Abstract
Fecal bacteria were studied in healthy elderly volunteers (age, 63 to 90 years; n = 35) living in the local community, elderly hospitalized patients (age, 66 to 103; n = 38), and elderly hospitalized patients receiving antibiotic treatment (age, 65 to 100; n = 21). Group- and species-specific primer sets targeting 16S rRNA genes were used to quantitate intestinal bacteria by using DNA extracted from feces and real-time PCR. The principal difference between healthy elderly volunteers and both patient cohorts was a marked reduction in the Bacteroides-Prevotella group following hospitalization. Reductions in bifidobacteria, Desulfovibrio spp., Clostridium clostridiiforme, and Faecalibacterium prausnitzii were also found in the hospitalized patients. However, total 16S rRNA gene copy numbers (per gram of wet weight of feces) were generally lower in the stool samples of the two groups of hospitalized patients compared to the number in the stool samples of elderly volunteers living in the community, so the relative abundance (percentage of the group- and species-specific rRNA gene copies in relation to total bacterial rRNA gene copies) of bifidobacteria, Desulfovibrio spp., C. clostridiiforme, and F. prausnitzii did not change. Antibiotic treatment resulted in further reductions in the numbers of bacteria and their prevalence and, in some patients, complete elimination of certain bacterial communities. Conversely, the numbers of enterobacteria increased in the hospitalized patients who did not receive antibiotics, and due to profound changes in fecal microbiotas during antibiotic treatment, the opportunistic species Enterococcus faecalis proliferated.
Degenerative transformations in gut physiology and function occur with aging and may be linked, in part, to changes in the composition and metabolic activities in the colonic ecosystem. As earlier studies have indicated, protective intestinal microorganisms such as bifidobacteria decrease in numbers, while putatively detrimental populations such as clostridia and enterobacteria increase in the elderly (16, 17, 19, 20, 22, 32, 33). The normal intestinal microbiota, however, provides an important natural defense mechanism against invading pathogens and prevents overgrowth of autochthonous opportunistic microorganisms, a process known as colonization resistance. The large intestinal ecosystem provides colonization resistance by a variety of mechanisms such as occupying adhesion sites in the gut and the production of antimicrobial agents. Host factors such as gut peristalsis and the secretion of saliva and gastric acid also contribute to the defense against invading pathogens. Detrimental changes in gastrointestinal ecology and function can have major consequences for the elderly, and a higher incidence of gastrointestinal infections is observed in older people (26). Diet is an important determinant of bowel function, and age-related changes in the amounts and types of food consumed caused by a decline in taste and smell, masticatory dysfunction, and swallowing difficulties (15) may be important factors contributing to alterations in the gut ecosystem in old age.
Malnutrition is also an important factor responsible for the impaired immune responses seen in elderly people (24). Immunologic changes with the onset of old age involve reductions in the efficiency of B- and T-cell-mediated responses (41), and the increase of circulating antibodies against commensal gut bacteria in older people has been associated with age-related changes such as decreased gastric acid secretion and increased mucosal permeability (35).
Old people are hospitalized twice as frequently as younger people, their length of stay is twice as long, and they receive twice as many medications as younger patients (13). It is estimated that at any given time, approximately 20% of the elderly patients in hospitals are receiving antibiotic treatment, principally for urinary and respiratory tract infections (M. E. T. McMurdo, unpublished data). Antibiotics can cause major disturbances in the intestinal microbiota, and important anaerobic species in the bowel such as bifidobacteria, lactobacilli, or bacteroides can be radically reduced, or even eradicated, during antibiotic treatment, while pathogenic bacteria can proliferate (44).
The commensal intestinal microbiota has been examined in detail in healthy adults, and a few investigations have dealt with the intestinal microbiota of healthy elderly people (16, 17, 19, 20, 22, 32, 33). However, the effects of antibiotic treatment on the gut bacteria in the elderly have been studied only on a very small group of volunteers (20), and little information is available on the effects of hospitalization on the gut bacteria of older people.
Most of the studies on the human gut bacteria have used cultivation techniques to monitor bacterial populations, although it has been reported that less then 25% of the intestinal species are cultivable (43). There is, consequently, a need for rapid, reliable, and specific methods to monitor and quantitate gut bacteria during the aging process. Molecular techniques based on the analysis of the 16S rRNA or rRNA genes are being increasingly used, and a variety of techniques such random cloning and sequencing (43), denaturing gel electrophoresis (46), fluorescence in situ hybridization (23), and dot blot hybridization (10) have been developed. Quantitative analysis of the fecal microbiota by employing species- or group-specific PCR has been done either by indirectly measuring PCR titers (45) or in combination with culture techniques (30), but only real-time PCR enabled dominant and subdominant intestinal populations to be directly quantitated by using DNA isolated from fecal material (29).
To overcome the methodological problems associated with culture techniques, real-time PCR was used in this investigation to monitor various bacterial groups in stool samples obtained from older people. Three different groups were recruited to analyze the effects of hospitalization and antibiotic treatment on the intestinal microbiota of elderly patients in comparison to the intestinal microbiota of a group of healthy older people living in the local community.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The strains listed below were obtained from the National Collection of Industrial and Marine Bacteria (NCIMB; Aberdeen, United Kingdom), the National Collection of Type Cultures (NCTC; London, United Kingdom), Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSM; Braunschweig, Germany), the American Type Culture Collection (ATCC; Manassas, Va.), and laboratory stock cultures at the University of Dundee (DUN). The following bacteria were used to evaluate the specificity of PCR primer sets (see Table 2): Bacteroides fragilis NCTC 9343, Bacteroides ovatus DUN 203, Bacteroides thetaiotaomicron NCTC 10582, Bacteroides vulgatus DUN 116, Bifidobacterium catenulatum NCIMB 702246, Bifidobacterium adolescentis NCIMB 702231, Bifidobacterium bifidum NCIMB 702715, Bifidobacterium breve NCTC 11815, Clostridium clostridiiforme DUN-120, Clostridium malenominatum DUN 102, Clostridium perfringens NCTC 8346, Clostridium butyricum NCIMB 7423, Clostridium bifermentans DUN-156, Clostridium perfringens NCTC 8533, Clostridium sordellii DUN 113, Clostridium tetani NCTC 5404, Clostridium difficile NCTC 11223, Clostridium histolyticum DSM 2158, Desulfovibrio desulfuricans NCIMB 12833, Desulfovibrio desulfuricans NCIMB 8307, Desulfovibrio vulgaris NCIMB 8303, Desulfococcus multivorans NCIMB 12965, Desulfobacter vibrioformis NCIMB 13525, Desulfobacter vibrioformis NCIMB 13525, Desulfotomaculum ruminis DSM 2154, Desulfotomaculum ruminis NCIMB 8452, Desulfotomaculum ruminis (laboratory strain provided by G. R. Gibson, University of Reading, United Kingdom), Escherichia coli ATCC 11775, E. coli NCTC 9001, Enterococcus faecalis ATCC 51299, Enterococcus faecalis NCTC 8131, Enterococcus faecium NCTC 12202, Eubacterium limosum DUN 112, Eubacterium rectale DUN-128, Eubacterium aerofaciens DUN 207, Eubacterium cylindroides DUN-119, Faecalibacterium prausnitzii ATCC 27768, Faecalibacterium prausnitzii NCIMB 13872, Lactobacillus plantarum DUN 145, Lactobacillus acidophilus DSM 20079, Peptostreptococcus anaerobius DUN 167, Ruminococcus flavefaciens 17 (laboratory strain provided by H. Flint, Rowett Research Institute, Aberdeen, United Kingdom), Ruminococcus albus SY3 (H. Flint), Ruminococcus productus DSM 2950, Shigella sonnei DUN-198, Shigella flexneri DUN 200, Salmonella enterica serovar Typhimurium DUN 217, and Veillonella parvula NCTC 12140.
TABLE 2.
Primer sets used in this study
| Target organism | Primer set | Sequence (5′ to 3′) | Product size (bp) | Annealing temp (°C) | Reference or source |
|---|---|---|---|---|---|
| All eubacteria | Uni331F | TCCTACGGGAGGCAGCAGT | 466 | 58 | 34 |
| Uni797R | GGACTACCAGGGTATCTATCCTGTT | 34 | |||
| Bacteroides-Prevotella group | Bac303F | GAAGGTCCCCCACATTG | 418 | 56 | 2a |
| Bac708R | CAATCGGAGTTCTTCGTG | 2 | |||
| Enterobacteriaceae | Eco1457F | CATTGACGTTACCCGCAGAAGAAGC | 195 | 63 | This study |
| Eco1652R | CTCTACGAGACTCAAGCTTGC | This study | |||
| Bifidobacterium genus | Bif164F | GGGTGGTAATGCCGGATG | 278 | 59 | 23 |
| Bif601R | TAAGCCATGGACTTTCACACC | 2a | |||
| Desulfovibrio genus | Dsv691F | CCGTAGATATCTGGAGGAACATCAG | 135 | 63 | 14 |
| Dsv826R | ACATCTAGCATCCATCGTTTACAGC | 14 | |||
| Faecalibacterium prausnitzii | Fprau223F | GATGGCCTCGCGTCCGATTAG | 199 | 58 | 45a |
| Fprau420R | CCGAAGACCTTCTTCCTCC | 45 | |||
| Ruminococcus albus | Ralb561F | CAGGTGTGAAATTTAGGGGC | 246 | 63 | This study |
| Ralb807R | GTCAGTCCCCCCACACCTAG | This study | |||
| Clostridium butyricum | Cbut825F | GTGCCGCCGCTAACGCATTAAGTAT | 213 | 72 | This study |
| Cbut1038R | ACCATGCACCACCTGTCTTCCTGCC | This study | |||
| Clostridium clostridiiforme | Cclos99F | AATCTTGATTGACTGAGTGGCGGAC | 148 | 62 | This study |
| Cclos247R | CCATCTCACACTACCGGAGTTTTTC | This study | |||
| Enterococcus faecalis | Efs130F | AACCTACCCATCAGAGGG | 360 | 57 | 31 |
| Efs490R | GACGTTCAGTTACTAACG | This study |
Modified from reference.
Desulfovibrios were grown in Postgate medium B in universal bottles (27); all other organisms were cultured on Wilkins-Chalgren agar plates or broth and incubated in an anaerobic chamber (in an atmosphere of 80% N2,10% CO2, and 10% H2) at 37°C.
DNA extraction from bacterial cultures.
Bacterial cells were collected and resuspended in 450 μl of deionized water to which 50 μl of lysozyme (50 mg ml−1) was added and incubated at 37°C for 30 min in a water bath. Twenty-five microliters of proteinase K solution (20 mg ml−1), 50 μl of 20% sodium dodecyl sulfate, 500 μl of aqua deionium, and 350 mg of 0.1-mm glass beads (BioSpec, Bartlesville, Okla.) were then added to mechanically destroy the cells by using a Mini-Beadbeater (BioSpec) for 2 min at maximum speed. After the suspension was heated at 70°C for 10 min, the bead-beating step was repeated, and the cell debris was removed by centrifugation (5,000 × g for 3 min). The supernatant containing bacterial DNA was purified by using a DNA purification kit (QIAGEN, Hilden, Germany) according to the manufacturer's instructions.
Subjects.
Three groups of elderly people (Table 1) from the Tayside area, East Scotland, with no history of gastrointestinal disease participated in this study, comprising healthy elderly volunteers (group H), hospitalized elderly patients (group P), and hospitalized elderly patients receiving antibiotic therapy (group PAB). Groups P and PAB were recruited from the Royal Victoria Hospital, Dundee, and stayed on a standard hospital diet. Group H volunteers lived in the local community and continued with their usual diet. Groups H and P had not received any antibiotics or laxatives within 2 months of the study. Group PAB received single antibiotics or combinations of drugs such as amoxicillin (29%), amoxicillin-clavulanic acid (35%), ciprofloxacin (12%), clarithromycin (6%), clindamycin (6%), flucloxacillin (12%), metronidazole (12%), rifampin (6%), and trimethoprim (12%) to treat various nongut infections. Group PAB patients diagnosed with C. difficile-associated diarrhea were excluded from the study. Written informed consent was obtained from each participant, and the study was approved by the Tayside Committee on Medical Research Ethics.
TABLE 1.
Age and sex of study group participants
| Group (no. of participants) | Mean age ± SD (yr) | Age range (yr) | Sex (no. male/ no. female) |
|---|---|---|---|
| H (35) | 71 ± 5.8 | 63-90 | 0/35 |
| P (38) | 81 ± 7.5a | 66-103 | 23/15 |
| PAB (21) | 81 ± 8.0b | 65-100 | 15/6 |
Indicates a significant difference (P < 0.05) between groups H and P.
Indicates a significant difference (P < 0.05) between groups H and PAB.
A total of 94 individuals took part in the study (Table 1). The two patient groups P and PAB were similar in terms of age range, mean age, and sex. Both groups P and PAB included higher numbers of men, with a mean age of 81 years. In contrast to these groups, group H volunteers were only female, and the mean age was 71 years.
Fecal slurries.
Stool samples were collected and processed within 1 h after defecation. Fresh feces were homogenized (10% wt/vol) in 0.1 M sodium phosphate buffer (pH 6.5) containing 80 mg of pectin liter−1 (citrus), 150 mg of starch liter−1 (soluble), 80 mg of xylan liter−1 (oatspelt), and 80 mg of mucin liter−1 (partially purified porcine gastric mucin) before being serially diluted 10-fold in half-strength peptone water.
DNA extraction from feces.
DNA was extracted from 2 ml of fecal slurry by using a QIAGEN stool kit with a modified protocol for cell lysis analogous to the one described for bacterial cells. Briefly, cells were collected by centrifugation at 20,000 × g (10 min) and resuspended in 1.4 ml of ASL lysis buffer. Glass beads (350 mg) were added, and the slurries were bead beaten for 2 min at maximum speed. After the suspension was heated for 5 min at 95°C, the bead-beating step was repeated, and the cell debris was removed by centrifugation (5,000 × g for 2 min). DNA-damaging substances and PCR inhibitors were removed from the supernatant containing the DNA by using the InhibitEX tablets provided in the kit. Protein digests and DNA purifications were done by using QIAamp spin columns, according to the manufacturer's instructions.
Primers and PCR amplification.
All primer sets used are listed in Table 2. Primer sets developed in this study were designed on the basis of 16S rRNA gene sequences available from the GenBank database and by means of the computer program Primer Premier for Windows, version 5.0 (Premier Biosoft International, Palo Alto, Calif.). The specificities of the oligonucleotide sequences for the target organisms were checked with an online tool provided by the Ribosomal Database Project (8). The primers were synthesized commercially by Invitrogen Life Technologies (Paisley, United Kingdom).
Each PCR mixture (50 μl) contained 10 mM Tris-HCl (pH 9.0), 50 mM KCl, 1.5 mM MgCl2, 0.25 mM concentrations of the deoxynucleoside triphosphates, 0.5 μM primer, 1 μl of bacterial template DNA, and 1 U of Taq DNA polymerase (Promega, Madison, Wis.). Determining the optimal annealing temperature for each primer set (Table 2) and testing the specificity of each primer set were done by using a Mastercycler gradient PCR machine (Eppendorf, Hamburg, Germany) and a Techne Genius PCR machine (Duxford, United Kingdom), respectively. The PCR program consisted of 35 cycles with a DNA denaturation step at 95°C (1 min), followed by an annealing step (1 min) and elongation step at 72°C (45 seconds). The PCR was completed with a final elongation step at 72°C (5 min).
Plasmid DNA standards.
PCR products of the different primer sets were purified by using a QIAquick spin PCR purification kit (QIAGEN), ligated into the pGEMT vector (Promega), and transformed into competent E. coli JM109 cells by heat shock at 42°C. Clones containing the correct plasmid insert were confirmed by PCR amplification and subcultured on Luria-Bertani medium supplemented with ampicillin (200 mg liter−1). Plasmid DNA was purified by using the Wizard Plus SV Minipreps DNA purification kit (Promega), and the plasmid DNA concentration was determined by electrophoresis and a comparison of band strengths against molecular marker DNA (2 log DNA Ladder N3200L; New England Biolabs Ltd., Hitchin, Hertfordshire, United Kingdom).
Real-time PCR.
An iCycler iQ apparatus (Bio-Rad, Hercules, Calif.), associated with iCycler Optical System Interface software (version 2.3; Bio-Rad), was used for the real-time PCR. Each reaction was done in duplicate in a volume of 20 μl with 96-well optical-grade PCR plates, sealed with optical sealing tape (Bio-Rad). Amplification reactions were done with iQ SYBR Green Supermix (Bio-Rad) containing 3 mM MgCl2, 20 mM Tris HCl (pH 8.4), 50 mM KCl, each deoxynucleoside triphosphate at a concentration of 200 μM, SYBR Green I, 10 nM fluorescein, 0.625 U of iTaq DNA polymerase mixed with the selected primer set at a concentration of 0.5 μM for each primer, and 1.6 μl of the respective template DNA or water. Amplifications were done with the following temperature profiles: one cycle at 95°C (3 min), 35 cycles of denaturation at 95°C (30 seconds), primer annealing (30 seconds), and one final cycle at 95°C (30 seconds). Finally, melt curve analyses were made by slowly heating the PCR mixtures from 55 to 95°C (1°C per cycle of 10 s) with simultaneous measurements of the SYBR Green I signal intensities. Quantitation was done by using standard curves made from known concentrations of plasmid DNA containing the respective amplicon for each set of primers.
Statistical analyses.
Logarithms of fecal 16S rRNA gene copy numbers were used to achieve normal distribution, and the mean ± standard deviation (SD) was calculated. SPSS for Windows, version 10.0.7, was employed for statistical analyses with a one-way analysis of variance and Bonferroni multiple comparisons to compare the three subject groups with each other. The percentage of bacterial group and species rRNA gene copies in relation to total rRNA gene copies (relative abundance) was calculated for each individual, and the median was determined for each subject group. A nonparametric Mann-Whitney U test was used for pairwise comparison of the subject groups.
RESULTS
Specificity of primers.
All primer sets were highly specific and gave positive results only for the corresponding target bacteria with the expected product size (Table 2). For the primer sets developed in this study, primer set Ecol475F/Ecol652R reacted with E. coli ATCC 11775, E. coli NCTC 9001, Shigella sonnei DUN-198, Shigella flexneri DUN 200, and Salmonella enterica serovar Typhimurium DUN 217; primer set Fprau223F/Fprau420R reacted with F. prausnitzii ATCC 27768 and F. prausnitzii NCIMB 13872; primer set Ralb561F/Ralb807R reacted with R. albus SY3; primer set Cbut825F/Cbut1038R reacted with C. butyricum NCIMB 7423; and primer set Cclos99F/Cclos247R reacted with C. clostridiiforme DUN 120.
Quantitation of bacterial populations in feces.
Real-time PCR showed major differences in the numbers of rRNA gene copies of various bacterial groups and species in stool samples from group H, group P, and group PAB participants (Table 3). A summary of all findings which are described in the following paragraphs can be found in Table 4. Total 16S rRNA gene copy numbers were significantly reduced by about one order of magnitude in both groups P and PAB compared to results for group H. Only a marginal reduction in total 16S rRNA gene copies was found in group PAB compared to group P.
TABLE 3.
Real-time PCR quantitation of 16S rRNA genes in feces of study group participantsa
| Group H (n = 35)
|
Group P (n = 38)
|
Group PAB (n = 21)
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | Min | Max | Prevalence (%) | Mean | SD | Min | Max | Prevalence (%) | Mean | SD | Min | Max | Prevalence (%) | |
| All eubacteria | 12.0 | 0.1 | 11.0 | 12.9 | 35 (100) | 11.1b | 0.7 | 9.4 | 12.4 | 38 (100%) | 10.8c | 0.7 | 9.6 | 12.0 | 21 (100) |
| Bacteroides-Prevotella group | 10.5 | 0.1 | 8.6 | 11.6 | 35 (100) | 8.8b | 1.2 | 6.6 | 11.0 | 33 (87%) | 8.4c | 1.2 | 6.9 | 10.6 | 19 (90) |
| Bifidobacterium genus | 9.5 | 0.1 | 7.6 | 10.7 | 35 (100) | 8.8 | 1.2 | 4.9 | 10.4 | 38 (100%) | 8.4c,d | 1.0 | 6.4 | 10.5 | 15 (71) |
| Desulfovibrio genus | 8.4 | 0.2 | 6.5 | 9.9 | 35 (100) | 7.7 | 1.2 | 5.6 | 9.2 | 36 (95%) | 7.0c,d | 1.1 | 5.2 | 9.4 | 15 (71) |
| Enterobacteriaceae | 7.3 | 0.3 | 5.0 | 9.4 | 34 (97) | 8.7 | 1.3 | 5.7 | 10.8 | 35 (92%) | 7.4 | 1.9 | 4.5 | 10.6 | 20 (95) |
| Faecalibacterium prausnitzii | 10.3 | 0.1 | 7.9 | 11.3 | 35 (100) | 9.4b | 0.8 | 7.4 | 10.6 | 37 (97%) | 8.3c,d | 1.0 | 6.3 | 10.2 | 20 (95) |
| Clostridium clostridiiforme | 10.2 | 0.1 | 9.1 | 11.0 | 35 (100) | 9.4b | 0.4 | 8.3 | 10.4 | 38 (100%) | 8.8c,d | 0.9 | 7.1 | 10.5 | 21 (100) |
| Clostridium butyricum | 7.5 | 0.3 | 5.9 | 8.9 | 34 (97) | 7.2 | 1.0 | 5.2 | 9.0 | 37 (97%) | 6.4c,d | 1.1 | 4.7 | 8.8 | 12 (57) |
| Ruminococcus albus | 5.7 | 0.3 | 4.2 | 9.2 | 32 (91) | 5.2 | 0.9 | 3.9 | 8.9 | 31 (82%) | 5.2c,d | 0.6 | 4.3 | 6.3 | 9 (43) |
| Enterococcus faecalis | 5.3 | 0.2 | 5.0 | 5.7 | 2 (6) | 6.8 | 0.5 | 6.0 | 7.8 | 7 (18%) | 6.9c | 0.9 | 5.4 | 8.4 | 8 (38) |
Results are expressed as log10 16S rRNA gene copies (per gram of wet weight). Min, minimum; Max, maximum.
Indicates a significant difference (P < 0.05) between groups H and P.
Indicates a significant difference (P < 0.05) between groups H and PAB.
Indicates a significant difference (P < 0.05) between groups P and PAB.
TABLE 4.
Summary of findings on fecal bacteria in groups of hospitalized patients compared to the control group
| Organism | Shifts observed from control group H to group P
|
Shifts observed from control group H to group PAB
|
||||
|---|---|---|---|---|---|---|
| 16S rRNA gene copy numbers | Relative abundance | Prevalence | 16S rRNA gene copy numbers | Relative abundance | Prevalence | |
| All eubacteria | Significant reduction | Not applicable | No change | Significant reduction | Not applicable | No change |
| Bacteroides-Prevotella group | Significant reduction | Significant reduction | Decrease | Significant reduction | Significant reduction | Decrease |
| Bifidobacterium genus | Nonsignificant reduction | Nonsignificant increase | No change | Significant reduction | No change | Decrease |
| Desulfovibrio genus | Nonsignificant reduction | Nonsignificant increase | No change | Significant reduction | Significant reduction | Decrease |
| Enterobacteriaceae | Nonsignificant increase | Significant increase | No change | No change | Significant reduction | No change |
| Faecalibacterium prausnitzii | Significant reduction | No change | No change | Significant reduction | Significant reduction | No change |
| Clostridium clostridiiforme | Significant reduction | No change | No change | Significant reduction | Nonsignificant reduction | No change |
| Clostridium butyricum | No change | Significant increase | No change | Significant reduction | Significant reduction | Decrease |
| Ruminococcus albus | Nonsignificant reduction | No change | Decrease | Significant reduction | Significant reduction | Decrease |
| Enterococcus faecalis | Nonsignificant increase | Significant increase | Increase | Significant increase | Significant increase | Increase |
Similar observations were made for the Bacteroides-Prevotella group (including species of the genera Bacteroides and Prevotella [2]), which was detected in high numbers in all stool samples from group H volunteers. However, in approximately 10% of the stool samples from both groups P and PAB, these organisms could not be detected (the detection limit was 104 16S rRNA gene copies per gram of wet weight), and the mean rRNA gene copy numbers were reduced significantly, by almost two orders of magnitude, compared to the results for group H. A nonsignificant reduction in rRNA genes was found in group PAB compared to group P.
A significant difference between the group H and P participants, who did not take antibiotics, and group PAB was found with bifidobacteria. Bifidobacteria were detected in only 71% of group PAB stool samples, and the mean 16S rRNA gene copy numbers were reduced significantly compared to results for both groups H and P. Bifidobacterial 16S rRNA genes were detected in every person in groups H and P, but the highest numbers were found in group H volunteers. Analogous observations were made with desulfovibrios. These organisms were detected in all fecal samples from group H and in 95% of the stool samples from group P, whose members had lower numbers of desulfovibrio-specific 16S rRNA genes. Species belonging to the genus Desulfovibrio were not detected in 29% of the group PAB patients, and a significant reduction in mean rRNA gene copy numbers was observed compared to the results for the other two groups.
Similarly, mean copy numbers of rRNA genes in C. butyricum and R. albus were highest in samples from group H and decreased slightly in group P. However, the most marked reductions were found in samples from group PAB.
The predominant fecal species F. prausnitzii and C. clostridiiforme were found in almost all samples, apart from one stool sample with no detectable F. prausnitzii found in both group P and group PAB. 16S rRNA genes of both species were almost one order of magnitude lower in samples from group P compared to group H, and they were further reduced by almost one order of magnitude in stool samples from group PAB.
Enterobacteriaceae were detected in almost all fecal samples. Numbers of rRNA genes belonging to these organisms were higher in samples from group P, although they were slightly less prevalent than in samples from groups H and PAB. Enterococcus faecalis was rarely detected by real-time PCR in stool samples from group H. These bacteria were found more often and in higher numbers in groups P and PAB, and a further increase in the prevalence of this species was observed in group PAB.
Relative abundance of rRNA genes in feces.
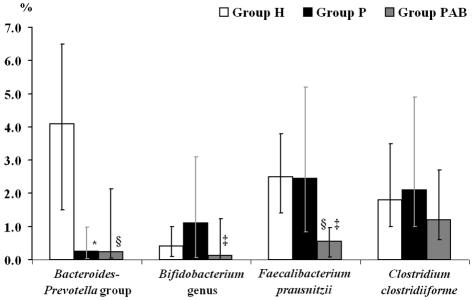
The percentages of specific rRNA genes were calculated for each individual, and the median of each subject group was determined to assess whether reductions in certain bacterial populations were linked with the decrease in total copy numbers of 16S rRNA genes in stool samples from groups P and PAB or whether shifts in relative abundances were occurring. The data revealed that the reduction in rRNA gene copy numbers of the Bacteroides-Prevotella group (Fig. 1) in stool samples from both group P and group PAB resulted in a significant decrease in the relative abundance of this population compared to the results for group H.
FIG. 1.
Percentage of 16S rRNA gene copies of the predominant fecal bacterial groups and species in relation to total bacterial 16S rRNA gene copies (relative abundance). The percentage was calculated for each individual, and the median was determined for each subject group. A nonparametric Mann-Whitney U test was used for pairwise comparison of the subject groups. Error bars represent the interquartile range. *, significant difference (P < 0.05) between groups H and P; §, significant difference (P < 0.05) between groups H and PAB; ‡, significant difference (P < 0.05) between groups P and PAB.
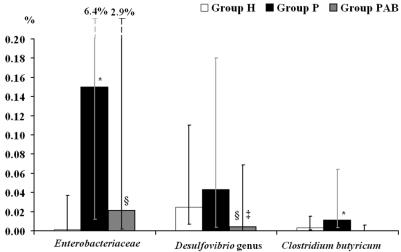
In contrast, the reduced numbers in 16S rRNA genes of other bacteria in samples from group P compared to group H samples did not lead to a decrease in relative abundance. Although bifidobacteria were present in the highest numbers in group H samples, the relative abundance of this genus was slightly higher in group P. The significant decrease in fecal bifidobacteria in samples from group PAB also caused a reduction in relative abundance. The abundance of desulfovibrios was also slightly higher in group P (Fig. 2), despite the elevated levels of rRNA genes in group H samples. However, the decline in these organisms in group PAB resulted in a significant reduction in their relative abundance. Similarly, mean copy numbers of 16S rRNA genes of F. prausnitzii and C. clostridiiforme were markedly reduced in the samples from groups P and PAB, but the abundance of both species was not significantly different between group P and group H, although it did decrease in group PAB (Fig. 1). Similar trends were observed with R. albus (data not shown).
FIG. 2.
Percentage of 16S rRNA gene copies of less abundant fecal bacterial groups and species in relation to total bacterial 16S rRNA gene copies (relative abundance). The percentage was calculated for each individual, and the median was determined for each subject group. A nonparametric Mann-Whitney U test was used for pairwise comparison of the subject groups. Error bars represent the interquartile range. *, significant difference (P < 0.05) between groups H and P; §, significant difference (P < 0.05) between groups H and PAB; ‡, significant difference (P < 0.05) between groups P and PAB.
C. butyricum 16S rRNA gene levels were relatively similar in samples from group H volunteers and groups P and PAB, but as a result of the reduced total numbers of bacterial 16S rRNA genes in group P samples, the relative abundance of this species increased significantly in these individuals (Fig. 2). However, a marked reduction in the relative abundance of C. butyricum in feces from group PAB was observed.
The abundance of Enterobacteriaceae was significantly higher in stool samples from both groups P and PAB compared to results for group H, but the percentage of these organisms was not as high in group PAB as in group P (Fig. 2). The relative abundance of Enterococcus faecalis in stool samples increased significantly from group H to group P to group PAB (data not shown).
DISCUSSION
Changes in intestinal bacterial community structure were observed among the three groups of elderly participants in this study. The highest total numbers of bacterial 16S rRNA genes occurred in stool samples from group H, and significantly lower numbers of 16S rRNA genes were detected in feces from groups P and PAB. Colonic transit has a major influence on large bowel function, and increased transit times are correlated with reduced fecal bacterial cell mass (42). Due to a lack of physical exercise and long periods of bed rest (1, 18, 36), intestinal transit times are often increased significantly in hospitalized patients (36), and this may have been a factor affecting population changes in groups P and PAB. Modified diets and eating patterns during hospitalization (37) may have also contributed.
The most marked difference between group H and groups P and PAB was seen with the Bacteroides-Prevotella group, with 16S rRNA gene copy numbers significantly lower in samples from groups P and PAB. Even though numbers of all fecal bacteria were generally reduced for groups P and PAB, the relative abundance of the Bacteroides-Prevotella group decreased over 10-fold in these subjects. Bacteroides are known to utilize a wide variety of carbon sources, and they account for the majority of polysaccharide digestion that occurs in the large intestine (28, 38). Marked reductions in such a nutritionally important bacterial group may have consequences for bacterial metabolism and the complex network of cross-feeding species that supports the colonic microbiota.
A previous study of culture techniques reported that the abundance of Bacteroides can be slightly reduced in older people compared to young adults but that the most marked reductions occurred in elderly hospitalized patients receiving antibiotics (20). However, the decrease in the abundance of the Bacteroides-Prevotella group in the present study correlated with hospitalization rather than with antibiotic treatment on its own. Antibiotics differ in their effects on the fecal microbiota (44), and in the present study, various drugs and drug combinations were given to group PAB. However, amoxicillin and/or amoxicillin-clavulanic acid were used most frequently, and they have been reported to have either no significant effect on fecal Bacteroides (12, 25) or to result in increased Bacteroides counts (7).
16S rRNA gene copies of other bacteria that play a role in carbohydrate breakdown and fermentation in the large intestine, such as bifidobacteria (9), F. prausnitzii (11), C. clostridiiforme (40), and R. albus (4), also decreased in stool samples from groups P and PAB. Reductions in the 16S rRNA gene copies found in group P samples correlated with the overall decrease in fecal bacteria, and the relative abundance of these organisms did not change significantly compared to results for group H. However, antibiotic therapy resulted in marked reductions in their relative abundance, and the prevalence of some bacteria was markedly reduced in group PAB stool samples. It is well established that antibiotics can cause major disturbances in the ecological balance between host and microorganism, which can lead to the proliferation of opportunistic invading species or pathogenic strains (44). For example, overgrowth of enterococci in the large bowel is common in antimicrobial therapy (44), and, accordingly, Enterococcus faecalis was found most frequently in group PAB stool samples. The reduction in important populations of carbohydrate-fermenting species in the large intestine seen in patients in groups P and PAB could also have major implications for the bacterial metabolism of the host. For example, F. prausnitzii (11), one of the most abundant species in the colon, is an important producer of butyrate, a major product of carbohydrate fermentation which is implicated in providing protection against colorectal cancer (3, 39) and ulcerative colitis (5, 6).
Bifidobacteria are regarded as being protective species in the large intestine, but there are indications that their numbers decline in older people, while the number of potentially harmful enterobacteria increases (16, 17, 19, 20, 22, 32, 33). Real-time PCR also demonstrated that bifidobacteria populations were lower and that enterobacteria populations were higher in the more frail and older patients in groups P and PAB. In our study, bifidobacteria were found in 100% of the fecal samples from groups H and P, in contrast to other studies, where these bacteria were isolated by using culture techniques and were found in only 50 to 86% of stool samples obtained from elderly subjects (16, 20, 32, 33), which highlights one of the advantages of cultivation-independent approaches in ecological studies of the large intestine.
However, one drawback of real-time PCR is that 16S rRNA gene copies cannot be directly converted into cell counts, because some bacteria can have a number of rrna operons, even within the same genus. For example, between two to five rrna operons have been found in different species belonging to the genus Bifidobacterium (21). Nevertheless, our results indicated that group H still had relatively high levels of fecal bifidobacteria. Individuals in group H were very health conscious and were living an active life. According to their statements, 25 out of 35 did regular exercises such as walking, gardening, dancing, gymnastics, cycling, or swimming. A similar observation was made in a previous study, in which a strong reduction in bifidobacteria was observed in a small group of elderly people, except for a 67-year-old, very health-conscious and fit man who had very high counts of fecal bifidobacteria, similar to those found in young adults (19).
Old age is often associated with increased ill health, which is leading to more frequent admissions to hospitals (13) and, together with a higher susceptibility to infection, to the increased use of antibiotics. The present study showed that members of groups P and PAB had abnormal intestinal microbiotas compared to members of group H and that antibiotic therapy contributed to further disturbances in the gut microbiota. Real-time PCR has been shown to provide a good quantitative tool for monitoring bacterial communities in the large bowel. This tool is needed to enable the development of therapeutic strategies, such as the use of pre- and probiotics, which may help to maintain a normal intestinal microbiota in elderly hospitalized patients and reduce proliferation of pathogenic microorganisms in the gut.
Acknowledgments
This work was supported by the Medical Research Council and The Health Foundation.
REFERENCES
- 1.Bazzochhi, G., J. Ellis, J. Villanueva-Meyer, J. Jing, S. N. Reddy, I. Mena, and W. J. Snape, Jr. 1990. Postprandial colonic transit and motor activity in chronic constipation. Gastroenterology 98:686-693. [DOI] [PubMed] [Google Scholar]
- 2.Bernhard, A. E., and K. G. Field. 2000. Identification of nonpoint sources of fecal pollution in coastal waters by using host-specific 16S ribosomal DNA genetic markers from fecal anaerobes. Appl. Environ. Microbiol. 66:1587-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradburn, D. M., J. C. Mathers, A. Gunn, J. Burn, P. D. Chapman, and I. D. Johnston. 1993. Colonic fermentation of complex carbohydrates in patients with familial adenomatous polyposis. Gut 34:630-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bryant, M. P. 1986. Genus Ruminococcus, p. 1093-1097. In P. H. A. Sneath, N. S. Mair, M. E. Sharpe, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 2. Williams & Wilkins, Baltimore, Md. [Google Scholar]
- 5.Chapman, M. A., M. F. Grahn, M. A. Boyle, M. Hutton, J. Rogers, and N. S. Williams. 1994. Butyrate oxidation is impaired in the colonic mucosa of sufferers of quiescent ulcerative colitis. Gut 35:73-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chapman, M. A., M. F. Grahn, M. Hutton, and N. S. Williams. 1995. Butyrate metabolism in the terminal ileal mucosa of patients with ulcerative colitis. Br. J. Surg. 82:36-38. [DOI] [PubMed] [Google Scholar]
- 7.Christensson, B., I. Nilsson-Ehle, and B. B. Ljungberg. 1991. A randomized multicenter trial to compare the influence of cefaclor and amoxicillin on the colonization resistance of the digestive tract in patients with lower respiratory tract infection. Infection 19:208-215. [DOI] [PubMed] [Google Scholar]
- 8.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Degan, B. A., and G. T. Macfarlane. 1991. Composition of carbohydrate substrate preferences in eight species of bifidobacteria. FEMS Microbiol. Lett. 84:151-156. [DOI] [PubMed] [Google Scholar]
- 10.Doré, J., A. Sghir, G. Hannequart-Gramet, G. Corthier, and P. Pochart. 1998. Design and evaluation of a 16S rRNA-targeted oligonucleotide probe for specific detection and quantitation of human faecal Bacteroides populations. Syst. Appl. Microbiol. 21:65-71. [DOI] [PubMed] [Google Scholar]
- 11.Duncan, S. H., G. L. Hold, H. J. Harmsen, C. S. Stewart, and H. J. Flint. 2002. Growth requirements and fermentation products of Fusobacterium prausnitzii, and a proposal to reclassify it as Faecalibacterium prausnitzii gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 52:2141-2146. [DOI] [PubMed] [Google Scholar]
- 12.Edlund, C., C. Stark, and C. E. Nord. 1994. The relationship between an increase in beta-lactamase activity after oral administration of three new cephalosporins and protection against intestinal ecological disturbances. J. Antimicrob. Chemother. 34:127-138. [DOI] [PubMed] [Google Scholar]
- 13.Feeney, E. T., M. P. Williams, and G. C. Doyle. 1986. Meeting the needs of hospitalized elderly. Nurs. Manage. 17:35-42. [PubMed] [Google Scholar]
- 14.Fite A., G. T. Macfarlane, J. H. Cummings, M. J. Hopkins, S. C. Kong, E. Furrie, S. Macfarlane. 2004. Identification and quantitation of mucosal and faecal desulfovibrios using real-time PCR. Gut 53:523-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gariballa, S. E., and A. J. Sinclair. 1998. Nutrition, ageing and ill health. Br. J. Nutr. 80:7-23. [DOI] [PubMed] [Google Scholar]
- 16.Gavini, F., C. Cayuela, J. M. Antoine, C. Lecoq, B. Levfebre, J. M. Membre, and C. Neut. 2001. Differences in the distribution of bifidobacterial and enterobacterial species in human faecal microflora of three different (children, adults, elderly) age groups. Microb. Ecol. Health Dis. 13:40-45. [Google Scholar]
- 17.Gorbach, S. L., L. Nahas, P. I. Lerner, and L. Weinstein. 1967. Studies of intestinal microflora. I. Effects of diet, age, and periodic sampling on numbers of fecal microorganisms in man. Gastroenterology 53:845-855. [PubMed] [Google Scholar]
- 18.Holdstock, D. J., and J. J. Misiewicz. 1970. Factors controlling colonic motility: colonic pressures and transit after meals in patients with total gastrectomy, pernicious anaemia and duodenal ulcer. Gut 11:100-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hopkins, M. J., R. Sharp, and G. T. Macfarlane. 2001. Age and disease related changes in intestinal bacterial populations assessed by cell culture, 16S rRNA abundance, and community cellular fatty acid profile. Gut 48:198-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hopkins, M. J., and G. T. Macfarlane. 2002. Changes in predominant bacterial populations in human faeces with age and with Clostridium difficile infection. J. Med. Microbiol. 51:448-454. [DOI] [PubMed] [Google Scholar]
- 21.Klappenbach, J. A., P. R. Saxman, J. R. Cole, and T. M. Schmidt. 2001. rrndb: the ribosomal RNA operon copy number database. Nucleic Acids Res. 29:181-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kleessen, B., B. Sykura, H. J. Zunft, and M. Blaut. 1997. Effects of inulin and lactose on fecal microflora, microbial activity, and bowel habit in elderly constipated persons. Am. J. Clin. Nutr. 65:1397-1402. [DOI] [PubMed] [Google Scholar]
- 23.Langendijk, P. S., F. Schut, G. J. Jansen, G. C. Raangs, G. R. Kamphuis, M. H. Wilkinson, and G. W. Welling. 1995. Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16S rRNA-targeted probes and its application in fecal samples. Appl. Environ. Microbiol. 61:3069-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lesourd, B. M., C. Laisney, R. Salvatore, S. Meaume, and R. Moulias. 1994. Decreased maturation of T-cell populations in the healthy elderly: influence of nutritional factors on the appearance of double negative CD4− CD8− CD2+ cells. Arch. Gerontol. Geriatr. 4:S149-S154. [DOI] [PubMed] [Google Scholar]
- 25.Lode, H., N. Von der Hoh, S. Ziege, K. Borner, and C. E. Nord. 2001. Ecological effects of linezolid versus amoxicillin/clavulanic acid on the normal intestinal microflora. Scand. J. Infect. Dis. 33:899-903. [DOI] [PubMed] [Google Scholar]
- 26.Lovat, L. B. 1996. Age related changes in gut physiology and nutritional status. Gut 38:306-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Macfarlane, G. T., and G. R. Gibson. 1991. Sulphate-reducing bacteria, p. 201-222. In P. N. Levett (ed.), Anaerobic microbiology-a practical approach. Oxford University Press, Oxford, England.
- 28.Macfarlane, G. T., and G. R. Gibson. 1991. Co-utilization of polymerized carbon sources in Bacteroides ovatus in a two-stage continuous culture system. Appl. Environ. Microbiol. 57:1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malinen, E., A. Kassinen, T. Rinttila, and A. Palva. 2003. Comparison of real-time PCR with SYBR Green I or 5′-nuclease assays and dot-blot hybridization with rDNA-targeted oligonucleotide probes in quantification of selected faecal bacteria. Microbiology 149:269-277. [DOI] [PubMed] [Google Scholar]
- 30.Matsuki, T., K. Watanabe, J. Fujimoto, Y. Miyamoto, T. Takada, K. Matsumoto, H. Oyaizu, and R. Tanaka. 2002. Development of 16S rRNA-gene-targeted group-specific primers for the detection and identification of predominant bacteria in human feces. Appl. Environ. Microbiol. 68:5445-5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meier, H., C. Koob, W. Ludwig, R. Amann, E. Frahm, S. Hoffmann, U. Obst, and K.-H. Schleifer. 1997. Detection of enterococci with rRNA targeted probes and their use for hygienic drinking water control. Water Sci. Technol. 35:437-444. [Google Scholar]
- 32.Mitsuoka, T., K. Hayakawa, and N. Kimura. 1974. Die Faekalflora bei Menschen II. Mitteilung: Die Zusammensetzung der Bifidobakterienflora der verschiedenen Altersgruppen. Zentbl. Bakteriol. Hyg. Abt. 1 Orig. A 226:469-478. [PubMed] [Google Scholar]
- 33.Mutai, M., and R. Tanaka. 1987. Ecology of Bifidobacterium in the human intestinal flora. Bifidobacteria and Microflora 6:33-41. [Google Scholar]
- 34.Nadkarni, M. A., F. E. Martin, N. A. Jacques, and N. Hunter. 2002. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology 148:257-266. [DOI] [PubMed] [Google Scholar]
- 35.Percival, R. S., P. D. Marsh, and S. J. Challacombe. 1996. Serum antibodies to commensal oral and gut bacteria vary with age. FEMS Immunol. Med. Microbiol. 15:34-42. [DOI] [PubMed] [Google Scholar]
- 36.Pontes, F. A., A. T. Silva, and A. C. Cruz. 1995. Colonic transit and the effect of lactulose or lactitol in hospitalized patients. Eur. J. Gastroenterol. Hepatol. 7:441-446. [PubMed] [Google Scholar]
- 37.Ross, D. G. 1993. Subjective data related to altered bowel elimination patterns among hospitalized elder and middle-aged persons. Orthop. Nurs. 12:25-32. [DOI] [PubMed] [Google Scholar]
- 38.Salyers, A. A. 1984. Bacteroides of the human lower intestinal tract. Annu. Rev. Microbiol. 38:293-313. [DOI] [PubMed] [Google Scholar]
- 39.Scheppach, W., H. P. Bartram, and F. Richter. 1995. Role of short-chain fatty acids in the prevention of colorectal cancer. Eur. J. Cancer 31A:1077-1080. [DOI] [PubMed] [Google Scholar]
- 40.Slade, A. P., G. M. Wyatt, C. E. Bayliss, and W. M. Waites. 1987. Comparison of populations of human faecal bacteria before and after in vitro incubation with plant cell wall substrates. J. Appl. Bacteriol. 62:231-240. [DOI] [PubMed] [Google Scholar]
- 41.Smith, J. L. 1998. Foodborne illnesses in the elderly. J. Food Prot. 61:1229-1239. [DOI] [PubMed] [Google Scholar]
- 42.Stephen, A. M., S. Wiggins, and J. H. Cummings. 1987. Effect of changing transit time on colonic microbial metabolism in man. Gut 28:601-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suau, A., R. Bonnet, M. Sutren, J. J. Godon, G. R. Gibson, M. D. Collins, and J. Doré. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65:4799-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sullivan, A., C. Edlund, and C. E. Nord. 2001. Effect of antimicrobial agents on the ecological balance of human microflora. Lancet Infect. Dis. 1:101-114. [DOI] [PubMed] [Google Scholar]
- 45.Wang, R. F., W. W. Cao, and C. E. Cerniglia. 1996. PCR detection and quantitation of predominant anaerobic bacteria in human and animal fecal samples. Appl. Environ. Microbiol. 62:1242-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zoetendal, E. G., A. D. Akkermans, and W. M. De Vos. 1998. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl. Environ. Microbiol. 64:3854-3859. [DOI] [PMC free article] [PubMed] [Google Scholar]