Abstract
A novel esterase gene (estI) of Lactobacillus casei CL96 was localized on a 3.3-kb BamHI DNA fragment containing an open reading frame (ORF) of 1,800 bp. The ORF of estI was isolated by PCR and expressed in Escherichia coli, the methylotrophic bacterium Methylobacterium extorquens, and the methylotrophic yeast Pichia pastoris under the control of T7, methanol dehydrogenase (PmxaF), and alcohol oxidase (AOX1) promoters, respectively. The amino acid sequence of EstI indicated that the esterase is a novel member of the GHSMG family of lipolytic enzymes and that the enzyme contains a lipase-like catalytic triad, consisting of Ser325, Asp516, and His558. E. coli BL21(DE3)/pLysS containing estI expressed a novel 67.5-kDa protein corresponding to EstI in an N-terminal fusion with the S · tag peptide. The recombinant L. casei CL96 EstI protein was purified to electrophoretic homogeneity in a one-step affinity chromatography procedure on S-protein agarose. The optimum pH and temperature of the purified enzyme were 7.0 and 37°C, respectively. Among the pNP (p-nitrophenyl) esters tested, the most selective substrate was pNP-caprylate (C8), with Km and kcat values of 14 ± 1.08 μM and 1,245 ± 42.3 S−1, respectively.
Microbial esterases and lipases are currently receiving considerable attention because of their potential applications in biotechnology for food processing, surfactant composition, detergents, paper, oil manufacture, diagnostics, and optically active drugs (27, 28). The enzymes that modify milk fat are lipases (triacylglycerol lipases; EC 3.1.1.3) and esterases (EC 3.1.1.1). Esterases are, by definition, enzymes that have the ability to hydrolyze ester substrates with short-chain fatty esters (≤C10), whereas lipases hydrolyze long-chain acylglycerols (≥C10) (54).
For many years, scientists in the dairy field have tried to shorten the maturation period and to reduce the inherent costs, as well as to enhance flavor intensity, for various cheeses. So far, lipolytic and proteolytic enzymes have been used extensively in the dairy and food processing industries for hydrolysis of milk fat with the purpose of flavor enhancement or acceleration of ripening processes in cheeses and cheese-related products, but bitterness and rancidity have precluded their use on a large scale. A correct blend of alkanoic acid with carbon chains from C6 to C10 appears to impart a cheese-like flavor (41). The release of long-chain (C10 to C16) fatty acids by microbial lipase produces a soapy flavor, whereas short-chain fatty acids from animal esterases produce unclean flavors (50). The free fatty acids, which are liberated by the action of lipases or esterases on milk fat, give many dairy products their typical flavor characteristics. Upon further breakdown of fatty acids, reactions with other components of the maturing cheeses and fermented dairy products, which may contribute to the formation of various flavor components, are likely to occur (51).
Esterases and lipases are widely present in various organisms from bacteria to higher eucaryotes. A common characteristic of these enzymes is that they contain a catalytic triad, composed of Ser, His, and Asp or Glu (15, 48). In addition, most of these enzymes have a structural motif, G-X-S-X-G, which contains the active-site serine residue (6). This motif is usually located between a β-strand and an α-helix, and it assumes an extremely sharp turn called a nucleophile elbow (42).
Although many reports on the cloning and expression of microbial lipases and esterases have been published (31, 32), very little has been reported on lactic acid bacteria as a source of enzymes. Recently, Fenster et al. (19, 20) reported the nucleotide sequences of arylesterase (EstB) and intracellular esterase (EstC) from Lactobacillus casei LILA. Amino acid sequence analysis revealed that the mature proteins possess the GXSXG motif at the active site, and EstB selectivity for para-nitrophenyl (pNP) esters was greatest for C5 and C6 compounds, whereas EstC was most selective for C3 and C4 compounds. Other studies showed that the esterase in L. casei CL96 (10) possesses strong esterolytic activity for C6 and C8 compounds. These properties are somewhat different from those of L. casei LILA EstB and EstC.
In this communication, we report the molecular properties and enzymatic characteristics of a novel esterase from L. casei CL96, and we also report the large-scale production of recombinant esterase in methylotrophic yeast (Pichia pastoris) and bacteria (Methylobacterium extorquens), since the Pichia expression system is well characterized (24) and M. extorquens (22, 23) has recently been shown to express high levels of recombinant protein in high-cell-density fed-batch culture.
MATERIALS AND METHODS
Strains, media, and plasmids.
The strains and plasmids used in this study and their properties are listed in Table 1. L. casei CL96 is a cheese isolate that was obtained from the Food Research and Development Centre, Agriculture and Agri-Food Canada (St. Hyacinthe, Quebec). Its identity was confirmed by 16S rRNA sequencing in our laboratory (2). The stock culture was maintained in MRS broth (14) containing glycerol (50%, vol/vol) at −70°C. Escherichia coli Top10F′ was used as a host strain for cloning and maintenance of plasmids. E. coli BL21(DE3)/pLysS was used as a host for expression of the estI gene under the control of the T7 promoter. E. coli transformants were grown at 37°C in Luria-Bertani (LB) broth containing, when required, 100 μg of ampicillin and 50 μg of kanamycin per ml. The pCWL70 vector (11; Y. J. Choi and B. H. Lee, Abstr. 101st Gen. Meet. Am. Soc. Microbiol., abstr. O107, p. 552, 2001), which has a positive selection mechanism based on a CCDB gene controlled by a lac promoter, was used for construction of the L. casei CL96 genomic library and as a cloning vector. For E. coli expression studies, pET29a (Novagen, Madison, Wis.) was used.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| Strains | ||
| E. coli Top10F′ | F′ [lacIq Tn10 (Tetr)] mcrA Δ(mrr-sdRMS-mcrBC) φ80 lacZ; cloning host | Invitrogen Canada |
| E. coli BL21(DE3)/pLysS | F−ompT hsdSB(rB− mB−) dcm (DE3); cloning and expression host for estI gene | Invitrogen Canada |
| L. casei CL96 | Wild type; source of estI gene | This study |
| M. extorquens ATCC 55366 | Wild type; cloning host for expression of estI gene | American Type Culture Collection |
| P. pastoris X-33 | Wild type; cloning host for expression of estI gene | Invitrogen Canada |
| Plasmids | ||
| pCWL70 | Positive selection vector; Ampr Emr | Choi and Lee, Abstr. 101st Gen. Meet. Am. Soc. Microbiol. 2001 |
| pCESTa | 3.3-kb BamHI fragment cloned into BamHI site of pCWL70; Ampr | This study |
| pET29a | E. coli expression vector; Kmr | Novagen |
| pCESTb | estI with BamHI and NotI cloned into BamHI-NotI sites of pET29a; Kmr | This study |
| pCM110 | M. extorquens expression vector; Tcr | 39 |
| pCESTc | estI with SpeI and ClaI cloned into SpeI-ClaI sites of pCM110; Tcr | This study |
| pPICZ B | Pichia expression vector; Zeor | Invitrogen Canada |
| pCESTd | estI with KpnI and NotI cloned into KpnI-NotI sites of pPICZ B; Zeor | This study |
Ampr, ampicillin resistant; Emr, erythromycin resistant; Kmr, kanamycin resistant; Tcr, tetracycline resistant; Zeor, zeocin resistant.
The methylotrophs M. extorquens and P. pastoris X-33 were used as host cells for heterologous gene expression. Plasmid pCM110, containing the homogeneous methanol dehydrogenase promoter (PmxaF) from M. extorquens AM1 (39), was used for estI expression in M. extorquens. The Pichia expression vector pPICZ B (Invitrogen Canada Inc., Burlington, Ontario, Canada) was used for yeast expression studies. The antibiotics tetracycline (30 μg/ml) and zeocin (100 μg/ml) were added to the medium, unless specified otherwise.
Medium composition for methylotrophs.
M. extorquens ATCC 55366 was grown in CHOI medium, as previously described (4), and 1% (vol/vol) methanol was used as the sole carbon source. P. pastoris was grown as specified in the Invitrogen manual; MD plates contained 1.34% (wt/vol) yeast nitrogen base, 1% (wt/vol) dextrose, and 2% (wt/vol) agar; BMGY contained 1% yeast extract, 2% peptone, 100 mM potassium phosphate (pH 7.0), 1.34% yeast nitrogen base, and 1% glycerol; BMMY contained 1% yeast extract, 2% peptone, 100 mM potassium phosphate (pH 7.0), 1.34% yeast nitrogen base, and 1% methanol.
DNA isolation and manipulations.
Plasmids from E. coli were prepared with a Mini and Midi-Prep system (Qiagen Inc., Mississauga, Ontario, Canada) according to the protocol provided by the company. Recombinant plasmids were constructed, and agarose gel electrophoresis was performed, according to the method of Sambrook and Russell (45). DNA fragments were isolated from agarose gels by using the QIAquick gel extraction system (Qiagen). T4 DNA ligase and other DNA-modifying enzymes were purchased from New England Biolabs Inc., GIBCO/BRL Life Technologies, Inc., or Pharmacia LKB Biotechnology and were used as recommended by the manufacturer. Electroporation was performed with a Gene-Pulser II electroporation apparatus (Bio-Rad Laboratories, Mississauga, Ontario, Canada).
Cloning of the esterase gene in E. coli.
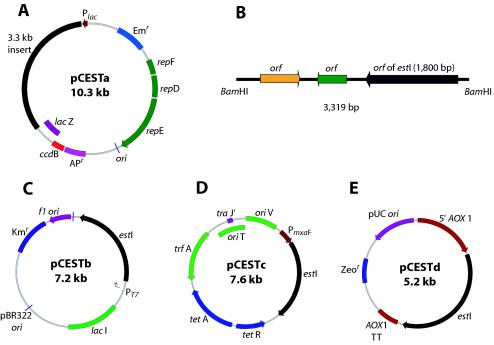
The esterase gene from L. casei CL96 was first cloned into pCWL70 at the BamHI site (pCESTa) (Fig. 1A). A positive clone was selected on the LA (LB broth with ampicillin) plate containing the chromogenic substrate 5-bromo-4-chloro-3-indolylcaprylate (X-caprylate), which showed a blue color because the esterase catalyzes the hydrolysis of X-caprylate, producing a blue precipitate. For expression of the mature estI gene, pCESTb (Fig. 1C) was constructed by cloning the estI gene into pET29a as follows. The forward primer 5′-GGA AGG ATC CAT GGA TCA ATC TAA AAC-3′ was designed to contain a BamHI site (underlined nucleotides) immediately upstream of the codon for aspartic acid (GAT), which is the first amino acid of mature esterase. The reverse primer 5′-GCA GCG GCC GCT TAT TTA TTT GTA ATA CCG-3′ was designed to contain a NotI site (underlined nucleotides) and tandem TAA stop codons immediately after the final lysine codon (AAA). PCR was performed with a GeneAmp PCR system 2400 (Perkin-Elmer, Boston, Mass.) by using the Taq PCR Core kit (Qiagen). The PCR product was digested with BamHI and NotI and was cloned into the same restriction enzyme sites in pET29a. The estI gene was expressed under the control of the T7 promoter by induction with isopropyl-β-d-thiogalactopyranoside (IPTG) (Roche Diagnostics, Laval, Quebec, Canada).
FIG. 1.
Schematic representation of plasmids pCESTa (A), pCESTb (C), pCESTc (D), and pCESTd (E) and location of estI in a 3.3-kb inserted fragment (B). Diagrams are not drawn to scale.
Cloning of the esterase gene in methylotrophs.
The E. coli expression vector pESTb, containing estI, served as a template for PCR. To express L. casei CL96 estI in M. extorquens, PCR primers (sense, 5′-CAA ACT AGT ATG GAT CAA TCT AAA AC-3′; antisense, 5′-CGG ATC GAT TTA TTT GTA ATA CCG-3′, containing SpeI and ClaI sites, respectively) were used to amplify the 1,800-bp estI fragment. This fragment was cloned into pCR2.1, and the estI fragment was excised with SpeI and ClaI and then ligated into pCM110 (39) to construct expression plasmid pCESTc (Fig. 1D). Possible constructs were confirmed by restriction digestion and sequencing.
To fuse the sequence encoding the mature esterase directly in frame to the cloning site in the Pichia expression vector pPICZ B, PCR was performed with the forward primer 5′-CCC GGT ACC ATG GAT CAA TCT AAA AC-3′, containing a KpnI site (underlined nucleotides), and the reverse primer 5′-GCA GCG GCC GCT TTA TTT GTA ATA CCG-3′, containing a NotI site (underlined nucleotides). The PCR product was digested with KpnI and NotI and then ligated into pPICZ B, linearized with the same enzymes, to produce pCESTd (Fig. 1E). After transformation of E. coli with pCESTd, zeocin-resistant clones were selected and analyzed by restriction digestion. For transformation of P. pastoris X-33, pCESTd was linearized with PmeI and used for electroporation according to the procedure of the manufacturer (Invitrogen).
DNA sequencing and sequence analysis.
The nucleotide sequences of both strands of pCESTa and other constructs were determined by AmpliTaq FS DNA polymerase fluorescent dye terminator reactions as recommended by the supplier (Perkin-Elmer). Sequencing products were detected by using an Applied Biosystems 373 stretch automated sequencer. Bioinformatics analyses were conducted on BioNavigator.com, provided by Entigen Corporation (http://www.bionavigator.com). Related sequences were obtained from database searches (SwissProt, PIR, EMBL, and GenBank) by using the BLASTP (version 2.1) and FASTA programs. Global multiple alignments between protein sequences were computed by using the CLUSTAL W program (52). A distance measure for the sequences was computed with the PROTDIST program of the PHYLIP package. The measures were used to construct a phylogenetic tree by the neighbor-joining method.
Enzyme and protein assays.
Esterase activity was determined by a spectrophotometric method using pNP-caprylate as the substrate. The rate of hydrolysis of pNP-caprylate at 37°C was measured in 50 mM sodium phosphate buffer (pH 7.0) at 420 nm according to the method of Kademi et al. (29, 30). One unit of activity was defined as the amount of enzyme that liberated 1 μmol of p-nitrophenol per min under the given assay conditions. The substrate specificity of the enzyme was determined on pNP esters of C2 to C18 fatty acids. The protein concentration was estimated by the method of Bradford (5) using the Bio-Rad protein assay kit with bovine serum albumin as a standard.
Purification of the recombinant esterase.
E. coli BL21(DE3)/pLysS harboring the pCESTb plasmid was used as a source of the recombinant enzyme. Cultivated cells were harvested at 12 h after induction with IPTG. The cell pellet was washed twice with 50 mM sodium phosphate buffer (pH 7.0) and then disrupted by using an ultrasonic disintegrator (Sonifier 450; Branson) for 10 min at 30-s intervals. The crude extract was separated from cell debris by centrifugation at 15,000 × g for 20 min at 4°C. The recombinant esterase with an S · tag was purified by using an S-protein agarose column. An S-protein agarose slurry (50%) was added to the cell lysate and mixed gently at 10°C for 1 h. The lysate containing the S-protein agarose mixture was loaded onto a column and washed twice with 5 ml of wash buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.1% Triton X-100). The washed S-protein agarose-containing target protein was resuspended in thrombin cleavage buffer (20 mM Tris-HCl [pH 8.4], 150 mM NaCl, 2.5 mM CaCl2), and the tag was removed by addition of biotinylated thrombin. The target protein released from the agarose no longer contained the S · tag peptide. The biotinylated thrombin was removed with streptavidin agarose according to the protocol provided by the company. The molecular mass of purified recombinant esterase was determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with 4% (wt/vol) stacking and 12% (wt/vol) separating gels as described by Laemmli (33). All the results presented in this report were obtained with the purified recombinant esterase.
Activity staining.
Nondenaturing gels were stained for esterase activity as described by Lambrechts and Galzy (34) with a minor modification. The gels were incubated in a solution (100 ml) of 50 mM phosphate buffer (pH 7.0) containing 0.001% (wt/vol) α-naphthylcaprylate dissolved in acetone and 0.03% (wt/vol) Fast Blue RR at room temperature. The reaction was stopped after 20 min by two rinses with distilled water.
Effects of metal ions and other inhibitor substances on enzyme activity.
The purified recombinant esterase was preincubated with various metal ions and reagents at a 1 mM concentration in 50 mM sodium phosphate buffer (pH 7.0) at 37°C for 30 min. The residual enzyme activity was assayed at 37°C. For the effects of inhibitors, different concentrations of diethyl pyrocarbonate (DEPC), phenylmethylsulfonyl fluoride (PMSF), phenylarsine oxide (PAO), and phenylglyoxal (PGO) were incubated with the enzyme for 30 min at 37°C. The reaction was stopped by chilling on ice, and aliquots were assayed for enzyme activity.
pH, temperature optimum, and stability.
The esterase activity for pNP-caprylate was measured over a pH range from 4.0 to 9.0 by using different buffers with 50 mM concentrations of sodium acetate (pH 4.0 to 5.0), sodium phosphate (pH 6.0 to 7.0), or Tris-HCl (pH 8.0 to 9.0). The temperature optimum was determined by performing pNP-caprylate assays at a temperature range from 20 to 70°C. For determination of stability, the enzyme was preincubated in 50 mM phosphate buffer (pH 7.0) at various temperatures for different periods and then assayed for residual activity at 37°C.
Nucleotide sequence accession number.
The nucleotide sequence data for estI have been deposited in GenBank under accession number AY251019.
RESULTS
Cloning of the L. casei CL96 esterase gene estI.
A genomic library was constructed by inserting BamHI fragments of L. casei CL96 chromosomal DNA into the pCWL70 vector. The ligation mixture was used to transform E. coli Top10F′ cells. Only 1 of the 15,000 transformant colonies screened turned blue in the enzymatic plate assay performed with X-caprylate as the substrate. The positive clone with the 3.3-kb insert was designated pCESTa (Fig. 1A) and was used for further characterization.
Southern hybridization was performed with total L. casei CL96 DNA digested with BamHI by using the PCR product of the inserted DNA fragment as a probe.
Sequence analysis and deduced amino acid sequence.
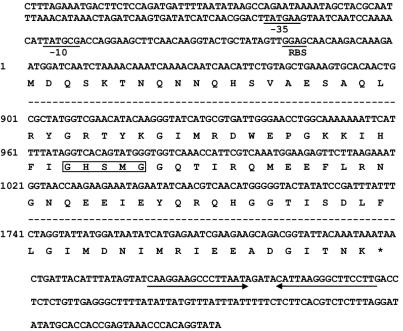
The full length of the 3.3-kb (estI/pCWL70) insert in plasmid pCESTa was sequenced. An open reading frame (ORF) search conducted in the six possible reading frames revealed the presence of three ORFs of 600 nucleotides or more. The deduced amino acid sequences from these three ORFs were subjected to protein sequence comparisons using the Bionavigator system. One of the three ORFs matched the known enzyme family that corresponds to the esterase gene. This ORF consists of 1,800 nucleotides encoding a protein of 599 amino acids and was designated estI (Fig. 2). A putative ribosomal binding site was located 8 bp upstream of the ATG start codon, and a potential −35/−10 consensus sequence was recognized. A rho-independent transcription terminator-like region was found 19 bp away from the termination codon of estI (Fig. 2). The G+C content was 35.27%, and the frequency of G or C at the third codon position was 35.4%, indicating a codon usage typical of lactobacilli (43). No potential signal sequence was found in estI, as revealed by the PSORT program (40).
FIG. 2.
Partial nucleotide sequence and deduced amino acid sequence of the estI gene from L. casei CL96. The proposed ribosomal binding site (RBS) and the −10 and −35 regions of the putative promoter are indicated. The stop codon is marked by an asterisk. A consensus amino acid sequence found in many lipolytic enzymes is boxed. The two inverted arrows at the end of the nucleotide sequence indicate a potential transcriptional termination site.
The deduced amino acid sequence of the potential esterase gene was compared with the known esterase and lipase amino acid sequences available from GenBank (NCBI database). EstI showed the highest homology to a lipase precursor of Staphylococcus epidermidis (54% identity) (37), a lipase precursor of Staphylococcus aureus (47% identity) (36), an esterase from Bacillus stearothermophilus (36% identity) (32), and a tributyrase from Geobacillus thermocatenulatus (35% identity) (46).
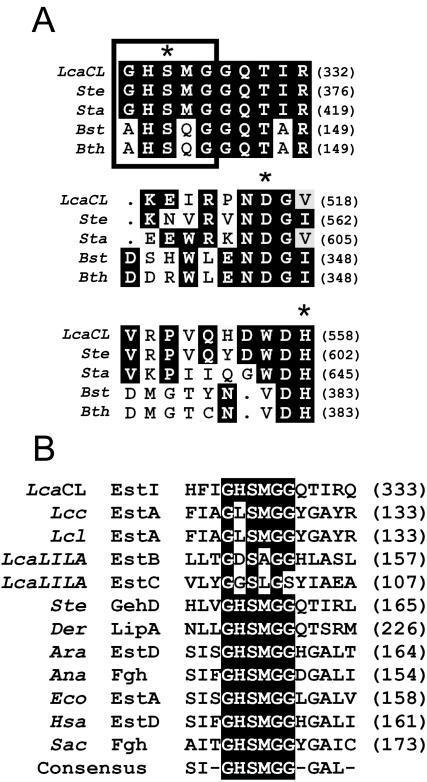
The alignment suggests that Ser325, Asp516, and His558 might be the catalytic triad (Fig. 3A), since these residues are highly conserved in most esterase and lipase groups (13, 31). The amino acid sequence GHSMG, starting at residue 323 (Fig. 3B), fits the GXSXG motif found in most bacterial and eucaryotic serine hydrolases, such as lipase and esterase, as well as in serine proteinases (6, 13). The sequence GHSMG in the L. casei CL96 esterase is also found in the lipases or esterases of S. epidermidis (37), Deinococcus radiodurans (56), and E. coli (3).
FIG. 3.
(A) Multiple alignment of the possible catalytic triad of L. casei CL96 EstI (LcaCL) (GenBank accession number AY251019) with lipase precursors from S. epidermidis (Ste) (AF090142) and S. aureus (Sta) (AAA26633), an esterase from B. stearothermophilus (Bst) (AF237623), and a tributyrin esterase from G. thermocatenulatus (Bth) (CAA64621). The residues of the catalytic triad are marked with asterisks. (B) Sequence alignment of the active-site consensus motif of EstI (LcaCL) with a tributyrin esterase from Lactococcus lactis subsp. cremoris (Lcc) (AF059739), an esterase from Lactococcus lactis (Lcl) (AF157601), an arylesterase (EstB) from L. casei LILA (LcaLILA) (AF494421), an esterase (EstC) from L. casei LILA (AF506279), a lipase precursor from S. epidermidis (Ste), esterases from D. radiodurans (Der) (AAF09912) and Arabidopsis thaliana (Ara) (AC002510), an FGH from Anabaena azollae (Ana) (AF035558), an esterase from E. coli (Eco) (AAC73458), esterase D (EstD) from Homo sapiens (Hsa) (NP001975), and FGH from Saccharomyces cerevisiae (Sac) (CAA84054). Highlighted residues are conserved in the majority of sequences aligned.
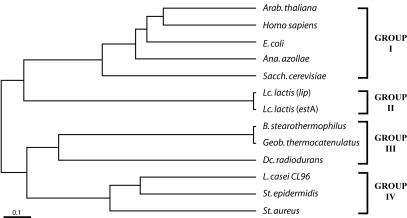
To verify the relationship of L. casei EstI to other known lipolytic proteins, as well as to S-formylglutathione hydrolases (FGH), we constructed a phylogenetic tree based on the amino acid sequences. The phylogenetic tree showed that the L. casei EstI protein belongs to group IV, which includes lipolytic proteins from Staphylococcus spp. (Fig. 4). The estI gene is separated from the esterolytic and lipolytic proteins of Lactococcus spp. (group II) (21), which suggests that the point of sequence divergence from lactic acid bacteria may be different for different genotypes.
FIG. 4.
Phylogenetic tree of lipolytic and esterolytic enzymes based on the alignment of EstI or EstI-like proteins. All of these enzymes contain the active-site motif GXSXG as described in the text. The proteins, as well as their accession numbers and sources, are described in the legend to Fig. 3.
The predicted EstI structure revealed that the active site of this enzyme was located in the known architecture of the α/β hydrolases (data not shown). The key nucleophile, Ser325, forms part of a catalytic triad with Asp516 and His558. This active-site architecture has also been observed in the structures of other lipases and esterases (16, 55).
Expression of recombinant esterase in E. coli.
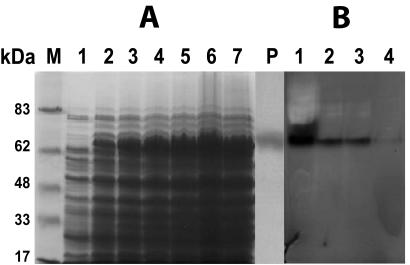
To ascertain that the estI gene indeed codes for an esterase, full-length estI was overexpressed in E. coli. Upon induction of E. coli BL21(DE3)/pLysS containing pET29a-estI with IPTG, a polypeptide with a molecular mass of about 68 kDa and with esterase activity was produced (Fig. 5); this result is in agreement with the calculated molecular mass of the predicted amino acid sequence (67,064 Da). The resulting recombinant protein should consist of the 599 amino acids with an N-terminal fusion of 36 amino acids corresponding to the S · tag epitope and a unique thrombin cleavage site, and its total molecular mass should be 67.5 kDa. Cultivation of E. coli BL21(DE3)/pLysS harboring pCESTb showed significant overexpression of the esterase gene (Fig. 6A). The relative quantity of recombinant protein appeared to increase up to 12 h after induction, and protein samples were prepared from cultures at this time. The presence of the S · tag epitope in the target protein was confirmed by Western blotting with the alkaline phosphatase-conjugated anti-S-protein antibody (data not shown). While the uninduced cell extracts showed slight esterase activity, no activity was found in crude extracts of the host strain, E. coli BL21(DE3)/pLysS.
FIG. 5.
(A) Overexpression of esterase from L. casei CL96 in E. coli BL21(DE3)/pLysS. Protein samples were separated in an SDS-12% polyacrylamide gel and stained with Coomassie brilliant blue R-250. Lanes: M, molecular mass standards; 1, E. coli BL21(DE3)/pLysS bearing vector pET29a; 2 to 7, E. coli BL21(DE3)/pLysS containing pET29a-estI after 2, 4, 6, 8, 10, and 12 h of induction, respectively; P, purified esterase (3 μg). (B) Staining of a 12% nondenaturing gel for activity of esterase from L. casei CL96 in E. coli BL21(DE3)/pLysS. Lanes 1 to 4, soluble fractions of crude lysates from 10, 8, 6, and 4 h after induction, respectively.
FIG. 6.
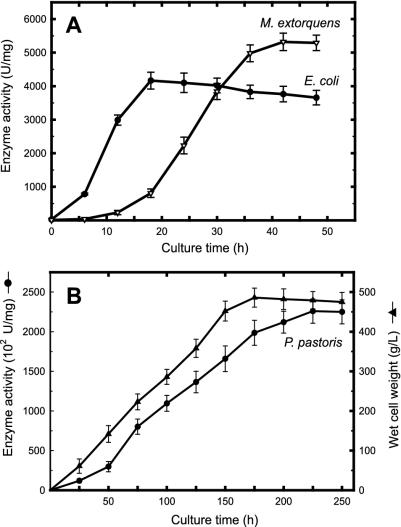
Total esterase activity profiles of recombinant E. coli and M. extorquens (A) and recombinant P. pastoris (B).
Expression of recombinant esterase in M. extorquens.
The estI gene under the control of the PmxaF promoter in M. extorquens provided a level of enzyme activity 1.2- to 1.5-fold greater than that found in recombinant E. coli. SDS-PAGE of cell extracts prepared from M. extorquens bearing pCESTc revealed the presence of recombinant esterase exhibiting a molecular mass of approximately 67 kDa, and zymogram analysis was then performed with native PAGE. The result was very similar to that seen with the recombinant esterase from E. coli (data not shown). The growth of recombinant M. extorquens carrying PmaxF-estI showed that the yield of recombinant esterase reached its maximum at late-exponential phase (about 5,320 U/mg) and then decreased slightly as the culture reached the early-stationary phase (Fig. 6A).
Expression of recombinant esterase in P. pastoris.
The pCESTd plasmid was digested with PmeI to generate a DNA fragment, which was integrated into the AOX1 locus of P. pastoris by homologous recombination. Electroporation of P. pastoris strain X-33 with this DNA fragment yielded about 102 zeocin-resistant transformants on selective plates containing 100 μg of zeocin/ml. Positive clones producing active esterase were screened on minimal agar plates containing X-caprylate, and expression was induced with vapor-phase methanol.
A positive P. pastoris clone was grown in shake flasks for 10 days with daily additions of methanol over a pH range from 4 to 8. Cell growth and enzyme production levels varied depending on the pH of the growth medium; a pH of 6.5 was optimal for enzyme production (data not shown).
High-cell-density culture was carried out in a bioreactor containing methanol in a fed-batch mode under optimum dissolved-oxygen, pH, and temperature conditions (Fig. 6B). When the recombinant Pichia strain was grown at pH 6.5 and 30°C, maximum cell density (about 468 g [wet weight] of cells/liter) was obtained after approximately 180 h. Production of recombinant esterase was initiated at 38 h after induction with methanol and continued to increase until the cells were harvested at 250 h. The final yield was 226 × 103 U/mg, which is more than 980-fold higher than that found in native cells.
Properties of recombinant esterase.
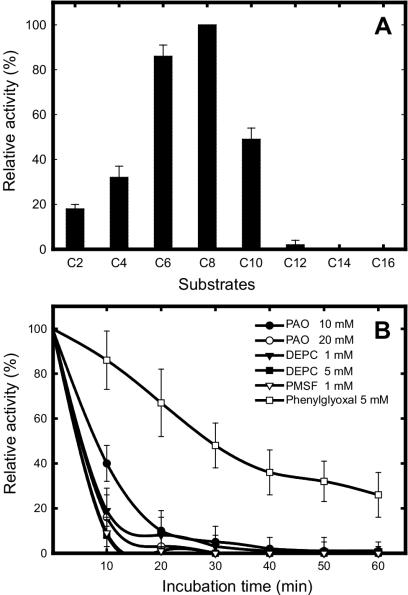
The substrate specificity of the purified enzyme from recombinant E. coli was determined with pNP esters of various acyl chain lengths (C2 to C16) at 37°C. Although the enzyme showed a wide range of substrate specificities, from C2 to C10, the highest activity was observed with pNP-caproate (C6) and pNP-caprylate (C8). However, activity toward longer pNP esters (C12 to C16) in the assay was also observed but was very low under the same assay conditions (Fig. 7A).
FIG. 7.
(A) Substrate specificity assay. Rates of hydrolysis were measured with fatty acids of various acyl chain lengths, from pNP-acetate to pNP-palmitate. The highest level of activity, observed with the C8 substrate, was taken as 100%. (B) Effects of inhibitors on recombinant esterase activity. Different concentrations of DEPC, PMSF, PAO, and PGO were incubated with the enzyme for 30 min at 37°C. The reaction was stopped by chilling on ice, and aliquots were assayed by the standard method.
Km and kcat values were calculated from Lineweaver-Burk plots by using a least-squares, best-fit Michaelis-Menten equation (Table 2). kcat values increased with increasing chain length, reaching a maximum with pNP-caprylate (C8), whereas Km values decreased with increasing acyl chain length, with the exceptions of pNP-caprate (C10) and pNP-laurate (C12). The specificity constant, kcat/Km, was higher for pNP-caproate (C6) and pNP-caprylate (C8) than for pNP-acetate (C2) and pNP-butyrate (C4), while pNP-caprylate (C8) showed maximum catalytic efficiency. These findings confirm that the enzyme is more specific for mid-chain fatty acids than for short-chain fatty acids, in agreement with the specific activity pattern shown in Fig. 7A.
TABLE 2.
Kinetic constants of purified recombinant esterase on pNP esters of fatty acidsa
| Substrate | Km (μM) | 102kcat (S−1) | kcat/Km (S−1 · μM−1) |
|---|---|---|---|
| pNP-acetate | 134 | 3.16 | 2.35 |
| pNP-butyrate | 97 | 9.88 | 10.18 |
| pNP-caproate | 19 | 11.94 | 62.80 |
| pNP-caprylate | 14 | 12.45 | 88.91 |
| pNP-caprate | 43 | 8.96 | 20.84 |
| pNP-laurate | 186 | 2.41 | 1.29 |
| pNP-myristate | — | — | — |
| pNP-palmitate | — | — | — |
Enzyme activity, determined at 37°C in 50 mM phosphate buffer (pH 7.0), is a mean from two or three independent experiments with less than 8% standard deviation. —, not detected.
Assays to determine the effects of metal ions on the esterase activity, by using pNP-caprylate as the substrate, showed that the enzyme was significantly inhibited by Hg2+, Ag+, and β-mercaptoethanol but was stimulated by Zn2+, Mg2+, and Ca2+ (data not shown). Incubation in the presence of Fe3+, Pb2+, or Cu2+ led to a slight inhibition, but the remaining ions did not show a significant effect. Assays with other inhibitors (Fig. 7B) showed that the enzyme was strongly inhibited by DEPC, a histidine modifier; PMSF, a serine protease inhibitor; and PAO, a cysteine modifier. However, PGO, an arginine modifier, caused only a slight loss of enzyme activity.
The enzyme was stable over a pH range of 6 to 8, and about 60% of enzyme activity was retained at 50°C (data not shown). The purified enzyme could be stored without significant loss of activity for several weeks at −20°C in a buffer (50 mM phosphate buffer, pH 7.0) supplemented with glycerol (10%, vol/vol).
DISCUSSION
The esterase of L. casei CL96 (EstI), which possesses unique genetic and biochemical properties, was characterized and overexpressed in various hosts: E. coli, M. extorquens, and P. pastoris. The fact that methylotrophs can grow on methanol, a relatively inexpensive compound, as a carbon and energy source led to the choice of host cells. The use of methanol as the sole substrate for fermentation processes has many advantages, such as high solubility in water and low toxicity.
Interestingly, the expression profile of the esterase in the methylotrophic bacterium M. extorquens ATCC 55366 under the control of the PmxaF promoter was very similar to that in E. coli, which was regulated by the T7 promoter. The large subunit of methanol dehydrogenase, MxaF, is expressed at high levels in M. extorquens strain AM1 and is tightly regulated by the PmxaF promoter (23). Nucleotide sequence comparisons of the PmxaF promoters and mxaF genes in M. extorquens strains AM1 and ATCC 55366 revealed 98% homology. Therefore, it is not surprising that the PmxaF promoter originating from strain AM1 drove the expression of estI in M. extorquens ATCC 55366 efficiently. The development of a strong and stable expression vector, pCM110 (39), and of a high-cell-density (110 g [dry weight]/liter) fed-batch fermentation process (4) renders M. extorquens an interesting biocatalyst for the expression of heterologous value-added proteins.
For expression of estI in the Pichia system, the AOX1 promoter was used to construct an expression cassette, and positive transformants were selected at low, medium, and high concentrations of zeocin (100, 500, and 1,000 μg/ml). Recombinant derivatives of plasmid pPICZ B, which had been linearized by digestion with PmeI, typically integrated at the AOX locus as a single copy, but multiple-copy integration occurred at a frequency of 1 to 10%. Resistance to higher levels of zeocin correlates with higher copy numbers of the integrated plasmid, which usually correlate with higher levels of expression of the recombinant target protein (44). The positive clone was found only on the selection plate containing 100 μg of zeocin/ml, indicating that a single copy was integrated. However, the rate of enzyme production by high-cell-density fed-batch culture was about 980-fold higher than that by native cells. Large-scale production of recombinant proteins is generally achieved by high-cell-density fed-batch cultivation, and it is known that the recombinant proteins produced by high-cell-density cultivation are often affected by many different parameters such as growth temperature and inducer concentration (57). Although the production of soluble proteins could be increased by cultivation at reduced temperatures (38), no apparent effect was found at 25°C in this study. Therefore, high-cell-density fed-batch culture of recombinant P. pastoris was carried out at 30°C.
The identity of L. casei strain CL96 was reconfirmed by 16S rRNA sequencing, since alignment of EstI with other esterolytic enzymes from lactic acid bacteria by using the BestFit program failed to reveal any significant homology. It is possible that the esterase gene (estI) may have arisen through horizontal transfer or evolution of genetic material among microorganisms. This may explain the high homology among esterases from different sources. Sequence analysis demonstrates that L. casei CL96 esterase may be classified as a group IV enzyme, as most of the highly conserved domains are found within this group. This group includes the Staphylococcus lipolytic enzymes (LipA), which have an active-site motif, GHSMG, identical to that of L. casei CL96 esterase. Thus, L. casei CL96 esterase is a true member of the lipolytic enzyme family, despite its lack of a conserved lid-like polypeptide chain, which is a pivotal sequence in lipases. The cloning and sequencing of the L. casei CL96 esterase gene is a logical step in the exploration of the molecular biology of L. casei. Gene isolation allows reverse-genetic analysis of the physiological significance of the enzyme; it becomes possible to knock out this gene in order to observe how this mutation would affect cell metabolism. Such an analysis may provide clues to the importance of EstI in L. casei CL96.
The crystal structures of many esterases and lipases (26, 55) show a common folding pattern called the α/β-hydrolase fold, with a catalytic site built by a conserved serine, an aspartate or a glutamate, and a histidine residue (12). Also, the highly conserved pentapeptide G-X-S-X-G forms a tight turn in a β-strand-turn-α-helix motif, called the nucleophilic elbow, which is the most strictly conserved structural motif of α/β-hydrolase (47). However, the active-site pentapeptides of lipolytic enzymes from Bacillus species show unique sequences (32). The distinct feature of these enzymes is the replacement of the first glycine residue in the canonical lipase consensus motif G-X-S-X-G by an alanine.
Sequence alignment of estI with other esterases and lipase precursors allowed the identification of the catalytic triad: the active-site serine (Ser325), Asp516, and His558, located near the carboxyl terminus.
Many esterases and lipases contain two or three conserved disulfide bridges, which are important for substrate binding or recognition (13). However, no potential disulfide bridge was found in EstI, which contains only one cysteine residue at position 502. Since this enzyme was significantly inhibited by the reducing agent β-mercaptoethanol, this cysteine residue might be involved in a catalytic mechanism. Similar characteristics have been observed for esterases from Lactobacillus plantarum 2739 (25), and Lactobacillus casei subsp. casei (7). These results were also supported by the observed inhibition of lactococcal esterase activity by Hg2+, which reacts with thiol groups (8, 53).
Inactivation of the L. casei CL96 esterase by PMSF and DEPC suggests the involvement of a histidine and a serine at the active site of the enzyme. The serine residue of the pentapeptide GXSXG is well known as the active catalytic residue that is modified by PMSF. Generally, lipases are only slightly inhibited by PMSF, because the lid structure covering the serine residue of the active site becomes inaccessible to the reagent.
The substrate specificity of L. casei CL96 esterase was studied by using the purified recombinant enzyme. The esterase specificity for pNP-caproate and pNP-caprylate was quite different from the specificities of other microbial esterases (9, 17), even esterases (EstB and EstC) from L. casei LILA (19, 20), which are mainly specific for short-chain (C2 to C6) fatty acids. Only the enzymes from B. stearothermophilus (32), Sulfolobus acidocaldarius (49), and Bacillus licheniformis (1) showed specificity patterns similar to that of EstI.
L. casei CL96 esterase was shown to be a good candidate for the production of mid-chain fatty acid esters and could play an important role in cheese ripening (35). It is known that fatty acids are important components in the flavors of many cheese types. Although butyric acid is considered to be an important cheese flavor, at high concentrations it imparts an undesirable flavor to cheese (18). The esterase from L. casei CL96 showed limited activity on C4 fatty acid esters relative to that of other esterases. The esterase purified in this study has been used for the production of enzyme-modified cheese, and the results indicate that the L. casei CL96 enzyme produces an intense cheese flavor comparable to that of extra-aged Cheddar within days (35); this performance is comparable to that of other commercial lipolytic or esterolytic enzymes used for cheese flavoring.
Acknowledgments
This work was supported by a strategic grant from the Natural Sciences and Engineering Research Council of Canada (NSERC-STRO202123) and in part by a European Community 5th Frame Work grant (QLK3-CT-2000-01528).
REFERENCES
- 1.Alvarez, M. E., M. V. Augier, and J. Baratti. 1999. Characterization of a thermostable esterase activity from the moderate thermophile Bacillus licheniformis. Biosci. Biotechnol. Biochem. 63:1865-1870. [DOI] [PubMed] [Google Scholar]
- 2.Bélanger, E. 1998. Genetic identification of Lactobacillus and Bifidobacteria species. M.Sc. thesis. McGill University, Montreal, Canada.
- 3.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, N. Gregor, G. F. Davis, H. A. Kirkpatrick, M. A. Goeden, J. D. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 4.Bourque, D., Y. Pomerleau, and D. Groleau. 1995. High-cell-density production of poly-β-hydroxybutyrate (PHB) from methanol by Methylobacterium extorquens: production of high-molecular-mass PHB. Appl. Microbiol. Biotechnol. 44:367-376. [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Brenner, S. 1988. The molecular evolution of genes and proteins: a tale of two serines. Nature 334:528-530. [DOI] [PubMed] [Google Scholar]
- 7.Castillo, I., T. Requena, P. Fernandez de Palencia, J. Fontecha, and M. Gobbetti. 1999. Isolation and characterization of an intracellular esterase from Lactobacillus casei subsp. casei IFPL731. J. Appl. Microbiol. 86:653-659. [Google Scholar]
- 8.Chich, J. F., K. Marchesseau, and J. C. Gripon. 1997. Intracellular esterase from Lactococcus lactis subsp. lactis NCDO 763: purification and characterization. Int. Dairy J. 7:169-174. [Google Scholar]
- 9.Choi, K. D., G. H. Jehon, J. S. Rhee, and O. J. Yoo. 1990. Cloning and nucleotide sequence of an esterase gene from Pseudomonas fluorescens and expression of the gene in Escherichia coli. Agric. Biol. Chem. 54:2039-2045. [PubMed] [Google Scholar]
- 10.Choi, Y. J., and B. H. Lee. 2001. Culture conditions for the production of esterase from Lactobacillus casei CL96. Bioprocess Biosystems Eng. 24:59-63. [Google Scholar]
- 11.Choi, Y. J., T. T. Wang, and B. H. Lee. 2002. Positive selection vectors. Crit. Rev. Biotechnol. 22:225-244. [DOI] [PubMed] [Google Scholar]
- 12.Cygler, M., and J. D. Schrag. 1997. Structure as basis for understanding interfacial properties of lipases. Methods Enzymol. 284:3-27. [DOI] [PubMed] [Google Scholar]
- 13.Cygler, M., J. D. Schrag, J. L. Sussman, M. Harel, L. Silman, M. K. Gentry, and B. P. Doctor. 1993. Relationship between sequence conservation and three-dimensional structure in a large family of esterases, lipases and related proteins. Protein Sci. 2:366-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Man, J. C., M. Rogosa, and M. Sharpe. 1960. A medium for the cultivation of lactobacilli. J. Appl. Bacteriol. 23:130-135. [Google Scholar]
- 15.Derewenda, Z. S. 1994. Structure and function of lipases. Adv. Protein Chem. 45:1-52. [DOI] [PubMed] [Google Scholar]
- 16.Derewenda, Z. S., and Y. Wei. 1995. A novel variant of the catalytic triad in the Streptomyces scabies esterase. J. Am. Chem. Soc. 117:2104-2105. [DOI] [PubMed] [Google Scholar]
- 17.Dupuis, C., C. Corre, and P. Voyaval. 1993. Lipase and esterase activities of Propionibacterium freudenreichii subsp. freudenreichii. Appl. Microbiol. Biotechnol. 59:4004-4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Engels, W. J. M., R. Dekker, C. de Jong, R. Neeter, and S. Visser. 1997. A comparative study of volatile compounds in the water-soluble fraction of various types of ripened cheese. Int. Dairy J. 7:255-264. [Google Scholar]
- 19.Fenster, K. M., K. L. Parkin, and J. L. Steele. 2003. Intracellular esterase from Lactobacillus casei LILA: nucleotide sequencing, purification, and characterization. J. Dairy Sci. 86:1118-1129. [DOI] [PubMed] [Google Scholar]
- 20.Fenster, K. M., K. L. Parkin, and J. L. Steele. 2003. Nucleotide sequencing, purification, and biochemical properties of an arylesterase from Lactobacillus casei LILA. J. Dairy Sci. 86:2547-2557. [DOI] [PubMed] [Google Scholar]
- 21.Fernandez, L., M. Beerthuyzen, J. Brown, R. J. Siezen, T. Coolbear, R. Holland, and O. P. Kuipers. 2000. Cloning, characterization, controlled overexpression, and inactivation of the major tributyrin esterase gene of Lactococcus lactis. Appl. Environ. Microbiol. 66:1360-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Figueira, M. M., L. Laramée, J. C. Murrell, D. Groleau, and C. B. Miguez. 2000. Production of green fluorescent protein by the methylotrophic bacterium Methylobacterium extorquens. FEMS Microbiol. Lett. 193:195-200. [DOI] [PubMed] [Google Scholar]
- 23.FitzGerald, K. A., and M. E. Lidstrom. 2003. Overexpression of a heterologous protein, haloalkane dehalogenase, in a poly-beta-hydroxybutyrate-deficient strain of the facultative methylotroph Methylobacterium extorquens AM1. Biotechnol. Bioeng. 81:263-268. [DOI] [PubMed] [Google Scholar]
- 24.Gellissen, G. 2000. Heterologous protein production in methylotrophic yeasts. Appl. Microbiol. Biotechnol. 54:741-750. [DOI] [PubMed] [Google Scholar]
- 25.Gobbetti, M., P. F. Fox, and L. Stepaniak. 1997. Isolation and characterization of a tributyrin esterase from Lactobacillus plantarum 2739. J. Dairy Sci. 80:3099-3106. [DOI] [PubMed] [Google Scholar]
- 26.Green, R., J. L. Schotte, L. Swenson, Y. Wei, and Z. S. Derewenda. 1992. Crystallization and preliminary crystallographic data of a Streptomyces scabies extracellular esterase. J. Mol. Biol. 227:569-571. [DOI] [PubMed] [Google Scholar]
- 27.Jaeger, K. E., B. W. Dijkstra, and M. T. Reetz. 1999. Molecular biology, three-dimensional structures, and biotechnological applications of lipases. Annu. Rev. Microbiol. 53:315-351. [DOI] [PubMed] [Google Scholar]
- 28.Jaeger, K. E., and M. T. Reetz. 1998. Microbial lipases form versatile tools for biotechnology. Trends Biotechnol. 16:396-403. [DOI] [PubMed] [Google Scholar]
- 29.Kademi, A., L. Fakhreddine, N. Abdelkader, and J. C. Baratti. 1999. Effect of culture condition on growth and esterase production by the moderate thermophile Bacillus circulans MAS2. J. Ind. Microbiol. Biotechnol. 23:188-193. [Google Scholar]
- 30.Kademi, A., N. Ait-Abdelkader, L. Fakhreddine, and J. C. Baratti. 1999. Thermostable esterase activity from newly isolated moderate thermophilic bacterial strains. Enzyme Microb. Technol. 24:332-338. [Google Scholar]
- 31.Khalameyzer, V., I. Fischer, U. T. Bornscheuer, and J. Altenbuchner. 1999. Screening, nucleotide sequence, and biochemical characterization of an esterase from Pseudomonas fluorescens with high activity towards lactones. Appl. Environ. Microbiol. 65:477-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim, H. K., S. Y. Park, J. K. Lee, and T. K. Oh. 1998. Gene cloning and characterization of thermostable lipase from Bacillus stearothermophilus L1. Biosci. Biotechnol. Biochem. 62:66-71. [DOI] [PubMed] [Google Scholar]
- 33.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 34.Lambrechts, C., and P. Galzy. 1995. Esterase activities of Brevibacterium sp. R312 and Brevibacterium linens 62. Biosci. Biotechnol. Biochem. 59:1464-1471. [Google Scholar]
- 35.Lee, B. H., S. Hailelassie, V. Yaylayan, and B. Stewart. 2001. Characterization of volatile flavors in enzyme-modified and natural cheddar cheese, p. 141-150. In A. Spanier, F. Shahidi, T. Parliament, C. Mussinan, C. T. Ho, and E. Contis (ed.), Food flavors and chemistry—advances of the new millenium. Royal Society of Chemistry, Cambridge, United Kingdom.
- 36.Lee, C. Y., and J. J. Iandolo. 1986. Lysogenic conversion of staphylococcal lipase is caused by insertion of the bacteriophage L54a genome into the lipase structural gene. J. Bacteriol. 166:385-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Longshaw, C. M., A. M. Farrell, J. D. Wright, and K. T. Holland. 2000. Identification of a second lipase gene, gehD, in Staphylococcus epidermidis: comparison of sequence with those of other staphylococcal lipases. Microbiology 146:1419-1427. [DOI] [PubMed] [Google Scholar]
- 38.Makrides, S. C. 1996. Strategies for achieving high-level expression of genes in Escherichia coli. Microbiol. Rev. 60:512-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marx, C. J., and M. E. Lidstrom. 2001. Development of improved versatile broad-host-range vectors for use in methylotrophs and other Gram-negative bacteria. Microbiology 147:2065-2075. [DOI] [PubMed] [Google Scholar]
- 40.Nakai, K., and P. Horton. 1999. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 1:34-36. [DOI] [PubMed] [Google Scholar]
- 41.Nelson, J. H., R. G. Jensen, and R. E. Pitas. 1977. Pregastric esterases and other oral lipases—a review. J. Dairy Sci. 60:327-362. [DOI] [PubMed] [Google Scholar]
- 42.Ollis, D. L., E. Cheah, M. Cygler, B. Dijkstra, F. Frolow, S. Franken, M. Harel, S. J. Remington, I. Silman, J. Schrag, J. L. Sussman, K. H. G. Verschueren, and A. Goldman. 1992. The α/β hydrolase fold. Protein Eng. 5:197-211. [DOI] [PubMed] [Google Scholar]
- 43.Pouwels, P. H., and J. A. M. Leunissen. 1994. Divergence in codon usage of Lactobacillus species. Nucleic Acids Res. 22:929-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Romanos, M. A., C. A. Scorer, K. Streekrishna, and J. Clare. 1998. The generation of multicopy recombinant strains. Methods Mol. Biol. 103:55-72. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 46.Schmidt-Dannert, C., M. L. Rua, S. Wahl, and R. D. Schmid. 1997. Bacillus thermocatenulatus lipase: a thermoalkalophilic lipase with interesting properties. Biochem. Soc. Trans. 25:178-182. [DOI] [PubMed] [Google Scholar]
- 47.Schrag, J. D., and M. Cygler. 1997. Lipases and α/β-hydrolase fold. Methods Enzymol. 284:85-107. [DOI] [PubMed] [Google Scholar]
- 48.Simons, F. A., J. W. Boots, M. P. Kats, A. J. Slotboom, M. R. Egmond, and H. M. Verheij. 1997. Dissecting the catalytic mechanism of staphylococcal lipases: chain length selectivity, interfacial activation and cofactor dependence. Biochemistry 36:14539-14550. [DOI] [PubMed] [Google Scholar]
- 49.Sobek, H., and H. Gorisch. 1989. Further kinetic and molecular characterization of an extremely heat stable carboxylesterase from the thermoacidophilic archaebacterium Sulfolobus acidocaldarius. Biochem. J. 261:993-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sood, V. K., and F. V. Kosikowski. 1979. Ripening changes and flavor development in microbial enzyme treated Cheddar cheese slurries. J. Food Sci. 44:1690-1694. [Google Scholar]
- 51.Stead, D. 1986. Microbial lipases; their characteristics, role in food spoilage and industrial uses. J. Dairy Res. 53:481-505. [DOI] [PubMed] [Google Scholar]
- 52.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsakalidou, E., and G. Kalantzopoulos. 1992. Purification and partial characterization of an esterase from Lactococcus lactis ssp. lactis strain ACA-DC 127. Lait 72:533-543. [DOI] [PubMed] [Google Scholar]
- 54.Verger, R. 1997. Interfacial activation of lipase: facts and artifacts. Trends Biotechnol. 15:32-38. [Google Scholar]
- 55.Wei, Y., J. A. Contreras, P. Sheffield, T. Osterlund, U. Derewenda, R. E. Kneusel, U. Matern, C. Holm, and Z. S. Derewenda. 1999. Crystal structure of brefeldin A esterase, a bacterial homolog of the mammalian hormone-sensitive lipase. Nat. Struct. Biol. 6:340-345. [DOI] [PubMed] [Google Scholar]
- 56.White, O., J. A. Eisen, J. F. Heidelberg, E. K. Hickey, J. D. Peterson, R. J. Dodson, D. H. Haft, M. L. Gwinn, W. C. Nelson, D. L. Richardson, K. S. Moffat, H. Qin, L. Jiang, W. Pamphile, M. Crosby, M. Shen, J. J. Vamathevan, P. Lam, L. McDonald, T. Utterback, C. Zalewski, K. S. Makarova, L. M. Aravind, M. J. Daly, K. W. Minton, R. D. Fleischmann, K. A. Ketchum, K. E. Nelson, S. Salzberg, H. O. Smith, J. C. Venter, and C. M. Fraser. 1999. Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science 286:1571-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wong, H. H., Y. C. Kim, S. Y. Lee, and H. N. Chang. 1998. Effect of post-induction nutrient feeding strategies on the production of bioadhesive protein in Escherichia coli. Biotechnol. Bioeng. 60:271-276. [DOI] [PubMed] [Google Scholar]