Abstract
Heterocyclic aromatic amines (HAAs) are carcinogens formed during the cooking of meats or arise in tobacco smoke. The genotoxic N-oxidized metabolites of HAAs bind to Cys residues of proteins to form arylsulfinamide adducts. However, these adducts are unstable and undergo hydrolysis during enzymatic digestion, and thus have been precluded as biomarkers of exposure to HAAs. Arylsulfinamide adducts of HAAs can undergo oxidation to form stable arylsulfonamide linkages, which are chemically stable and amenable for analysis. 2-Amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) is a carcinogen present in cooked meat. We established a quantitative MS-based method to measure the sulfinamide adduct of PhIP formed at the cysteine34 (Cys34) residue of human serum albumin (SA), following chemical oxidation of PhIP-modified SA with m-chloroperoxybenzoic acid. Different enzyme systems (trypsin; chymotrypsin; trypsin/chymotrypsin; proteinase K; pronase E; and pronase E/leucine aminopeptidase/prolidase) were evaluated for their proficiency of digestion of SA modified with PhIP. The strongest signal was observed for the L31QQC*PFEDHVK41 peptide, by ultraperformance liquid chromatography and ion trap MS. A limit of quantification value was 0.3 fmol of LQQC*PFEDHVK per μg SA, or 2.5 adducts per 105 SA molecules, when assaying 0.75 μg of SA.
Biological significance
This article describes a mass spectrometric based method to characterize and measure human serum albumin (SA) adducts of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP), a carcinogenic heterocyclic aromatic amine formed in cooked meats and tobacco smoke. PhIP undergoes metabolic activation to form reactive N-oxidized intermediates that bind to DNA and proteins. N-oxidized PhIP metabolites bind to the Cys34 residue of SA to form a sulfinamide linkage. However, the linkage undergoes hydrolysis during proteolysis, precluding the employment of this adduct as a biomarker in human studies. We have shown that the sulfinamide linkage undergoes oxidation to form the [cysteine-S-yl-PhIP]-S-dioxide, a sulfonamide linked adduct which is stable toward proteolysis. The specificity and efficiency of several different proteases toward the digestion of the SA-Cys34-PhIP adduct were examined. The combination of trypsin and chymotrypsin produced the single-missed cleaved peptide LQQC*PFEDHVK in high yield. Moreover, denaturation and chemical reduction of the internal Cys disulfide bonds of SA were not required for the recovery of LQQC*PFEDHVK. The novel chemistry and proteomic approaches developed in this study may be applied to monitor biologically reactive N-oxidized intermediates of arylamines through their adduction products formed at nucleophilic Cys residues of proteins.
Keywords: Biomarkers, Protein adducts, Food carcinogens
1. Introduction
Hemoglobin (Hb) and serum albumin (SA) are the two major proteins in blood, and both proteins form adducts with different classes of environmental and dietary genotoxicants, therapeutics, and endogenously produced electrophiles [1–4]. Several of these protein adducts have been employed as biomarkers in human studies. Early studies employed gas chromatography coupled to mass spectrometry to measure several types of protein adducts [1,5]; however, liquid chromatography/mass spectrometry has emerged as the method of choice for the analysis of many protein toxicant adducts [6–8].
Aromatic amines and heterocyclic aromatic amines (HAAs) are carcinogens formed during the combustion of tobacco or during the high-temperature cooking of meats, poultry, or fish [9,10]. The genotoxic N-oxidized metabolites of these procarcinogens bind to Cys residues of proteins to form a sulfinamide adduct [1,4]. The N-oxidized arylamine metabolites are thought to be formed in the liver, exported to blood, and undergo a co-oxidation reaction with Hb to form sulfinamide adducts at the Hb-Cys93β residue [11,12]. These sulfur–nitrogen linked adducts are labile and undergo hydrolysis during proteolysis [1,13,14]. The labile nature of the Hb arylsulfinamide bond has been exploited to monitor occupational and environmental exposures to some monocyclic and bicyclic aromatic amines following mild acid or base hydrolysis of Hb and liberation of the arylamines [1,15,16]. However, the measurement of the arylamines does not identify a specific Hb adduct, and it is inferred that adduct formation occurred at the Hb-Cys93β residue [14,17]. The structure of the putative Hb sulfinamide adduct formed with 4-aminobiphenyl was proven by X-ray crystallography [14,17]. Nevertheless, the possibility that some arylamines form labile covalent adducts at other nucleophilic sites or by non-covalent binding to Hb cannot be excluded.
The employment of Hb arylsulfinamide adducts as a dosimeter has some limitations. A number of aromatic amines and HAAs do not form Hb sulfinamide adducts in vivo, perhaps because alternative metabolic pathways predominate or because the N-oxidized metabolites do not efficiently penetrate the erythrocyte or do not react with the heme moiety to form the nitrosoarene intermediates [1]. The Cys34 residue of human SA is highly nucleophilic and known to scavenge a variety of environmental and endogenous electrophiles [3,4,6–8]. The Cys34 of SA reacts with N-oxidized metabolites of arylamines and HAAs to form putative arylsulfenamide or arylsulfinamide adducts [18–22]. The formation of these arylamine adducts with N-acetyltransferase has also been inferred by MS analysis of the intact protein; however, these adducts likewise undergo hydrolysis during proteolysis [23–26].
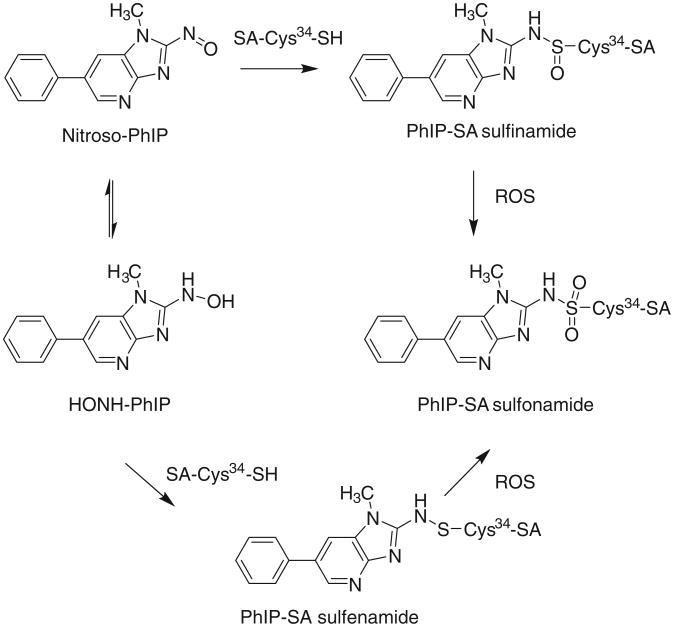
We recently discovered that the SA-Cys34 sulfinamide adduct of the cooked meat carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) undergoes oxidation with m-chloroperoxybenzoic acid (m-CPBA) to form [cysteine-S-yl-PhIP]-S-dioxide, a stable arylsulfonamide-SA linkage (Fig. 1) [27]. The same SA adduction product occurs for the tobacco carcinogen 4-aminobiphenyl [27]. The SA-arylsulfonamide adduct is stable toward acid, heat or reducing reagents commonly used in proteolytic digestion, and the intact adducted peptide can be characterized by liquid chromatography–tandem mass spectrometry (LC–MS/MS), following proteolysis [27].
Fig. 1. Reaction of N-oxidized PhIP metabolites with Cys34 of SA adducts and formation of the sulfonamide adduct by chemical oxidation with reactive oxygen species (ROS).
The biomonitoring of SA arylsulfonamide adducts in humans is challenging because there are many non-modified peptides of SA present in vast excess to the adducted peptide in the protein digest [8,28]. Proficient enzymatic digestion of the chemically modified SA, combined with selective enrichment and chromatographic separations of the adducted peptide is crucial in the development of a validated biomarker. Proteases of different specificities can digest proteins to polypeptide chains of different lengths. Combinations of several proteases can further break down proteins to smaller peptides or even monoamino acids, and different digestion strategies have been employed for digestion and quantification of proteins [29–31]. However, the adduction of a bulky carcinogen to a protein can impact the efficacy of a protease, resulting in the occurrence of missed-cleaved peptides. For example, SA adducts of HAAs, therapeutics, acetaminophen, mustards or lipid peroxidation products formed at the Cys34 residue undergo digestion with pronase, trypsin, or a combination of trypsin and chymotrypsin to produce C*PF, ALVLIAFAQYLQQC*PFEDHVK, LQQC*PF, or missed cleavage peptide LQQC*PFEDHVK [3,6,28,32,33].
The goals of our study were to establish a robust method to measure the arylsulfinamide adduct of PhIP formed with Cys34 of human SA, following its conversion to the stable arylsulfonamide structure, and to examine the proficiencies of different proteases to recover PhIP-adducted Cys34 peptides for measurement by ion trap MS.
2. Experimental section
2.1. Caution
PhIP is a carcinogen and should be handled in a well-ventilated fume hood with the appropriate protective clothing.
2.2. Chemicals and materials
PhIP and 2-amino-1-[2H3C]-methyl-6-phenylimidazo[4,5-b] pyridine ([2H3]-PhIP, 99% isotopic purity) were purchased from Toronto Research Chemicals (Toronto, ON, Canada). 2-Amino-1-methyl-6-[2H5]-phenylimidazo[4,5-b]pyridine ([2H5]-PhIP, 99% isotopic purity) was a gift from Dr. Mark Knize and Dr. Kristen Kulp (Lawrence Livermore National Laboratory, Livermore, CA). Human SA, β-mercaptoethanol (βME), iodoacetamide (IAA), dithiothreitol (DTT), and m-CPBA were obtained from Sigma-Aldrich (St. Louis, MO). Human plasma was purchased from Bioreclamation LLC (Hicksville, NY) and SA was purified with a HiTrap Blue affinity column (GE Healthcare, Piscataway, NJ) [26]. LQQCPFEDHVK was purchased from Biosynthesis (Lewisville, TX) and LQQCPˆFEDHVK (Pˆ: 13C and 15N universally-labeled proline) was from New England Peptide (Gardner, MA). Sequence grade trypsin and chymotrypsin were purchased from Promega (Madison, WI), and biochemical grade trypsin, chymotrypsin, proteinase K, pronase E, leucine aminopeptidase, and prolidase were purchased from Sigma (St. Louis, MO). All solvents were high-purity B&J Brand from Honeywell Burdick and Jackson (Muskegon, MI). Isolute C18 solid phase extraction (SPE) columns (25 mg) were from Biotage (Charlotte, NC).
2.2.1. Synthesis of 2-hydroxyamino-1-methyl-6-phenylimidazo[4,5-b]pyridine (HONH-PhIP) and 2-Nitroso-1-methyl-6-phenylimidazo[4,5-b]pyridine (NO-PhIP)
HONH-PhIP and HONH-[2H5]-PhIP were prepared by reduction of their respective nitroderivatives with hydrazine, using Pd/C as a catalyst [34]. NO-PhIP was obtained by oxidation of HONH-PhIP with K3Fe(CN)6 [26].
2.2.2. Syntheses of the PhIP sulfonamide adduct of LQQC*PFEDHVK,LQQC*PFEDHVK sulfonic acid, and IAA alkylated LQQC*PFEDHVK
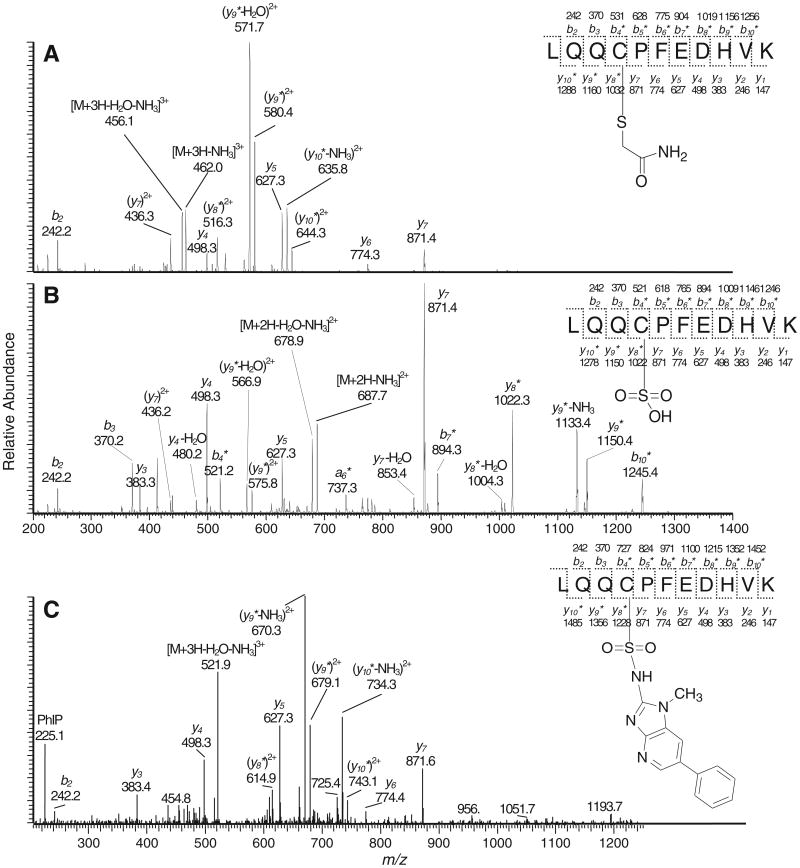
LQQCPFEDHVK (15 nmol) was reacted with NO-PhIP (45 nmol) in 10 mM phosphate buffer (0.4 mL, pH 8.5) and CH3CN (0.4 mL) at 5 °C for 1 h. The mixture was then acidified topH 5.5 by the addition of 50 mM potassium phosphate buffer (0.5 mL, pH 3.6), and incubated at 37 °C for 1 h to form the PhIP sulfinamide LQQC*PFEDHVK (C-[S=O]-PhIP). The sulfonamide LQQC*PFEDHVK (C-[SO2]-PhIP) was obtained by direct oxidation of the mixture with m-CPBA at a molar ratio of 1:10 [27]. The sulfonic acid of LQQC*PFEDHVK (C-[SO3H]) was synthesized by treatment of LQQCPFEDHVK (7.4 nmol) with oxone (44.4 nmol) in 0.1 M potassium phosphate buffer (0.25 mL, pH 7.4) for 3 h at room temperature. IAA alkylated LQQC*PFEDHVK (C-[S–CH2CONH2]) was prepared by the reaction of LQQCPFEDHVK (7.4 nmol) in 0.1 M potassium phosphate buffer (0.25 mL, pH 7.4) with excess IAA (480 nmol) in the dark for 2 h at room temperature. Thereafter, LQQC*PFEDHVK (C-[SO2]-PhIP), LQQC*PFEDHVK (C-[SO3H]) and LQQC*PFEDHVK (C-[S–CH2CONH2]) were purified by HPLC (Acquasil C18, 5 μm, 4.6 × 250 mm, Thermo, Bellefonte, PA) using a linear gradient starting from 100% A (0.1% HCO2H) to 55% B (95% CH3CN containing 4.9% H2O and 0.1% HCO2H) over 15 min, at a flow rate of 1 mL/min. The UV was monitored at 210 nm for LQQC*PFEDHVK (C-[SO3H]) and (C-[S–CH2CONH2]) peptides and 320 nm for the (C-[SO2]-PhIP) peptide. The formation of LQQC*PFEDHVK (C-[SO3H]) and (C-[S–CH2CONH2]) was quantitative based upon the HPLC UV analysis. The amount of the (C-[SO2]-PhIP) adduct was based upon its UV chromophore at 320 nm, employing the molar extinction coefficient of PhIP [27]. The structures of the LQQC*PFEDHVK peptides were confirmed by MS/MS spectra (Figs. 2 and S-1).
Fig. 2. ESI/MS product ion spectra of (A) LQQC*PFEDHVK (C-[S-CH2CONH2]) ([M + 3H]3+ at m/z 467.6) and (B) LQQC*PFEDHVK (C-[SO3H]) ([M + 2H]2+ at m/z 696.3), and (C) LQQC*PFEDHVK (C-[SO2]-PhIP sulfonamide) ([M + 3H]3+ at m/z 533.2). Asterisk (*) labeled ions are PhIP adducted.
2.2.3. Modification of human SA with N-oxidized derivatives of PhIP
Human SA was pretreated with β-mercaptoethanol to reduce mixed disulfides formed at Cys34 [19]. Following PD-10 gel chromatography in 0.1 M potassium phosphate buffer (pH 7.4), the sulfhydryl content of SA was determined using Ellman's reagent [35]. The SA reduced with β-mercaptoethanol contained a sulfhydryl content of 0.95 mol –SH/mol SA. SA (2 mg, 30 nmol) was reacted with a solution of HONH-PhIP and HONH-[2H5]-PhIP (15 nmol of each in 40 μL C2H5OH) in 0.1 M potassium phosphate buffer (pH 7.4, 1 mL) at 37 °C for 18 h. Alternatively, SA (1 mg, 15 nmol) was reacted with unlabeled HONH-PhIP (15 nmol) under the same reaction conditions. Thereafter, unbound carcinogen was removed by solvent extraction with ethyl acetate, and the modified SA was run through a PD-10 gel column in 0.1 M phosphate buffer (pH 7.4). Unmodified or PhIP-modified SA (0.5 mg, 7.5 nmol) was oxidized with m-CPBA for 4 h in 0.1 M phosphate buffer (pH 7.4, 1 mL) with a protein:m-CPBA molar ratio of 1:50. The samples were then incubated for 1 h with a Branson 2510 model sonicator. Thereafter, the SA was run through a PD-10 gel column in 50 mM ammonium bicarbonate (pH 8.5) [27].
2.2.4. Proteolytic digestion
Enzyme solutions were prepared as described previously [26]. Five different proteolytic conditions were employed to digest SA adducts modified by HONH-PhIP.
Pronase E digestion: PhIP-modified SA (5 μg, 75 pmol) in 50 μL 50 mM ammonium bicarbonate buffer (pH 8.5) containing1 mM CaCl2 was treated with pronase E at a protease: protein ratio of 1:2 and 1:10 (w/w) for 20 h at 37 °C, followed by adduct enrichment with SPE [26,28].
Pronase E/leucine aminopeptidase/prolidase digestion: PhIP-modified SA (5 μg, 75 pmol) in 50 μL 50 mM ammonium bicarbonate buffer (pH 8.5) containing MnCl2 (1 mM) and CaCl2 (1 mM) was treated with pronase E at a protease:protein ratio of 1:2 (w/w), leucine aminopeptidase at a protease: protein ratio of 1:30 (w/w), and prolidase at a protease:protein ratio of 1:8 (w/w). The digestion was conducted at 37 ° C for 20 h, followed by adduct enrichment with SPE [27].
Proteinase K digestion: PhIP-modified SA (5 μg, 75 pmol) in 50 μL 50 mM ammonium bicarbonate buffer (pH 8.5) was digested with proteinase K at a protease:protein ratio of 1:20 (w/w). The digestion was conducted at 37 °C or 55 °C for 18 h, followed by adduct enrichment with SPE.
Trypsin/chymotrypsin digestion: The PhIP-modified SA (5 μg, 75 pmol) in 50 μL 50 mM ammonium bicarbonate buffer (pH 8.5) containing CaCl2 (1 mM) was digested with trypsin at a protease:protein ratio of 1:50 (w/w) and chymotrypsin at a protease:protein ratio of 1:25 (w/w) at 37 °C for 20 h, followed by adduct enrichment with SPE [26,28].
Chymotrypsin digestion: The PhIP-modified SA (5 μg, 75 pmol) in 50 μL 50 mM ammonium bicarbonate buffer (pH 8.5) containing CaCl2 (1 mM) was incubated with chymotrypsin at a protease:protein ratio of 1:2.5 (w/w) at 25 °C for 20 h, followed by adduct enrichment with SPE.
2.2.5. Ultraperformance liquid chromatography-electrospray ionization/multistage scan mass spectrometry (UPLC-ESI/MSn)
Chromatography of the peptide digests was conducted with a NanoAcquity UPLC system (Waters Corp., Milford, MA) equipped with a Magic C18AQ column (0.3 × 150 mm, 3 μm particle size, MichromBioresource Inc., Auburn, CA). The peptides were resolved with a linear gradient starting from 90% A solvent (4.99% CH3CN, 0.01% HCO2H, 95% H2O) to 60% B solvent (95% CH3CN containing, 4.99% H2O, and 0.01% HCO2H) over 15 min, at a flow rate of 5 μL/min. For data-dependent analyses, a longer gradient of 60 min was employed [28].
MS spectra were acquired at positive ionization mode with a linear quadrupole ion trap mass spectrometer (LTQ, Thermo Fisher, San Jose, CA) equipped with an Advance CaptiveSpray™ source from Michrom Bioresource Inc. The temperature of capillary tube was set at 200 °C; the spray voltage was 1.5 kV; and the in-source fragmentation was10 V. There was no sheath or auxiliary gas. The automatic gain control (AGC) settings were full MS target 30,000 and MSn target 10,000; the maximum injection time was 10 ms, and one microscan was used for data acquisition. The isolation width m/z = 2.0, activation Q = 0.25, activation time = 15 ms, and a normalized CE = 25% were employed for all peptides. Xcalibur version 2.07 software was used for data manipulations. The tandem mass spectra of peptides were interpreted manually and facilitated by online software (Protein Prospector, Univ. of California, San Francisco, http://us.expasy.org/proteomics).
2.2.6. Estimates PhIP-Cys34 SA adducts
The total ion counts obtained by MS/MS of the peptides were used to estimate adduct levels of PhIP-modified SA. LQQC*PˆFEDHVK (C-[SO2]-PhIP) sulfonamide (2.2 pmol) was employed as the internal standard and added to PhIP-modified SA (5 μg protein) prior to proteolysis. The PhIP adducts were monitored as LQQC*PFEDHVK ([M + 3H]3+ at m/z 533.7), LQQC*PF ([M + H]+ at m/z 989.4), QC*PF ([M + H]+ at m/z 748.2), C*PF ([M + H]+ at m/z 620.2) or QC*PF (or QQC*PF ([M + H]+ at m/z 876.2)) for proteolytic digestions conducted, respectively, with trypsin/chymotrypsin, chymotrypsin, pronase E, pronase E/leucine aminopeptidase/prolidase or proteinase K (vide infra). For studies determining the lower limit of quantification of the LQQC*PFEDHVK peptide, five prominent product ions were employed and reported in Fig. 7.
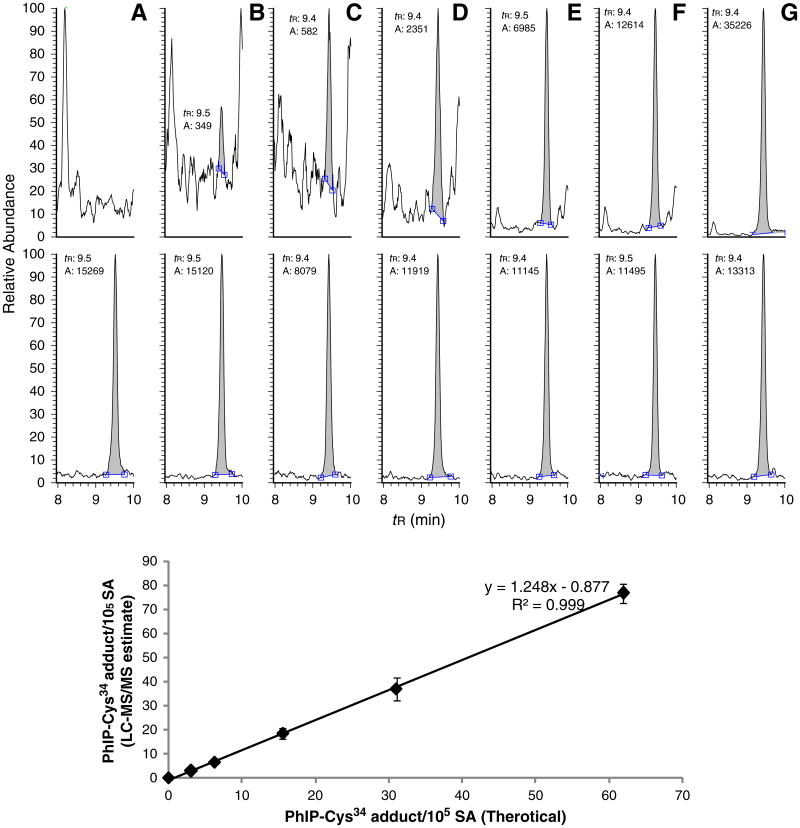
Fig. 7.
(A) UPLC–ESI/MS/MS chromatograms and estimates of LQQC*PFEDHVK (C-[SO2]-PhIP) sulfonamide ([M + 3H]3+ at m/z 533.7) recovered from trypsin/chymotrypsin digestion of a serial dilution of m-CPBA oxidized HONH-PhIP-modified SA (5 μg, 75 pmol) with unmodified SA: (A) 0, (B) 1.6, (C) 3.1, (D) 6.2, (E) 15.5, (F) 31.0, and (G) 62.0 adducts per 105 SA molecules (upper panel) and 0.75 μg of SA digest was employed for MS/MS analysis. The theoretical and target values of adduct levels are shown in the lower panel. The internal standard LQQC*PˆFEDHVK (C-[SO2]-PhIP) sulfonamide was added at a level of 14.9 pmol. Each value represents the average and standard deviation of three individual experiments.
2.2.7. Impact of denaturation of SA on estimates of Cys34 SA adducts of PhIP, Cys34-[SO3H] and Cys34 (C-[S–CH2CONH2])
The effect of denaturation on the efficacy of proteolytic digestion of SA with trypsin or trypsin/chymotrypsin was examined by determining the sequence coverage of the digested SA employing data dependent MS/MS scanning (described below), and by estimates of Cys34 containing peptide adducts recovered from trypsin/chymotrypsin digest. Five microgram (7.5 pmol) of IAA-alkylated SA, m-CPBA oxidized SA, or m-CPBA oxidized HONH-PhIP-modified SA underwent enzymatic digestion without or with prior denaturation, reduction and alkylation of the SA [28]. The total ion counts obtained by MS/MS of LQQC*PFEDHVK (C-[SO3H], [M + 2H]2+ at m/z 696.3), LQQC*PF (C-[S-CH2CONH2], [M + H]+ at m/z 792.3), LQQC*PFEDHVK (C-[SO2]-PhIP, [M + 3H]3+ at m/z 533.7) and their internal standards LQQC*PˆFEDHVK (C-[SO3H], [M + 2H]2+ at m/z 699.3, 14.9 pmol), LQQC*PF (C-[S-CH2CONH2]), [M + H]+ at m/z 798.3, 14.9 pmol) and LQQC*PˆFEDHVK (C-[SO2]-PhIP) ([M + 3H]3+ at m/z 535.7, 4.5 pmol) were added to the chemically-treated SA prior to proteolysis.
2.2.8. Data-dependent MS/MS
A full scan MS was obtained for the eluting peptides of SA in the range of 300–2000 Da, followed by three data-dependent MS/MS scans [28]. The MS/MS spectra of peptides were obtained using dynamic exclusion of previously analyzed precursors for 180 s with a repeat of 3 and a repeat duration of 60 s. The MS/MS spectra were obtained by CID of the peptide ions at a normalized collision energy of 35% to generate a series of b and y ions as major fragments. Fully automated data analysis on peptide adducts was conducted by converting RAW data to mz5 format in the ProteoWizard ms Convert tool [36]. Spectra were identified by the MyriMatch algorithm [28,37], version 2.1.111, using a 31 protein subset database containing SA from an initial search against the RefSeq human protein database, version 37.3. MyriMatch was configured to have Cys to contain carbamidomethyl (+57.0 Da) for IAA treatment, or (+32.0 or 48.0 Da) for oxidation of Cys to the sulfinic or sulfonic acids, and to allow for the possibility of oxidation (+16.0 Da) on methionines and deamidation (−17.0 Da) of N-terminal glutamines. Peptides modified with HONH-PhIP and HONH-[2H5]-PhIP were allowed to have dynamic modifications at [C,K,Y,S,T,W,H] of 222.1 (HONH-PhIP–H2O) and 227.1 Da (HONH-[2H5]-PhIP–H2O), or dynamic modifications at [C] of 238.1 and 243.1 Da for adduction with NO-PhIP (sulfinamide adducts) and 254.1 and 259.1 Da (sulfonamide adducts). Candidate peptides were allowed to have trypsin cleavages or protein termini at one or both termini (semitryptic search), and up to two missed cleavages were permitted. The precursor error was set at 1.25 m/z, but fragmentions were required to match within 0.5 m/z. Analyses of modified SA were also performed with trypsin/chymotrypsin digests, employing the same configurations. The IDPicker algorithm v3.0.420 filtered the identifications for each spectrum with a 5% identification false discovery rate at the peptide spectrum match level.
Isotopic data-dependent analyses of PhIP and [2H5]-PhIP modified SA were conducted as previously described [28]. The survey full scan mass spectra of the peptides were acquired on the characteristic isotopic pattern of unlabelled PhIP/[2H5]-PhIP at a partner intensity ratio of 65–100%, which triggered the acquisition of the product ion spectra of the adducts. The mass to charge differences were set at m/z 5.00, 2.50, or 1.67, respectively, for singly, doubly, or triply charged peptide species.
3. Results and discussion
3.1. Identification of major Cys34 peptide adducts from different proteolytic digestions of PhIP-modified SA
Proteolytic digestion of SA produces polypeptides of various lengths according to the specificity of the enzymes employed. Missed cleavages of peptides can occur during the digestion due to the solvent buffer conditions, post-translational modifications of proteins, non-specific chemical or enzymatic cleavage of proteins, or by the introduction of bulky molecules, such as PhIP into SA, which can affect the efficiency of enzyme digestion [38]. We previously reported that the N-oxidized metabolites of PhIP, HONH-PhIP and NO-PhIP, selectively reacted at the Cys34 residue of human SA [26,28]. The peptides recovered by proteolysis of m-CPBA-oxidized SA containing the Cys34 sulfonamide adducts of PhIP and [2H5]-PhIP (C-[SO2]-PhIP) in a 1:1 ratio were identified by the characteristic isotopic pattern of the adducts, which triggered the acquisition of MS/MS spectra [28]. The major adducted peptide was the single-missed cleavage peptide LQQC*PFEDHVK, when digestion was done with trypsin/chymotrypsin; the fully digested adducted peptide LQQC*PF was recovered in a much lower amount [28]. In this study, we have examined the proficiency of digestion of the SA-Cys34 adducts of PhIP with pronase E, or a mixture of pronase E/leucine aminopeptidase/prolidase, proteinase K, chymotrypsin, and by treatment with trypsin/chymotrypsin.
The major PhIP peptide adducts recovered from the different proteases are reported in Table 1. The digestion of the m-CPBA-oxidized PhIP-SA with pronase E produced the QC*PF sulfonamide as the major adduct rather than the fully digested C* (C-[SO2]-PhIP) sulfonamide). The recovery of the incompletely digested peptide adduct could be due to the steric effect of the bulky PhIP moiety adducted to the Cys34 and/or by the Pro35 residue, which may have hindered the effectiveness of proteolysis. Increasing the ratio of protease: SA (from 1:10 to 1:2) produced less QC*PF sulfonamide, in association with the appearance of the C*PF sulfonamide. The C*PF sulfonamide adduct, along with only minor amounts of C*P and C* sulfonamide adducts, was also detected following proteolytic digestion with a 3-enzyme mixture containing pronase E, leucine aminopeptidase, and prolidase (Fig. S-2).
Table 1.
Proteolytic peptide adducts containing Cys34, which were derived from different enzymatic systems. (Bold and italicized peptides were major peptide adducts for each proteolytic digestion system.)
| Enzyme(s) | Peptide adducts | Charge | m/z |
|---|---|---|---|
| Pronase E/leucine | C* | 1 | 376.1 |
| Aminopeptidase/prolidase | C*P | 1 | 473.1 |
| C*PF | 1 | 620.2 | |
| Pronase E (1:10) | QC*PF | 1 | 748.2 |
| Proteinase K (40 °C) | QC*PF | 1 | 748.2 |
| QQC*PF | 1 | 876.2 | |
| Proteinase K (55 °C) | QC*PF | 1 | 748.2 |
| QQC*PF | 1 | 876.2 | |
| Trypsin/chymotrypsin | LQQC*PF | 1 | 989.4 |
| LQQC*PFEDHVK | 3 | 533.2 | |
| Chymotrypsin | LQQC*PF | 1 | 989.4 |
| LQQC*PFEDHVK | 3 | 533.2 |
Remark: C* refers to C-[SO2]-PhIP with a mass addition of 254.1 Da to Cys.
Adduction products of mustards, acrylamide, and acetaminophen formed at the Cys34 of human SA are also resistant toward enzymatic hydrolysis and recovered principally as C*PF or QC*PF, following digestion with pronase [3,32,39]. QC*PF was the predominant adduct of PhIP recovered with proteinase K, but the signal was much lower than the signal obtained from any of the other proteases. The major peptide adduct obtained from the enzymatic digestion of PhIP-SA with trypsin/chymotrypsin occurred as the single-missed cleavage LQQC*PFEDHVK (C-[SO2]-PhIP) sulfonamide adduct. The double-missed cleavage peptide, LQQCPFEDHVKLVNEVTEF was not detected (L. Peng, unpublished observations). Employing larger quantities of chymotrypsin increased the amount of the fully cleaved LQQC*PF peptide; however, the signal of response was still much weaker than the missed cleavage peptide adduct. LQQC*PF was obtained as the major peptide only when highly elevated quantities of chymotrypsin (chymotrypsin/SA: 1/2.5) were employed for digestion. The digestion of PhIP-SA with chymotrypsin at 25 °C did produce several-fold more LQQC*PF sulfonamide than did the digestion at 37 °C.
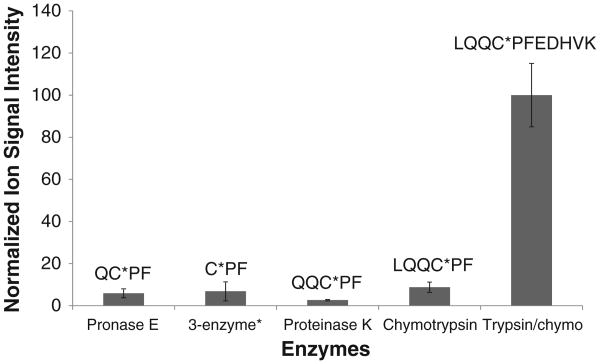
The signal of response of PhIP-SA peptides is determined by the efficiency of enzyme digestion, the ionization efficiency of the adducted peptide, and by any potential ion suppression matrix effects. The corresponding chromatograms of the MS ion signals for major peptide adducts recovered from the different proteases are shown in Fig. 3. The enzymatic digestion of SA with trypsin/chymotrypsin provided the highest signal of all the PhIP adducted peptides. The intensity of the LQQC*PFEDHVK (C-[SO2]-PhIP) signal obtained from trypsin/chymotrypsin digestion of SA was 10-fold or greater than the signal of the PhIP peptide adducts recovered following digestion of SA with other proteases (Fig. 4).
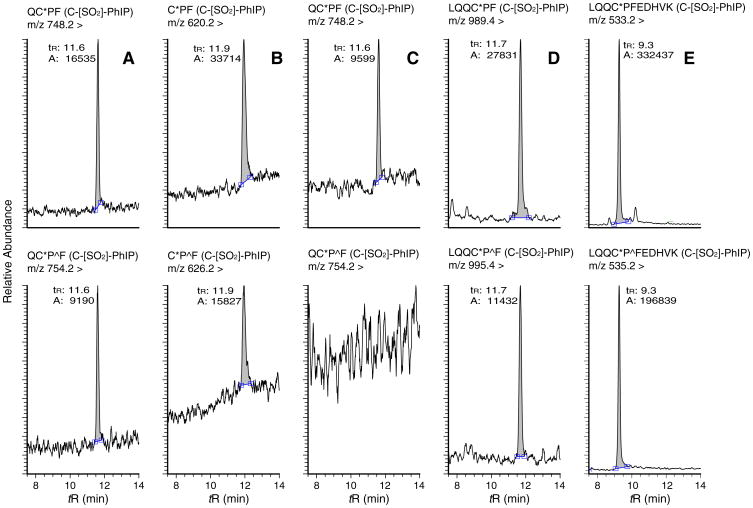
Fig. 3.
UPLC-ESI/MS2 chromatograms of Cys34 peptide adducts recovered from m-CPBA oxidized HONH-PhIP-modified SA digests (upper panel) and corresponding isotopically labeled peptide adducts recovered from the internal standard LQQC*PˆFEDHVK (C-[SO2]-PhIP) sulfonamide (bottom panel), following (A) pronase E; (B) pronase E/leucine aminopeptidase/prolidase; (C) chymotrypsin; and (D) trypsin/chymotrypsin digestions. Pˆ is 13C15N universally labeled proline. All adducts and internal standards were measured at m/z [M + H]+ except for LQQC*PFEDHVK, which was monitored at m/z [M + 3H]3+. ‘A’ refers to the area.
Fig. 4.
Comparison of digestion efficiency and recovery of the SA-Cys34 containing PhIP adducts (C-[SO2]-PhIP), by different proteases. Each value represents the average and standard deviation of three individual experiments. The response of the adduct recovered from digestion with proteinase K was poor and no further studies were conducted. * The 3 enzyme systems contained pronase E, leucine aminopeptidase and prolidase.
We examined the ionization efficiencies and signals of response of several synthetic peptides. The signal of response of the unmodified synthetic triply charged peptide LQQCPFEDHVK at ([M + 3H]3+) at m/z 448.6 was ∼2-fold greater than response of the singly protonated peptide LQQCPF at ([M + H]+) m/z 735.3, both in full and MS/MS scan modes (Fig. S-3). However, the signal of the triply protonated synthetic peptide adduct LQQC*PFEDHVK(C-[SO2]-PhIP) ([M + 3H]3+ at m/z 533.7) was ∼6 and 11-fold greater than the signal of the singly protonated peptide LQQC*PF(C-[SO2]-PhIP) ([M + H]+ at m/z 989.5), respectively in full and MS2 scan modes. These data signify that the charge state and not matrix effects are responsible for the high ionization efficiency and elevated response of the LQQC*PFEDHVK (C-[SO2]-PhIP) sulfonamide adduct over the singly protonated C*-[SO2]-PhIP adducts, and thereby explains the large differences in response of signals of the different PhIP peptide adducts obtained by digestion of SA with different proteases (Fig. 4).
3.2. Estimates of PhIP-SA Cys34 sulfonamide adducts as a function of different proteolytic digestion conditions of PhIP-modified SA
LQQC*PˆFEDHVK (C-[SO2]-PhIP) sulfonamide adduct was employed as an internal standard and added to the PhIP-modified SA prior to proteolytic digestion. The stability of LQQC*PFEDHVK: LQQC*PˆFEDHVK (4.5 pmol, 1:1) toward the different proteases (trypsin, trypsin/chymotrypsin, pronase E, or pronase E, leucine aminopeptidase and prolidase) was examined by employing the same quantities of enzymes that were used for the digestion of PhIP-modified SA (5 μg) described above. The abilities of the different proteases to digest the 1-missed cleaved tryptic/chymotryptic peptide (C-[SO2]-PhIP) sulfonamide homologues were assessed by isotopic data-dependent scanning and monitoring mass to charge differences set at m/z 6.0, 3.0, or 2.0, respectively, for singly, doubly, or triply charged peptide species. Trypsin/chymotrypsin did not digest LQQC*PFEDHVK: LQQC*PˆFEDHVK at F, and the fully cleaved product LQQC*PF: LQQC*PˆF was not detected. In contrast, the unlabeled and the labeled (C-[SO2]-PhIP) sulfonamide peptide did undergo digestion with elevated levels of chymotrypsin, or by digestion with pronase E, or by treatment with pronase E/leucine aminopeptidase/prolidase to produce the same smaller peptides observed in the different proteolytic digests of PhIP-modified SA(Table 1). Thus, LQQC*PˆFEDHVK (C-[SO2]-PhIP) was selected as an internal standard to measure the amount of (C-[SO2]-PhIP) sulfonamide in SA digested by these different proteases.
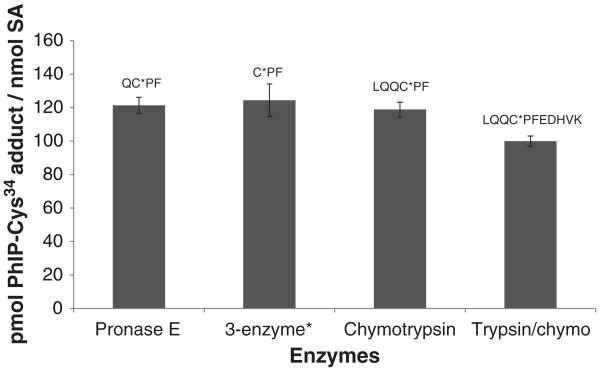
The estimates of the sulfonamide adducts were obtained by measurement of LQQC*PFEDHVK, LQQC*PF, QC*PF or C*PF in m-CPBA oxidized PhIP-modified SA, following digestion with, respectively, trypsin/chymotrypsin, chymotrypsin, pronase Eor pronase/leucine aminopeptidase/prolidase (Fig. 5). The SA was not denatured prior to enzyme digestion. The mean levels of adducts recovered from the different digests were within 20% of each other. The level of LQQC*PFEDHVK (C-[SO2]-PhIP) obtained from the PhIP-modified SA digested with trypsin/chymotrypsin of accounted for greater than 65% of the PhIP bound to SA, on the basis of UV measurements and recovery of PhIP following mild acid hydrolysis of the presumed sulfinamide of PhIP [26]. Because of high signal of response, the LQQC*PFEDHVK PhIP sulfonamide adduct was selected as the target peptide, following proteolysis of SA with trypsin/chymotrypsin.
Fig. 5.
UPLC–ESI/MS/MS estimates of peptide adducts containing Cys34 recovered from m-CPBA-oxidized HONH-PhIP-modified SA with different proteases. Isotopic labeled LQQC*PˆFEDHVK (C-[SO2]-PhIP) sulfonamide was employed as the internal standard. Each value represents the average and standard deviations of three individual experiments. *The 3 enzyme system contained pronase E, leucine aminopeptidase and prolidase.
3.3. The influence of denaturation of SA on the efficacy of enzyme digestion and recovery of PhIP-Cys34 SA-Cys34SO3H and SA-Cys34S-CH2CONH2 adducts
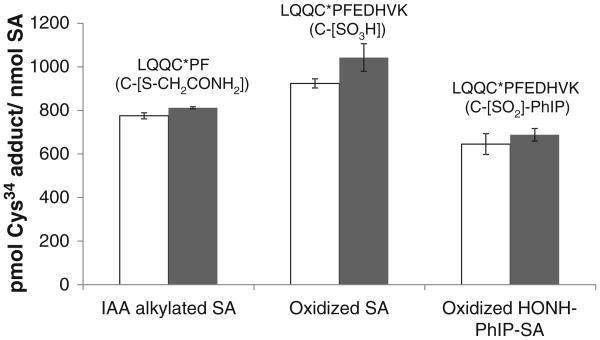
Cys34 is the only reduced sulfhydryl residue present in human SA, which contains 17 intrachain disulfide bonds, and the resistance of native SA to denaturation is well known [40,41]. Like many proteins, SA is commonly denatured by heat treatment and chemical reduction of the internal disulfide bonds in the presence of 6 M guanidine – HCl, followed by alkylation of the newly formed sulfhydryl groups with IAA, to expose as many sites as possible prior to proteolytic digestion [38]. However, comparable amounts of LQQC*PFEDHVK (C-[SO2]-PhIP) were recovered from native and denatured SA, following digestion with trypsin/chymotrypsin (Fig. 6) [27]. Moreover, similar amounts of LQQC*PFEDHVK (C-[SO2]-PhIP) were recovered irrespective of whether the digestion SA was conducted by treatment with trypsin/chymotrypsin simultaneously at 37 °C; or by incubation with trypsin alone at 37 °C, followed by incubation with chymotrypsin at its optimal temperature of 25 °C (L. Peng, unpublished observations).
Fig. 6.
Estimates of modified peptides containing Cys34 recovered from trypsin/chymotrypsin digestion of SA either alkylated with IAA, oxidized with m-CPBA, or modified with HONH-PhIP followed by m-CPBA oxidation. Adduct levels of modified SA were determined without (open bar) or with predenaturation of SA (filled bar). Isotopically labeled LQQC*PˆF (C-[S-CH2CONH2]), LQQC*PˆFEDHVK (C-[SO3H]) and LQQC*PˆFEDHVK (C-[SO2]-PhIP) sulfonamide were employed as internal standards. Each value represents the average and standard deviations of three individual experiments. The level of LQQC*PˆFEDHVK (C-[SO2]-PhIP) was multiplied by 30× to be visualized on the same scale with the other adducts.
The Cys34 sulfonic acid (C-[SO3H]) of SA prepared by m-CPBA-mediated oxidation of SA (pre-reduced with βME to obtain 0.95 mol “Cys34-SH” per mol SA) and the carbamido-methylated Cys34 (C-[S-CH2CONH2]) of pre-reduced SA prepared by alkylation of SA with IAA, were also recovered in high yield, irrespective of denaturation of SA (Fig. 6). The efficacy of enzyme digestion was based on the quantitation of LQQC*PFEDHVK (C-[SO3H]) from m-CPBA-oxidized SA and LQQC*PF (C-[S-CH2CONH2]) from IAA-treated SA. The levels of LQQC*PFEDHVK (C-[SO3H]) recovered from the trypsin/chymotrypsin digests of native and denatured SA samples were greater than 90% of the target value. In contrast to SA-Cys34 modified as C-[SO3H] or C-[SO2]-PhIP, the SA-Cys34 alkylated with IAA underwent digestion to form primarily LQQC*PF (C-[S-CH2CONH2]) rather than LQQC*PFEDHVK (C-[S-CH2-CONH2]) (Fig. S-4). The internal standard LQQC*PˆFEDHVK was also quantitatively digested at the F residue and recovered as LQQC*PˆF (C-[S-CH2CONH2]) ([M + H]+ at m/z 798.3). The amount of LQQC*PF (C-[S-CH2CONH2]) recovered was ∼80% of the target value for both native and denatured SA. These results demonstrate that L31QQC*PFEDHVK41 and L31QQC*PF36 peptides are efficiently recovered in high yield irrespective of prior denaturation of SA. The accessibility of the peptide sequence containing the Cys34 in native SA may be attributed to its position in subdomain lA, which is at the N-terminal portion of SA; this subdomain is flexible and distant from the first of the 17 intrachain disulfide linkages which commence between Cys53 and Cys62 of SA [41].
3.4. Comparison of the specificities of sequence and biochemical grade trypsin and chymotrypsin to digest SA and recover PhIP-SA Cys34 adducts
The price of sequence grade trypsin and chymotrypsin is more than 1000-fold greater than the cost of biochemical grade enzyme per unit activity. With the low level of PhIP modification of SA that reportedly occurs in humans [42,43], we anticipate that 100 μg up to several mgs of SA will be required for chemical analysis. The high cost of sequence grade enzymes would preclude extensive biomonitoring studies in humans. Therefore, we sought to determine whether less expensive biochemical grade trypsin and chymotrypsin could be employed to efficiently digest PhIP-modified SA and recover PhIP peptides with the same specificity as the sequence grade enzymes. The efficacy of digestion was determined by the sequence coverage of native or denatured commercial SA or SA obtained from plasma, following purification by HiTrap Blue affinity chromatography. Sequence coverage was done by data dependent MS/MS scanning and analysis of the peptide digest with the MyriMatch algorithm parameters described in Chemicals and materials. The sequence coverage of SA was greater than 87% and 94%, respectively, when sequence grade trypsin (Promega) and biochemical grade trypsin (Sigma) were employed for digestion. As expected, the sequence coverage of non-denatured, non-reduced SA, by both grades of enzymes, was decreased to 55%. Under the identical MS/MS scanning parameters, the sequence coverage of m-CPBA-oxidized SA, IAA alkylated SA, or m-CPBA-oxidized HONH-PhIP-modified SA digested with trypsin/chymotrypsin accounted for 60–65% of the protein sequence. The lower level of sequence coverage is attributed in part to the higher percentage of hydrophilic peptides of low molecular weight (di-, tri- or tetra-peptides) which were lost during SPE processing, or the sparse number of y and b ions present in the MS/MS spectra of some small molecular weight peptides may have been insufficient for recognition by the MyriMatch algorithm.
We then compared the efficacy and specificity of sequence (Promega) and biochemical (Sigma) grades of trypsin and chymotrypsin to digest and recover LQQC*PFEDHVK (C-[SO2]-PhIP) from PhIP-modified SA. The adduct was recovered with the same efficiency from PhIP-modified SA digested with both grades of proteases (Fig. S-5), Thus, an economical and quantitative method was established to measure LQQC*PFEDHVK (C-[SO2]-PhIP) employing biochemical grade trypsin/chymotrypsin for proteolytic digestion of m-CPBA-oxidized PhIP-SA without denaturation of SA.
The intraday and interday measurements of the mean adduct levels were determined on three different days over a time period of two weeks and estimated at: 19.7, 20.3, and 19.6 pmol PhIP/nmol SA, when 5 μg SA was assayed. The within-day coefficient of variation (%) and between-day coefficient variation were: 9.8 and 9.1%. Then, we examined the efficacy of biochemical grade trypsin/chymotrypsin to digest variable amounts of PhIP-modified SA, by digesting 5 μg (73 pmol), 25 μg (365 pmol) or 125 μg (1825 pmol) of SA with a constant amount of trypsin/chymotrypsin/SA ratio (1/2/50) in the presence of 6.7 pmol of LQQC*PˆFEDHVK (C-[SO2]-PhIP) as the internal standard in a final volume of 0.25 mL of ammonium bicarbonate buffer (pH 8.5). The estimates of LQQC*PFEDHVK (pmol PhIP/nmol SA) obtained from 3 independent measurements ranged from (mean ± standard deviation): 19.6 ± 1.1 (5 μg of SA), 24.5 ± 1.7 (25 μg of SA) and 21.9 ± 1.3 (125 μg of SA), demonstrating that variable quantities of PhIP-modified SA can be digested with similar efficiencies.
3.5. Lower limit of quantification of PhIP-SA Cys34 adducts
Serial dilutions of m-CPBA oxidized HONH-PhIP-modified SA with unmodified SA were subjected to digestion with trypsin/chymotrypsin. The limit of detection and limit of quantification (LOQ) were ∼0.15 fmol and 0.3 fmol of LQQC*PFEDHVK (C-[SO2]-PhIP) adduct/μg SA, or approximately 1 and 2 adducts per 105 SA molecules, when 0.75 μg of SA digest was employed for MS/MS (Fig. 7). The LOQ may be further decreased by assaying more SA, or by selective enrichment of target peptide adducts prior to MS/MS analysis, or by employment of a high resolution accurate MS instrument.
4. Conclusion
N-Oxidized metabolites of HAAs and aromatic amines bind to Cys34 of rodent SA or human SA in vitro to form putative labile arylsulfenamide or arylsulfinamide adducts [18,19,22,44]. However, the adducts are unstable and undergo hydrolysis during proteolytic digestion, precluding the employment of these types of protein carcinogen adducts as biomarkers to assess human exposure. PhIP adduct formation occurs with SA in humans [21,42,43]. At least one adduct underwent hydrolysis under mildly acidic pH to produce PhIP, suggesting that a portion of the PhIP had formed a sulfur–nitrogen linked adduct at the Cys34 residue of SA [21]. The chemistry that we describe to oxidize labile arylsulfenamide and arylsulfinamide protein adducts to stable arylsulfonamide linkages represents a novel approach to measure labile arylsulfinamide protein adducts [27]. Studies are ongoing to improve the selective enrichment LQQC*PFEDHVK (C-[SO2]-PhIP) adduct from the complex SA digest matrix and the sensitivity of adduct detection by high resolution and accurate mass measurements of the SA-PhIP sulfonamide adduct, which is expected to occur at levels of several adducts per 107 SA molecules in humans [42,43]. With these improvements in analytical methodology, the SA arylsulfonamide adducts of PhIP, other HAAs, or aromatic amines, which are prominently formed well-done cooked meats or tobacco smoke, may serve as biomarkers in molecular epidemiology that seek to address the role of these chemicals in diet or tobacco associated cancers.
Supplementary Material
Acknowledgments
We would like to thank the Dr. David Tabb Laboratory at the Department of Biomedical Informatics, and the Department of Biochemistry and Mass Spectrometry Research Center, Vanderbilt University, for the introduction to and use of the MyriMatch software program.
This research was supported by Grant R01 CA122320 (R.J.T.)
Footnotes
Transparency document: The Transparency document associated with this article can be found, in the online version.
Conflict of interest: The authors declare no conflict of interest pertaining to this research.
Appendix A. Supplementary data: Additional information as noted in text (Figs. S-1–S-5). Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.jprot.2014.03.023.
Contributor Information
Lijuan Peng, Email: lijuan_peng@hotmail.com.
Robert J. Turesky, Email: Rturesky@umn.edu.
References
- 1.Skipper PL, Tannenbaum SR. Protein adducts in the molecular dosimetry of chemical carcinogens. Carcinogenesis. 1990;11:507–18. doi: 10.1093/carcin/11.4.507. [DOI] [PubMed] [Google Scholar]
- 2.Rappaport SM. Biomarkers intersect with the exposome. Biomarkers. 2012;17:483–9. doi: 10.3109/1354750X.2012.691553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Noort D, Hulst AG, de Jong LP, Benschop HP. Alkylation of human serum albumin by sulfur mustard in vitro and in vivo: mass spectrometric analysis of a cysteine adduct as a sensitive biomarker of exposure. Chem Res Toxicol. 1999;12:715–21. doi: 10.1021/tx9900369. [DOI] [PubMed] [Google Scholar]
- 4.Liebler DC. Proteomic approaches to characterize protein modifications: new tools to study the effects of environmental exposures. Environ Health Perspect. 2002;110(Suppl. 1):3–9. doi: 10.1289/ehp.02110s113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tornqvist M, Fred C, Haglund J, et al. Protein adducts: quantitative and qualitative aspects of their formation, analysis and applications. J Chromatogr B Analyt Technol Biomed Life Sci. 2002;778:279–308. doi: 10.1016/s1570-0232(02)00172-1. [DOI] [PubMed] [Google Scholar]
- 6.Aldini G, Gamberoni L, Orioli M, et al. Mass spectrometric characterization of covalent modification of human serum albumin by 4-hydroxy-trans-2-nonenal. J Mass Spectrom. 2006;41:1149–61. doi: 10.1002/jms.1067. [DOI] [PubMed] [Google Scholar]
- 7.Colzani M, Aldini G, Carini M. Mass spectrometric approaches for the identification and quantification of reactive carbonyl species protein adducts. J Proteomics. 2013;92:28–50. doi: 10.1016/j.jprot.2013.03.030. [DOI] [PubMed] [Google Scholar]
- 8.Rappaport SM, Li H, Grigoryan H, et al. Adductomics: Characterizing exposures to reactive electrophiles. Toxicol Lett. 2012;213:83–90. doi: 10.1016/j.toxlet.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sugimura T, Wakabayashi K, Nakagama H, Nagao M. Heterocyclic amines: Mutagens/carcinogens produced during cooking of meat and fish. Cancer Sci. 2004;95:290–9. doi: 10.1111/j.1349-7006.2004.tb03205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turesky RJ, Le Marchand L. Metabolism and biomarkers of heterocyclic aromatic amines in molecular epidemiology studies: lessons learned from aromatic amines. Chem Res Toxicol. 2011;24:1169–214. doi: 10.1021/tx200135s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kiese M, Taeger K. The fate of phenylhydroxylamine in human red cells. Naunyn Schmiedebergs Arch Pharmacol. 1976;292:59–66. doi: 10.1007/BF00506490. [DOI] [PubMed] [Google Scholar]
- 12.Kiese M. The biochemical production of ferrihemoglobin-forming derivatives from aromatic amines, and mechanisms of ferrihemoglobin formation. Pharmacol Rev. 1966;18:1091–161. [PubMed] [Google Scholar]
- 13.Neumann HG. Biomonitoring of aromatic amines and alkylating agents by measuring hemoglobin adducts. Int Arch Occup Environ Health. 1988;60:151–5. doi: 10.1007/BF00378690. [DOI] [PubMed] [Google Scholar]
- 14.Green LC, Skipper PL, Turesky RJ, et al. In vivo dosimetry of 4-aminobiphenyl in rats via a cysteine adduct in hemoglobin. Cancer Res. 1984;44:4254–9. [PubMed] [Google Scholar]
- 15.Gan J, Skipper PL, Gago-Dominguez M, et al. Alkylaniline-hemoglobin adducts and riskof non-smoking-related bladder cancer. J Natl Cancer Inst. 2004;96:1425–31. doi: 10.1093/jnci/djh274. [DOI] [PubMed] [Google Scholar]
- 16.Bryant MS, Vineis P, Skipper PL, Tannenbaum SR. Hemoglobin adducts of aromatic amines: associations with smoking status and type of tobacco. Proc Natl Acad Sci U S A. 1988;85:9788–91. doi: 10.1073/pnas.85.24.9788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ringe D, Turesky RJ, Skipper PL, Tannenbaum SR. Structure of the single stable hemoglobin adduct formed by 4-aminobiphenyl in vivo. Chem Res Toxicol. 1988;1:22–4. doi: 10.1021/tx00001a003. [DOI] [PubMed] [Google Scholar]
- 18.Skipper PL, Obiedzinski MW, Tannenbaum SR, et al. Identification of the major serum albumin adduct formed by 4-aminobiphenyl in vivo in rats. Cancer Res. 1985;45:5122–7. [PubMed] [Google Scholar]
- 19.Turesky RJ, Skipper PL, Tannenbaum SR. Binding of 2-amino-3-methylimidazo[4,5-f]quinoline to hemoglobin and albumin in vivo in the rat Identification of an adduct suitable for dosimetry. Carcinogenesis. 1987;8:1537–42. doi: 10.1093/carcin/8.10.1537. [DOI] [PubMed] [Google Scholar]
- 20.Lynch AM, Murray S, Boobis AR, et al. The measurement of MeIQx adducts with mouse haemoglobin in vitro and in vivo: implications for human dosimetry. Carcinogenesis. 1991;12:1067–72. doi: 10.1093/carcin/12.6.1067. [DOI] [PubMed] [Google Scholar]
- 21.Magagnotti C, Orsi F, Bagnati R, et al. Effect of diet on serum albumin and hemoglobin adducts of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) in humans. Int J Cancer. 2000;88:1–6. doi: 10.1002/1097-0215(20001001)88:1<1::aid-ijc1>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 22.Chepanoske CL, Brown K, Turteltaub KW, Dingley KH. Characterization of a peptide adduct formed by N-acetoxy-2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP), a reactive intermediate of the food carcinogen PhIP. Food Chem Toxicol. 2004;42:1367–72. doi: 10.1016/j.fct.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 23.Liu L, Wagner CR, Hanna PE. Isoform-selective inactivation of human arylamine N-acetyltransferases by reactive metabolites of carcinogenic arylamines. Chem Res Toxicol. 2009;22:1962–74. doi: 10.1021/tx9002676. [DOI] [PubMed] [Google Scholar]
- 24.Liu L, Wagner CR, Hanna PE. Human arylamine N-acetyl-transferase 1: in vitro and intracellular inactivation by nitrosoarene metabolites of toxic and carcinogenic arylamines. Chem Res Toxicol. 2008;21:2005–16. doi: 10.1021/tx800215h. [DOI] [PubMed] [Google Scholar]
- 25.Guo Z, Wagner CR, Hanna PE. Mass spectrometric investigation of the mechanism of inactivation of hamster arylamine N-acetyltransferase 1 by N-hydroxy-2-acetylaminofluorene. Chem Res Toxicol. 2004;17:275–86. doi: 10.1021/tx030045o. [DOI] [PubMed] [Google Scholar]
- 26.Peng L, Turesky RJ. Mass spectrometric characterization of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine N-oxidized metabolites bound at Cys34 of human serum albumin. Chem Res Toxicol. 2011;24:2004–17. doi: 10.1021/tx2003504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng L, Turesky RJ. Capturing labile sulfenamide and sulfinamide serum albumin adducts of carcinogenic arylamines by chemical oxidation. Anal Chem. 2013;85:1065–72. doi: 10.1021/ac3028273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peng L, Dasari S, Tabb DL, Turesky RJ. Mapping serum albumin adducts of the food-borne carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine by data-dependent tandem mass spectrometry. Chem Res Toxicol. 2012;25:2179–93. doi: 10.1021/tx300253j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baxter JH, Lai CS, Phillips RR, et al. Direct determination of methionine sulfoxide in milk proteins by enzyme hydrolysis/high-performance liquid chromatography. J Chromatogr A. 2007;1157:10–6. doi: 10.1016/j.chroma.2007.04.035. [DOI] [PubMed] [Google Scholar]
- 30.Tsao M, Otter DE. Quantification of glutamine in proteins and peptides using enzymatic hydrolysis and reverse-phase high-performance liquid chromatography. Anal Biochem. 1999;269:143–8. doi: 10.1006/abio.1998.3091. [DOI] [PubMed] [Google Scholar]
- 31.Burkhart JM, Schumbrutzki C, Wortelkamp S, et al. Systematic and quantitative comparison ofdigest efficiency and specificity reveals the impact of trypsin quality on MS-based proteomics. J Proteomics. 2012;75:1454–62. doi: 10.1016/j.jprot.2011.11.016. [DOI] [PubMed] [Google Scholar]
- 32.Damsten MC, Commandeur JN, Fidder A, et al. Liquid chromatography/tandem mass spectrometry detection of covalent binding of acetaminophen to human serum albumin. Drug Metab Dispos. 2007;35:1408–17. doi: 10.1124/dmd.106.014233. [DOI] [PubMed] [Google Scholar]
- 33.Li F, Chordia MD, Woodling KA, Macdonald TL. Irreversible alkylation of human serum albumin by zileuton metabolite 2-acetylbenzothiophene-S-oxide: a potential model for hepatotoxicity. Chem Res Toxicol. 2007;20:1854–61. doi: 10.1021/tx7001417. [DOI] [PubMed] [Google Scholar]
- 34.Turesky RJ, Lang NP, Butler MA, et al. Metabolic activation of carcinogenic heterocyclic aromatic amines by human liver and colon. Carcinogenesis. 1991;12:1839–45. doi: 10.1093/carcin/12.10.1839. [DOI] [PubMed] [Google Scholar]
- 35.Riener CK, Kada G, Gruber HJ. Quick measurement of protein sulfhydryls with Ellman's reagent and with 4,4′-dithiodipyridine. Anal Bioanal Chem. 2002;373:266–76. doi: 10.1007/s00216-002-1347-2. [DOI] [PubMed] [Google Scholar]
- 36.Kessner D, Chambers M, Burke R, et al. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics. 2008;24:2534–6. doi: 10.1093/bioinformatics/btn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tabb DL, Fernando CG, Chambers MC. MyriMatch: highly accurate tandem mass spectral peptide identification by multivariate hypergeometric analysis. J Proteome Res. 2007;6:654–61. doi: 10.1021/pr0604054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kinter M, Sherman NE. Protein Sequencing and Identification Using Tandem Mass Spectrometry. New York: Wiley-Interscience; 2000. [Google Scholar]
- 39.Noort D, Fidder A, Hulst AG. Modification of human serum albumin by acrylamide at cysteine-34: a basis for a rapid biomonitoring procedure. Arch Toxicol. 2003;77:543–5. doi: 10.1007/s00204-003-0484-5. [DOI] [PubMed] [Google Scholar]
- 40.Peters T., Jr Serum albumin. Adv Protein Chem. 1985;37:161–245. doi: 10.1016/s0065-3233(08)60065-0. [DOI] [PubMed] [Google Scholar]
- 41.Sugio S, Kashima A, Mochizuki S, et al. Crystal structure of human serum albumin at 2.5 A resolution. Protein Eng. 1999;12:439–46. doi: 10.1093/protein/12.6.439. [DOI] [PubMed] [Google Scholar]
- 42.Dingley KH, Curtis KD, Nowell S, et al. DNA and protein adduct formation in the colon and blood of humans after exposure to a dietary-relevant dose of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine. Cancer Epidemiol Biomarkers Prev. 1999;8:507–12. [PubMed] [Google Scholar]
- 43.Garner RC, Lightfoot TJ, Cupid BC, et al. Comparative biotransformation studies of MeIQx and PhIP in animal models and humans. Cancer Lett. 1999;143:161–5. doi: 10.1016/s0304-3835(99)00118-4. [DOI] [PubMed] [Google Scholar]
- 44.Lynch AM, Murray S, Zhao K, et al. Molecular dosimetry of the food-borne carcinogen MeIQx using adducts of serum albumin. Carcinogenesis. 1993;14:191–4. doi: 10.1093/carcin/14.2.191. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.