Abstract
Rhizodegradation of organic pollutants, such as polycyclic aromatic hydrocarbons, is based on the effect of root-produced compounds, known as exudates. These exudates constitute an important and constant carbon source that selects microbial populations in the plant rhizosphere, modifying global as well as specific microbial activities. We conducted an experiment in two-compartment devices to show the selection of bacterial communities by root exudates and phenanthrene as a function of distance to roots. Using direct DNA extraction, PCR amplification, and thermal gradient gel electrophoresis screening, bacterial population profiles were analyzed in parallel to bacterial counts and quantification of phenanthrene biodegradation in three layers (0 to 3, 3 to 6, and 6 to 9 mm from root mat) of unplanted-polluted (phenanthrene), planted-polluted, and planted-unpolluted treatments. Bacterial community differed as a function of the distance to roots, in both the presence and the absence of phenanthrene. In the planted and polluted treatment, biodegradation rates showed a strong gradient with higher values near the roots. In the nonplanted treatment, bacterial communities were comparable in the three layers and phenanthrene biodegradation was high. Surprisingly, no biodegradation was detected in the section of planted polluted treatment farthest from the roots, where the bacterial community structure was similar to those of the nonplanted treatment. We conclude that root exudates and phenanthrene induce modifications of bacterial communities in polluted environments and spatially modify the activity of degrading bacteria.
Polycyclic aromatic hydrocarbons (PAHs) are hydrophobic organic pollutants, composed of two or more fused aromatic rings. They are produced by organic matter combustion, naturally during forest fires or anthropogenically during fossil hydrocarbon burning. Due to their carcinogenic properties, their recalcitrance to degradation, and their widespread occurrence in contaminated industrial wastelands, the methods of eliminating PAHs from heavily polluted environments have been amply investigated. Numerous soil microorganisms have been shown to biotransform and to mineralize PAH (3, 17). Microbially based remediation techniques consist mainly of improving such activities in situ with the optimization of soil parameters such as pO2 or addition of mineral nutrients.
The use of plants to improve remediation of recalcitrant organic pollutants, such as PAHs, includes rhizodegradation, which combines physical-chemical modifications in the rhizosphere affecting pollutant bioavailability and stimulating effects on microbial processes. Roots indeed increase soil porosity and, by creating oriented fluxes, cause a decrease of water and nutrient levels, especially nitrogen and phosphorus (29). On the other hand, roots exude a high diversity and quantity of organic carbon compounds into the surrounding soil (28). Exudation is a dynamic phenomenon that qualitatively and quantitatively varies depending on plant species, physiological status such as stage development (25), nutritional conditions (10, 32), and the presence of microorganisms (19). By controlling C balance in the rhizosphere (18), root exudation is likely the most selective force affecting microbial communities. The rhizosphere effect is indeed well known to increase microbial density and activity compared to those for bulk soil and to select specific microorganisms (20). As a consequence, any variation in root exudation could induce variations of microbial communities.
Using compartment devices with sand and pot experiments with polluted soil, we have previously shown rhizospheric gradients of both bacterial numbers and PAH biodegradation (7, 14). However, to our knowledge, how PAH and root exudates affect bacterial community structure in a polluted rhizosphere has not been investigated. The objectives of this work were to verify if microbial communities and their degrading activity were affected by (i) soluble root exudates, (ii) the distance to roots, and (iii) the presence of PAHs. A compartment device was designed to obtain a dense root mat simulating an active exuding surface and laterally replenishing a compartment with C from exudates, in the presence or absence of phenanthrene (PHE) (7). Using such devices, we analyzed microbial community structure; quantified heterotrophic and PAH-degrading bacteria and PHE biodegradation, at three distances from the roots; and compared planted-polluted, planted-unpolluted, and unplanted-polluted devices. Total genomic DNA was extracted from the three layers of the lateral compartment, and bacterial 16S ribosomal DNA (rDNA) was amplified by universal primers to compare profiles of communities by thermal gradient gel electrophoresis (TGGE) analysis.
MATERIALS AND METHODS
Experimental design.
Compartmented pots, previously described in the work of Corgié et al. (7), consisted of a vertical polyvinyl chloride tube forming a central root compartment (RC) and a horizontal rhizosphere compartment (RHC) separated from the central compartment by a 37-μm-pore-size nylon mesh. Three treatments were carried out with four replicates per treatment: planted pots with unpolluted sand in RHC (P−PHE), planted pots with polluted sand in RHC (P+PHE), and nonplanted pots with polluted sand in RHC (NP+PHE). Briefly, a germinated seed of ryegrass (Lolium perenne L. cv. Barclay) was planted in RC and grown for 4 weeks. Washed quartz sand (0.125- to 2-mm-diameter grains) was spiked with PHE, as described in the work of Corgié et al. (7), to a final concentration of 500 μg · g of sand−1. A microbial suspension from a PAH-contaminated soil (14), prepared and filtrated to 0.2 μm as previously described (7), was added to polluted and unpolluted sand to a final concentration of approximately 105 cells g of sand−1. Polluted and unpolluted inoculated sands were filled in RHC and watered with nutrient solution until saturation. RHCs were immediately inserted into the RC, and pots were incubated for another 4 weeks in a growth chamber (24 and 20°C day and night temperatures, respectively; 16-h light period; 60 to 70% relative humidity; 350 μmol of PAR [photosynthetically active radiation] m−2 s−1). At harvest, lateral compartments were removed and the sand was cut in 3-mm layers from the separating mesh: 0 to 3, 3 to 6, and 6 to 9 mm, called S1, S2, and S3, respectively. For each sample, 1 g was immediately used for microbial counts, 1 g was frozen at −80°C prior to DNA extraction, and the remaining sand was air dried overnight and stored at 4°C prior to chemical analysis.
PAH quantification and microbial enumeration.
The PHE concentration was analyzed using gas chromatography-mass spectrometry (SARM, CRPG-CNRS, Vandoeuvre-lès-Nancy, France) after Soxhlet extraction with chloroform on 2 g of dried samples (2).
The number of culturable microorganisms was determined using a most probable number (MPN) procedure in 96-well microtiter plates (1) from serial dilutions (10−2 to 10−5) of initial suspension (1 g of sand in 10 ml of 0.85% NaCl) and inoculated into 40 wells per dilution. Culturable heterotrophic microorganisms were cultivated (1 week, 28°C) in nutrient broth (Difco). PAH-degrading bacteria were cultivated (3 weeks, 28°C) in a mineral medium (Bushnel Haas; Difco) supplemented with a PAH mixture (PHE, fluorene, anthracene, and fluoranthene). Absorbance was measured at 620 nm for heterotrophs and at 405 to 620 nm for PAH degraders. A computer program using standard McCrady standards was used to calculate MPNs.
DNA extraction, 16S rDNA amplification, and TGGE profiling.
Isolation of total DNA, based on a bead beating lysis and a phenol-chloroform purification, was modified from the method of Picard et al. (26) as follows. Sand particles of RHC were used to ensure physical lysis of bacterial cells and to homogenize solvent and aqueous phases. Plasticware was certified DNase and RNase free, and all solutions were autoclaved before use. Briefly, 800 μl of extraction buffer (100 mM Tris, 100 mM EDTA, 100 mM NaCl, 1% [wt/vol] polyvinylpolypyrrolidone, 2% [wt/vol] sodium dodecyl sulfate, pH 8) was added to each sand sample (approximately 1 g) in a 2-ml centrifuge tube and beaten on a horizontal grinder (Retsch) at maximum speed (approximately 1 m s−1) for 1 min. A phenol solution (800 μl) (phenol-chloroform-isoamyl alcohol, 25/24/1) was added, and the mixture was homogenized again for 30 s. After centrifugation at 14,000 × g for 2 min, the supernatant was transferred to a new microcentrifuge tube with 800 μl of chloroform solution (chloroform-isoamyl alcohol, 24/1, vol/vol). The mixture was gently homogenized and centrifuged at 14,000 × g for 5 min, and the supernatant was transferred to a fresh microtube. After the previous step was repeated, DNA was precipitated with an equal volume (800 μl) of isopropanol at 4°C for 15 min and centrifuged at 14,000 × g for 20 min. The supernatant was discarded, and the DNA pellet was rinsed twice with ethanol (70%, 4°C) and vacuum dried. The pellet was then resuspended in 100 μl of sterile ultrapure water and stored at −80°C for further analysis.
The eubacterial primer set, 968f and 1401r, was used to amplify a 475-bp fragment of the 16S rDNA (8, 12). A 40-bp GC clamp was attached to the 5′ end of the primer 968f to avoid complete separation of DNA strands during denaturing electrophoresis. For each amplification reaction, 1 μl of crude DNA extract was added to 49 μl of PCR mix consisting of 50 mM buffer, 3 mM MgCl2, 0.2 mM deoxynucleoside triphosphates, 2 μM (each) primer, and 2 U of Taq polymerase (Fastart; Roche Diagnostic). DNA was amplified in an iCycler (Bio-Rad Laboratories) with the following amplification program: 94°C for 5 min (1 cycle); 94°C for 40 s, 56°C for 30 s, and 72°C for 40 s (38 cycles); and 72°C for 5 min (1 cycle). PCR products, stained with ethidium bromide, were visualized under UV light after electrophoresis in a 1% (wt/vol) agarose gel to verify the size of amplified DNA.
TGGE was performed with a Dcode universal mutation detection system (Bio-Rad Laboratories). An exhaustive thermal gradient was previously determined, using MacMelt software provided by the manufacturer, on 43 bacterial 16S rDNA sequences (α, β, γ, and δ divisions of the Proteobacteria and genus of Eubacteria) from GenBank, targeted with the primer set used. The polyacrylamide gels (6% [wt/vol] acrylamide, 0.21% [wt/vol] bisacrylamide, 8 M urea, 1.25× Tris-acetate-EDTA, and 0.2% [vol/vol] glycerol) were allowed to polymerize for 2 h. DNA samples (5 μl) were separated by electrophoresis in 1.25× Tris-acetate-EDTA at a constant voltage (100 V), with a temperature gradient from 57 to 67°C (temperature increment of 0.7°C/h). After electrophoresis, gels were stained with ethidium bromide and numerized under UV light. Bands of interest were extracted from the gel and DNA amplified with the previous primer set (nonclamped) prior to sequencing (AB BioGen). One-strand sequences (435 bp) obtained were compared to sequences available in the GenBank database (Blastn software).
The presence-absence of bands is generally well suited for comparing profiles when they are really different with enough species not shared between the samples. The relative intensity of detectable species enabled an increase in the sensitivity of population profiling methods (24). Quantification of band mobility and band intensity was performed using Kodak 1D 3.5.2 software. For each profile, the intensity of the bands was summed and the relative intensity of each band was calculated, so that the relative band intensity reflected the abundance of the species in the initial sample (23). Community similarities between TGGE profiles, based on the relative band intensity, were analyzed by principal component analysis (33) with ADE-4 software (http://pbil.univ-lyon1.fr/ADE-4/). Analyses of variance were performed on ordination plots for the two principal components to determine the effect of PHE and distance to roots on bacterial communities. Kruskal-Wallis tests were performed to compare mean values of bacterial numbers and remaining PHE from each section for the three treatments.
RESULTS
Bacterial numbers and PHE degradation.
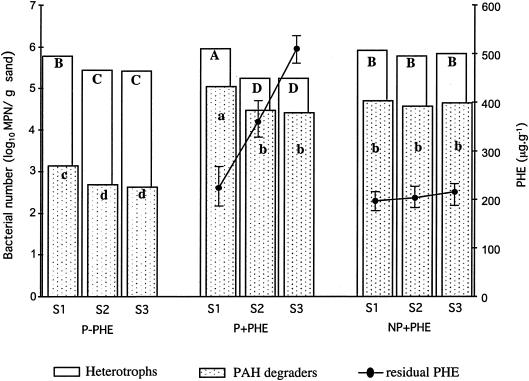
No significant difference in the number of culturable heterotrophic bacteria and PAH-degrading bacteria was observed among the three sections of the unplanted treatment (Fig. 1). In planted treatments, the microbial numbers of both heterotrophic and PAH-degrading bacteria were higher in S1 than in S2 and S3. The highest numbers of heterotrophic bacteria and PAH-degrading bacteria were observed in section S1 of the planted and polluted treatment. The lowest numbers of PAH-degrading bacteria were found in the unpolluted treatment. In planted treatment PHE concentration significantly decreased with the distance to the roots (P < 0.05) with higher degradation in S1 (approximately 58%). In S2, PHE degradation reached 28%, whereas in S3 no degradation was observed. In the nonplanted treatment, PHE degradation did not significantly differ between sections (approximately 60%).
FIG. 1.
Numbers (log10 of MPN) of culturable heterotrophs and PAH degraders and PHE concentration as a function of the distance to the roots (S1, S2, and S3 correspond to the three consecutive sections from the RC) for planted and nonpolluted (P−PHE), planted and polluted (P+PHE), and nonplanted polluted (NP+PHE) treatments. Capital letters for total heterotrophs and small letters for PAH degraders indicate significant differences between treatments and section (P < 0.05).
Bacterial community structure.
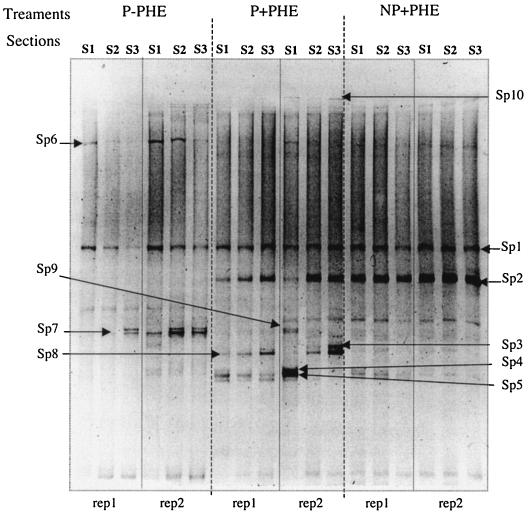
In planted treatments, community profiles clearly indicated qualitative changes in bacterial species and quantitative changes in their band intensity as a function of the distance to the roots (Fig. 2). In contrast, in the nonplanted treatment, community profiles appeared identical for all the sections. Some species were present in all the samples, such as Sp1, whose sequence showed 100% homology with Prosthecobacter fusiformis (sequence accession no. U60015). Other species appeared only in the presence of PHE, such as Sp2 (unidentified) and Sp3, Sp4, and Sp5, showing 100% homology with Caulobacter sp. (Sp3 sequence accession no., AF301221; Sp4 and Sp5 sequence accession, no. AJ227755). Sp4 and Sp5 occurrence was higher in S1 sections of the planted and polluted treatment, whereas Sp3 was higher in S3 sections. Sp6 (unidentified) was found only in planted treatments, and its occurrence decreased with increasing distance from the roots. In contrast Sp7, close to a Pseudomonas sp. (100% homology, sequence accession no. AJ419674), was more abundant in S3 sections. Sp8, close to an unidentified and unculturable bacterium (96% homology, sequence accession no. AF226221), and Sp9, belonging to the Burkholderiales (94% homology, sequence accession no. AY154366), were detected only in planted treatments. Sp10, common to polluted treatments, showed 93% homology with an unidentified Rhodocyclaceae member (sequence accession no. AF351219).
FIG. 2.
TGGE gel of bacterial 16S rDNA stained with ethidium bromide. DNA was amplified from sample DNA extracts for the three sections (S1, S2, and S3 correspond to the three consecutive sections from the RC) for planted and nonpolluted (P−PHE), planted and polluted (P+PHE), and nonplanted polluted (NP+PHE) treatments (rep 1 and rep 2 correspond to two of the four replicates performed for each treatment).
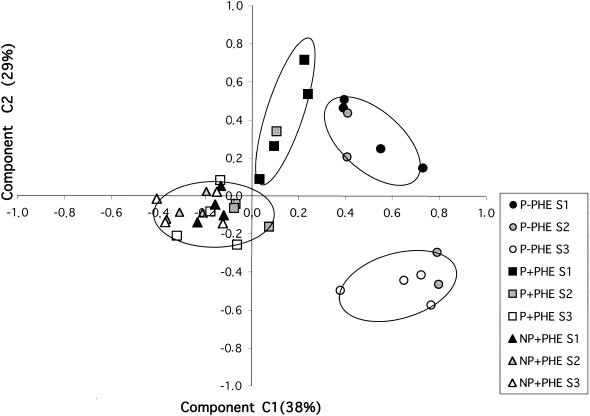
To compare all the profiles, a principal component analysis was performed on the occurrence data calculated from relative band intensities on the TGGE gels. Analysis of variance, performed on plot ordinates for the three main component axes, showed that the C1 axis was significantly correlated with the presence-absence of PHE whereas the C2 axis was correlated with the distance to the roots (Fig. 3). Based on the similarity of occurrence patterns between profiles, four main groups were significantly separated by these two parameters. For the nonpolluted treatment, S1-type profiles and S3-type profiles were significantly separated on the C2 axis, indicating different communities as a function of the distance to the roots. S2 community profiles grouped with S1- or S3-type profiles. In contrast, samples of the NP+PHE treatment formed a homogenous group and few differences in bacterial communities within the sections were detected. For the P+PHE treatment, profiles of the S1 section were significantly different from the profiles of the S2 and S3 sections that grouped with the NP+PHE samples. The S1 profiles of the P+PHE treatment were significantly different from the S1 profiles of the P−PHE treatment.
FIG. 3.
Ordination plots, by treatments and sections, of bacterial communities generated by principal component analysis of bacterial species occurrence from 16S rDNA TGGE profiles. Values on the axes indicate percentages of total variation explained by the axes.
DISCUSSION
Our results confirm the previous findings (6) that biodegradation of PAH and bacterial numbers in the rhizosphere are a function of distance to roots. Using pot experiments with industrial polluted soils, Joner et al. (14) also found a similar gradient of PHE degradation in the rhizosphere, confirming that our compartment device is a simplified system well suited to studying rhizosphere processes. However, the extent to which root exudates affect microbial processes depends on experimental conditions and on soil characteristics (16) and cannot be extrapolated from results obtained in such devices to the situation in real soil. The low number of species detected in all treatments may be due to the staining procedure with ethidium bromide as well as the amplification protocol revealing only the most abundant species. The initial inoculum also had low species diversity and was dominated by six species detected on TGGE gels (data not shown), suggesting that this limited number of species may also be partly due to the preparation of the bacterial inoculum. The comparable number of species in the initial inoculum and the samples collected after incubation shows that diversity has been maintained during the incubation period.
Individual effects of exudates or PHE.
In the nonpolluted treatment, where root exudates were the only source of carbon, the number of culturable bacteria was highest in the section closest to the roots. Two specific bacterial communities were significantly separated, the first one corresponding to the more rhizospheric section, S1, and the second corresponding to the furthest section, S3. The communities corresponding to the intermediary layer S2 grouped with one of the two other communities. Communities were then spatially selected and gradually changed from a rhizospheric to a nonrhizospheric community structure. Gradients of populations showed differences in species composition and occurrence. Some majority species differing between treatments and sections were extracted prior to sequencing. The occurrence of Sp1, Sp6, and Sp7 decreased with distance to roots. Pseudomonas species were detected in planted treatments but were also present in nonplanted ones. Some species of this family, which is usually found at higher density in the rhizosphere (21), are also well known for their capabilities to ensure the primary steps of PAH catabolism, such as dioxygenation or ring cleavage (9, 27). Some species remained unidentified but matched species found in the plant environment such as Sp8, which was highly similar to an unknown bacterium found in the rhizosphere of maize (5). Other species were found only in polluted treatments, such as Sp2 or Sp10, both unidentified. The distribution of the three Caulobacter species detected was dependent on the distance to roots. It is the first time, to our knowledge, that this genus has been reported in a PAH-contaminated rhizosphere and that the species distribution is shown to vary with distance to roots. The Caulobacter genus belongs to the alpha subgroup of the Proteobacteria and is characterized by the presence of a prostheca. This wall extension of the cytoplasm confers an adaptation advantage in oligotrophic growth conditions by favoring nutrient uptake or cell adsorption onto solids. These species are reported to have a narrow range of C sources, but none of them have been described as PAH degraders even though their presence was reported in chlorophenol-contaminated aquifers (22). P. fusiformis belongs to the Verrumicrobiales order (11) and also has prosthecae. It has been found in many environments, including soils with different amendments of fertilizers or organic matter (30), and is present in all the samples of the three treatments. The abundance of these dominant species could be due to the low carbon content in the lateral compartment, which created an oligotrophic and highly selective environment.
In the unplanted treatment, in which PHE was the only source of carbon, no variation in the number of culturable bacteria was detected between layers. In parallel, no variations were observed in the composition and species abundance of communities. The microbial inoculum came from a polluted soil in which PHE was the more abundant PAH. Previous studies with the same soil showed that PAH degraders represented approximately 15% of total culturable bacteria and that they were able to degrade three-, four-, and five-ring PAH (14). At harvest, PAH degraders represented about 50% of total bacteria in all the sections of the lateral compartments. This consortium formed a homogenous group that most probably was already selected in the initial soil sample and that had been overselected by growing only on PHE in the compartment experiment. Biodegradation was equal in all the layers and reached about 60% in 4 weeks, confirming the efficiency of a complex microbial community adapted to PAHs (4, 34).
Combined effects of exudates and PHE.
In the planted and polluted treatment, numbers of total heterotrophs and PAH degraders were higher in the first layer, as in the unpolluted treatment. Two communities were distinguished, the first one corresponding to the S1 section and the second one grouping the sections S2 and S3. The latter grouped with the profiles of the nonplanted treatment, suggesting that microbial communities were less affected by root exudates than by the presence of PHE. In comparing the two planted treatments, the S1 groups were separated, indicating that root exudates on one hand and PHE on the other hand indeed selected communities in the rhizosphere. Using phospholipid fatty acid profile analysis, Joner et al. (15) also found different microbial communities when they compared planted and nonplanted pots, spiked or not with PAH. PHE degradation presented a strong spatial gradient as a function of the distance to the roots and reached 58% in the first section. This effect of distance to roots on PAH degradation was previously observed in compartmented devices (6). Similar gradients of PAH disappearance were observed in pot experiments with PAH-contaminated industrial soils in which biodegradation was higher close to the roots but was equal or inferior to that of the nonplanted soil at a further distance from roots (>0.6 to 0.9 mm) (16). The results of the present experiment actually confirm that the rhizosphere can improve but also reduce or inhibit the biodegradation of PAH at a certain distance from the roots. As mentioned by Joner and Leyval (13), the mechanisms behind biodegradation in the rhizosphere are probably more complex than in bulk soil. Biodegradation may be affected by rhizosphere-induced changes in microbial activity and diversity, depletion of nutrients, and water consumption but also by possible adsorption of PAH to roots or cell debris. Although adsorption of PAH to roots or cell debris was limited in the compartment device, as roots were not directly in contact with PHE, the physicochemical behavior of PAH in the lateral compartments may have been affected by the presence of the root mat. Therefore, comparisons of biodegradation rates between planted and nonplanted devices do not reflect the intrinsic biodegradation efficiency of rhizosphere communities but the entire environmental modifications induced by the rhizosphere.
Biodegradation in the rhizosphere should depend mainly on the quantity of C from exudates or on specific compounds from exudates such as phenolic acids (31) that are able to stimulate PAH biodegradation by cometabolism (6). Root exudates, principally sugars and organic acids, have a short life span in soils (18) and are quickly mineralized or transformed, so that root exudate-mediated biodegradation could be time dependent. In the compartment device, this organic C pool should be freshly renewed on a daily basis, in contrast to the fixed amount of carbon brought as PHE. The decrease and the inhibition of PHE biodegradation in the second and third sections of the planted and polluted treatment could also be transitory before the extension of the rhizosphere. Such a hypothesis will be investigated in further studies with time course experiments. Community profiles based on DNA indicate the structure of microbial populations at harvest and the selection of species that leads to this specific community. However, to provide complementary information on biodegradation processes in the rhizosphere and relate the observed spatial selection of bacterial communities in the rhizosphere to PHE biodegradation, other techniques such as RNA-based PCR and hybridization with degradation gene probes could be used.
REFERENCES
- 1.Binet, P., J. M. Portal, and C. Leyval. 2000. Dissipation of 3-6-ring polycyclic aromatic hydrocarbons in the rhizosphere of ryegrass. Soil Biol. Biochem. 32:2011-2017. [Google Scholar]
- 2.Binet, P., J. M. Portal, and C. Leyval. 2000. Fate of polycyclic aromatic hydrocarbons (PAH) in the rhizosphere and mycorrhizosphere of ryegrass. Plant Soil 227:207-213. [Google Scholar]
- 3.Bouchez, M., D. Blanchet, V. Bardin, F. Haeseler, and J.-P. Vandecasteele. 1999. Efficiency of defined strains and of soil consortia in the biodegradation of polycyclic aromatic hydrocarbon (PAH) mixtures. Biodegradation 10:429-435. [DOI] [PubMed] [Google Scholar]
- 4.Bouchez, M., D. Blanchet, and J.-P. Vandecasteele. 1995. Degradation of polycyclic aromatic hydrocarbons by pure strains and by defined strain association: inhibition phenomena and cometabolism. Appl. Microbiol. Biotechnol. 43:156-164. [DOI] [PubMed] [Google Scholar]
- 5.Chelius, M. K., and E. W. Triplett. 2001. The diversity of Archae and bacteria in association with roots of Zea mays L. Microb. Ecol. 41:252-263. [DOI] [PubMed] [Google Scholar]
- 6.Chen, S. H., and M. D. Aitken. 1999. Salicylate stimulates the degradation of high-molecular weight polycyclic aromatic hydrocarbons by Pseudomonas saccharophila P15. Environ. Sci. Technol. 33:435-439. [Google Scholar]
- 7.Corgié, S. C., E. Joner, and C. Leyval. 2003. Rhizospheric degradation of phenanthrene is a function of proximity to roots. Plant Soil 275:143-150. [Google Scholar]
- 8.Felske, A., A. D. L. Akkermans, and W. M. De Vos. 1998. Quantification of 16S rRNAs in complex bacterial communities by multiple competitive reverse transcription-PCR in temperature gradient gel electrophoresis fingerprints. Appl. Environ. Microbiol. 64:4581-4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foght, J. M., and D. W. S. Westlake. 1988. Degradation of polycyclic aromatic hydrocarbons and aromatic heterocycles by a Pseudomonas species. Can. J. Microbiol. 34:1135-1141. [DOI] [PubMed] [Google Scholar]
- 10.Gransee, A., and L. Wittenmayer. 2000. Qualitative and quantitative analysis of water-soluble root exudates in relation to plant species and development. J. Plant Nutr. Soil Sci. 163:381-385. [Google Scholar]
- 11.Hedlund, B., J. Gosink, and J. Staley. 1996. Phylogeny of Prosthecobacter, the fusiform caulobacters: members of a recently discovered division of the Bacteria. Int. J. Syst. Bacteriol. 46:960-966. [DOI] [PubMed] [Google Scholar]
- 12.Heuer, H., K. Hartung, G. Wieland, I. Kramer, and K. Smalla. 1999. Polynucleotide probes that target a hypervariable region of 16S rRNA genes to identify bacterial isolates corresponding to bands of community fingerprints. Appl. Environ. Microbiol. 65:1045-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joner, E., and C. Leyval. 2003. Phytoremediation of organic pollutants using mycorrhizal plants: a new aspect of rhizosphere interactions. Agronomie 23:495-502. [Google Scholar]
- 14.Joner, E. J., S. C. Corgie, N. Amellal, and C. Leyval. 2002. Nutritional constraints to degradation of polycyclic aromatic hydrocarbons in a simulated rhizosphere. Soil Biol. Biochem. 34:859-864. [Google Scholar]
- 15.Joner, E. J., A. Johansen, A. P. Loibner, M. A. de la Cruz, O. H. Szolar, J. M. Portal, and C. Leyval. 2001. Rhizosphere effects on microbial community structure and dissipation and toxicity of polycyclic aromatic hydrocarbons (PAHs) in spiked soil. Environ. Sci. Technol. 35:2773-2777. [DOI] [PubMed] [Google Scholar]
- 16.Joner, E. J., and C. Leyval. 2003. Rhizosphere gradients of polycyclic aromatic hydrocarbon (PAH) dissipation in two industrial soils and the impact of arbuscular mycorrhiza. Environ. Sci. Technol. 37:2371-2375. [DOI] [PubMed] [Google Scholar]
- 17.Kanaly, R. A., and S. Harayama. 2000. Biodegradation of high-molecular-weight polycyclic aromatic hydrocarbons by bacteria. J. Bacteriol. 182:2059-2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuzyakov, Y., H. Ehrensberger, and K. Stahr. 2001. Carbon partitioning and below-ground translocation by Lolium perenne. Soil Biol. Biochem. 33:61-74. [Google Scholar]
- 19.Laheurte, F., C. Leyval, and J. Berthelin. 1990. Root exudates of maize, pine and beech seedlings influenced by mycorrhizal and bacterial inoculation. Symbiosis 9:111-116. [Google Scholar]
- 20.Lynch, J. M. (ed.). 1990. The rhizosphere, vol. 1. John Wiley & Sons, Ltd., Chichester, United Kingdom.
- 21.Mahaffee, W. F., and J. W. Kloepper. 1997. Temporal changes in the bacterial communities of soil, rhizosphere, and endorhiza associated with field-grown cucumber (Cucumis sativus L.). Microb. Ecol. 34:210-223. [DOI] [PubMed] [Google Scholar]
- 22.Männistö, M. K., M. A. Tiirola, M. S. Salkinoja-Salonen, M. S. Kulomaa, and J. A. Puhakka. 1999. Diversity of chlorophenol-degrading bacteria isolated from contaminated boreal groundwater. Arch. Microbiol. 171:189-197. [DOI] [PubMed] [Google Scholar]
- 23.Marschner, P., and K. Baumann. 2003. Changes in bacterial community structure induced by mycorrhizal colonisation in spilt-root maize. Plant Soil 251:279-289. [Google Scholar]
- 24.Marschner, P., C.-H. Yang, R. Lieberei, and D. E. Crowley. 2001. Soil and plant specific effects on bacterial community composition in the rhizosphere. Soil Biol. Biochem. 33:1437-1445. [Google Scholar]
- 25.Meharg, A. A., and K. Killham. 1990. Carbon distribution within the plant and rhizosphere in laboratory and field-grown Lolium perenne at different stages of development. Soil Biol. Biochem. 22:471-477. [Google Scholar]
- 26.Picard, C., C. Ponsonnet, E. Paget, X. Nesme, and P. Simonet. 1992. Detection and enumeration of bacteria in soil by direct DNA extraction and polymerase chain reaction. Appl. Environ. Microbiol. 58:2717-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ringelberg, D. B., J. W. Talley, E. J. Perkins, S. G. Tucker, R. G. Luthy, E. J. Bouwer, and H. L. Fredrickson. 2001. Succession of phenotypic, genotypic, and metabolic community characteristics during in vitro bioslurry treatment of polycyclic aromatic hydrocarbon-contaminated sediments. Appl. Environ. Microbiol. 67:1542-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rovira, A. D. 1969. Plant root exudates. Bot. Rev. 35:35-57. [Google Scholar]
- 29.Schilling, G., A. Gransee, A. Deubel, G. Lezovic, and S. Ruppel. 1998. Phosphorus availability, root exudates, and microbial activity in the rhizosphere. Z. Pflanzernernähr. Bodenkd. 161:465-478. [Google Scholar]
- 30.Sessitsch, A., A. Weilharter, M. H. Gerzabek, H. Kirchmann, and E. Kandeler. 2001. Microbial population structures in soil particle size fractions of a long-term fertilizer field experiment. Appl. Environ. Microbiol. 67:4215-4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shim, H., S. Chauhan, D. Ryoo, K. Bowers, S. M. Thomas, K. A. Canada, J. G. Burken, and T. K. Wood. 2000. Rhizosphere competitiveness of trichloroethylene-degrading, poplar-colonizing recombinant bacteria. Appl. Environ. Microbiol. 66:4673-4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Warembourg, F. R., and H. D. Estelrich. 2000. Towards a better understanding of carbon flow in the rhizosphere: a time-dependent approach using carbon-14. Biol. Fertil. Soils 30:528-534. [Google Scholar]
- 33.Wikström, P., A. C. Andersson, and M. Forsman. 1999. Biomonitoring complex microbial communities using random amplified polymorphic DNA and principal component analysis. FEMS Microbiol. Lett. 28:131-139. [Google Scholar]
- 34.Yuan, S. Y., S. H. Wei, and B. V. Chang. 2000. Biodegradation of polycyclic aromatic hydrocarbons by a mixed culture. Chemosphere 41:1463-1468. [DOI] [PubMed] [Google Scholar]