3; see also Table S1, column 5). Among the three B. pinnipedialis strains tested, strains M163/99/10 and M292/94/1, isolated from a hooded and a common seal, respectively, were clearly GAD positive, whereas strain B2/94, isolated from a common seal, was only weakly positive (yellow/green) despite its putatively intact GadB protein.
To evaluate the specificity of the Rice test, 16 strains of the genus Ochrobactrum (O. anthropi and O. intermedium), the closest phylogenetic neighbor of Brucella, were also analyzed. In fact, in the genome sequences of the two reference strains of these species (LMG 3331 and LMG 3301), the two genes gadB and gadC of the GAD system are not present. A GAD-negative result was found for all 16 strains tested (see Table S1 in the supplemental material, notes). In contrast, the E. coli strains tested were all GAD positive except for CC118. These results indicated that the Rice test may be useful to distinguish environmental and atypical strains of Brucella from those of Ochrobactrum, for which they are sometimes mistaken using standard biochemical tests.
To confirm the functionality of the GAD system among Brucella strains, the activity of the GadC antiporter was assayed by measurement of GABA levels in the medium at pH 3.5, as described previously (23). GABA was detected in all strains with a potentially functional GAD system (i.e., intact GadB and GadC proteins), with the exception of B. pinnipedialis strain B2/94, for which results of the Rice test were found to be only weakly positive (Fig. 1 and Table 1), thereby justifying a very limited or null GABA export.
GAD-positive and GABA-exporting Brucella strains are resistant to extreme acid pH in the presence of glutamate.
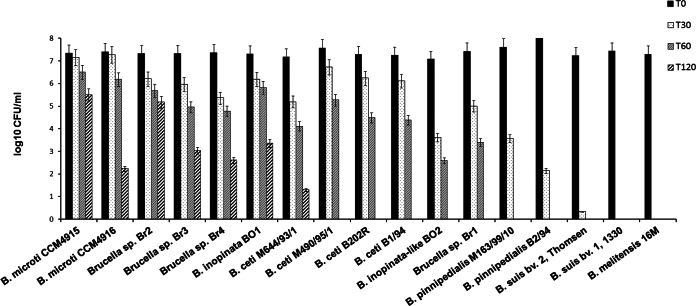
To identify possible glutamate-dependent AR phenotypes among Brucella strains, a variety of Brucella GAD-positive and -negative strains were subjected to acid shock in modified GMM at pH 2.5 for 30, 60, and 120 min in the presence/absence of Glu (1.5 mM). After 30 min, viable bacteria were undetectable for all classical Brucella species tested (B. abortus, B. canis, B. melitensis, B. neotomae, B. ovis, and B. suis), whereas a significant reduction in viability (≥104-fold) was observed for the B. pinnipedialis strains (Fig. 2).
FIG 2.
Survival of representative Brucella strains in minimal medium at pH 2.5 with 1.5 mM Glu. Decrease of viability during the acid shock (at the indicated time, T) was expressed as a log10 of the number of CFU (CFU/ml) at each time point (30, 60, and 120 min) versus bacteria present at time zero. B. abortus S19 and ATCC 23488 (544), B. neotomae 5K33, and B. ovis ATCC 25840 strains have the same acid-sensitive phenotypes as B. suis bv. 1, 1330 and B. melitensis 16M.
In contrast, the GAD-positive and GABA-producing strains (B. microti CCM4915 and CCM4916, three strains of Brucella isolated from bullfrogs, B. inopinata BO1, B. inopinata-like BO2, and four strains of B. ceti) were acid resistant, with a reduction in viability varying from 102- to 104-fold. Some of these strains maintained viability even after 2 h of incubation at this extreme pH (Fig. 2). Incubation in modified GMM at pH 2.5 without Glu for 30 min resulted in killing of all strains, suggesting an important role of the GAD system in acid resistance. Even though we could not detect any clear correlation between a strong AR phenotype and levels of exported GABA, it was nevertheless evident that a GABA export of at least 0.12 mM/h was necessary to support AR for 1 h at extreme acid pH (Table 1, rightmost column), the only exception being B. inopinata-like BO2.
Heterologous expression of gadBC of Brucella species in E. coli.
To compare the functioning of the GAD system of representative strains of Brucella species in a unique background genotype, their gadBC regions were overexpressed in an acid-sensitive triple mutant strain of E. coli MG1655, specifically constructed to be devoid of the GAD system (i.e., deleted in gadA and gadBC) (Table 2). In addition, four strains derived from E. coli MG1655 were constructed as controls: a wild-type strain and a triple mutant strain, carrying the pBBR1MCS nonrecombinant plasmid, and two triple mutant strains, carrying pBBR1MCS with the gadBC operon of E. coli MG1655 or B. microti CCM4915.
TABLE 2.
GAD test, GAD activity, and GABA export in overnight cultures of E. coli strains grown in LBG, before assaying survival following exposure to extreme acid
| Description of E. coli MG1655 wild type and gadABC triple mutant (Δ) and complemented strainsa | GAD Rice test result | GAD activity (U/mgb) | Concn (mM) of GABAc | % survivald |
|---|---|---|---|---|
| Wild type_p | + | 1.29 ± 0.03 | 0.86 ± 0.35 | 86.6 ± 12.2 |
| Δ_p | − | 0.03 ± 0.03 | 0.00 ± 0.00 | 0.0 ± 0.0 |
| Δ_ pgadBC (E. coli MG1655) | + | 2.34 ± 0.36 | 3.17 ± 0.21 | 70.3 ± 23.7 |
| Δ_ pgadBC (B. ceti M644/93/1) | + | 0.45 ± 0.01 | 0.28 ± 0.15 | 72.5 ± 16.7 |
| Δ_ pgadBC (B. ceti B1/94) | + | 0.36 ± 0.07 | 0.40 ± 0.14 | 99.2 ± 1.2 |
| Δ_ pgadBC (B. inopinata BO1) | + | 0.54 ± 0.10 | 0.16 ± 0.06 | 60.2 ± 16.4 |
| Δ_ pgadBC (B. inopinata-like BO2) | + | 0.64 ± 0.08 | 0.25 ± 0.15 | 83.6 ± 18.4 |
| Δ_ pgadBC (B. microti CCM4915) | + | 0.55 ± 0.01 | 0.31 ± 0.12 | 79.2 ± 22.3 |
| Δ_ pgadBC (B. microti CCM4916) | + | NDe | 0.31 ± 0.11 | 95.3 ± 8.0 |
| Δ_ pgadBC (B. pinnipedialis M163/99/10) | + | 0.51 ± 0.03 | 0.54 ± 0.13 | 70.7 ± 24.8 |
| Δ_ pgadBC (B. pinnipedialis B2/94) | + | ND | 0.18 ± 0.11 | 62.0 ± 30.4 |
| Δ_ pgadBC (Brucella sp. Br2) | + | 0.86 ± 0.14 | 0.20 ± 0.09 | 69.0 ± 22.0 |
| Δ_ pgadBC (B. suis bv. 1, 1330) | − | 0.03 ± 0.03 | 0.00 ± 0.00 | 0.0 ± 0.0 |
| Δ_pgadBC (B. suis bv. 2, Thomsen) | + | 0.49 ± 0.05 | 0.10 ± 0.03 | 0.0 ± 0.0 |
p, pBBR1MCS plasmid, either nonrecombinant (p only) or carrying gadBC (pgadBC) of E. coli MG1655 or Brucella strains (name of strain).
Micromoles of GABA produced per minute at 37°C per mg of total protein.
Concentration of GABA exported in spent medium was measured using GABase (Sigma).
Bacterial survival was expressed as the percentage of viable bacteria present in EG at pH 2.5 in the presence of 1.5 mM Glu after 2 h of incubation. Mean values ± SD are from 3 to 8 independent experiments.
ND, not determined.
As expected, on the basis of our previous work with ΔgadC and ΔgadA ΔgadB simple and double mutant strains (23), the E. coli triple mutant strain (carrying [or not] the pBBR1MCS control plasmid) was GAD and GABA negative and sensitive to extreme acid stress, with no survivors after 2 h at pH 2.5 in EG in the presence of Glu. In contrast, homologous and heterologous complementations of the same strain with the gadBC operons of E. coli or B. microti CCM4915 resulted in GAD activity. Hence, these strains were able to export GABA in LBG and displayed AR in EG at pH 2.5 in the presence of Glu to a level identical to that of the E. coli wild-type strain (Table 2). The presence of the plasmid pBBR1MCS per se did not affect the AR phenotype of the strains investigated.
Similarly, the E. coli triple mutant strains carrying the gadBC operons of various GAD-positive and GABA-positive Brucella strains were able to express a functional GadB protein and export GABA (B. ceti B1/94, B. ceti M644/93/1, B. inopinata BO1, B. inopinata-like BO2, B. microti CCM4916, B. pinnipedialis M163/99/10, and Brucella sp. Br2). These strains were as acid resistant as the E. coli triple mutant strains complemented with the operon of E. coli or B. microti CCM4915 (Table 2). In contrast, the E. coli triple mutant strain complemented with the gadBC operon of B. suis 1330 was devoid of GAD activity, unable to export GABA, and sensitive to acid challenge (Table 2). The mutant strain complemented with the gadBC region of the B. suis bv. 2 strain (Thomsen) expressed a functional GadB protein but was unable to export GABA and was also very sensitive to pH 2.5 even in the presence of Glu. As mentioned above, this strain possesses a gadC gene encoding a truncated GadC, very likely inactive.
Western blot analysis with crude lysates of recombinant E. coli strains grown to stationary phase showed that the GadB protein was detectable with anti-GadB (E. coli) antibodies only if the gadBC operon was derived from GAD-positive Brucella strains (data not shown). Truncated GadB proteins were not detected by Western blotting for the E. coli strains complemented with gadBC from GAD-negative strains, probably because of instability of the synthesized polypeptide. As a matter of fact, in B. suis ATCC 23445 bv. 2 (Thomsen), possessing intact GadB and truncated GadC, GadB was detectable by Western blotting and activity assays (Table 2 and data not shown). This suggests that the normal regulatory machinery for expression was still intact in this otherwise acid-sensitive strain.
The results of the complementation experiments corroborated the finding that the more acid-resistant strain was not necessarily the one exporting more GABA and that an extracellular GABA concentration of 0.16 mM was sufficient to support full expression of AR.
DISCUSSION
As unicellular organisms, bacteria are more vulnerable to sudden physicochemical environmental changes. To bypass the acid stress encountered in food or in the gastrointestinal tract of animal and human hosts, orally acquired bacteria apply various mechanisms to cope with this stress, among which the hydrolysis of urea by ureases (e.g., in Helicobacter pylori, Klebsiella, and all Brucella species except B. ovis) and the decarboxylation of different amino acids are the most effective (18, 29, 30). Based on the decarboxylation of Glu by the cytosolic enzyme glutamate decarboxylase (Gad) and the Glu/GABA antiport by the inner membrane protein GadC, the GAD system is the most efficient amino acid-dependent AR system in E. coli, S. flexneri, L. monocytogenes, and Lactobacillus lactis (18). In recent work, it was demonstrated that the GAD system is also functional as a gadBC operon in the newly described species B. microti but not in strain B. suis 1330, contributing to extreme AR in vitro and to the success of orally acquired infection in mice (23).
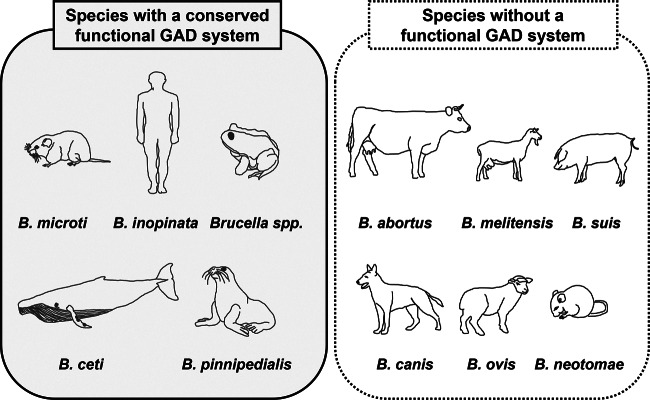
In the present work, we studied the functionality of the GAD system among a collection of strains of the genus Brucella. Our in silico analysis revealed that all Brucella species for which genome sequences are available possessed a gadBC locus homologous to those of B. microti and E. coli. However, only a definite group of strains may possess a potentially functional GAD system on the basis of the putative Gad proteins (Fig. 3). Experimental data confirmed that all existing isolates of B. microti (n = 12), B. inopinata BO1, B. inopinata-like BO2, and also several B. ceti, B. pinnipedialis, and Brucella species strains isolated from bullfrogs were GAD positive and exported GABA. Representative strains of these species were acid resistant at pH 2.5 in the presence of Glu. When the gadBC locus of these strains was used to complement a triple mutant strain of E. coli MG1655 deleted of its endogenous GAD system, glutamate-dependent AR was restored in this otherwise acid-sensitive strain. Among the B. pinnipedialis strains investigated in this study, the B2/94 strain, isolated from a common seal, was only weakly GAD positive and unable to export GABA, despite the presence of a putatively intact gadBC locus; mutations in as yet unidentified transcriptional regulators might be responsible for the observed GAD-negative phenotype. According to their gadBC sequences, Brucella strain 83/13 and Brucella sp. NF2653, isolated from Australian rodents (12), are also expected to have a functional GAD system.
FIG 3.
Distribution of Brucella species based on the functionality of the GAD system.
In agreement with their putative GAD sequences, the representative strains of the terrestrial classical Brucella species tested (B. abortus, B. canis, B. melitensis, B. neotomae, and B. suis) were found to have a nonfunctional GAD system, i.e., were GAD negative, unable to export GABA, and highly sensitive to extreme acidity even after short-time incubation (Fig. 3). A nonfunctional GAD system was also confirmed for the B. ovis ATTC 25840 (63/290) and B. suis bv. 2 (Thomsen) strains. According to their putative GadB proteins, these strains were GAD positive, but due to their truncated GadC proteins, they did not export GABA, which finally makes them acid sensitive, comparable to the other classical Brucella strains. Genome analysis of 3 recently sequenced B. suis bv. 2 strains suggests identical Gad phenotypes (31).
Therefore, the intriguing question is the following: why was the functional GAD system lost in these host-adapted and important human food-borne pathogens, although it might have conferred an advantage regarding survival in food and during passage through the gastrointestinal tract? The oral route is considered the most common route of infection in humans, which are only accidental hosts of Brucella; in livestock and domesticated animals, the pathogenic strains are preferentially transmitted by sexual or vertical routes, where the GAD system does not play a significant role. In fact, experimental infections of mice with Brucella spp. by the oral route succeed only with high doses of living bacteria >109 (CFU) (23, 32, 33). Thus, stomach acidity may significantly affect the survival of brucellae despite the presence of several factors allowing resistance to the acid conditions, such as urease and HdeA (19, 32–35). In addition, it is controversial that the primary invasion after ingestion of classical pathogenic Brucella spp. occurs through the intestine. In fact, several observations indicate that the mucosae of the face area (tonsils, lymph nodes, and conjunctival and respiratory mucosae), are the normal sites of entry of Brucella (36), which is consistent with the absence of a functional GAD system in most pathogenic Brucella species.
Our previous results with B. microti showed that the loss of a functional GAD system significantly reduces the number of living bacteria recovered from the target organs liver and spleen after oral infection of mice (33). Based on our present work, it seems conceivable that Brucella strains that have conserved this system may directly infect their hosts via the gastrointestinal tract. However, it must be stressed that the natural hosts and ecological niches of these strains are still unknown and the verification of this hypothesis is not straightforward, since appropriate animal models cannot be readily defined.
Our data suggested that the state of the GAD system may reflect the adaptive evolution of Brucella species for their respective environments and hosts. These agree with results of phylogenic tree analysis of the genus Brucella: five main groups of host-associated Brucella species rapidly diverged from a likely free-living ancestor close to B. microti, resulting in explosive radiation (37, 38). Our hypothesis is that, starting from a GAD-positive ancestor, all the strains adapted to specific hosts have lost this system, except those isolated from marine mammals, among which the gad locus is still conserved, though not equally expressed. Multiple alignments of the available sequences of gadB and gadC of Brucella strains included in this study (see Fig. S1 and S2 in the supplemental material) indicate that point mutations arose independently in each group, leading to truncated proteins. The genotypes of the two strains B. ovis ATTC 25840 and B. suis bv 2 (Thomsen) (gadB intact and gadC interrupted) indicate that in both groups of strains of B. ovis and B. suis, the loss of the GAD system is still in progress.
In several pathogenic bacteria, this evolutionary step can be accompanied by a loss or inactivation of specific genes which are not essential or even detrimental for the interaction with a host. These gene modifications, referred as “pathoadaptive mutations”, may occur by point mutations, microdeletion, or insertional inactivation, as described for the lysine-cadaverine AR system of S. flexneri (39). However, there is no evidence indicating that this also applies to the GAD system of Brucella; in fact, the behaviors of the wild type and a ΔgadBC mutant strain of B. microti were identical during cellular and intraperitoneal murine infections. In addition, a mutant strain of B. suis 1330 expressing the functional GAD system of B. microti CCM4915 is neither advantaged nor affected in the cellular model of infection compared to the parental wild-type strain (23).
Because gadBC protects the bacterium from acid pH levels of <2.5, it may confer to marine and “new and atypical” Brucella species an adaptive advantage for survival in particularly acidic niches encountered in various environments, such as water, food, soil, and the gastrointestinal tract of hosts. In the case of B. microti, the bacterium was recovered from soil several years after its first isolation from voles (6). The other new and atypical Brucella species or strains may be also considered more environmental than the classical pathogenic species of the genus. Moreover, it cannot be excluded that a functional GAD system is conserved because it may play a role in the protection against other, as yet unidentified stresses. For example, it has been shown that GadC plays a role in protection against oxidative stress in Francisella tularensis (40) and that GadB is part of a compensatory pathway (GABA shunt) that partially overcomes metabolic defect in the interrupted TCA cycle, as in L. monocytogenes and Mycobacterium tuberculosis (41, 42).
In conclusion, our results showed that the GAD system has been lost in classical pathogenic Brucella species, which are most adapted to livestock and human hosts, and it is conserved in new and in atypical strains of ancestral origin as well as in marine pathogenic Brucella species, which may have retained their ability to persist in acidic external and/or gastrointestinal host environments.
In addition, our data indicate that GadB activity may be useful to discriminate newly described or atypical species of Brucella which are GAD positive (B. microti, B. inopinata BO1, B. inopinata-like BO2, and strains from bullfrogs) from closely related Ochrobactrum species and also from classical terrestrial pathogenic species of Brucella, which are GAD negative. Thus, the simple, easy-to-handle, and inexpensive colorimetric Rice test may be added to other conventional and rapid biochemical tests used for the identification of new or atypical Brucella species. A more complete GAD database has to be established by increasing the variety and number of strains tested, including more atypical Brucella isolates.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grant 1329-485 from the Federal Institute for Risk Assessment, Germany (to A.O. and S.K.), and by funding from Sapienza University of Rome and Fondazione Roma (to D.D.B.). Travels and research stays of A.O., D.D.B., M.A.D., and D.B. were supported by the 2011-2012 Galilée program of Egide (Hubert Curien program), no. 25960UE, from the French and Italian Ministry of Foreign and Europeans Affairs. M.A.D. was a recipient was awarded a postdoctoral fellowship from the foundation Infectiopôle Sud, and A.O. was awarded a Congés pour Recherches ou Conversions Thématiques (CRCT) from Montpellier University.
We thank E. Bianchini for her participation in genetic constructions of E. coli strains, B. Saadeh for preliminary GABA tests, C. Göllner for GAD tests, P. Bouhours for preparation of bacterial media, and I. Jacques and D. O'Callaghan for their generous gifts of several marine strains and of B. inopinata BO1 and B. inopinata-like BO2 strains, respectively.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02928-14.
REFERENCES
- 1.Pappas G, Papadimitriou P, Akritidis N, Christou L, Tsianos EV. 2006. The new global map of human brucellosis. Lancet Infect Dis 6:91–99. doi: 10.1016/S1473-3099(06)70382-6. [DOI] [PubMed] [Google Scholar]
- 2.Pappas G. 2010. The changing Brucella ecology: novel reservoirs, new threats. Int J Antimicrob Agents 36(Suppl 1):S8–S11. doi: 10.1016/j.ijantimicag.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 3.Scholz HC, Kämpfer P, Cloeckaert A. 2012. Brucella: relationship to other Alphaproteobacteria, current taxonomy and the emergence of new species. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 4.Scholz HC, Hofer E, Vergnaud G, Le Fleche P, Whatmore AM, Al Dahouk S, Pfeffer M, Kruger M, Cloeckaert A, Tomaso H. 2009. Isolation of Brucella microti from mandibular lymph nodes of red foxes, Vulpes vulpes, in lower Austria. Vector Borne Zoonotic Dis 9:153–156. doi: 10.1089/vbz.2008.0036. [DOI] [PubMed] [Google Scholar]
- 5.Scholz HC, Hubalek Z, Sedlacek I, Vergnaud G, Tomaso H, Al Dahouk S, Melzer F, Kampfer P, Neubauer H, Cloeckaert A, Maquart M, Zygmunt MS, Whatmore AM, Falsen E, Bahn P, Gollner C, Pfeffer M, Huber B, Busse HJ, Nockler K. 2008. Brucella microti sp. nov., isolated from the common vole Microtus arvalis. Int J Syst Evol Microbiol 58:375–382. doi: 10.1099/ijs.0.65356-0. [DOI] [PubMed] [Google Scholar]
- 6.Scholz HC, Hubalek Z, Nesvadbova J, Tomaso H, Vergnaud G, Le Fleche P, Whatmore AM, Al Dahouk S, Kruger M, Lodri C, Pfeffer M. 2008. Isolation of Brucella microti from soil. Emerg Infect Dis 14:1316–1317. doi: 10.3201/eid1408.080286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Al Dahouk S, Hofer E, Tomaso H, Vergnaud G, Le Fleche P, Cloeckaert A, Koylass MS, Whatmore AM, Nockler K, Scholz HC. 2012. Intraspecies biodiversity of the genetically homologous species Brucella microti. Appl Environ Microbiol 78:1534–1543. doi: 10.1128/AEM.06351-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De BK, Stauffer L, Koylass MS, Sharp SE, Gee JE, Helsel LO, Steigerwalt AG, Vega R, Clark TA, Daneshvar MI, Wilkins PP, Whatmore AM. 2008. Novel Brucella strain (BO1) associated with a prosthetic breast implant infection. J Clin Microbiol 46:43–49. doi: 10.1128/JCM.01494-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scholz HC, Nockler K, Gollner C, Bahn P, Vergnaud G, Tomaso H, Al Dahouk S, Kampfer P, Cloeckaert A, Maquart M, Zygmunt MS, Whatmore AM, Pfeffer M, Huber B, Busse HJ, De BK. 2010. Brucella inopinata sp. nov., isolated from a breast implant infection. Int J Syst Evol Microbiol 60:801–808. doi: 10.1099/ijs.0.011148-0. [DOI] [PubMed] [Google Scholar]
- 10.Tiller RV, Gee JE, Lonsway DR, Gribble S, Bell SC, Jennison AV, Bates J, Coulter C, Hoffmaster AR, De BK. 2010. Identification of an unusual Brucella strain (BO2) from a lung biopsy in a 52 year-old patient with chronic destructive pneumonia. BMC Microbiol 10:23. doi: 10.1186/1471-2180-10-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schlabritz-Loutsevitch NE, Whatmore AM, Quance CR, Koylass MS, Cummins LB, Dick EJ Jr, Snider CL, Cappelli D, Ebersole JL, Nathanielsz PW, Hubbard GB. 2009. A novel Brucella isolate in association with two cases of stillbirth in non-human primates—first report. J Med Primatol 38:70–73. doi: 10.1111/j.1600-0684.2008.00314.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tiller RV, Gee JE, Frace MA, Taylor TK, Setubal JC, Hoffmaster AR, De BK. 2010. Characterization of novel Brucella strains originating from wild native rodent species in North Queensland, Australia. Appl Environ Microbiol 76:5837–5845. doi: 10.1128/AEM.00620-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eisenberg T, Hamann HP, Kaim U, Schlez K, Seeger H, Schauerte N, Melzer F, Tomaso H, Scholz HC, Koylass MS, Whatmore AM, Zschock M. 2012. Isolation of potentially novel Brucella spp. from frogs. Appl Environ Microbiol 78:3753–3755. doi: 10.1128/AEM.07509-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cotter PD, Gahan CG, Hill C. 2001. A glutamate decarboxylase system protects Listeria monocytogenes in gastric fluid. Mol Microbiol 40:465–475. doi: 10.1046/j.1365-2958.2001.02398.x. [DOI] [PubMed] [Google Scholar]
- 15.De Biase D, Tramonti A, Bossa F, Visca P. 1999. The response to stationary-phase stress conditions in Escherichia coli: role and regulation of the glutamic acid decarboxylase system. Mol Microbiol 32:1198–1211. doi: 10.1046/j.1365-2958.1999.01430.x. [DOI] [PubMed] [Google Scholar]
- 16.Castanie-Cornet MP, Penfound TA, Smith D, Elliott JF, Foster JW. 1999. Control of acid resistance in Escherichia coli. J Bacteriol 181:3525–3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanders JW, Leenhouts K, Burghoorn J, Brands JR, Venema G, Kok J. 1998. A chloride-inducible acid resistance mechanism in Lactococcus lactis and its regulation. Mol Microbiol 27:299–310. doi: 10.1046/j.1365-2958.1998.00676.x. [DOI] [PubMed] [Google Scholar]
- 18.Lund P, Tramonti A, De Biase D. 2 July 2014. Coping with low pH: molecular strategies in neutralophilic bacteria. FEMS Microbiol Rev doi: 10.1111/1574-6976.12076. [DOI] [PubMed] [Google Scholar]
- 19.De Biase D, Pennacchietti E. 2012. Glutamate decarboxylase-dependent acid resistance in orally acquired bacteria: function, distribution and biomedical implications of the gadBC operon. Mol Microbiol 86:770–786. doi: 10.1111/mmi.12020. [DOI] [PubMed] [Google Scholar]
- 20.Tsai MF, McCarthy P, Miller C. 2013. Substrate selectivity in glutamate-dependent acid resistance in enteric bacteria. Proc Natl Acad Sci U S A 110:5898–5902. doi: 10.1073/pnas.1301444110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Audic S, Lescot M, Claverie JM, Scholz HC. 2009. Brucella microti: the genome sequence of an emerging pathogen. BMC Genomics 10:352. doi: 10.1186/1471-2164-10-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jimenez de Bagues MP, Ouahrani-Bettache S, Quintana JF, Mitjana O, Hanna N, Bessoles S, Sanchez F, Scholz HC, Lafont V, Kohler S, Occhialini A. 2010. The new species Brucella microti replicates in macrophages and causes death in murine models of infection. J Infect Dis 202:3–10. doi: 10.1086/653084. [DOI] [PubMed] [Google Scholar]
- 23.Occhialini A, Jimenez de Bagues MP, Saadeh B, Bastianelli D, Hanna N, De Biase D, Kohler S. 2012. The glutamic acid decarboxylase system of the new species Brucella microti contributes to its acid resistance and to oral infection of mice. J Infect Dis 206:1424–1432. doi: 10.1093/infdis/jis522. [DOI] [PubMed] [Google Scholar]
- 24.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rice EW, Johnson CH, Dunnigan ME, Reasoner DJ. 1993. Rapid glutamate decarboxylase assay for detection of Escherichia coli. Appl Environ Microbiol 59:4347–4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 27.De Biase D, Tramonti A, John RA, Bossa F. 1996. Isolation, overexpression, and biochemical characterization of the two isoforms of glutamic acid decarboxylase from Escherichia coli. Protein Expr Purif 8:430–438. doi: 10.1006/prep.1996.0121. [DOI] [PubMed] [Google Scholar]
- 28.Capitani G, De Biase D, Aurizi C, Gut H, Bossa F, Grutter MG. 2003. Crystal structure and functional analysis of Escherichia coli glutamate decarboxylase. EMBO J 22:4027–4037. doi: 10.1093/emboj/cdg403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mobley HL, Island MD, Hausinger RP. 1995. Molecular biology of microbial ureases. Microbiol Rev 59:451–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith LD, Ficht TA. 1990. Pathogenesis of Brucella. Crit Rev Microbiol 17:209–230. doi: 10.3109/10408419009105726. [DOI] [PubMed] [Google Scholar]
- 31.Ferreira AC, Tenreiro R, Correa de Sa MI, Dias R. 2014. Complete genome sequences of three Iberian Brucella suis biovar 2 strains isolated from wild boars. Genome Announc 2:e00618-14. doi: 10.1128/genomeA.00618-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sangari FJ, Seoane A, Rodriguez MC, Aguero J, Garcia Lobo JM. 2007. Characterization of the urease operon of Brucella abortus and assessment of its role in virulence of the bacterium. Infect Immun 75:774–780. doi: 10.1128/IAI.01244-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paixao TA, Roux CM, den Hartigh AB, Sankaran-Walters S, Dandekar S, Santos RL, Tsolis RM. 2009. Establishment of systemic Brucella melitensis infection through the digestive tract requires urease, the type IV secretion system, and lipopolysaccharide O antigen. Infect Immun 77:4197–4208. doi: 10.1128/IAI.00417-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bandara AB, Contreras A, Contreras-Rodriguez A, Martins AM, Dobrean V, Poff-Reichow S, Rajasekaran P, Sriranganathan N, Schurig GG, Boyle SM. 2007. Brucella suis urease encoded by ure1 but not ure2 is necessary for intestinal infection of BALB/c mice. BMC Microbiol 7:57. doi: 10.1186/1471-2180-7-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Valderas MW, Alcantara RB, Baumgartner JE, Bellaire BH, Robertson GT, Ng WL, Richardson JM, Winkler ME, Roop RM II. 2005. Role of HdeA in acid resistance and virulence in Brucella abortus 2308. Vet Microbiol 107:307–312. doi: 10.1016/j.vetmic.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 36.Gorvel JP, Moreno E, Moriyon I. 2009. Is Brucella an enteric pathogen? Nat Rev Microbiol 7:250. doi: 10.1038/nrmicro2012-c1 (Reply, 7:250. doi:). [DOI] [PubMed] [Google Scholar]
- 37.Wattam AR, Foster JT, Mane SP, Beckstrom-Sternberg SM, Beckstrom-Sternberg JM, Dickerman AW, Keim P, Pearson T, Shukla M, Ward DV, Williams KP, Sobral BW, Tsolis RM, Whatmore AM, O'Callaghan D. 2014. Comparative phylogenomics and evolution of the Brucellae reveal a path to virulence. J Bacteriol 196:920–930. doi: 10.1128/JB.01091-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Audic S, Lescot M, Claverie JM, Cloeckaert A, Zygmunt MS. 2011. The genome sequence of Brucella pinnipedialis B2/94 sheds light on the evolutionary history of the genus Brucella. BMC Evol Biol 11:200. doi: 10.1186/1471-2148-11-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prosseda G, Di Martino ML, Campilongo R, Fioravanti R, Micheli G, Casalino M, Colonna B. 2012. Shedding of genes that interfere with the pathogenic lifestyle: the Shigella model. Res Microbiol 163:399–406. doi: 10.1016/j.resmic.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 40.Ramond E, Gesbert G, Rigard M, Dairou J, Dupuis M, Dubail I, Meibom K, Henry T, Barel M, Charbit A. 2014. Glutamate utilization couples oxidative stress defense and the tricarboxylic acid cycle in Francisella phagosomal escape. PLoS Pathog 10:e1003893. doi: 10.1371/journal.ppat.1003893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feehily C, O'Byrne CP, Karatzas KA. 2013. Functional gamma-aminobutyrate shunt in Listeria monocytogenes: role in acid tolerance and succinate biosynthesis. Appl Environ Microbiol 79:74–80. doi: 10.1128/AEM.02184-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tian J, Bryk R, Itoh M, Suematsu M, Nathan C. 2005. Variant tricarboxylic acid cycle in Mycobacterium tuberculosis: identification of alpha-ketoglutarate decarboxylase. Proc Natl Acad Sci U S A 102:10670–10675. doi: 10.1073/pnas.0501605102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.