Abstract
We report that the odd-skipped related 1 (OSR1) gene encoding a zinc-finger transcription factor was preferentially methylated in gastric cancer by genome-wide methylation screening. OSR1 expression was frequently silenced or down-regulated in gastric cancer cell lines. OSR1 expression was also significantly down-regulated at both mRNA and protein levels in primary gastric cancer tissues compared with adjacent normal tissues. The silencing or down-regulation of OSR1 was closely associated with promoter hypermethylation. Overexpression of OSR1 significantly inhibited cell growth, arrested the cell cycle, and induced apoptosis in the gastric cancer cell lines AGS, MKN28, and MGC803. Conversely, knockdown of OSR1 by OSR1-short hairpin RNA significantly enhanced cell growth, promoted the cell cycle, and inhibited apoptosis in the normal gastric epithelial cell line GES1. The dual-luciferase reporter assay revealed that OSR1 activated p53 transcription and repressed the T-cell factor (TCF)/lymphoid enhancer factor (LEF). Complementary DNA expression array and western blotting showed that OSR1 increased the expression of nuclear p53, p21, Fas, and death receptor-5, and suppressed the expression of cyclin D1 and cyclin-dependent kinase 4 in the p53 signalling pathway. In addition, OSR1 suppressed the expression of cytoplasmic β-catenin, TCF-1, and LEF1 in the Wnt/β-catenin signalling pathway. OSR1 methylation was detected in 51.8% of primary gastric cancer patients (85 of 164) by bisulphite genomic sequencing. Multivariate Cox regression analysis showed that OSR1 methylation was an independent predictor of poor survival. Kaplan–Meier survival curves revealed that OSR1 methylation was associated with shortened survival in TNM stage I–III patients. In conclusion, OSR1 acts as a functional tumour suppressor through the transcriptional activation of p53 and repression of TCF/LEF in gastric cancer. Detection of OSR1 methylation may serve as a potential biomarker of the early stage of gastric cancer. © 2014 The Authors. The Journal of Pathology published by John Wiley & Sons Ltd on behalf of Pathological Society of Great Britain and Ireland.
Keywords: odd-skipped related 1 (OSR1), methylation, tumour suppressor, prognosis, gastric cancer
Introduction
Gastric cancer is one of the most common cancers world-wide, especially in Asia 1. There is accumulating evidence that aberrant DNA methylation is a hallmark of gastric cancer 2. The identification of novel tumour suppressor genes repressed by DNA methylation will provide us with insights into the functional roles in gastric carcinogenesis and will be useful in discovering potential biomarkers 3,4.
We used methylated DNA immunoprecipitation-chip (MeDIP-chip) to identify the novel methylated genes on a genome-wide scale 5. The principle of MeDIP-chip is the use of an antibody specific for methylcytosine to immunoprecipitation of methylated DNA for downstream applications. Promoter methylated genes are determined and compared between cancer cell lines and normal tissues. From our preliminary data, we discovered that the promoter of the odd-skipped related 1 (OSR1) gene was preferentially methylated in gastric cancer cell lines. The OSR1 gene is located on human chromosome 2p24.1 encoding a zinc-finger transcription factor 6. OSR1 plays important roles in the development of intermediate mesoderm, and it is also essential for embryonic heart and urogenital formation 7–9. Although the role of OSR1 in cancer development is unknown, hypermethylated CpG islands associated with OSR1 have been reported in lung cancer 10. This suggests that OSR1 has some kind of function in cancer development.
It is known that the Wnt/β-catenin pathway and p53 pathway play important roles in gastric carcinogenesis 11–13. In the Wnt/β-catenin canonical pathway, Wnt ligands bind to the transmembrane receptors Frizzled and lipoprotein receptor-related protein (LRP) 5 and 6, which leads to the accumulation of β-catenin in the cytoplasm 14,15. The accumulated β-catenin is translocated from cytoplasm to the nucleus, where it forms a complex with T-cell factor (TCF)/lymphoid enhancer factor (LEF) to mediate the transcription of target genes 16–18. The Wnt/β-catenin canonical pathway becomes an oncogenic pathway in cancer development. On the other hand, p53 is a known tumour suppressor and activates the transcription of the downstream target genes involved in cell cycle arrest and apoptosis 19,20. Notably, Huang suggested that OSR1 was one of the agonists of the p53 network by genome-wide functional analysis 21. In the present study, we have investigated the expression and promoter methylation profile of OSR1.
Materials and methods
Cell lines
The gastric cancer cell lines (AGS, MKN28, MKN45, SNU1, SNU16, SNU638, SNU719, MGC803, and NCI-N87) and immortalized normal human gastric epithelial cell line (GES1) were purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA) and the Cancer Research Institute of Beijing University, China. The cell lines were cultured in RPMI-1640 medium (Gibco; Life Technologies, Carlsbad, CA, USA) with 10% fetal bovine serum (FBS) in 5% CO2 at 37 °C.
The colon cancer cell line HCT116 p53-wild type cells were purchased from ATCC and HCT116 p53-knockout cells were kindly provided by Professor Bert Vogelstein from the Ludwig Center and the Howard Hughes Medical Institute at the Johns Hopkins University School of Medicine. HCT116 p53-wild type cells were cultured in McCoy's 5A medium (Sigma-Aldrich, St Louis, MO, USA) with 10% FBS and HCT116 p53-knockout cells were cultured in Dulbecco's modified Eagle's medium (Gibco; Life Technologies) with 10% FBS in 5% CO2 at 37 °C.
Gastric tissue samples
Paired biopsy tissues from primary gastric cancer and adjacent normal mucosa were obtained during endoscopy at the Prince of Wales Hospital of the Chinese University of Hong Kong. Primary gastric cancer tissues were collected during operation from gastric cancer patients diagnosed at the First Affiliated Hospital of Sun Yat-sen University in Guangzhou, China. In addition, age-matched biopsy specimens of normal gastric mucosa were obtained as controls. Written consent was obtained prior to sample collection, and the study protocol was approved by the Clinical Research Ethics Committee of the Chinese University of Hong Kong and the Sun Yat-sen University of Medical Sciences.
Genome-wide profiling by MeDIP-chip
We determined the DNA methylation profile of gastric cancer cells by MeDIP-chip using the Agilent Custom Human CpG Island Microarray and Human Promoter Array (Agilent Technologies, Santa Clara, CA, USA) as reported previously 5.
Semi-quantitative reverse transcriptase-PCR and quantitative real-time PCR analyses
Semi-quantitative reverse transcriptase (RT)-PCR was performed by Go-Taq DNA polymerase (Promega, Madison, WI, USA). Quantitative real-time PCR was performed by SYBR Green PCR Master Mix (Applied Biosystems; Life Technologies) on a 7500HT Fast Real-Time PCR System (Applied Biosystems; Life Technologies). The primer sequences are listed in Supplementary Table 1.
Treatment with 5-aza-2'-deoxycytidine and trichostatin A
After 24 h of cell seeding, cells were treated with 2 µm of the demethylating agent 5-aza-2'-deoxycytidine (5-Aza; Sigma-Aldrich) for 96 h. Some cell lines were further treated with 300 nm of the histone deacetylase inhibitor trichostatin A (TSA) for an additional 24 h.
Methylation-specific PCR and bisulphite genomic sequencing
Methylation-specific PCR (MSP) and bisulphite genomic sequencing (BGS) were performed to amplify the bisulphite-modified DNA by TaKaRa Taq HS (TaKaRa, Ohtsu, Japan). The MSP region was selected from −19 to +83 bp of the transcription start site (TSS) and the BGS region was selected from −55 to +202 bp of the TSS (Figure 1D). Primer sequences for MSP and BGS are listed in Supplementary Table 1. PCR products of BGS were submitted to the Research Center of BGI, Hong Kong for sequencing. The methylation percentage of each CpG site was calculated according to the formula methylation % = HC/(HC + HT) × 100% (HC = height of peak C and HT = height of peak T).
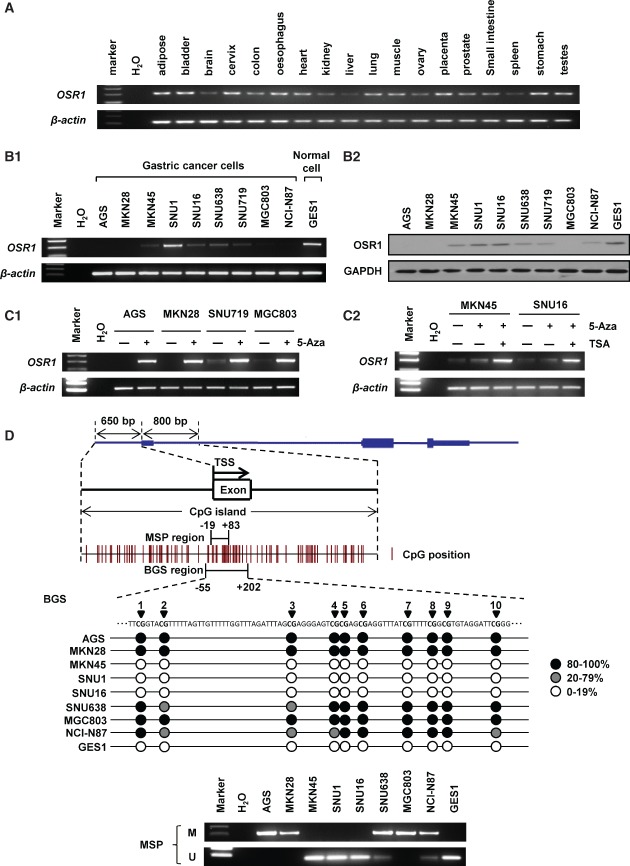
Figure 1.
OSR1 is silenced or down-regulated by promoter methylation in gastric cancer cells. (A) Expression of OSR1 mRNA in human organ tissues was determined by semi-quantitative reverse transcriptase (RT)-PCR. (B1) Expression of OSR1 mRNA in gastric cancer cell lines and normal gastric epithelial cell line. (B2) Expression of OSR1 protein in gastric cancer cell lines and normal gastric epithelial cell line. (C1) Treatment with the demethylating agent 5-aza-2'-deoxycytidine (5-Aza) restored OSR1 expression in gastric cancer cell lines. (C2) Combination treatment with 5-Aza and the histone deacetylase inhibitor trichostatin A (TSA) restored OSR1 expression in MKN45 and SNU16. (D) A typical CpG island spans the promoter region of OSR1. The transcription start site (TSS) and the regions of bisulphite genomic sequencing (BGS) and methylation-specific PCR (MSP) are indicated. Methylation status of OSR1 in the gastric cancer cell lines was determined by BGS and MSP (M, methylated; U, unmethylated).
Immunohistochemistry
Immunohistochemistry was performed by Histostain-Plus Kits (Broad Spectrum) and an Invitrogen 2nd Generation LAB-SA Detection System (Invitrogen; Life Technologies). Slides were incubated with a primary antibody to OSR1 (ab74003; Abcam, Cambridge, MA, USA) followed by a biotinylated secondary antibody and enzyme conjugate, stained with 3,3'-diaminobenzidine, and finally counterstained with haematoxylin. The extent of OSR1 staining was scored by assigning the percentage of positive tumour cells (0, none; 1, < 20% of positive staining cells; 2, 20–50% of positive staining cells; 3, > 50% of positive staining cells).
Western blotting
Protein blots were detected with primary antibodies to OSR1 (sc-376545; Santa Cruz Biotechnology, Santa Cruz, CA, USA), p21 (sc-6246; Santa Cruz Biotechnology), cyclin D1 (sc-8396; Santa Cruz Biotechnology), cyclin-dependent kinase 4 (CDK4; #2906; Cell Signaling Technology, Danvers, MA, USA), Fas (#4233; Cell Signaling Technology), cleaved caspase-3 (#9661; Cell Signaling Technology), cleaved poly-(ADP-ribose) polymerase (PARP; #5625; Cell Signaling Technology), cyclin B1 (sc-245; Santa Cruz Biotechnology), followed by a secondary antibody, and developed with enhanced chemiluminescence (Amersham, Arlington Heights, IL, USA). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH; sc-32233; Santa Cruz Biotechnology) served as a loading control.
Nuclear protein and cytoplasmic protein were extracted by NE-PER Nuclear and Cytoplasmic Extraction Reagents (Pierce; Thermo Fisher Scientific, Rockford, IL, USA). Nuclear protein was detected with p53 (sc-126; Santa Cruz Biotechnology) antibody and lamin A/C (612162; BD Biosciences, San José, CA, USA) was used as a loading control. Cytoplasmic protein was detected with β-catenin (#9582; Cell Signaling Technology) antibody and actin (sc-1615; Santa Cruz Biotechnology) was used as a loading control.
Construction of the OSR1 expression plasmid
The complete open reading frame of human OSR1 (NM_145260.2) was amplified by PCR from normal human stomach complementary DNA (cDNA). The inserts were subcloned into pcDNA3.1 (+) expression vector (Invitrogen; Life Technologies) by HindIII and XbaI. The primer sequences are listed in Supplementary Table 1. Plasmid DNA was purified from transformed E. coli cultures by a Pure Yield Plasmid Midiprep System (Promega). Cells were transfected with pcDNA3.1-OSR1 or empty pcDNA3.1 by Lipofectamine 2000 (Invitrogen; Life Technologies).
OSR1 knockdown
Cells were transfected with short hairpin RNA (shRNA) pGFP-V-RS-shOSR1 (TG302745, Origene) or shRNA control vector by Lipofectamine 2000.
Cell viability, colony formation, and cell proliferation assay
After 48 h of transfection, cells were cultured in the medium with G418 (Roche Applied Science, Mannheim, Germany) for pcDNA3.1 or puromycin (Gibco; Life Technologies) for the shRNA vector. After 14 days of selection to establish stable clones, cells were seeded for 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulphophenyl)-2H-tetrazolium (MTS; Promega) assay to measure the cell viability and colony formation assay counting the colonies (≥50 cells per colony) after staining with 5% crystal violet. Cell proliferation was monitored by the xCELLigence system (Roche Applied Science).
Cell cycle assay
Stably transfected cells were fixed in 70% ethanol and stained with PI/RNase Staining Buffer (BD Biosciences). The cells were sorted by a fluorescence activated cell sorting (FACS) Calibur system (BD Biosciences) and analysed by ModFit 3.0 software (Verity Software House, Topsham, ME, USA).
Apoptosis assay
Stably transfected cells were stained with annexin V-APC (BD Biosciences) and 7-aminoactinomycin (7-AAD). The cells were sorted by a FACS Calibur system and annexin V-positive cells were counted as apoptotic cells.
Dual-luciferase reporter assay
Stably transfected cells in a 24-well plate were co-transfected with luciferase reporter plasmid (p53-Luc or TOPflash; 200 ng/well) and pRL-cytomegalovirus vector (5 ng/well) by Lipofectamine 2000. After 48 h of transfection, luciferase activities were analysed by a Dual-Luciferase Reporter Assay System (Promega) and normalized to the control Renilla.
Mutational analysis of β-catenin
Genomic DNA was extracted using PureLink Genomic DNA Kits (Invitrogen; Life Technologies) and amplified by PCR using TaKaRa Ex Taq (TaKaRa). β-Catenin is encoded by the CTNNB1 gene in humans 22. We examined exon 3 of CTNNB1 using the primers previously reported by Sasaki 23. PCR products of BGS were submitted to the Research Center of BGI, Hong Kong for sequencing.
cDNA expression array
Gene expression profiles in stably transfected cells were analysed by the Human p53 Signaling Pathway RT2 Profiler PCR Array (PAHS-027Z; Qiagen, Hilden, Germany) and the Human Wnt Signaling Pathway RT2 Profiler PCR Array (PAHS-043Z; Qiagen). Gene expression changes ≥ 1.5-fold or ≤ 0.67-fold were considered to be of biological significance.
Genome-wide DNA methylation and microsatellite instability analyses
Genome-wide DNA methylation status in 43 gastric cancer tissues was detected by a 5-mC DNA ELISA Kit (Zymo Research, Irvine, CA, USA). Microsatellite instability (MSI) was examined by an MSI Analysis System, Version 1.2 (Promega).
Statistics
Values are expressed as mean ± standard deviation (SD). The independent Student t-test was used to compare the difference between two groups. One-way analysis of variance (ANOVA) was used to compare the difference between multiple groups, and the results were analysed by Tukey's multiple comparisons test. The χ2 test was used to compare the clinicopathological characteristics of gastric cancer patients and OSR1 methylation. Univariate and multivariate Cox regression analyses were performed to assess the prognostic value of OSR1 methylation. Overall survival in relation to methylation status was evaluated by Kaplan–Meier survival curve and log-rank test. Differences with p < 0.05 were considered to be statistically significant.
Results
OSR1 expression is epigenetically down-regulated in gastric cancer cells
We examined the expression of OSR1 mRNA through semi-quantitative RT-PCR. In human normal tissues including normal gastric tissue, expression of OSR1 mRNA was broadly detected (Figure 1A). In contrast, expression of OSR1 mRNA was silenced or down-regulated in most of the gastric cancer cells, and high expression of OSR1 mRNA was found in the normal gastric epithelial cell line GES1 (Figure 1B1). In keeping with the mRNA expression, silence or down-regulation of OSR1 protein in gastric cancer cell lines was demonstrated by western blotting (Figure 1B2). To confirm whether OSR1 expression was repressed by promoter methylation, the demethylating agent 5-Aza was employed. The results showed that 5-Aza treatment could restore OSR1 expression in AGS, MKN28, SNU719, and MGC803 (Figure 1C1). Although the restoration of OSR1 by 5-Aza was insufficient in MKN45 and SNU16, combination treatment with 5-Aza and the histone deacetylase inhibitor TSA markedly restored OSR1 expression (Figure 1C2).
The methylation status of OSR1 was further evaluated by BGS and MSP analysis. Full methylation was detected in AGS, MKN28, and MGC803, and partial methylation was found in SNU638 and NCI-N87. No methylation was detected in MKN45, SNU1, SNU16, and GES1. The MSP results were confirmed to be consistent with the BGS results (Figure 1D).
OSR1 expression is down-regulated in primary gastric cancer tissues
Expression of OSR1 mRNA in paired primary gastric cancer tissues was evaluated by quantitative real-time PCR. The expression levels of OSR1 mRNA were significantly down-regulated by 0.39-fold in 20 gastric cancers compared with adjacent normal tissues (p < 0.01; Figure 2A).
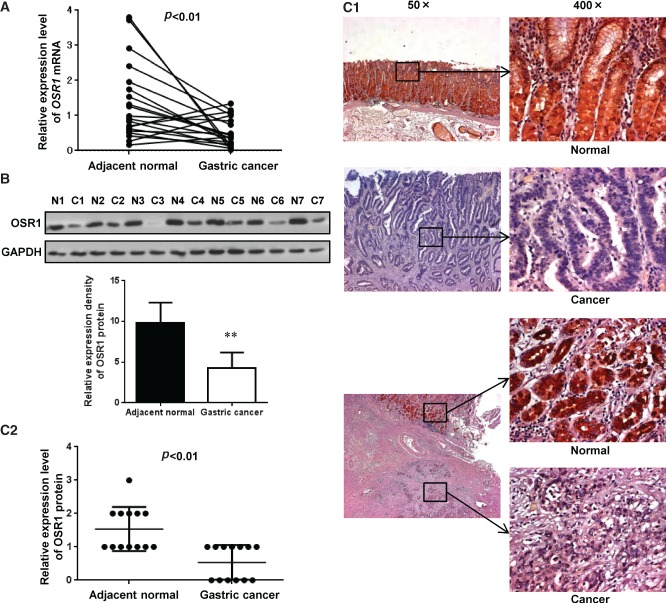
Figure 2.
OSR1 is down-regulated in primary gastric cancer tissues. (A) Expression levels of OSR1 mRNA in 20 paired primary gastric cancer tissues were determined by quantitative real-time PCR. (B) Expression levels of OSR1 protein in seven paired primary gastric cancer tissues were determined by western blotting. Quantitative analysis of the relative expression density is shown in the lower panel. Each column represents the mean ± standard deviation (SD). **p < 0.01 (N, normal; C, cancer). (C1) Expression levels of OSR1 protein in 13 paired primary gastric cancer tissues were determined by immunohistochemistry. Expression of OSR1 protein in normal gastric tissues and gastric cancer tissues is shown. The lower three images indicate cancer cell infiltration under the adjacent normal epithelial mucosa. (C2) Immunohistochemistry scoring was performed according to the percentage of positive tumour cells (0, none; 1, < 20%; 2, 20–50%; 3, > 50%). Each bar represents the mean ± SD.
Expression of OSR1 protein in paired gastric cancer tissues was further examined by western blotting. The expression levels of OSR1 protein were significantly down-regulated by 0.44-fold in seven gastric cancers compared with adjacent normal tissues (p < 0.01; Figure 2B). Expression of OSR1 protein was also evaluated by immunohistochemical analysis. OSR1 protein was strongly expressed in both the nucleus and the cytoplasm in adjacent normal tissues. However, OSR1 signal was much reduced in primary gastric cancer tissues (Figure 2C1). To quantify the extent of OSR1 staining, we scored the rate of positive tumour cells. The results showed that the expression levels of OSR1 protein were significantly down-regulated by 0.35-fold in 13 gastric cancers compared with adjacent normal tissues (p < 0.01; Figure 2C2).
OSR1 inhibits cell growth
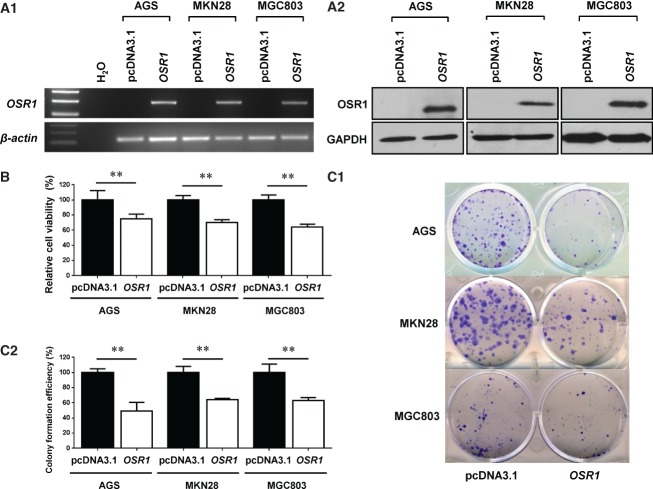
We examined the effect of OSR1 overexpression on cell growth in AGS, MKN28, and MGC803, which showed complete methylation and silencing of OSR1. Re-expression of OSR1 mRNA and protein was confirmed by semi-quantitative RT-PCR (Figure 3A1) and western blotting (Figure 3A2). Ectopic expression of OSR1 significantly inhibited cell viability to 74.8% in AGS (p < 0.01), 70.4% in MKN28 (p < 0.01), and 64.1% in MGC803 (p < 0.01), as revealed by MTS assay compared with empty pcDNA3.1 (Figure 3B). The colony numbers of OSR1-transfected cells were significantly decreased to 49.4% in AGS (p < 0.01), 64.0% in MKN28 (p < 0.01), and 63.1% in MGC803 (p < 0.01) by colony formation assay compared with empty pcDNA3.1-transfected cells (Figures 3C1 and 3C2).
Figure 3.
Overexpression of OSR1 inhibits cell growth. (A1) Overexpression of OSR1 mRNA was revealed by semi-quantitative RT-PCR. (A2) Overexpression of OSR1 protein was confirmed by western blotting. (B) Effect of OSR1 overexpression on cell viability was determined by MTS assay. Each column represents the mean ± SD. **p < 0.01. (C1) Effect of OSR1 overexpression on colony numbers was determined by colony formation assay. (C2) Quantitative analysis of colony formation efficiency (%) is shown. Each column represents the mean ± SD. **p < 0.01.
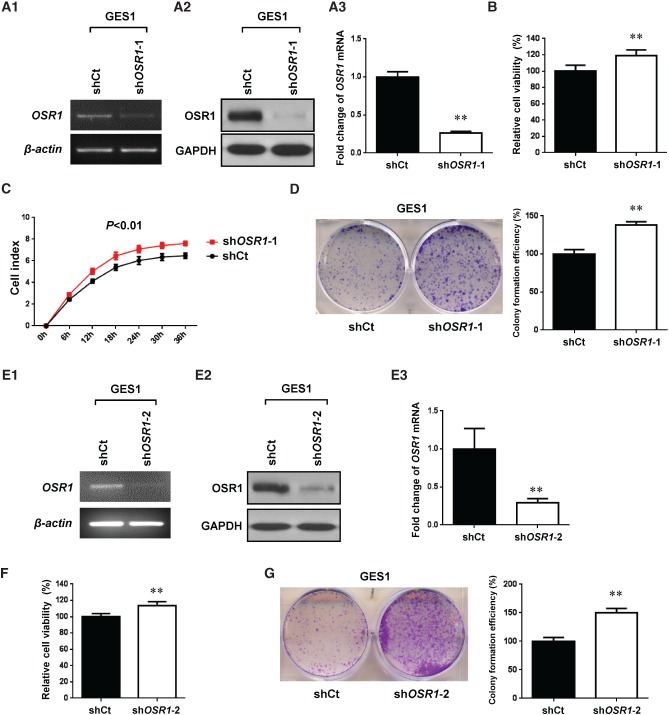
We further investigated the effect of OSR1 knockdown by OSR1-shRNA-1 on cell growth in GES1. Knockdown of OSR1 was confirmed by semi-quantitative RT-PCR (Figure 4A1), western blotting (Figure 4A2), and quantitative real-time PCR (p < 0.01; Figure 4A3). Knockdown of OSR1 by OSR1-shRNA-1 significantly enhanced cell viability to 119.1% (p < 0.01; Figure 4B), cell proliferation (p < 0.01; Figure 4C), and colony numbers to 137.7% (p < 0.01; Figure 4D). We validated the results of the knockdown assays using OSR1-shRNA-2 (Figures 4E–4G).
Figure 4.
Knockdown of OSR1 by OSR1-shRNA-1 (A–D) and OSR1-shRNA-2 (E–G) enhances cell growth. (A1) Knockdown of OSR1 mRNA was revealed by semi-quantitative RT-PCR (shCt, shRNA control vector; shOSR1-1, OSR1-shRNA-1). (A2) Knockdown of OSR1 protein was confirmed by western blotting. (A3) Knockdown efficiency of OSR1 mRNA was examined by quantitative real-time PCR. Each column represents the mean ± SD. **p < 0.01. (B) Effect of OSR1 knockdown on cell viability was determined by MTS assay. Each column represents the mean ± SD. **p < 0.01. (C) Effect of OSR1 knockdown on cell proliferation was determined by xCELLigence analysis. Data are shown as mean ± SD. (D) Effect of OSR1 knockdown on colony numbers was determined by colony formation assay. Quantitative analysis of colony formation efficiency (%) is shown in the right panel. Each column represents the mean ± SD. **p < 0.01. (E1) Knockdown of OSR1 mRNA was revealed by semi-quantitative RT-PCR (shCt, shRNA control vector; shOSR1-2, OSR1-shRNA-2). (E2) Knockdown of OSR1 protein was confirmed by western blotting. (E3) Knockdown efficiency of OSR1 mRNA was examined by quantitative real-time PCR. Each column represents the mean ± SD. **p < 0.01. (F) Effect of OSR1 knockdown on cell viability was determined by MTS assay. Each column represents the mean ± SD. **p < 0.01. (G) Effect of OSR1 knockdown on colony numbers was determined by colony formation assay. Quantitative analysis of colony formation efficiency (%) is shown in the right panel. Each column represents the mean ± SD. **p < 0.01.
OSR1 induces cell cycle arrest
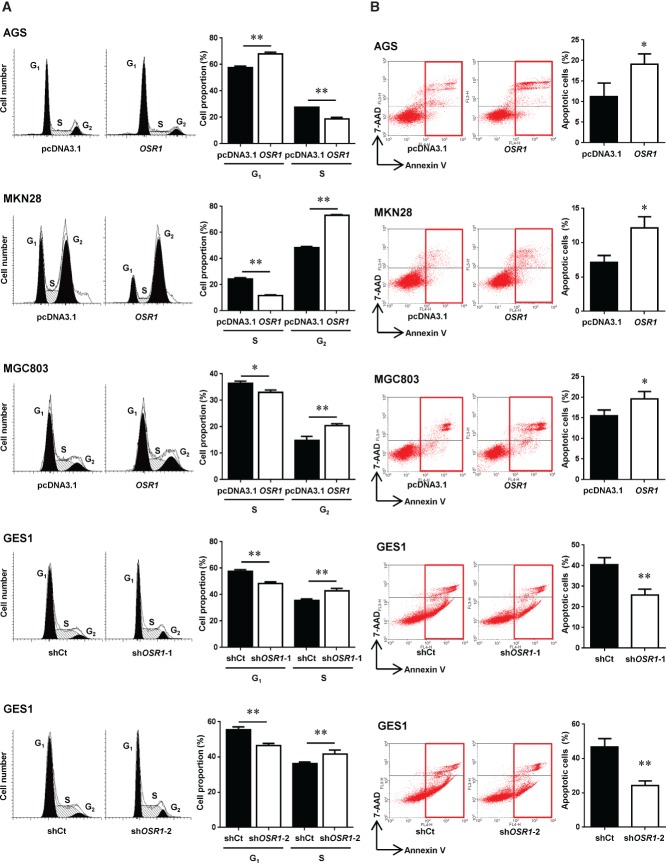
Overexpression of OSR1 significantly increased cells at the G1 phase (57.7 ± 1.1% versus 68.2 ± 1.1%, p < 0.01) and decreased cells at the S phase (27.4 ± 0.18% versus 18.7 ± 1.2%, p < 0.01) in AGS. On the other hand, overexpression of OSR1 significantly reduced cells at the S phase (24.6 ± 0.78% versus 11.8 ± 0.48%, p < 0.01) and elevated cells at the G2 phase (48.2 ± 1.1% versus 73.1 ± 0.60%, p < 0.01) in MKN28. Overexpression of OSR1 also significantly decreased cells at the S phase (36.3 ± 0.94% versus 33.1 ± 0.84%, p < 0.05) and increased cells at the G2 phase (14.9 ± 1.6% versus 20.3 ± 0.86%, p < 0.01) in MGC803. Knockdown of OSR1 by OSR1-shRNA-1 and OSR1-shRNA-2 significantly decreased cells at the G1 phase (57.6 ± 1.3% versus 48.3 ± 1.3%, p < 0.01; 55.6 ± 1.5% versus 46.5 ± 1.2%, p < 0.01) and increased cells at the S phase (35.8 ± 0.91% versus 42.7 ± 2.1%, p < 0.01; 36.3 ± 0.91% versus 41.6 ± 2.4%, p < 0.01) in GES1 (Figure 5A).
Figure 5.
OSR1 arrests the cell cycle and induces apoptosis. (A) Effects of OSR1 overexpression and knockdown on the cell cycle were determined by fluorescence activated cell sorting (FACS) analysis. Quantitative analysis of cell proportion (%) is shown in the right panel. Each column represents the mean ± SD. *p < 0.05; **p < 0.01 (shCt, shRNA control vector; shOSR1-1, OSR1-shRNA-1; shOSR1-2, OSR1-shRNA-2). (B) Effects of OSR1 overexpression and knockdown on apoptosis were determined by FACS analysis after the dual staining with annexin V-APC and 7-aminoactinomycin (7-AAD). Quantitative analysis of apoptotic cells (%) is shown in the right panel. Each column represents the mean ± SD. *p < 0.05; **p < 0.01.
OSR1 induces apoptosis
Overexpression of OSR1 significantly increased the numbers of apoptotic cells in AGS (11.2 ± 3.3% versus 19.1 ± 2.5%, p < 0.05), MKN28 (7.1 ± 1.1% versus 12.2 ± 1.6%, p < 0.05), and MGC803 (15.6 ± 1.3% versus 19.6 ± 1.8%, p < 0.05). Knockdown of OSR1 in GES1 by OSR1-shRNA-1 (40.5 ± 3.4% versus 25.8 ± 2.8%, p < 0.01) and OSR1-shRNA-2 (46.7 ± 4.9% versus 24.4 ± 2.6%, p < 0.01) significantly decreased the numbers of apoptotic cells (Figure 5B).
OSR1 activates p53 transcription and represses TCF transcription
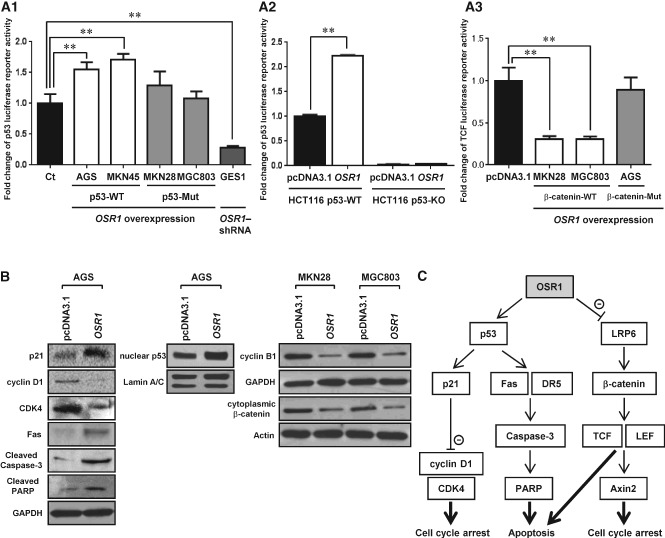
To clarify the tumour suppressive pathways modulated by OSR1, we measured p53 luciferase reporter activity. Overexpression of OSR1 significantly increased p53 activity by 1.5-fold in AGS (p < 0.01) and 1.7-fold in MKN45 (p < 0.01). Conversely, knockdown of OSR1 by OSR1-shRNA significantly repressed p53 activity by 0.28-fold in GES1 (p < 0.01). In contrast, OSR1 transfection had no effect on p53 activity in the p53-mutant cell lines MKN28 and MGC803 (Figure 6A1). In the colon cancer cell line HCT116, overexpression of OSR1 significantly elevated p53 activity by 2.2-fold in HCT116 p53-wild type cells, but p53 activity could not be detected in HCT116 p53-knockout cells (Figure 6A2).
Figure 6.
OSR1 activates the transcription of p53 and represses the transcription of T-cell factor (TCF). (A1) Effect of OSR1 overexpression and knockdown on p53 luciferase reporter activity was determined in gastric cancer cell lines (WT, wild type; Mut, mutant). Each column represents the mean ± SD. **p < 0.01. (A2) Effect of OSR1 overexpression on p53 luciferase reporter activity was determined in a colon cancer cell line (WT, wild type; KO, knockout). Each column represents the mean ± SD. **p < 0.01. (A3) Effect of OSR1 overexpression on TCF luciferase reporter activity was determined in gastric cancer cell lines (WT, wild type; Mut, mutant). Each column represents the mean ± SD. **p < 0.01. (B) Effect of OSR1 overexpression on the expression levels of candidates in the p53 and Wnt/β-catenin signalling pathway was validated by western blotting. (C) Schematic diagram showing the mechanism of OSR1 tumour suppressive function.
TCF luciferase reporter activity was also examined using TOPflash. Overexpression of OSR1 significantly repressed TCF activity by 0.30-fold in MKN28 (p < 0.01) and 0.31-fold in MGC803 (p < 0.01). In contrast, overexpression of OSR1 did not affect TCF activity in AGS (Figure 6A3). In mutation analysis of β-catenin, there were no mutations in MKN28 and MGC803, but there was one mutation at position 88 bp of exon 3 (GGA to GAA) and the protein sequence was also changed (Gly to Glu) in AGS (Supplementary Table 2).
Identification of genes modulated by OSR1
We employed the cDNA expression array to examine the molecular mechanism by which OSR1 exerted tumour suppressive activities. Fold changes of OSR1-modulated downstream genes in the p53 signalling pathway were measured in AGS and GES1. Overexpression of OSR1 significantly increased the expression levels of CDKN1A (p21Cip1; 3.3-fold), Fas (14.2-fold), death receptor-5 (DR5; 3.9-fold), and Bax (1.5-fold), and suppressed the expression level of CDK4 (0.66-fold) in AGS. Conversely, knockdown of OSR1 significantly reduced the expression levels of p21Cip1 (0.57-fold), Fas (0.66-fold), and DR5 (0.59-fold), and elevated the expression level of CDK4 (1.5-fold) in GES1.
Fold changes of OSR1-modulated downstream genes in the Wnt/β-catenin signalling pathway were also measured in MKN28 and MGC803. Overexpression of OSR1 significantly suppressed the levels of LRP6 (0.41-fold), CTNNB1 (0.47-fold), TCF-1 (0.44-fold), LEF1 (0.46-fold), and Axis inhibition protein 2 (Axin2; 0.36-fold) in MKN28. Overexpression of OSR1 significantly suppressed the levels of LRP6 (0.41-fold), CTNNB1 (0.66-fold), TCF-1 (0.49-fold), LEF1 (0.43-fold), and Axin2 (0.66-fold) in MGC803 (Supplementary Table 3).
The results from the cDNA expression array were validated by western blotting. Overexpression of OSR1 enhanced the protein expression of p21, Fas, cleaved caspase-3, cleaved PARP, and nuclear p53, and suppressed the protein expression of cyclin D1 and CDK4 in AGS. Overexpression of OSR1 suppressed the protein expression of cyclin B1 and cytoplasmic β-catenin in MKN28 and MGC803 (Figure 6B). A schematic diagram for the mechanism of OSR1 tumour suppressive function derived from the cDNA expression array and western blotting is shown in Figure 6C.
Clinicopathological features of OSR1 methylation in gastric cancer patients
We evaluated OSR1 methylation in 164 primary gastric cancer tissues by BGS. Methylated OSR1 was detected in 51.8% of primary gastric cancers (85 of 164). We next determined the association between OSR1 methylation and clinicopathological features such as age, gender, Helicobacter pylori infection, Lauren type, differentiation, and tumour–nodes–metastasis (TNM) stage, but no correlations were found (Supplementary Table 4).
In addition, we examined the association of OSR1 promoter methylation with genome-wide DNA methylation or MSI, respectively. Global DNA hypomethylation was found in 81.4% of gastric cancers (35 of 43), and MSI was detected in 18.6% of gastric cancers (8 of 43). Neither genome-wide DNA methylation nor MSI had a correlation with OSR1 promoter methylation (Supplementary Table 5).
OSR1 methylation is associated with poor survival of gastric cancer patients
We further examined the prognostic factors of gastric cancer patients. From the results of univariate Cox regression analysis, OSR1 methylation was found to be associated with a significant increased risk of cancer-related death. The relative risk (RR) was 2.32, with 95% confidence interval (CI) ranging from 1.45 to 3.70 (p < 0.01). TNM stage was also a significant predictor of outcome as expected. After the adjustment for potential confounding factors, multivariate Cox regression analysis showed that OSR1 methylation was an independent predictor of poorer survival of gastric cancer patients (RR 1.70, 95% CI 1.04–2.80; p < 0.05). TNM stage was another independent predictor, and patients in TNM stages I–III had a significantly better survival compared with patients in stage IV (Table1).
Table 1.
Univariate and multivariate Cox regression analyses of prognostic factors in gastric cancer patients
| Univariate | Multivariate | |||
|---|---|---|---|---|
| Variable | RR (95% CI) | p value | RR (95% CI) | p value |
| Age | 1.00 (0.98–1.02) | 0.75 | 1.01 (0.99–1.02) | 0.53 |
| Gender | ||||
| Male | 0.99 (0.63–1.55) | 0.97 | 0.91 (0.57–1.44) | 0.68 |
| Female | 1.00 | 1.00 | ||
| H. pylori | ||||
| Positive | 1.05 (0.56–1.97) | 0.88 | ||
| Negative | 1.00 | |||
| Lauren | ||||
| Intestinal | 0.67 (0.38–1.18) | 0.17 | ||
| Diffuse/mixed | 1.00 | |||
| Differentiation | ||||
| Poor | 1.24 (0.72–2.13) | 0.44 | ||
| Moderate/well | 1.00 | |||
| TNM stage | ||||
| I | 0.09 (0.03–0.28) | < 0.01 | 0.10 (0.03–0.31) | < 0.01 |
| II | 0.15 (0.07–0.32) | < 0.01 | 0.16 (0.07–0.34) | < 0.01 |
| III | 0.26 (0.15–0.45) | < 0.01 | 0.29 (0.16–0.50) | < 0.01 |
| IV | 1.00 | 1 | ||
| OSR1 methylation | ||||
| Methylated | 2.32 (1.45–3.70) | < 0.01 | 1.70 (1.04–2.80) | 0.04 |
| Unmethylated | 1.00 | 1.00 | ||
RR = relative risk; CI = confidence interval.
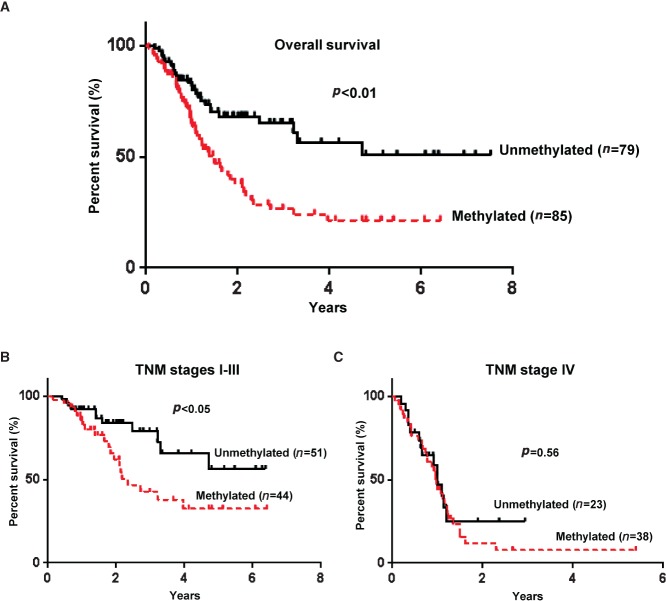
Kaplan–Meier survival curves showed that gastric cancer patients with OSR1 methylation had a significantly poorer overall survival than patients without methylation based on the log-rank test (p < 0.01; Figure 7A). Patients with OSR1 methylation in TNM stages I–III showed a significantly poorer survival than patients without methylation (p < 0.05; Figure 7B), but patients in TNM stage IV did not differ significantly (p = 0.56; Figure 7C).
Figure 7.
OSR1 methylation is associated with poor survival of patients at the early stage of gastric cancer. (A) Kaplan–Meier curves of gastric cancer patients. (B) Kaplan–Meier curves of gastric cancer patients in tumour–nodes–metastasis (TNM) stages I–III. (C) Kaplan–Meier curves of gastric cancer patients in TNM stage IV.
Discussion
In this study, we demonstrated that OSR1 was frequently silenced or down-regulated in gastric cancer cell lines. In addition, we found that OSR1 was also significantly down-regulated at both mRNA and protein levels in primary gastric cancer tissues compared with their adjacent normal tissues. The silencing or down-regulation of OSR1 was closely associated with promoter hypermethylation, as confirmed by treatment with the demethylating agent 5-Aza and methylation analysis by BGS and MSP, inferring that promoter methylation was the principal regulatory mechanism of OSR1 inactivation in gastric cancer. There were low levels of methylated gastric cancer cell lines with silencing or down-regulation of OSR1 expression. We demonstrated that another transcription regulating mechanism such as histone modification also contributed to the gene silencing in these cell lines.
Down-regulation of OSR1 in gastric cancer suggested that OSR1 could function as a potential tumour suppressor. We therefore investigated the biological functions of OSR1 by gain and loss of OSR1 in human gastric cancer cells. Overexpression of OSR1 significantly inhibited cell growth in the gastric cancer cell lines AGS, MKN28, and MGC803. Conversely, knockdown of OSR1 significantly enhanced cell growth in GES1. Flow cytometry analysis showed that OSR1 induced cell cycle arrest at the G1 phase and apoptosis in AGS and GES1. On the other hand, OSR1 induced cell cycle arrest at the G2 phase and apoptosis in MKN28 and MGC803. The tumour suppressive mechanism in AGS and GES1 was likely to be different in MKN28 and MGC803. Collectively, these observations indicate for the first time that OSR1 functions as a novel tumour suppressor in gastric cancer.
We investigated the signalling pathways by dual-luciferase reporter assay. OSR1 showed a tumour suppressive effect through the transcriptional activation of p53 in the p53-wild type cell lines AGS, MKN45, and GES1, and through the transcriptional repression of TCF in MKN28 and MGC803. OSR1 did not repress TCF luciferase reporter activity in AGS, and we confirmed that it was due to the β-catenin mutation in AGS. This mutation may make the Wnt/β-catenin signalling pathway constitutively active, and AGS cells are independent of upstream Wnt signals 24. Therefore, overexpression of OSR1 could not repress TCF luciferase reporter activity in AGS.
We also studied the molecular mechanisms by which OSR1 exerted tumour suppressive activity in gastric cancer using the cDNA expression array and western blotting. We found that OSR1 stabilized p53 in the nucleus, enhanced p21, suppressed cyclin D1 and CDK4 which drove the G1 to S phase transition, and stimulated Fas and DR5 receptors, which in turn induced apoptosis through caspase-3 25–29. We also found that OSR1 suppressed the expression of LRP6, cytoplasmic β-catenin, TCF-1, LEF1, and Axin2 in the Wnt/β-catenin signalling pathway. Cytoplasmic β-catenin is considered to separate the centrosome for spindle formation in mitosis 30,31. The level of cytoplasmic β-catenin and TCF transcriptional activity was the highest at the G2/M phase as was the peak of cyclin B1, which drives the G2 to M phase transition 32–34. In addition, Axin2 reaches a peak at the G2/M phase and provides negative feedback to oscillate the β-catenin levels 35–37. Therefore, the components of the Wnt/β-catenin signalling may play a crucial role during mitosis.
Although promoter hypermethylation of CpG islands leads to the inactivation of tumour suppressor genes, global DNA hypomethylation is often found in cancers simultaneously 38. We found that OSR1 promoter methylation occurred independently of global DNA methylation and MSI status. We further evaluated the clinical significance and prognostic value of OSR1 promoter methylation in primary gastric cancer patients who had known survival data. In multivariate Cox regression survival analysis, OSR1 methylation was significantly correlated with a shorter survival, independent of other clinicopathological features. This means that OSR1 promoter methylation can be an independent prognostic marker for gastric cancer patients. As the TNM stage is a highly important predictor of disease recurrence, we used Kaplan–Meier curves stratified by both methylation status and TNM stage. The results showed that OSR1 methylation was significantly associated with shorter survival for TNM stage I–III gastric cancer patients. This result implies that the propensity for OSR1 promoter methylation may specifically predict the most aggressive and fatal types of gastric cancer at an early stage.
In conclusion, we suggest that OSR1 is a novel tumour suppressor gene commonly silenced or down-regulated by promoter methylation in gastric cancer. OSR1 shows the tumour suppressive function of inhibiting cell growth, arresting the cell cycle, and inducing apoptosis through the transcriptional activation of p53 and the transcriptional repression of TCF/LEF. OSR1 methylation is an independent predictor of poor survival of patients at the early stage of gastric cancer.
Acknowledgments
This project was supported by research funds of a China 863 Program (2012AA02A203; 2012AA02A504), a China 973 Program (2010CB529305), and Hong Kong ITF (ITS/214/12).
Author contribution statement
KO, YD, XL, JL, NZ, LX, and MYYG performed the experiments. EKWN provided material support. TA, FKLC, and JJYS commented on this study and provided technical support. KO designed, analysed data, and drafted the paper. JY designed, supervised this study, and revised the paper.
Abbreviations
7-AAD, 7-aminoactinomycin; Axin2, Axis inhibition protein 2; 5-Aza, 5-aza-2'-deoxycytidine; BGS, bisulphite genomic sequencing; CDK4, cyclin-dependent kinase 4; DR5, death receptor-5; FACS, fluorescence activated cell sorting; LEF, lymphoid enhancer factor; LRP, lipoprotein receptor-related protein; MeDIP-chip, methylated DNA immunoprecipitation-chip; MSI, microsatellite instability; MSP, methylation-specific PCR; MTS, 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulphophenyl)-2H-tetrazolium; OSR1, odd-skipped related 1; PARP, poly-(ADP-ribose) polymerase; RR, relative risk; TCF, T-cell factor; TNM, tumour–nodes–metastasis; TSA, trichostatin A; TSS, transcription start site
SUPPORTING INFORMATION ON THE INTERNET
The following supporting information may be found in the online version of this article.
Table S1. List of primer sequences.
Table S2. The sequence of β-catenin exon 3 in gastric cancer cell lines.
Table S3. Fold changes of OSR1-modulated downstream genes in the p53 signalling pathway and Wnt/β-catenin signalling pathway were determined by cDNA expression array.
Table S4. The association between clinicopathological features and OSR1 methylation in gastric cancer patients.
Table S5. The association between global DNA methylation/MSI and OSR1 methylation in gastric cancer patients.
Supporting Information
List of primer sequences
The sequence of β-catenin exon 3 in gastric cancer cell lines
Fold changes of OSR1-modulated downstream genes in the p53 signalling pathway and Wnt/β-catenin signalling pathway were determined by cDNA expression array
The association between clinicopathological features and OSR1 methylation in gastric cancer patients
The association between global DNA methylation/MSI and OSR1 methylation in gastric cancer patients
References
- Ferlay J, Shin HR, Bray F. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- Otani K, Li X, Arakawa T. Epigenetic-mediated tumor suppressor genes as diagnostic or prognostic biomarkers in gastric cancer. Expert Rev Mol Diagn. 2013;13:445–455. doi: 10.1586/erm.13.32. [DOI] [PubMed] [Google Scholar]
- Kang GH, Lee HJ, Hwang KS. Aberrant CpG island hypermethylation of chronic gastritis, in relation to aging, gender, intestinal metaplasia, and chronic inflammation. Am J Pathol. 2003;163:1551–1556. doi: 10.1016/S0002-9440(10)63511-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang GH, Lee S, Kim JS. Profile of aberrant CpG island methylation along multistep gastric carcinogenesis. Lab Invest. 2003;83:519–526. doi: 10.1097/01.lab.0000064704.53132.65. [DOI] [PubMed] [Google Scholar]
- Zhao J, Liang Q, Cheung KF. Genome-wide identification of Epstein–Barr virus-driven promoter methylation profiles of human genes in gastric cancer cells. Cancer. 2013;119:304–312. doi: 10.1002/cncr.27724. [DOI] [PubMed] [Google Scholar]
- Katoh M. Molecular cloning and characterization of OSR1 on human chromosome 2p24. Int J Mol Med. 2002;10:221–225. [PubMed] [Google Scholar]
- Wang Q, Lan Y, Cho ES. Odd-skipped related 1Odd 1) is an essential regulator of heart and urogenital development. Dev Biol. 2005;288:582–594. doi: 10.1016/j.ydbio.2005.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James RG, Kamei CN, Wang Q. Odd-skipped related 1 is required for development of the metanephric kidney and regulates formation and differentiation of kidney precursor cells. Development. 2006;133:2995–3004. doi: 10.1242/dev.02442. [DOI] [PubMed] [Google Scholar]
- Tena JJ, Neto A, de la Calle-Mustienes E. Odd-skipped genes encode repressors that control kidney development. Dev Biol. 2007;301:518–531. doi: 10.1016/j.ydbio.2006.08.063. [DOI] [PubMed] [Google Scholar]
- Rauch TA, Wang Z, Wu X. DNA methylation biomarkers for lung cancer. Tumour Biol. 2012;33:287–296. doi: 10.1007/s13277-011-0282-2. [DOI] [PubMed] [Google Scholar]
- Ganesan K, Ivanova T, Wu Y. Inhibition of gastric cancer invasion and metastasis by PLA2G2A, a novel beta-catenin/TCF target gene. Cancer Res. 2008;68:4277–4286. doi: 10.1158/0008-5472.CAN-07-6517. [DOI] [PubMed] [Google Scholar]
- Du R, Xia L, Sun S. URG11 promotes gastric cancer growth and invasion by activation of beta-catenin signalling pathway. J Cell Mol Med. 2010;14:621–635. doi: 10.1111/j.1582-4934.2008.00622.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahara E. Molecular mechanism of stomach carcinogenesis. J Cancer Res Clin Oncol. 1993;119:265–272. doi: 10.1007/BF01212724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- Bhanot P, Brink M, Samos CH. A new member of the frizzled family from Drosophila functions as a Wingless receptor. Nature. 1996;382:225–230. doi: 10.1038/382225a0. [DOI] [PubMed] [Google Scholar]
- Mao J, Wang J, Liu B. Low-density lipoprotein receptor-related protein-5 binds to Axin and regulates the canonical Wnt signaling pathway. Mol Cell. 2001;7:801–809. doi: 10.1016/s1097-2765(01)00224-6. [DOI] [PubMed] [Google Scholar]
- Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- Linke SP, Clarkin KC, Wahl GM. p53 mediates permanent arrest over multiple cell cycles in response to gamma-irradiation. Cancer Res. 1997;57:1171–1179. [PubMed] [Google Scholar]
- Midgley CA, Owens B, Briscoe CV. Coupling between gamma irradiation, p53 induction and the apoptotic response depends upon cell type in vivo. J Cell Sci. 1995;108:1843–1848. doi: 10.1242/jcs.108.5.1843. [DOI] [PubMed] [Google Scholar]
- Huang Q, Raya A, DeJesus P. Identification of p53 regulators by genome-wide functional analysis. Proc Natl Acad Sci U S A. 2004;101:3456–3461. doi: 10.1073/pnas.0308562100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus C, Liehr T, Hulsken J. Localization of the human beta-catenin gene (CTNNB1) to 3p21: a region implicated in tumor development. Genomics. 1994;23:272–274. doi: 10.1006/geno.1994.1493. [DOI] [PubMed] [Google Scholar]
- Sasaki Y, Morimoto I, Kusano M. Mutational analysis of the beta-catenin gene in gastric carcinomas. Tumour Biol. 2001;22:123–130. doi: 10.1159/000050606. [DOI] [PubMed] [Google Scholar]
- Li J, Mo ML, Chen Z. HSulf-1 inhibits cell proliferation and invasion in human gastric cancer. Cancer Sci. 2011;102:1815–1821. doi: 10.1111/j.1349-7006.2011.02024.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el-Deiry WS, Tokino T, Velculescu VE. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- He G, Siddik ZH, Huang Z. Induction of p21 by p53 following DNA damage inhibits both Cdk4 and Cdk2 activities. Oncogene. 2005;24:2929–2943. doi: 10.1038/sj.onc.1208474. [DOI] [PubMed] [Google Scholar]
- Quelle DE, Ashmun RA, Shurtleff SA. Overexpression of mouse D-type cyclins accelerates G1 phase in rodent fibroblasts. Genes Dev. 1993;7:1559–1571. doi: 10.1101/gad.7.8.1559. [DOI] [PubMed] [Google Scholar]
- Musgrove EA, Lee CS, Buckley MF. Cyclin D1 induction in breast cancer cells shortens G1 and is sufficient for cells arrested in G1 to complete the cell cycle. Proc Natl Acad Sci U S A. 1994;91:8022–8026. doi: 10.1073/pnas.91.17.8022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupt S, Berger M, Goldberg Z. Apoptosis – the p53 network. J Cell Sci. 2003;116:4077–4085. doi: 10.1242/jcs.00739. [DOI] [PubMed] [Google Scholar]
- Huang P, Senga T, Hamaguchi M. A novel role of phospho-beta-catenin in microtubule regrowth at centrosome. Oncogene. 2007;26:4357–4371. doi: 10.1038/sj.onc.1210217. [DOI] [PubMed] [Google Scholar]
- Bahmanyar S, Kaplan DD, Deluca JG. β-Catenin is a Nek2 substrate involved in centrosome separation. Genes Dev. 2008;22:91–105. doi: 10.1101/gad.1596308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson G, Niehrs C. Emerging links between CDK cell cycle regulators and Wnt signaling. Trends Cell Biol. 2010;20:453–460. doi: 10.1016/j.tcb.2010.05.002. [DOI] [PubMed] [Google Scholar]
- Orford K, Orford CC, Byers SW. Exogenous expression of beta-catenin regulates contact inhibition, anchorage-independent growth, anoikis, and radiation-induced cell cycle arrest. J Cell Biol. 1999;146:855–868. doi: 10.1083/jcb.146.4.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmeda D, Castel S, Vilaro S. Beta-catenin regulation during the cell cycle: implications in G2/M and apoptosis. Mol Biol Cell. 2003;14:2844–2860. doi: 10.1091/mbc.E03-01-0865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jho EH, Zhang T, Domon C. Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol Cell Biol. 2002;22:1172–1183. doi: 10.1128/MCB.22.4.1172-1183.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd-Lewis B, Fletcher AG, Dale TC. Toward a quantitative understanding of the Wnt/beta-catenin pathway through simulation and experiment. Wiley Interdiscip Rev Syst Biol Med. 2013;5:391–407. doi: 10.1002/wsbm.1221. [DOI] [PubMed] [Google Scholar]
- Hadjihannas MV, Bernkopf DB, Bruckner M. Cell cycle control of Wnt/beta-catenin signalling by conductin/axin2 through CDC20. EMBO Rep. 2012;13:347–354. doi: 10.1038/embor.2012.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M. DNA hypomethylation, cancer, the immunodeficiency, centromeric region instability, facial anomalies syndrome and chromosomal rearrangements. Journal Nutr. 2002;132:2424S–2429S. doi: 10.1093/jn/132.8.2424S. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of primer sequences
The sequence of β-catenin exon 3 in gastric cancer cell lines
Fold changes of OSR1-modulated downstream genes in the p53 signalling pathway and Wnt/β-catenin signalling pathway were determined by cDNA expression array
The association between clinicopathological features and OSR1 methylation in gastric cancer patients
The association between global DNA methylation/MSI and OSR1 methylation in gastric cancer patients