Abstract
Magnetotactic bacteria synthesize magnetosomes, which cause them to orient and migrate along magnetic field lines. The analysis of magnetotaxis and magnetosome biomineralization at the molecular level has been hindered by the unavailability of genetic methods, namely the lack of a means to introduce directed gene-specific mutations. Here we report a method for knockout mutagenesis by homologous recombination in Magnetospirillum gryphiswaldense. Multiple flagellin genes, which are unlinked in the genome, were identified in M. gryphiswaldense. The targeted disruption of the flagellin gene flaA was shown to eliminate flagella formation, motility, and magnetotaxis. The techniques described in this paper will make it possible to take full advantage of the forthcoming genome sequences of M. gryphiswaldense and other magnetotactic bacteria.
The capability of magnetotaxis in magnetotactic bacteria (MTB) results from their active motility in combination with the presence of magnetosomes, which cause the cell to passively align along magnetic field lines. Magnetotaxis is thought to function as a navigational mechanism by interacting with the Earth's magnetic field, such that a magnetotactic cell effectively acts as a “self-propelled magnetic compass needle” (9). By interaction with other taxis mechanisms such as aerotaxis and phototaxis, magnetotaxis thereby facilitates the orientation in chemically stratified habitats such as aquatic sediments (5).
Magnetotaxis and magnetosome formation have attracted broad interdisciplinary interest for several reasons. Magnetosome biomineralization is a well-established example of controlled mineral formation by bacteria in aquatic sediments on Earth (22, 24, 40), and magnetosome characteristics have been recently considered for use as biosignatures to identify presumptive Martian magnetofossils (21). Because of the unique characteristics of bacterial magnetite crystals, there is considerable biotechnological interest in magnetosome biomineralization (1, 35). In addition, MTB provide a simple model for studying magnetoreception, which might be useful for an understanding of this phenomenon in more complex systems such as higher animals (15). However, both magnetotaxis and magnetosome biomineralization have remained poorly understood at the molecular level, mostly because of the lack of appropriate genetic tools due to past difficulties in culturing and transforming these fastidious organisms.
Here we report a method for knockout mutagenesis by homologous recombination in Magnetospirillum gryphiswaldense, which has recently emerged as a model system to study magnetotaxis and magnetosome biomineralization (11, 32, 34). The biochemical and proteomic analysis of the magnetosome membrane in M. gryphiswaldense in combination with reverse genetics has led to the identification of a number of genes encoding magnetosome membrane proteins, which are organized in the genome in several different operons (7, 8, 10). The isolation and characterization of spontaneous nonmagnetic mutants revealed a large magnetosome island harboring most of the magnetosome membrane protein genes as well as numerous further mam genes with an implicated role in biomineralization but yet unknown function (7, 8, 31, 33). Although a conjugative system for random Tn5-based mutagenesis has been reported in M. gryphiswaldense and Magnetospirillum sp. strain AMB-1 (20, 36, 42), so far there has been no means for site-directed mutagenesis, which is particularly desirable with the increasing availability of genome sequence data from M. gryphiswaldense and other MTB (reference 7 and http://www.jgi.doe.gov/tempweb/JGI_microbial/html/index.html.) This lack has been a major impediment to elucidating the gene functions involved in magnetotaxis and biomineralization and was the impetus for the present study.
Motility is a key factor in magnetotaxis, but nearly nothing is known about its structural and molecular components. In order to establish gene disruption, we analyzed the flaA gene encoding a flagellin homologue, whose mutagenesis should yield a predictable and easily detectable phenotype. Flagellin is the principal constituent of bacterial flagellar filaments, which consist of an assembly of about 20,000 flagellin subunits (19, 28). In this study, the targeted disruption of the flagellin gene flaA was found to eliminate flagella formation and motility. The genetic technique described herein will allow future exploitation of the substantial genome data that have become available for M. gryphiswaldense.
MATERIALS AND METHODS
Strains and growth conditions.
Characteristics of the strains used in this study are shown in Table 1. For Escherichia coli strains, the culture conditions were as previously described (26). Liquid cultures of M. gryphiswaldense strains were grown microaerobically at 28°C in flask standard medium (FSM) containing 50 μM ferric citrate essentially as previously described (11). Single colonies were grown on activated charcoal agar medium (ACAM) that was incubated microaerobically at 28°C as described elsewhere (36). Selection against the sacB gene was performed by the addition of 5% sucrose to ACAM. Transconjugants with the gusA gene were incubated on ACAM containing 50 μg of 5-bromo-4-chloro-3-indoxyl-β-d-glucuronide per ml. M. gryphiswaldense strain R3/S1, which is resistant to both rifampin and streptomycin by spontaneous mutation, was isolated similarly as described before (36).
TABLE 1.
Bacterial strains, plasmids, and primers
| Strain, plasmid, or primer | Description | Source or reference |
|---|---|---|
| Strains | ||
| E. coli S17-1 | thi pro hsdR recA with RP4-2 (Tc::Mu, Km::Tn7) | 39 |
| M. gryphiswaldense MSR-1 DSMZ 6361 | Wild type | 30 |
| M. gryphiswaldense R3/S1 | Rifr, Smr spontaneous mutant | This work |
| M. gryphiswaldense Da136 | M. gryphiswaldense ΔflaA | This work |
| Plasmids | ||
| pK19mobsacB | Kmr, sacB modified from B. subtilis, lacZα | 27 |
| pBBR1MCS2 | Kmr, lacZα | 17 |
| pBBR1MCS5 | Gmr, lacZα | 17 |
| pGEM-T Easy | Ampr, lacZα, PCR cloning vector | Promega |
| pDa87 | pGEM-T Easy, containing a 2.5-kb PCR fragment with the flaA gene of M. gryphiswaldense | This work |
| pDa102 | pGEM-T Easy, containing a PCR fragment with PstI linker and the Gmr of pBBR1MCS5 | This work |
| pDa103 | pK19mobsacB, containing the 2.5-kb fragment with the flaA gene | This work |
| pDa115 | pDa103 with Gmr insertion in the PstI cutting site (flaA::Gm) | This work |
| pDa116 | pK19mobGII with EcoRI fragment containing flaA::Gm | This work |
| Primersa | ||
| MCS5foPstI | CTGCAGGACGCACACCGTGGAAA | |
| MCS5rwPstI | CTGCAGGCGGCGTTGTGACAATTT | |
| flaAUp3 | TTGTCGGGGAAACGGAAGC | |
| flaALo3 | CATCAGCCGCCAGAAAGGAC | |
| flaRTfo | TAGCGACTTGACCACCCGTAAG | |
| flaRTrw | ACCTTCCTTCAGAGCGTTCACG | |
| SondeflAfo2 | GCTTCACCTATGGTGCCGC | |
| SondeflArw2 | GCCGTCATACCCACGAAAGC |
Sequence 5′ to 3′. PstI restriction sites are underlined.
DNA techniques.
DNA isolation, digestion, ligation and transformation essentially followed standard methods (26). For Southern hybridization, DNA was isolated, digested with restriction enzymes, electrophoresed, and blotted on a Hybond N membrane (Amersham). Probe DNA was labeled with digoxigenin-dUTP by using a PCR labeling kit (Roche, Mannheim, Germany) and the primers sondeflAfo2 and sondeflArw2. Prehybridization and hybridization were carried out at 68°C. Signals were detected with anti-digoxigenin-alkaline phosphatase and CDP-Star (Roche).
Primers used for PCR are listed in Table 1. PCR amplification was performed with the Mastercycler gradient (Eppendorf, Hamburg, Germany) by using standard protocols.
RT-PCR.
The isolation of the total RNA from M. gryphiswaldense was performed by standard techniques (26). Isolated RNA was treated with DNase (MBI Fermentas) and then used in a reverse transcriptase (RT) reaction (Moloney murine leukemia virus reverse transcriptase; MBI Fermentas) with a hexanucleotide primer mix (Roche Molecular Biochemicals). For negative control reverse transcriptase was omitted from the reaction mixture. The obtained cDNA was amplified by PCR by using PCR Master Mix (Promega) and primers flaRTfo and flaRTrw, which amplify a 577-bp fragment of the flaA gene.
Biparental conjugation.
Recombinant plasmids were introduced into the recipient strain M. gryphiswaldense R3/S1 by biparental conjugation with E. coli S17-1 as a donor as described previously (36). For selection of homologous recombination events, up to 1010 cells were mixed and incubated microaerobically on ACAM for 8 h. Cells were flushed from the agar surface into sterile liquid medium containing 50 μg of streptomycin to counterselect against the E. coli donor. To increase the ratio of homologous recombination events, the cells were incubated in this medium overnight before they were plated onto ACAM with rifampin (150 μg/liter) and streptomycin (50 μg/liter) and the appropriate antibiotic for plasmid selection.
Construction of flaA insertion mutations.
A 2.5-kb fragment was amplified by PCR by using primers flaAUp3 and flaALo3 and then subcloned into the pGEM-T Easy vector (Promega, Mannheim, Germany). The fragment was excised with EcoRI and ligated with the pK19mobsacB vector (the PstI restriction site of the vector was eliminated before) containing the sacB gene (27) as a counterselectable suicide marker. The gentamicin resistance cassette of the broad-host-range plasmid pBBR1MCS5 was amplified by PCR (each primer with a PstI linker) and subcloned into the pGEM-T Easy vector. The resulting plasmid, pDa102, was digested with PstI, and the purified gentamicin cassette was ligated into the PstI restriction site of the 2.5-kb fragment inside the flaA gene to yield plasmid pDa115. The construct was excised from pDa115 by EcoRI digestion and ligated into the suicide vector pK19mobGII containing the gusA gene (14) as a chromogenic marker. Correct insertion into the M. gryphiswaldense chromosome by single and double crossovers was confirmed by PCR with primers flaAFo3 and flaALo3 as well as by Southern hybridization with probe I.
Analysis of DNA and protein sequence data.
Genome sequence data from M. gryphiswaldense MSR-1 were used from the whole genome shotgun in progress (R. Reinhardt, MPI Molecular Genetics, Berlin-Dahlem, Germany), at the present stage of eightfold sequencing coverage. The basic analysis of DNA and protein sequences was done by the MacVector 7.0 software package (Oxford Molecular Ltd.). Sequence alignments were carried out by using the ClustalW algorithm (41), which is part of the same software. Protein sequences were compared to the GenBank, EMBL, and SwissProt databases. Preliminary sequence data for Magnetospirillum magnetotacticum MS-1 was obtained from the U.S. Department of Energy Joint Genome Institute at http://www.jgi.doe.gov/tempweb/JGI_microbial/html/index.html.
Electron microscopy.
Cells were adsorbed on carbon-coated copper grids and negatively stained with 2% (wt/vol) uranyl acetate. Samples were viewed and recorded with a Philips CM12 transmission electron microscope at an accelerating voltage of 120 kV.
SDS-PAGE.
Whole-cell extracts of M. gryphiswaldense were prepared by boiling the cells in sodium dodecyl sulfate (SDS) sample buffer for 10 min and then were separated by one-dimensional SDS-polyacrylamide gel electrophoresis (PAGE) (18). Approximately 20 μg of protein per lane was loaded onto a 12% polyacrylamide gel. The gels were digitized and analyzed by using ImageMaster 1D software (Amersham Pharmacia).
Motility assays.
Swarm plate assays were done by stabbing cells into semisolid 0.25% FSM agar and incubating the plate at 28°C under microaerobic conditions for 72 h. For motility assays in oxygen gradient tubes (1.5 by 15 cm), the FSM medium with 0.3% agar was inoculated with the cells and incubated for 48 h exposed to the air in the absence of an external magnetic field. In order to demonstrate magnetotaxis, a ferrite plate magnet (10 by 10 by 2.5 cm) was applied close to the tubes to generate a horizontal magnetic field, which covered the whole area of the length of the tube.
Nucleotide sequence accession numbers.
The nucleotide sequences of the M. gryphiswaldense flaA, flaB, and flaC genes have been deposited in the GenBank, EMBL, and DDJB libraries under the accession numbers CR354386, CR354387, and CR354388, respectively.
RESULTS AND DISCUSSION
Identification and sequence analysis of genes encoding flagellin homologues in M. gryphiswaldense and M. magnetotacticum.
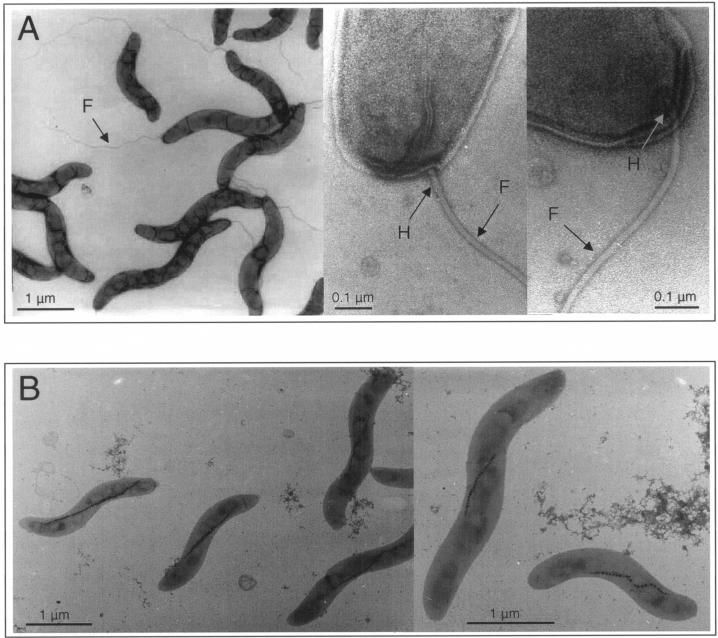
M. gryphiswaldense is highly motile by means of a single flagellum at each pole, which is slightly subterminally inserted into the cell body. The filament, which has a length of up to 5 μm and a diameter of approximately 20 nm, appears to be connected to the cell wall by a hook-like structure (Fig. 1A).
FIG. 1.
Electron micrographs of M. gryphiswaldense cells. (A) Wild type. The filament (F) appears to be connected to the cell wall by a hook-like structure (H). (B) A nonflagellated mutant strain, Da136.
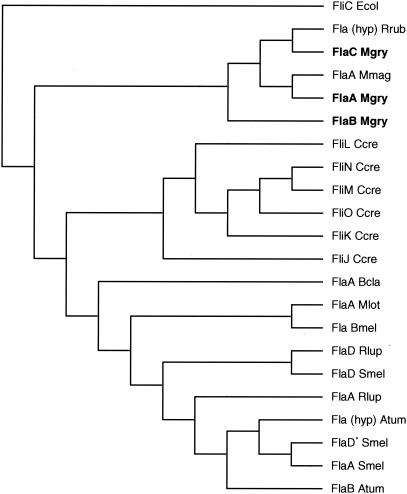
Inspection of the preliminary genome assembly (draft analysis; http://www.jgi.doe.gov/tempweb/JGI_microbial/html/index.html) of the closely related M. magnetotacticum identified several open reading frames (ORFs) with similarity to flagellin-related genes. One of them (flaA, gene 11 on contig 3879) encoding a putative 552-amino-acid (aa) protein with a predicted mass of 56.54 kDa and a pI of 8.55 was used in homology searches against the preliminary genome assembly of M. gryphiswaldense. A highly similar ORF was identified as the top hit, which was accordingly assigned to flaA of M. gryphiswaldense. The deduced amino acid sequence of M. gryphiswaldense FlaA indicated a protein of 295 aa residues with a mass of 31.36 kDa and a pI of 8.19. Similarity searches in the preliminary genome of M. gryphiswaldense identified two additional genes dubbed flaB and flaC with high similarity to flaA (E values of 2e-28 to 2e-36) and which apparently are unlinked on the chromosome, as well as several ORFs with lower similarity (E values of 0.048 to 2e-06; data not shown). This indicates that the filament, which has been described before for a number of other bacteria (4, 12, 28), is likely composed of multiple flagellin proteins in M. gryphiswaldense. FlaA, FlaB, and FlaC of M. gryphiswaldense have the highest similarity to a number of hypothetical and identified flagellin proteins from other α-proteobacteria (Fig. 2). All putative Fla proteins from both M. magnetotacticum and M. gryphiswaldense display the characteristic three-domain organization of bacterial flagellins with conserved N- and C-terminal domains and a variable central domain (19, 28). Interestingly, the N- and C-terminal amino acid sequences of the FlaA proteins are very similar for the two Magnetospirillum strains, whereas the central domains are highly divergent and have different lengths. In contrast, the hypothetical FlaA protein of Rhodospirillum rubrum displays extensive sequence similarity (64% similarity; 45% identity) to M. gryphiswaldense FlaA over its whole length.
FIG. 2.
Dendrogram showing the sequence similarity of selected full-length flagellin proteins from various α-proteobacteria. Abbreviations (with proteins and accession numbers): Rrub, Rhodospirillum rubrum [Fla(hyp), ZP_00013883]; Mgry, M. gryphiswaldense, Mmag, M. magnetotacticum (FlaA ZP_00056435); Ccres, Caulobacter crescentus (FliL, AAC35988; FliN, AAB95381.2; FliM, AAB95380.1; FliO, AAB95382.2; FliK, NP_420274; FliJ, P02969), Bcla, Bartonella clarridgeiae (FlaA, CAB64773); Mlot, Mesorhizobium loti (FlaA, NP_104151); Bmel, Brucella melitensis (FlaB, NP_541127); Rlup, Rhizobium lupini (FlaD, AAG14366; FlaA, AAG14364); Smel, Sinorhizobium meliloti (FlaD, AAB81422; putative FlaD*, NP_384777; FlaA, NP_384775); Atum, Agrobacterium tumefaciens [Fla(hyp), NC_003062]. Flagellin sequences determined in this study are in bold. The E. coli (Ecol) flagellin (FliC, NC_000913) was used as an outgroup marker. The multiple alignment and dendrogram were constructed by using the ClustalW program of the MacVector 7.0 software. Branch lengths are not to scale.
Construction of a flagellin mutant by gene replacement.
Homologous recombination is a versatile tool, allowing the creation of marked or unmarked insertion or deletion mutations in selected genes. Derivatives of the mobilizable pK19mob vector (pMB-1-replicon [27]) were selected for insertional mutagenesis experiments because of the vector's general inability to replicate in bacteria outside the enterobacterial group. As expected, all conjugation experiments with this vector failed to yield antibiotic-resistant transformants, indicating that plasmids harboring the pMB-1-replicon do not replicate in M. gryphiswaldense and can, therefore, be used as suicide vectors to introduce mutations into the chromosome.
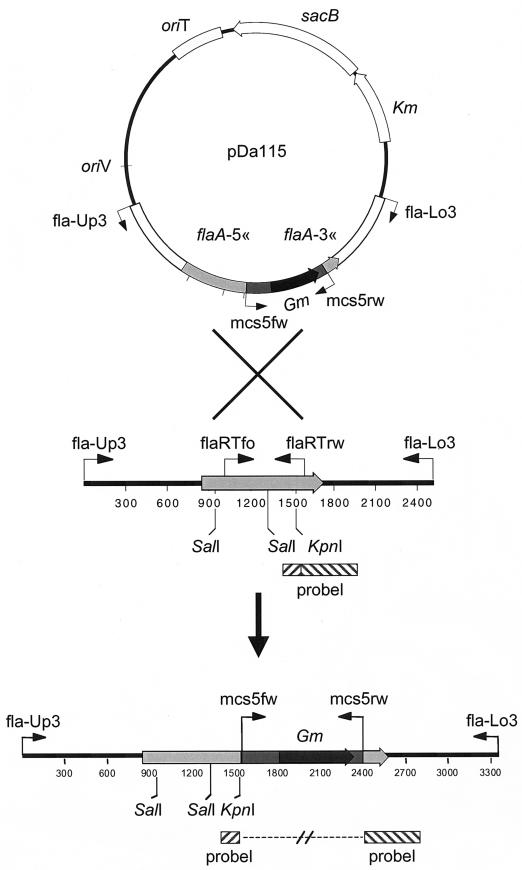
The isolation of rare double recombinants can often be greatly facilitated by the use of markers, which are easily screenable or counterselectable (2, 23). To test if this is an applicable strategy for study of M. gryphiswaldense, we constructed suicide plasmids based on either the pK19mobGII vector (14) harboring the gusA gene, which encodes the enzyme β-glucuronidase, or the pK19mobsacB vector (27), which harbors the genetically modified allele of the sacB gene of Bacillus subtilis (38) coding for the levansucrase enzyme that confers a lethal phenotype to many gram-negative bacteria in the presence of sucrose. The resulting plasmids pDa115 (sacB) (Fig. 3) and pDa116 (gusA) both contained the flaA gene with the gentamicin cassette inserted (flaA::Gm). The extent of homologous M. gryphiswaldense sequences present before and after the gentamicin marker was 1,549 and 979 bp, respectively. Conjugation with plasmid pDa116 resulted in numerous gentamicin-resistant colonies with a frequency of approximately 10−6 colonies per recipient cell. Every one of the 96 examined colonies resulted from a single crossover event, as detected by PCR and sensitivity to both kanamycin and gentamicin. Several clones harboring single crossovers were further propagated in liquid medium containing gentamicin, but lacking kanamycin. However, we repeatedly failed to identify double-crossover mutants by replica plating on ACAM containing either kanamycin or gentamicin for the loss of the plasmid-borne kanamycin marker. Several of the colonies eventually turned blue after prolonged incubation on 5-bromo-4-chloro-3-indoxyl-β-d-glucuronide-containing ACAM plates. However, color development was not reproducible and was unstable during serial transfers, and no clear correlation between decolorization and a particular genotype (loss of the inserted vector) could be detected. The potential use of gusA as a screenable marker in M. gryphiswaldense thus requires further elaboration.
FIG. 3.
Scheme of construction of double crossovers. Plasmid pDa115 represents the suicide vector used to inactivate the M. gryphiswaldense flaA gene. The primers used are shown as arrows. Restriction sites and the probe used in the Southern blot analysis are indicated.
The growth of M. gryphiswaldense clones harboring the sacB gene was inhibited on ACAM plates by the presence of sucrose (MIC, 5%), whereas the wild-type control lacking the sacB gene grew at sucrose concentrations up to 7.5 to 10%. Therefore, we concluded that sacB can be used as a marker to counterselect for the rare, gene-replacing, second recombination.
By using plasmid pDa115, numerous gentamicin-resistant colonies were obtained in conjugation experiments on ACAM plates containing 5% sucrose. As revealed by PCR and Southern blot analysis, all colonies resulted from homologous recombination with the M. gryphiswaldense flaA locus. Three classes of mutants could be distinguished based on their different genotypes. Two classes of single-crossover mutants resulted from a single-crossover event with either the left arm (5′-insertion) or the right arm (3′-insertion). The ratio of the number of left- to the number of right-arm insertion mutants was approximately 1:20. As these mutants were still found to contain the inserted plasmid pDa115, we concluded that the gained insensitivity to sucrose was due to the loss of the sacB function by spontaneous mutation, which has repeatedly been observed before (3, 6, 13). Both 5′ and 3′ single-crossover insertion mutants displayed motility, which was virtually indistinguishable from the wild-type strain. This finding indicates that the insertion of the plasmid, which results in cells that are merodiploid for flaA, has no polar effects on the expression of downstream genes that might putatively affect motility and flagellar assembly.
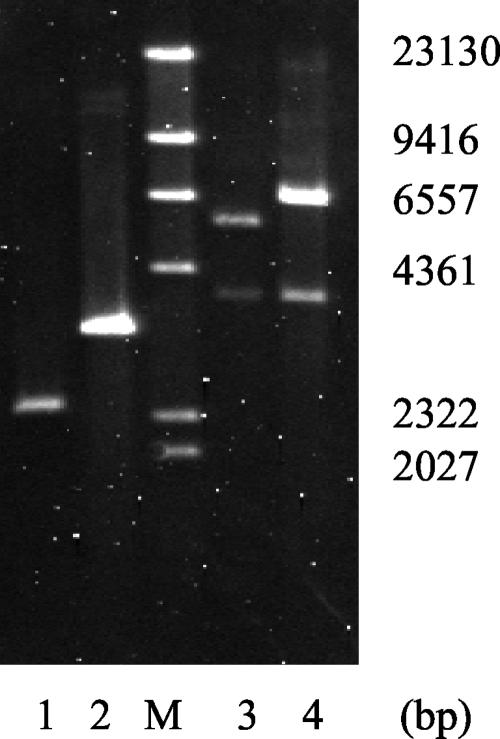
Approximately 1% of the mutants, however, were found to represent a third class of mutants, which was due to reciprocal crossover events (Fig. 3) as revealed by replica plating and Southern blot analysis (Fig. 4). One clone, dubbed strain Da136, was selected for further analysis.
FIG. 4.
Confirmation of flaA disruption by Southern blot analysis of genomic DNA from the wild type (lanes 1 and 3) and strain Da136 (lanes 2 and 4). DNA was digested with SalI (lanes 1 and 2) and KpnI (lane 3 and 4). Lane M, molecular weight markers. The blot was hybridized with probe I (shown in Fig. 3), which overlaps the KpnI restriction site. Digesting with SalI revealed a larger band for Da136 because of the insertion of the gentamicin cassette. As expected, digesting with KpnI yielded two signals for the wild type and strain Da136. The smaller hybridizing fragments of the wild-type and Da136 DNA have identical sizes, while the positions of the larger bands differ by the size of the inserted gentamicin cassette.
Phenotypic analysis of the mutant strain Da136.
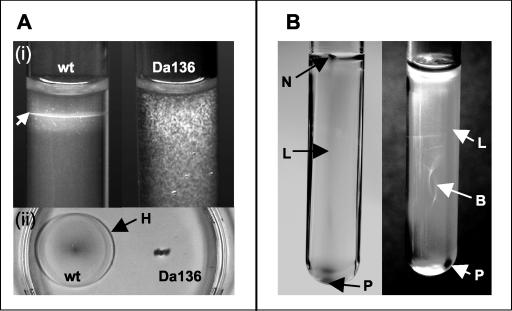
Microscopic analysis of strain Da136 revealed a total loss of motility. In the absence of a magnetic field, wild-type bacteria grew as sharp microaerophilic bands in oxygen gradients, while growth of the mutant had a fuzzy appearance. Likewise, the mutant failed to form chemotactic halos in semisolid swarm plates (Fig. 5A). The appearance of the growth and the distribution of mutant cells in semisolid oxygen gradient tubes were unaffected by external magnetic fields, although cells contained magnetosomes and passively aligned along magnetic fields. In contrast, the wild-type cells formed characteristic polar, three-dimensional magnetotactic patterns in the presence of a horizontal magnetic field (Fig. 5B). Specifically, cells accumulated at the wall facing the magnetic South pole as a line leading into a nose-like tip close to the agar surface, while at the opposite side (distal from the magnetic South pole) a cell pellet was visible close to the bottom of the tube.
FIG. 5.
Motility assays in semisolid agar. (A) Growth in the absence of a magnetic field. At the top (i), wild-type (wt) M. gryphiswaldense grew as sharp microaerophilic bands (arrow) in oxygen gradients, while growth of the nonmotile mutant strain Da136 was fuzzy. At the bottom (ii), bacteria were stabbed into motility agar in a petri dish. The wild type formed a large chemotactic swarming halo (H) with a diameter of approximately 4 cm after 48 h, while no spreading of mutant strain Da136 was visible. (B) Magnetotactic patterns of wild-type M. gryphiswaldense in the presence of a horizontal magnetic field. On the left side of the panel, at the wall facing the magnetic South pole, cells accumulated as a line (L) leading into a nose-like tip (N) close to the agar surface. On the right side of the panel, at the opposite side (distal to the South pole of the magnet), a cell pellet (P) was visible close to the bottom of the same tube. In addition, a spherical pattern resembling a bubble (B) was formed in the center of the tube.
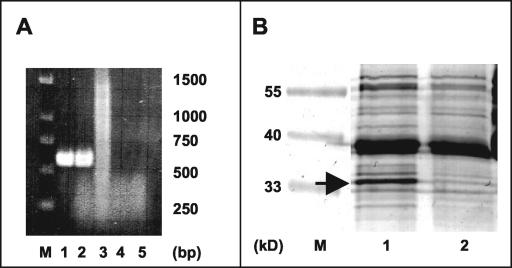
Electron microscopy confirmed that the loss of motility was in fact due to the lack of flagellar filaments, resulting in a bald phenotype (Fig. 1B). In addition, we failed to detect any remaining rod-like structures or appendages, which were occasionally observed in flagellin mutants of other bacteria (16, 37). While the RT-PCR revealed transcription of the flaA gene in the wild type, a transcript was no longer detectable in the mutant strain as expected (Fig. 6A). The disappearance of the FlaA protein in the null mutant strain Da136 was anticipated since the flagellar proteins represent a significant proportion of the total cellular protein (25). Figure 6B shows the Coomassie blue-stained polypeptide profiles from the wild-type and mutant strains. An abundant band (approximately 15% of total protein) at 33.2 kDa was present in whole-cell extracts of the wild type, while in the mutant strain only a faint band was visible at the same position, which was equivalent to less than 4% of the total cell protein. We concluded that the 33.2-kDa band in the wild type corresponds to the FlaA protein, whereas the faint band in strain Da135 is likely to represent an unrelated protein with an electrophoretic mobility coincidentally resembling the FlaA band. The slightly higher apparent molecular mass of the FlaA band compared to its predicted mass might be explained by glycosylation of the flagellin protein, as there is increasing evidence that protein glycosylation is involved in the flagellar assembly process in a number of bacteria (29). In summary, from these results it can be concluded that the specific knockout of flaA function results in a deficiency in the assembly of flagella and, consequently, in the loss of magnetotaxis.
FIG. 6.
(A) Analysis of the expression of the flaA gene in M. gryphiswaldense wild type and mutant strain Da136 by RT-PCR using various DNA templates. Lane 1, genomic DNA used as a template (positive control); lane 2, cDNA of the wild type; lane 3, cDNA of strain Da136; lane 4, wild-type RNA, reverse transcriptase omitted (negative control); lane 5, Da136 RNA, reverse transcriptase omitted (negative control). DNA was amplified by using primers flaRTfo and flaRTrw. (B) Analysis of the flagellin synthesis by SDS-12% PAGE. Whole-cell extracts of the wild type (lane 1) and the mutant Da136 (lane 2) were stained with Coomassie brilliant blue. A putative flagellin polypeptide was present in the wild-type strain (arrow) but absent from the mutant strain. M, molecular weight markers.
Acknowledgments
This study was supported by the BMBF Biofuture program and the Max Planck Society.
We thank Katja Junge and Susanne Ullrich for help and Katja Schmidt for excellent technical assistance. We are grateful to Harald Engelhardt and Günter Pfeifer (MPI f. Biochemistry, Martinsried, Germany) for help and access to the electron microscope. We thank Richard Reinhardt (MPI f. Molecular Genetics, Berlin-Dahlem, Germany) for access to genomic sequencing data. Douglas Bartlett (La Jolla, Calif.), Matthias Keller, Alfred Pühler (Bielefeld, Germany), and Gerrit Voordouw (Calgary, Canada) are acknowledged for their kind gifts of plasmids and bacterial strains. Preliminary sequence data for M. magnetotacticum MS-1 was obtained from the U.S. Department of Energy Joint Genome Institute at http://www.jgi.doe.gov/tempweb/JGI_microbial/html/index.html.
REFERENCES
- 1.Bäuerlein, E. 2003. Biomineralization of unicellular organisms: an unusual membrane biochemistry for the production of inorganic nano- and microstructures. Angew. Chem. Int. Ed. Engl. 42:614-641. [DOI] [PubMed] [Google Scholar]
- 2.Bitan-Banin, G., R. Ortenberg, and M. Mevarech. 2003. Development of a gene knockout system for the halophilic archaeon Haloferax volcanii by use of the pyrE gene. J. Bacteriol. 185:772-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cai, Y. P., and C. P. Wolk. 1990. Use of a conditionally lethal gene in Anabaena sp. strain PCC 7120 to select for double recombinants and to entrap insertion sequences. J. Bacteriol. 172:3138-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ely, B., T. W. Ely, W. B. Crymes, Jr., and S. A. Minnich. 2000. A family of six flagellin genes contributes to the Caulobacter crescentus flagellar filament. J. Bacteriol. 182:5001-5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frankel, R. B., D. A. Bazylinski, M. S. Johnson, and B. L. Taylor. 1997. Magneto-aerotaxis in marine coccoid bacteria. Biophys. J. 73:994-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fu, R., and G. Voordouw. 1997. Targeted gene-replacement mutagenesis of dcrA, encoding an oxygen sensor of the sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough. Microbiology 143:1815-1826. [DOI] [PubMed] [Google Scholar]
- 7.Grünberg, K., E. C. Müller, A. Otto, R. Reszka, D. Linder, M. Kube, R. Reinhardt, and D. Schüler. 2004. Biochemical and proteomic analysis of the magnetosome membrane in Magnetospirillum gryphiswaldense. Appl. Environ. Microbiol. 70:1040-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grünberg, K., C. Wawer, B. M. Tebo, and D. Schüler. 2001. A large gene cluster encoding several magnetosome proteins is conserved in different species of magnetotactic bacteria. Appl. Environ. Microbiol. 67:4573-4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guell, D. C., H. Brenner, R. B. Frankel, and H. Hartman. 1988. Hydrodynamic forces and band formation in swimming magnetotactic bacteria. J. Theor. Biol. 135:525-542. [Google Scholar]
- 10.Handrick, R., S. Reinhardt, D. Schultheiss, T. Reichart, D. Schüler, and D. Jendrossek. 2004. Unraveling the function of the Rhodospirillum rubrum activator of polyhydroxybutyrate (PHB) degradation: the activator is a PHB-granule-bound protein (phasin). J. Bacteriol. 186:2466-2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heyen, U., and D. Schüler. 2003. Growth and magnetosome formation by microaerophilic Magnetospirillum strains in an oxygen-controlled fermentor. Appl. Microbiol. Biotechnol. 61:536-544. [DOI] [PubMed] [Google Scholar]
- 12.Josenhans, C., A. Labigne, and S. Suerbaum. 1995. Comparative ultrastructural and functional studies of Helicobacter pylori and Helicobacter mustelae flagellin mutants: both flagellin subunits, FlaA and FlaB, are necessary for full motility in Helicobacter species. J. Bacteriol. 177:3010-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaniga, K., I. Delor, and G. R. Cornelis. 1991. A wide-host-range suicide vector for improving reverse genetics in gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene 109:137-141. [DOI] [PubMed] [Google Scholar]
- 14.Katzen, F., A. Becker, M. V. Ielmini, C. G. Oddo, and L. Ielpi. 1999. New mobilizable vectors suitable for gene replacement in gram-negative bacteria and their use in mapping of the 3′ end of the Xanthomonas campestris pv. campestris gum operon. Appl. Environ. Microbiol. 65:278-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kirschvink, J. L. 1997. Magnetoreception: homing in on vertebrates. Nature 390:339-341. [Google Scholar]
- 16.Klose, K. E., and J. J. Mekalanos. 1998. Differential regulation of multiple flagellins in Vibrio cholerae. J. Bacteriol. 180:303-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop II, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 18.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 19.Macnab, R. M. 2003. How bacteria assemble flagella. Annu. Rev. Microbiol. 57:77-100. [DOI] [PubMed] [Google Scholar]
- 20.Matsunaga, T., C. Nakamura, J. Burgess, and S. Sode. 1992. Gene transfer in magnetic bacteria: transposon mutagenesis and cloning of genomic DNA fragments required for magnetosome synthesis. J. Bacteriol. 174:2748-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nealson, K. H., and B. L. Cox. 2002. Microbial metal-ion reduction and Mars: extraterrestrial expectations? Curr. Opin. Microbiol. 5:296-300. [DOI] [PubMed] [Google Scholar]
- 22.Petersen, N., T. Von Dobeneck, and H. Vali. 1986. Fossil bacterial magnetite in deep-sea sediments from the South Atlantic Ocean. Nature 320:611-615. [Google Scholar]
- 23.Reyrat, J. M., V. Pelicic, B. Gicquel, and R. Rappuoli. 1998. Counterselectable markers: untapped tools for bacterial genetics and pathogenesis. Infect. Immun. 66:4011-4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson, S. G., J. T. S. Sahota, and F. Oldfield. 2000. Early diagenesis in North Atlantic abyssal plain sediments characterized by rock-magnetic and geochemical indices. Marine Geol. 163:77-107. [Google Scholar]
- 25.Rosey, E. L., M. J. Kennedy, D. K. Petrella, R. G. Ulrich, and R. J. Yancey, Jr. 1995. Inactivation of Serpulina hyodysenteriae flaA1 and flaB1 periplasmic flagellar genes by electroporation-mediated allelic exchange. J. Bacteriol. 177:5959-5970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Schäfer, A., A. Tauch, W. Jäger, J. Kalinowski, G. Thierbach, and A. Pühler. 1994. Small mobilizable multipurpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 28.Scharf, B., H. Schuster-Wolff-Buhring, R. Rachel, and R. Schmitt. 2001. Mutational analysis of the Rhizobium lupini H13-3 and Sinorhizobium meliloti flagellin genes: importance of flagellin A for flagellar filament structure and transcriptional regulation. J. Bacteriol. 183:5334-5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schirm, M., E. C. Soo, A. J. Aubry, J. Austin, P. Thibault, and S. M. Logan. 2003. Structural, genetic and functional characterization of the flagellin glycosylation process in Helicobacter pylori. Mol. Microbiol. 48:1579-1592. [DOI] [PubMed] [Google Scholar]
- 30.Schleifer, K., D. Schüler, S. Spring, M. Weizenegger, R. Amann, W. Ludwig, and M. Köhler. 1991. The genus Magnetospirillum gen. nov., description of Magnetospirillum gryphiswaldense sp. nov. and transfer of Aquaspirillum magnetotacticum to Magnetospirillum magnetotacticum comb. nov. Syst. Appl. Microbiol. 14:379-385. [Google Scholar]
- 31.Schübbe, S., M. Kube, A. Scheffel, C. Wawer, U. Heyen, A. Meyerdierks, M. Madkour, F. Mayer, R. Reinhardt, and D. Schüler. 2003. Characterization of a spontaneous nonmagnetic mutant of Magnetospirillum gryphiswaldense reveals a large deletion comprising a putative magnetosome island. J. Bacteriol. 185:5779-5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schüler, D. 2002. The biomineralization of magnetosomes in Magnetospirillum gryphiswaldense. Int. Microbiol. 5:209-214. [DOI] [PubMed] [Google Scholar]
- 33.Schüler, D. 2004. Molecular analysis of a subcellular compartment: the magnetosome membrane in Magnetospirillum gryphiswaldense. Arch. Microbiol. 1-7. 181: [DOI] [PubMed]
- 34.Schüler, D., and E. Bäuerlein. 1998. Dynamics of iron uptake and Fe3O4 biomineralization during aerobic and microaerobic growth of Magnetospirillum gryphiswaldense. J. Bacteriol. 180:159-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schüler, D., and R. B. Frankel. 1999. Bacterial magnetosomes: microbiology, biomineralization and biotechnological applications. Appl. Microbiol. Biotechnol. 52:464-473. [DOI] [PubMed] [Google Scholar]
- 36.Schultheiss, D., and D. Schüler. 2003. Development of a genetic system for Magnetospirillum gryphiswaldense. Arch. Microbiol. 179:89-94. [DOI] [PubMed] [Google Scholar]
- 37.Schuster, S. C., and E. Bäuerlein. 1992. Location of the basal disk and a ringlike cytoplasmic structure, two additional structures of the flagellar apparatus of Wolinella succinogenes. J. Bacteriol. 174:263-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Selbitschka, W., S. Niemann, and A. Pühler. 1993. Construction of gene replacement vectors for gram-negative bacteria using a genetically modified sacRB gene as a positive selection marker. Appl. Microbiol. Biotechnol. 38:615-618. [Google Scholar]
- 39.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilisation system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology. 1:784-791. [Google Scholar]
- 40.Stolz, J. 1990. Biogenic magnetite and the magnetization of sediments. J. Geophys. Res. 95:4355-4361. [Google Scholar]
- 41.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wahyudi, A. T., H. Takeyama, and T. Matsunaga. 2001. Isolation of Magnetospirillum magneticum AMB-1 mutants defective in bacterial magnetic particle synthesis by transposon mutagenesis. Appl. Biochem. Biotechnol. 91-93:147-154. [DOI] [PubMed] [Google Scholar]