Abstract
Marine sponges (Porifera) harbor large amounts of commensal microbial communities within the sponge mesohyl. We employed 16S rRNA gene library construction using specific PCR primers to provide insights into the phylogenetic identity of an abundant sponge-associated bacterium that is morphologically characterized by the presence of a membrane-bound nucleoid. In this study, we report the presence of a previously unrecognized evolutionary lineage branching deeply in the domain Bacteria that is moderately related to the Planctomycetes, Verrucomicrobia, and Chlamydia lines of decent. Because members of this lineage showed <75% 16S rRNA gene sequence similarity to known bacterial phyla, we suggest the status of a new candidate phylum, named “Poribacteria”, to acknowledge the affiliation of the new bacterium with sponges. The affiliation of the morphologically conspicuous sponge bacterium with the novel phylogenetic lineage was confirmed by fluorescence in situ hybridization with newly designed probes targeting different sites of the poribacterial 16S rRNA. Consistent with electron microscopic observations of cell compartmentalization, the fluorescence signals appeared in a ring-shaped manner. PCR screening with “Poribacteria”-specific primers gave positive results for several other sponge species, while samples taken from the environment (seawater, sediments, and a filter-feeding tunicate) were PCR negative. In addition to a report for Planctomycetes, this is the second report of cell compartmentalization, a feature that was considered exclusive to the eukaryotic domain, in prokaryotes.
Sponges (Porifera) are ancient metazoans dating back more than 580 million years. They populate tropical and subtropical benthic marine habitats but are also found at higher latitudes and even in freshwater lakes and streams. So far an estimated 15,000 species have been described, but the true diversity is probably much higher (20). Particularly the tropical sponges are known for their colorful appearances and their morphological plasticity, encompassing encrusting, rope, ball, and vase shapes ranging in size from a few millimeters to >1 m. Sponges are diploblast metazoans that lack true tissues or organs. In spite of their simple organization, genome sequencing has revealed genes encoding functions that are highly homologous to those of their vertebrate analogs (4, 31, 32). As sessile filter feeders, they pump large volumes of water through a specialized canal system, termed the aquiferous system. The filtration capacities of sponges are remarkably efficient (up to 24,000 liters kg−1 day−1), leaving the expelled water essentially sterile (36, 39, 44, 54). Food particles such as unicellular algae and bacteria are removed from the seawater and translocated into the mesohyl interior. The mesohyl is a collagen scaffold that constitutes much of the sponge body. Single amoeboid sponge cells, termed archaeocytes, move freely through the mesohyl matrix and digest food particles by phagocytosis (3, 5).
The presence of large amounts of microorganisms within the mesohyl of many demosponges is well documented (45, 46, 51, 52, 53; for recent reviews, see references 19 and 24). Bacteria can contribute up to 40% of the sponge biomass (equal to about 108 to 109 bacteria g of tissue−1) and are probably permanently associated with the host sponge unless they are disturbed by external stress factors (13, 43, 48, 50). Several recent studies have sought to address the phylogenetic diversity of microbial communities associated with marine sponges by using 16S rRNA gene sequence analysis (1, 18, 27, 38, 49). A comprehensive analysis showed that sponges from different oceans contain phylogenetically complex, yet highly specific, microbial signatures (18, 19). In particular, representatives of the poorly characterized phyla Chloroflexi (formerly green nonsulfur bacteria), Acidobacteria, and Actinobacteria as well as Alpha-, Gamma-, and Deltaproteobacteria are abundant in gene libraries. It is noteworthy that none of these sponge-specific bacteria have so far been cultured or are related to cultured bacteria.
Very early on researchers noted that similar bacterial morphotypes occur in taxonomically different sponges (46, 53). One particular morphotype has gained attention because of a nucleoid-like structure within the microbial cell (16, 17). Six different morphological variants were described that resembled the general appearance of sponge-associated morphotype E according to Vacelet (45) and Friedrich et al. (12) and morphotype 4 according to Wilkinson (53). Prokaryotic cells do not generally have cell compartmentalization, with the only exception being members of the phylum Planctomycetes (26). Because of the presence of nucleoid-containing bacteria in the sponge mesohyl and because fluorescence in situ hybridization (FISH) yielded Planctomycetes-specific signals in tissue cryosections (13, 49), this study aimed to target these microorganisms by using molecular methods in order to provide insights into their phylogeny, abundance, and distribution among tropical and subtropical sponge species.
MATERIALS AND METHODS
Sponge collection.
The sponge Aplysina aerophoba was collected by scuba diving in Banyuls sur Mer, France (42°29′N, 03°08′E), at depths of 5 to 15 m in April 2000. The sponges Aplysina fistularis, Aplysina insularis, Aplysina lacunosa, Verongula gigantea, and Smenospongia aurea were collected by scuba diving offshore of Little San Salvador Island, Bahamas (24°34.39′N, 75°58.00′W), at depths of 5 to 15 m in July 2000. Individual specimens were placed separately into plastic bags and brought to the surface. After collection, sponge tissues were cut into pieces and stored at −80°C until use.
Amplification of 16S rRNA genes and library construction.
Genomic DNAs were extracted from sponge tissues frozen in liquid nitrogen by using Fast DNA spin kits for soil (Q-Biogene, Heidelberg, Germany) according to the manufacturer's protocol. Amplification of the 16S rRNA genes was accomplished with the primers PLA46f (5′-GGA TTA GGC ATG CAA GTC-3′) and 1390r (5′-CGG GCG GTG TCT ACA A-3′) (34, 57). A second library was constructed from A. aerophoba, with PLA46f as a forward primer and PLA886 (5′-GGG AGT ATG GTC GCA AGG C-3′) as a reverse primer (34). The resulting sequences from this library are identified by the prefix “Q.” PCR cycling conditions were as follows: initial denaturation (2 min at 96°C); 30 cycles of denaturation (1 min at 96°C), primer annealing (1 min at 56°C), and elongation (1.5 min at 72°C); and a final extension step (10 min at 72°C). Genomic DNA from Pirellula marina was used as a positive control. PCR products were ligated into the pGEM-T-Easy vector (Promega) and transformed into CaCl2-competent Escherichia coli DH5α. Plasmid DNAs were isolated by standard miniprep protocols. Correct insert sizes were verified by EcoRI restriction digestion and agarose gel electrophoresis (41).
Design and application of “Poribacteria”-specific PCR primers for screening.
In order to determine whether the novel candidate phylum “Poribacteria” is specific to sponges, we designed the primers POR389f (5′ ACG ATG CGA CGC CGC GTG 3′) and POR1130r (5′ GGC TCG TCA CCA GCG GTC 3′) and used them to probe environmental samples from within the vicinity of sponges. Seawater was collected from both dive locations (Banyuls sur Mer and Little San Salvador Island) on the same dives but prior to the removal of sponges from the reef surface. In order to compensate for the estimated 100 to 1,000 times higher bacterial concentration in sponge tissues than in seawater, we collected 1 to 3 liters of seawater, prefiltered it through folded filter papers, and then filtered it through 0.2-μm-pore-size bottle-top SFCA membrane filters (Nalgene, Rochester, N.Y.). Genomic DNAs were extracted from the filters after thermal lysis in a boiling water bath for 10 min and subsequent precipitation in ethanol. Also, coarse and fine sediment samples taken within a 1-m radius of an A. fistularis sponge and tissues of a filter-feeding tunicate (Ecteinascidia turbinata) were probed for the presence of the candidate phylum “Poribacteria”. Several DNA template dilutions (1:1, 1:10, 1:100, and 1:1,000) were added to PCRs that were performed at an annealing temperature of 68°C. As a positive control, genomic DNAs from frozen sponge tissues and “Poribacteria”-positive clones from 16S ribosomal DNA libraries were used. PCR amplification with the universal eubacterial primers 27f and 1492r was performed to validate the quality of the environmental DNAs. Furthermore, the resulting PCR amplicons were used in a nested PCR approach using the primers POR389f and POR1130r. As a negative control, 1 μl of H2O instead of DNA was always added to a PCR. The resulting PCR products were analyzed in 1% agarose gels.
In order to assess the distribution of the novel candidate phylum “Poribacteria,” we probed several other sponges by PCR. These were Aplysina cavernicola from Elba (Italy) (43), Theonella swinhoei from the Western Caroline Islands (Palau) (18), Agelas wiedenmayeri from the Florida Keys (30), A. fistularis from offshore of San Diego, Calif. (unpublished), and Aplysina cauliformis, Aplysina archeri, Pseudoceratina crassa, Xestospongia muta, Chondrilla nucula, Agelas wiedenmayeri, Agelas cerebrum, Ptilocaulis sp., and Ectyoplasia ferox from around Little San Salvador Island.
Restriction fragment length polymorphism, sequencing, and phylogenetic analyses.
PCR products were characterized by single digestion with the restriction endonucleases HaeIII and AvaI. Clones with similar restriction patterns were grouped together, and random clones from each group were chosen for sequence analysis. Sequencing was performed on a LiCor 4200 automated sequencer (LiCor Inc., Lincoln, Nebr.) and an ABI 377XL automated sequencer (Applied Biosystems). Sequence data were edited with ABI Prism Autoassembler, v. 2.1, software (Perkin-Elmer) and then checked for chimera formation (CHECK_CHIMERA online analysis of Ribosomal Database Project II [RDP-II] [6]). Phylogenetic trees including 16S rRNA gene sequences of approximately 1.3 kb were constructed with ARB software (http://www.arb-home.de). Trees were calculated by using the neighbor-joining (Jukes-Cantor correction), maximum parsimony, and maximum likelihood methods implemented in ARB. Partial sequences (about 600 bp) were analyzed with the basic local alignment search tool (BLASTn) algorithm for initial identification. Affiliations of partial sequences were confirmed by adding them to a tree consisting of full-length sequences only without changing the tree topology by use of the ARB “parsimony interactive” method.
FISH.
The inner core (about 15 g) of freshly collected A. aerophoba was cut into pieces and disintegrated in 50 ml of ice-cold Ca2+- and Mg2+-free artificial seawater (400 mM NaCl, 27.6 mM Na2SO4, 2.3 mM NaHCO3, 8.9 mM KCl, 0.8 mM KBr, 0.4 mM H3BO3, 0.15 mM SrCl2, 0.07 mM NaF) (40) by use of a mortar and pestle. The suspension was vortexed for 10 min, prefiltered through folded filter papers to remove tissue fibers, and centrifuged at 4°C for 30 min at 100 × g to remove the remaining sponge cells. Bacteria were obtained from the supernatant by centrifugation at 4°C at 12,000 × g for 30 min to ensure complete microbial recovery and washed three times in ice-cold Ca2+- and Mg2+-free artificial seawater. The bacterium-containing fractions were fixed overnight in 0.2-μm-pore-size-filtered seawater containing 4% paraformaldehyde, resuspended in 50% ethanol-phosphate-buffered saline (PBS), and stored at −20°C.
For FISH analyses, aliquots were heat fixed at 40°C onto microscope slides, dehydrated in an aqueous ethanol series (50, 70, and 96%) for 3 min at each step, and air dried. FISH was performed in an isotonically equilibrated humid chamber at 46°C in hybridization buffer (0.9 M NaCl, 20 mM Tris-HCl [pH 7.4], 0.01% sodium dodecyl sulfate) and various amounts of formamide (10 to 30%) for 4 h. Probes were used at a concentration of 2 ng/μl. The hybridization buffer was gently rinsed off and the slides were incubated at 48°C in wash buffer (0.7 M NaCl, 20 mM Tris-HCl [pH 7.4], 50 mM EDTA, 0.01% sodium dodecyl sulfate) for 20 min. The wash buffer was rinsed off with ice-cold distilled water, and the slides were air dried in the dark and mounted in Citifluor (Citifluor Ltd., London, United Kingdom). Visualization was achieved with an Axiolab microscope (MC 80; Zeiss). Each hybridization was performed at least five times in parallel and three times independently. The hybridization conditions for the new “Poribacteria”-specific probes were adjusted with an increasing formamide series and evaluated visually. Table 1 lists the FISH probes used for this study.
TABLE 1.
FISH probes used for this studya
| Probe | % FA | Sequence (5′-3′) | Specificity | Range of positionsb | Reference |
|---|---|---|---|---|---|
| EUB338Ic | 35 | GCTGCCTCCCGTAGGAGT | Bacteria, including Planctomycetes, Verrucomicrobia, Chloroflexi, and candidate division OP11 | 338-355 | 2 |
| EUB338IIc | 35 | GCAGCCACCCGTAGGTGT | 338-355 | 7 | |
| EUB338IIIc | 35 | GCTGCCACCCGTAGGTGT | 338-355 | 7 | |
| GNS934 | 30 | ACCACACGCTCCGCTGCTTGT | Chloroflexi sponge cluster I | 934-955 | This study |
| SRB385 | 35 | CGGCGTCGCTGCGTCAGG | Deltaproteobacteria | 385-402 | 2 |
| PLA46 | 30 | GGATTAGGCATGCAA | Planctomycetes | 46-63 | 34 |
| PLA886 | 35 | GCCTTGCGACCATACTCCC | Planctomycetes | 886-904 | 34 |
| Competitor | GCCTTGCGACCGTACTCCC | ||||
| POR1130 | 30 | GGCTCGTCACCAGCGGTC | Candidate phylum “Poribacteria” | 1130-1148 | This study |
| POR389 | 20 | CACGCGGCGTCGCATCGT | Candidate phylum “Poribacteria” | 389-407 | This study |
| POR600 | 10 | CCGAACCCTTTCACGTCT | Candidate phylum “Poribacteria” | 600-618 | This study |
New probe sequences were deposited at probeBase (http://www.probebase.net; 28).
Target site on 16S rRNA according to E. coli numbering.
The probes EUB338I to EUB338III were used as a probe mix.
Transmission electron microscopy.
A. aerophoba was fixed in 2.5% glutaraldehyde-PBS for 12 h, rinsed 3 times for 20 min each in 1× PBS, and postfixed in 2% osmium tetroxide-1× PBS for 12 h. The pieces were dehydrated in an ethanol series (30, 50, 70, and 90% and three times at 100%), incubated three times for 20 min each in propylene oxide, and polymerized in Epon 812 (Serva) for 4 days at 60°C. The embedded sponge pieces were sectioned with an ultramicrotome (OM U3; C. Reichert, Vienna, Austria). For contrast, 70- to 80-nm-thick sections were poststained with 0.5% uranyl acetate in methanol for 10 min and with Reynold's lead citrate for 5 min. The resulting sections were investigated by electron microscopy (EM 10; Zeiss).
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences obtained in this study were deposited in EMBL/GenBank/DDBJ under accession numbers AY485280 to AY485299.
RESULTS
Phylogenetic analysis.
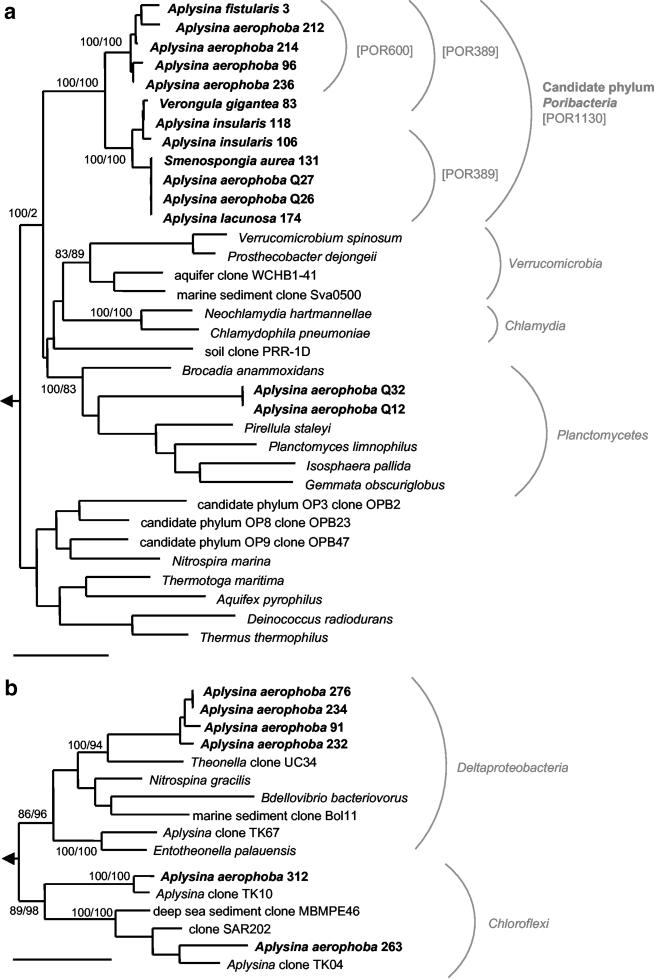
Amplification of the 16S rRNA genes from the total sponge-derived DNA yielded fragments of the expected size of 1,300 or 800 bp, depending on which PCR primer set was used. Altogether, 600 clones were compared by restriction fragment length polymorphism analyses, resulting in six major restriction patterns. A total of 157 representative clones were chosen for sequencing. Thirteen chimeric sequences were excluded from further analyses. A large portion (n = 41 [29%]) of the recovered sequences, with an average of only 74.6% 16S rRNA sequence similarity to members of known phyla, formed a novel evolutionary lineage branching deeply in the domain Bacteria in all treeing methods applied (Fig. 1a). Based on the phylogenetically distant position of this phylum, the candidate phylum “Poribacteria” was proposed. A moderate relationship of this evolutionary lineage to the Planctomycetes, Verrucomicrobia, and Chlamydia lines of descent was suggested by the monophyletic grouping of these phyla in maximum likelihood and neighbor-joining trees, while parsimony trees showed no consistent branching pattern (Fig. 1a). All near-full-length sequences belonging to the candidate phylum “Poribacteria” were conserved in 13 of 15 (87%) signature nucleotides that are characteristic of the Planctomycetes (15, 25, 42, 55). The average in-cluster similarity (92.7%) from sponge-derived sequences is high compared to that for other phyla (9). The remaining 103 16S rRNA gene clones consisted of sequences affiliated with (i) the Deltaproteobacteria (n = 41 [28%]), (ii) the Chloroflexi (n = 51 [35%]), (iii) the Planctomycetes (n = 4 [3%]), and (iv) a subset of sequences with various phylogenetic positions (n = 7 [5%]) (Fig. 1b).
FIG. 1.
Phylogenetic maximum likelihood dendrograms calculated with nearly complete 16S rRNA gene sequences (>1,300 bp) of the candidate phylum “Poribacteria” (a) and of the Deltaproteobacteria and Chloroflexi (b) that were recovered from marine verongid and dictyoceratid sponges. Sequences obtained in this study are indicated in bold. The specificities of newly designed, “Poribacteria”-specific FISH probes POR389, POR600, and POR1130 are indicated in square brackets. Neighbor-joining or maximum parsimony (100 resamplings) bootstrap values are provided for relevant groups. Bar, 10% divergence. The arrow indicates the outgroup.
To determine the specificity of “Poribacteria” and their distribution within sponges, we applied “Poribacteria”-specific PCR primers to various environmental samples and to other sponge species. A positive PCR product amplified with the universal bacterial primers 27f and 1492r served as an internal control for the validity of each DNA sample. The seawater samples from both dive locations, coarse and fine sediments taken near a sponge colony, and a filter-feeding tunicate were PCR negative with the “Poribacteria”-specific primers POR389f and POR1130r in conventional and nested PCRs. Positive “Poribacteria”-specific PCR products were obtained from all tested sponges of the order Verongida (A. aerophoba, A. cavernicola [Elba, Italy], A. lacunosa, A. cauliformis, A. fistularis [San Diego, Calif.], A. insularis, A. archeri, V. gigantea, and Pseudoceratina crassa) independent of their geographic location. Positive PCR products amplified with “Poribacteria”-specific primers were also recovered from S. aurea (Dictyoceratida), X. muta (Haplosclerida), and T. swinhoei (Lithistida). Negative “Poribacteria”-specific PCR results were obtained with the sponges C. nucula (Hadromerida), Agelas wiedenmayeri and Agelas cerebrum (both Agelasida), Ptilocaulis sp. (Halichondria), and Ectyoplasia ferox (Poecilosclerida).
FISH.
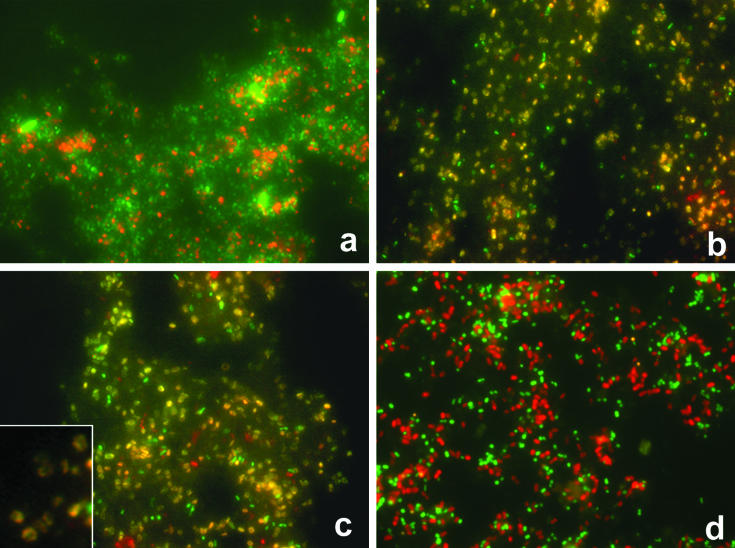
In order to visualize the presence of the “Poribacteria” and to assess their relative abundance within the microbial consortium of A. aerophoba, we designed a set of specific FISH probes, using the probe design and probe match tool of the ARB software package (Table 1). Hybridization with the EUB338 probe mix revealed a very high signal abundance. An estimation of the ratio of FISH signals to DAPI-stained cells revealed >98% detection when the EUB338 probe mix was applied (data not shown). The application of the bacterial probe mix EUBI-III together with the “Poribacteria”-specific probe POR1130 revealed a high abundance of this lineage within the microbial consortium of A. aerophoba (Fig. 2a). In fact, “Poribacteria”-specific signals were visibly more abundant than all previously applied specific FISH probes (12, 13).
FIG. 2.
FISH of bacterial preparations from A. aerophoba. (a) Hybridization with EUB338 probe mix (FITC-labeled) and “Poribacteria”-specific probe POR1130 (Cy3-labeled). (b and c) Hybridization with probe POR1130 (FITC-labeled) and probes POR389 (b) and POR600 (c) (Cy3-labeled), specific for the “Poribacteria” subgroups. Yellow signals result from cohybridization. Note the ring-shaped fluorescent signal in the inset in panel c. (d) Hybridization with probe GNS934, specific for the Chloroflexi I cluster (FITC-labeled), and probe POR1130 (Cy3-labeled).
The identity of the “Poribacteria” was further confirmed by the application of probes that were specific to the two major subgroups of this lineage. Cohybridization is visible when a red signal (Cy3) colocalizes with a green signal (fluorescein isothiocyanate [FITC]), resulting in a yellow signal. Cohybridization was indeed observed after the application of the probe POR1130 (FITC labeled) together with either probe POR389 or POR600 (Cy3 labeled) (Fig. 2b and c). Because these probes differ in their target sites on the corresponding 16S rRNA, their colocalization confirms the authenticity of the “Poribacteria” lineage. It is noteworthy that the FISH signals appeared in a ring-shaped manner that is consistent with the presence of a nucleoid in this particular cell type (Fig. 2b and c).
Because sequences from other lineages were also recovered from the gene libraries, cohybridization reactions were performed to assess the relative abundance of the different lineages. After cohybridization of the Chloroflexi I cluster-specific probe GNS934 with POR1130, green and red signals were visible as independent spots in about equal amounts (Fig. 2d). Cohybridization of the probe POR1130 with probe SRB385, targeting many Deltaproteobacteria, revealed that the “Poribacteria” are more abundant than the Deltaproteobacteria (data not shown). After the application of Planctomycetes-specific probes PLA46 and PLA886 to microbial preparations, signals were detected only rarely (data not shown).
Transmission electron microscopy.
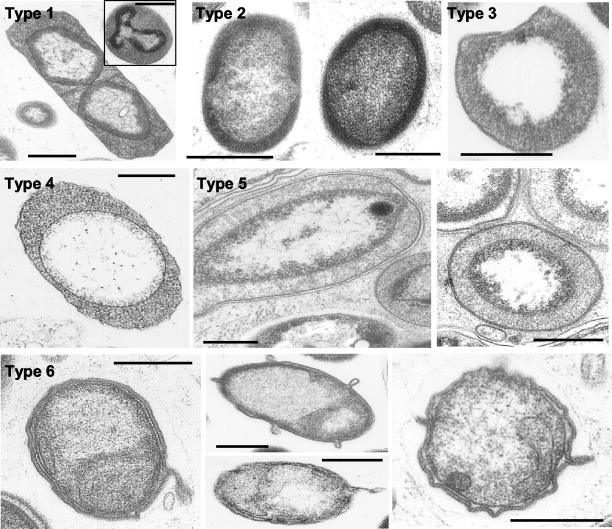
Electron microscopy was performed to provide insights into the morphology of the nucleoid-containing microorganisms of A. aerophoba. Altogether, six different morphotypes were found that closely resemble the different subtypes of Australian Great Barrier Reef sponges (Fig. 3; Table 2) (16, 17). Five of the morphotypes contain a membrane-bound organelle that occupies a central position in the cell. The organelle contains fibrillar DNA that is surrounded by an electron-dense riboplasm. The morphotypes vary in cell shape (short and long rods and D-shaped cells), membrane structure (gram-negative and S-layer types), and density of the cytoplasm. A sixth morphotype shows a reverse topology in that the nuclear material lies in the cytoplasm, whereas the membrane-bound compartment is DNA-free. Morphotype 6 can easily be distinguished by bleb-like structures of the outer cell membrane.
FIG. 3.
Transmission electron microscopy of bacteria associated with A. aerophoba. The nucleoid-containing bacterial morphotypes, according to the work of Fuerst et al. (17), are described in Table 2.
TABLE 2.
Characteristics of sponge-associated bacterial morphotypes containing membrane-bound nucleoidsa
| Morphotype | Morphological characteristics |
|---|---|
| 1 | Long rods with membrane-bound nuclear region and S-layer-type outer membrane, cell division by septation |
| 2 | Short fat rods with membrane-bound nuclear region and electron-dense cytoplasm |
| 3 | D-shaped cells with clear membrane-bound nuclear region and S-layer-type outer membrane |
| 4 | Rods with clear membrane-bound nuclear region and gram-negative outer membrane |
| 5 | Rods with clear, membrane-bound nuclear region and S-layer-type outer membrane |
| 6 | Rods with membrane-bound compartment that is devoid of DNA; the compartment contains most of the RNA, while the DNA is restricted to the cytoplasm; the outer membrane contains budding appendages; there is an unusually wide periplasmic space and a phenotypic resemblance to G. aurantiaca (56) |
The data were summarized from the work of Fuerst et al. (17).
DISCUSSION
16S rRNA gene-based studies have enormously enhanced our knowledge about the phylogenetic diversity of environmental microbial communities. More than 40 different phyla are now recognized for the domain Bacteria, about half of which contain cultured representatives. Even though databases encompassing >78,000 rRNA gene sequences have been established (RDP-II preview [http://rdp.cme.msu.edu/html/analyses_preview.html]), the extent of prokaryotic diversity is still underestimated even at the phylum level. A striking example of novel microbial diversity is the description of an entirely new phylum, Gemmatimonadetes (formerly candidate phylum BD [33] or KS-B [29]), which was recently added to the domain Bacteria (56). Moreover, the OP candidate phylum from hot springs (23), the WS6, TM7, and SC1-SC4 candidate phyla from soil communities (9, 10, 11, 21, 22), and the BRC1 candidate phylum from rice roots (8) have been described in recent years. The term “candidate phylum” is reserved for phylogenetic lineages that show <75% sequence similarity to known phyla but that lack cultivated representatives or contain too few sequences for a precise phylogenetic placement.
In this study, we report the discovery of a novel, deeply rooting candidate phylum of bacteria in marine sponges. This study was originated because electron microscopic data suggested the presence of unusual bacteria with membrane-bound nuclear bodies in several sponges (12, 16, 17, 45). Because cell compartmentalization has so far only been observed in the phylum Planctomycetes (14, 26), the Planctomycetes-specific FISH probes Pla46 and Pla886 were used as PCR primers. After library construction and phylogenetic analysis, a novel lineage was identified that showed an average of 74.6% sequence similarity to any of the known bacterial phyla (Fig. 1a). The phylogenetic lineage was reproducibly monophyletic and stable after the application of different tree-building algorithms. Because of the lack of relatedness to known bacterial phyla and because bacterial cultivation has so far been unsuccessful (37), the status of candidate phylum and the name “Poribacteria” are proposed. This name was chosen to acknowledge the affiliation of these bacteria with sponges (phylum Porifera). The poribacterial lineage currently encompasses 12 nearly complete 16S rRNA gene sequences and 29 partial sequences from five tropical and one temperate verongid species. The average in-cluster similarity of this lineage is high (92.7%) compared to those of other published phyla (9). If this value reflects the limited resources that have been sampled so far (verongid sponges), then one would expect this value to increase as more sequences become available. Alternatively, if the “Poribacteria” lineage has been affiliated with sponges for evolutionarily long periods of time, reduced diversity as a result of adaptation would be expected.
The “Poribacteria” lineage is moderately related to the Planctomycetes, Chlamydia, and Verrucomicrobia lines of descent. Also, the lineage shows an affiliation with the soil clone PRR-1D, which was recovered in a PCR screening assay (the PV assay) that led to the discovery of considerable bacterial diversity in the Planctomycetes-Verrucomicrobia branch of the tree (8). The question arises regarding why the “Poribacteria” lineage has not been recovered in any previous environmental 16S rRNA gene library. One possible explanation is that there may be mismatches in the target region of the “Poribacteria” 16S rRNA genes in the universal primers 27f and/or 1492r, as has been reported for Planctomycetes (47) and some spirochetes (35). Similarly, the FISH probe EUB338, originally thought to target all bacteria, also did not hybridize with Planctomycetes and Verrucomicrobia, a flaw that has been compensated for by the construction of a more comprehensive probe mix, EUBI-III (7).
The 16S rRNA gene libraries made for this study also contained sequences of specific phylogenetic clusters of the Chloroflexi and Deltaproteobacteria that had previously been reported from marine sponges (Fig. 2b) (18, 49). Their amplification was most likely due to nonspecific primer binding, as there are three mismatches each between the Pla46 primer sequence and the corresponding 16S rRNA gene sequence of the closely related Chloroflexi and Deltaproteobacteria. However, their recovery is still noteworthy, as it extends their documented distribution to five additional tropical sponge species. Surprisingly, only 2 of 157 sequenced clones were affiliated with the Planctomycetes even though group-specific primers were used. We concluded, for the following reasons, that the Planctomycetes probably play a minor role in the microbial community structure of marine sponges: (i) with the exception of clones Q12 and Q32 from this study, planctomycete sequences have not been recovered from any sponge-derived 16S ribosomal DNA library to date; (ii) Planctomycetes have been cultivated from Aplysina sponges, but the application of FISH probes revealed that they are only minor components of the sponge-associated microbial community (37); and (iii) the application of the FISH probes Pla46 and Pla886 produced fluorescent signals on sponge tissue cryosections (13), but not when bacterial preparations were used (data not shown), indicating either nonspecific binding of the FISH probes to the host extracellular matrix or the lysis of planctomycetes during the bacterial extraction procedure.
In order to visualize members of the novel candidate phylum “Poribacteria” within sponge tissues, we designed a set of “Poribacteria”-specific FISH probes (Table 1). Cohybridization with the universal bacterial probe mix EUBI-III showed that the “Poribacteria” lineage is an abundant member of the sponge-associated microbial community (Fig. 2a). As expected, the application of the phylum-specific probe POR1130 with either the subphylum probe POR389 or POR600 resulted in the cohybridization of most cells (Fig. 2b and c). The remaining green and red signals reflect either a certain degree of nonspecific binding to nontarget microorganisms or the presence of as yet unrecognized poribacterial subgroups. It is also noteworthy that the resulting fluorescence forms a ring-shaped signal (Fig. 2b and c). This finding is consistent with the presence of a membrane-bound compartment in the target microorganisms (Table 2; Fig. 3). Simultaneous hybridization with a Chloroflexi-specific probe and a “Poribacteria”-specific probe resulted in more signal abundance than was reported previously for other phylum- and subphylum-level probes (Fig. 2d) (12).
Electron microscopic studies were performed to characterize the nucleoid-containing bacterial morphotypes (Fig. 2). The structural plasticity level was high with respect to cell shape, membrane structure, and cytosol density (Table 2). The cell shapes represented included short and long rods as well as D-shaped cells. The cell walls represented were typical cell walls of gram-negative bacteria but also the S-layer-type cell walls of Archaea. Morphotype 6 contains characteristic bleb-like structures. Previous reports showed that in morphotypes 1 to 5, the fibrillar DNA is located in the nucleoid, while in morphotype 6, the RNA is contained in the nucleoid and the DNA is located in the cytoplasm (17). Morphotype 6 shows some resemblance to Gemmatimonas aurantiaca, the first cultivated representative of the novel phylum Gemmatimonadetes (56). Both are rod-shaped cells with a gram-negative cell envelope. The characteristic outer cell wall is wavy, contains budding structures, and is separated from the cytosol by an atypically wide periplasm. Interestingly, Gemmatimonadetes 16S rRNA gene sequences have been recovered from the sponge T. swinhoei from Palau, suggesting that sponges may indeed be a reservoir for this novel bacterial phylum (56).
The results of this study did not permit us to correlate the electron microscopic data with the molecular analysis results with absolute certainty. However, the following lines of evidence suggest that the “Poribacteria” candidate phylum does represent the nucleoid-containing morphotypes. (i) The “Poribacteria” lineage is moderately related to the Planctomycetes lineage, which is the only other phylum known for cell compartmentalization. (ii) The abundance of FISH signals correlated well with the abundance of the nucleoid-containing morphotypes as examined by electron microscopy. In both studies, the “Poribacteria” amounted to an estimated one-fourth to one-third of the total microbial community. (iii) Of all the FISH probes tested in this and previous studies, only the “Poribacteria”-specific FISH probes fluoresced in a ring-shaped manner. This is consistent with immunogold labeling studies by Fuerst et al. (17), who showed that, at least for morphotypes 1 to 5, the RNA is absent from the nucleoid. Whether morphotype 6 belongs to the “Poribacteria” or to another lineage such as the Gemmatimonadetes remains to be investigated.
The candidate phylum “Poribacteria” appears to be specifically affiliated with marine sponges, as samples taken from the immediate environment (seawater, sediments, and a filter-feeding invertebrate) were PCR negative. However, these results do not necessarily mean that the “Poribacteria” are absent from the environment; it suggests, rather, that the “Poribacteria” are concentrated in sponges. Negative PCR results can be explained if the concentrations in the environment are exceedingly low, if bacteria are only seasonally present, or if other seawater phylogenetic lineages mask the amplification of the poribacterial 16S rRNA gene sequences. PCR screening with specific primers indicated the presence of the “Poribacteria” in other sponge species (X. muta and T. swinhoei) and in sponges from different geographic regions (Palau [T. swinhoei], offshore California [A. fistularis], and by electron microscopic evidence, the Great Barrier Reef [17]). However, not all sponges are equally well suited for the maintenance of the “Poribacteria,” as some species gave negative PCR results (C. nucula, Agelas sp., and Ecytoplasia ferox).
The distribution of the candidate phylum “Poribacteria” is consistent with the general microbial pattern that has been described for several marine sponges (18, 19). Marine sponges have a unique microbial signature that consists of specific sequence clusters belonging to at least seven different known bacterial phyla. As a result of this study, the candidate phylum “Poribacteria” has been added to the list. To our knowledge, this is only the second report of cell compartmentalization for a prokaryotic lineage. With “Poribacteria”-specific PCR primers and FISH probes in hand, it is now possible to explore the abundance and distribution patterns of this elusive, sponge-specific candidate phylum.
Acknowledgments
We gratefully acknowledge the marine operations personnel at the Laboratoire Arago (Banyuls-sur-Mer, France) and the captain and crew of the R/V Seaward Johnson (HBOI, Fort Pierce, Fla.) for their expert help during sponge collection. We thank Christine Gernert (Universität Würzburg) for excellent technical assistance, J. Pawlik (UNC Wilmington) for cruise organization, and Feras Lafi (University of Queensland) for stimulating discussions during the 6th Sponge Conference.
Financial support for this work was provided by grant SFB567 to U.H. from the DFG.
REFERENCES
- 1.Althoff, K., C. Schütt, R. Steffen, R. Batel, and W. E. G. Müller. 1998. Evidence for a symbiosis between bacteria of the genus Rhodobacter and the marine sponge Halichondria panicea: harbor also for putatively toxic bacteria? Mar. Biol. 130:529-536. [Google Scholar]
- 2.Amann, R., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergquist, P. R. 1978. Sponges. University of California Press, Berkeley, Calif.
- 4.Böhm, M., H. C. Schroder, I. M. Muller, W. E. Muller, and V. Gamulin. 2000. The mitogen-activated protein kinase p38 pathway is conserved in metazoans: cloning and activation of p38 of the SAPK2 subfamily from the sponge Suberites domuncula. Biol. Cell 92:95-104. [DOI] [PubMed] [Google Scholar]
- 5.Brusca, R. C., and G. J. Brusca. 1990. Phylum Porifera: the sponges, p. 181-210. In A. D. Sinauer (ed.), Invertebrates. Sinauer Press, Sunderland, Mass.
- 6.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daims, H., A. Bruhl, R. Amann, K. H. Schleifer, and M. Wagner. 1999. The domain-specific probe EUB338 is insufficient for the detection of all bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434-444. [DOI] [PubMed] [Google Scholar]
- 8.Derakshani, M., T. Lukow, and W. Liesack. 2001. Novel bacterial lineages at the (sub)division level as detected by signature nucleotide-targeted recovery of 16S rRNA genes from bulk soil and rice roots of flooded rice microcosms. Appl. Environ. Microbiol. 67:623-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dojka, M. A., J. K. Harris, and N. R. Pace. 2000. Expanding the known diversity and environmental distribution of an uncultured phylogenetic division of bacteria. Appl. Environ. Microbiol. 66:1617-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dojka, M. A., P. Hugenholtz, S. K. Haack, and N. R. Pace. 1998. Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl. Environ. Microbiol. 64:3869-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunbar, J., S. Takala, S. M. Barns, J. A. Davis, and C. R. Kuske. 1999. Levels of bacterial community diversity in four arid soils compared by cultivation and 16S rRNA gene cloning. Appl. Environ. Microbiol. 65:1662-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedrich, A. B., H. Merkert, T. Fendert, J. Hacker, P. Proksch, and U. Hentschel. 1999. Microbial diversity in the marine sponge Aplysina cavernicola (formerly Verongia cavernicola) analyzed by fluorescence in situ hybridisation (FISH). Mar. Biol. 134:461-470. [Google Scholar]
- 13.Friedrich, A. B., J. Hacker, I. Fischer, P. Proksch, and U. Hentschel. 2001. Temporal variations of the microbial community associated with the Mediterranean sponge Aplysina aerophoba. FEMS Microbiol. Ecol. 38:105-113. [Google Scholar]
- 14.Fuerst, J. A. 1995. The planctomycetes: emerging models for microbial ecology, evolution and cell biology. Microbiology 141:1493-1506. [DOI] [PubMed] [Google Scholar]
- 15.Fuerst, J. A., H. G. Gwilliam, M. Lindsay, A. Lichanska, C. Belcher, J. E. Vickers, and P. Hugenholtz. 1997. Isolation and molecular identification of planctomycete bacteria from postlarvae of the giant tiger prawn, Penaeus monodon. Appl. Environ. Microbiol. 63:254-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuerst, J. A., R. I. Webb, M. J. Garson, L. Hardy, and H. M. Reiswig. 1998. Membrane-bounded nucleoids in microbial symbionts of marine sponges. FEMS Microbiol. Lett. 166:29-34. [Google Scholar]
- 17.Fuerst, J. A., R. I. Webb, M. J. Garson, L. Hardy, and H. M. Reiswig. 1999. Membrane-bounded nuclear bodies in a diverse range of microbial symbionts of Great Barrier Reef sponges. Memoirs Queensland Museum 44:193-203. [Google Scholar]
- 18.Hentschel, U., J. Hopke, M. Horn, A. B. Friedrich, M. Wagner, and B. S. Moore. 2002. Molecular evidence for a uniform microbial community in sponges from different oceans. Appl. Environ. Microbiol. 68:4431-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hentschel, U., L. Fieseler, M. Wehrl, C. Gernert, M. Steinert, J. Hacker, and M. Horn. 2003. Microbial diversity of marine sponges, p. 60-88. In W. E. G. Müller (ed.), Molecular marine biology of sponges. Springer-Verlag, Heidelberg, Germany. [DOI] [PubMed]
- 20.Hooper, J. N. A., and R. W. M. van Soest. 2002. Systema Porifera. A guide to the classification of sponges, vol. 1. Plenum Publishers, New York, N.Y.
- 21.Hugenholtz, P., G. W. Tyson, R. I. Webb, A. M. Wagner, and L. L. Blackall. 2001. Investigation of candidate division TM7, a recently recognized major lineage of the domain Bacteria with no known pure-culture representatives. Appl. Environ. Microbiol. 67:411-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hugenholtz, P., C. Pitulle, K. L. Hershberger, and N. R. Pace. 1998. Novel division level bacterial diversity in a Yellowstone hot spring. J. Bacteriol. 180:366-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Imhoff, J. F., and R. Stöhr. 2003. Sponge-associated bacteria: general overview and special aspects of bacteria associated with Halichondria panicea, p. 35-56. In W. E. G. Müller (ed.), Molecular marine biology of sponges. Springer-Verlag, Heidelberg, Germany. [DOI] [PubMed]
- 25.Liesack, W., and E. Stackebrandt. 1992. Occurrence of novel groups of the domain Bacteria as revealed by analysis of genetic material isolated from an Australian terrestrial environment. J. Bacteriol. 174:5072-5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lindsay, M. R., R. I. Webb, and J. A. Fuerst. 1997. Pirellulosomes: a new type of membrane-bounded cell compartment in planctomycete bacteria of the genus Pirellula. Microbiology 143:739-748. [DOI] [PubMed] [Google Scholar]
- 27.Lopez, J. V., P. J. McCarthy, K. E. Janda, R. Willoughby, and S. A. Pomponi. 1999. Molecular techniques reveal wide phylogenetic diversity of heterotrophic microbes associated with Discodermia spp. (Porifera: Demospongiae). Memoirs Queensland Museum 44:329-341. [Google Scholar]
- 28.Loy, A., M. Horn, and M. Wagner. 2003. probeBase—an online resource for rRNA-targeted oligonucleotide probes. Nucleic Acids Res. 31:514-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Madrid, V. M., J. Y. Aller, R. C. Aller, and A. Y. Chistoserdov. 2001. High prokaryote diversity and analysis of community structure in mobile mud deposits off French Guiana: identification of two new bacterial candidate divisions. FEMS Microbiol. Ecol. 37:197-209. [Google Scholar]
- 30.Meinhold, M. 2003. M.S. thesis. Universität Würzburg, Würzburg, Germany.
- 31.Müller, W. E. G. 1998. Molecular phylogeny of eumetazoa: genes in sponges (Porifera) give evidence for the monophyly of animals. Prog. Mol. Subcell. Biol. 9:89-126. [DOI] [PubMed] [Google Scholar]
- 32.Müller, W., M. Bohm, V. Grebenjuk, A. Skorokhod, I. Muller, and V. Gamulin. 2002. Conservation of the positions of metazoan introns from sponges to humans. Gene 295:299-309. [DOI] [PubMed] [Google Scholar]
- 33.Mummey, D. L., and P. D. Stahl. 2003. Candidate division BD: phylogeny, distribution and abundance in soil ecosystems. Syst. Appl. Microbiol. 26:228-235. [DOI] [PubMed] [Google Scholar]
- 34.Neef, A., R. Amann, H. Schlesner, and K. H. Schleifer. 1998. Monitoring a widespread bacterial group: in situ detection of planctomycetes with 16S rRNA-targeted probes. Microbiology 144:3257-3266. [DOI] [PubMed] [Google Scholar]
- 35.Paster, B. J., F. E. Dewhirst, W. G. Weisburg, L. A. Tordoff, G. J. Fraser, R. B. Hespell, T. B. Stanton, L. Zablen, L. Mandelco, and C. R. Woese. 1991. Phylogenetic analysis of the spirochetes. J. Bacteriol. 173:6101-6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pile, A. J. 1997. Finding Reiswig's missing carbon: quantification of sponge feeding using dual-beam flow cytometry, p. 1403-1410. In H. A. Lessios and I. G. Macintyre (ed.), Proceedings of the 8th International Coral Reef Symposium, vol. 2. Smithsonian Tropical Research Institute, Balboa, Panama. [Google Scholar]
- 37.Pimentel-Elardo, S., M. Wehrl, A. B. Friedrich, P. J. Jensen, and U. Hentschel. 2003. Isolation of planctomycetes from Aplysina sponges. Aquat. Microb. Ecol. 33:239-245. [Google Scholar]
- 38.Preston, C. M., K. Y. Wu, T. F. Molinski, and E. F. DeLong. 1996. A psychrophilic crenarchaeon inhabits a marine sponge: Cenarchaeum symbiosum gen. nov., sp. nov. Proc. Natl. Acad. Sci. USA 93:6241-6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reiswig, H. 1974. Water transport, respiration and energetics of three tropical marine sponges. J. Exp. Mar. Biol. Ecol. 14:231-249. [Google Scholar]
- 40.Rottmann, M., H. C. Schroeder, M. Gramzow, K. Renneisen, B. Kurelec, A. Dorn, U. Friese, and W. E. Mueller. 1987. Specific phosphorylation of proteins in pore complex-laminae from the sponge Geodia cydonium by the homologous aggregation factor and phorbol ester. Role of protein kinase C in the phosphorylation of DNA topoisomerase II. EMBO J. 6:3939-3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 42.Schmid, M., U. Twachtmann, M. Klein, M. Strous, S. Juretschko, M. Jetten, J. W. Metzger, K. H. Schleifer, and M. Wagner. 2000. Molecular evidence for genus level diversity of bacteria capable of catalyzing anaerobic ammonium oxidation. Syst. Appl. Microbiol. 23:93-106. [DOI] [PubMed] [Google Scholar]
- 43.Thoms, C., M. Horn, W. Wagner, U. Hentschel, and P. Proksch. 2003. Monitoring microbial diversity and natural products profiles of the sponge Aplysina cavernicola following transplantation. Mar. Biol. 142:685-692. [Google Scholar]
- 44.Turon, X., J. Galera, and M. J. Uriz. 1997. Clearance rates and aquiferous systems in two sponges with contrasting life-history strategies. J. Exp. Zool. 278:22-36. [Google Scholar]
- 45.Vacelet, J. 1975. Étude en microscopie électronique de l'association entre bactéries et spongiaires du genre Verongia (Dictyoceratida). J. Microsc. Biol. Cell 23:271-288. [Google Scholar]
- 46.Vacelet, J., and C. Donadey. 1977. Electron microscope study of the association between some sponges and bacteria. J. Exp. Mar. Ecol. 30:301-314. [Google Scholar]
- 47.Vergin, K. L., E. Urbach, J. L. Stein, E. F. DeLong, B. D. Lanoil, and S. J. Giovannoni. 1998. Screening of a fosmid library of marine environmental genomic DNA fragments reveals four clones related to members of the order Planctomycetales. Appl. Environ. Microbiol. 64:3075-3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Webster, N., and R. T. Hill. 2001. The culturable microbial community of the Great Barrier Reef sponge Rhopaloeides odorabile is dominated by an α-proteobacterium. Mar. Biol. 138:843-851. [Google Scholar]
- 49.Webster, N. S., K. J. Wilson, L. L. Blackall, and R. T. Hill. 2001. Phylogenetic diversity of bacteria associated with the marine sponge Rhopaloeides odorabile. Appl. Environ. Microbiol. 67:434-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Webster, N. S., R. I. Webb, M. J. Ridd, R. T. Hill, and A. P. Negri. 2001. The effects of copper on the microbial community of a coral reef sponge. Environ. Microbiol. 31:19-31. [DOI] [PubMed] [Google Scholar]
- 51.Wilkinson, C. R. 1978. Microbial associations in sponges. I. Ecology, physiology and microbial populations of coral reef sponges. Mar. Biol. 49:161-167. [Google Scholar]
- 52.Wilkinson, C. R. 1978. Microbial associations in sponges. II. Numerical analysis of sponge and water bacterial populations. Mar. Biol. 49:169-176. [Google Scholar]
- 53.Wilkinson, C. R. 1978. Microbial associations in sponges. III. Ultrastructure of the in situ associations in coral reef sponges. Mar. Biol. 49:177-185. [Google Scholar]
- 54.Wehrl, M. 2001. M.S. thesis. Universität Würzburg, Würzburg, Germany.
- 55.Woese, C. R. 1987. Bacterial evolution. Microbiol. Rev. 51:221-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang, H., Y. Sekiguchi, S. Hanada, P. Hugenholtz, H. Kim, Y. Kamagata, and K. Nakamura. 2003. Gemmatimonas aurantiaca gen. nov., sp. nov., a gram-negative, aerobic, polyphosphate-accumulating micro-organism, the first cultured representative of the new bacterial phylum Gemmatimonadetes phyl. nov. Int. J. Syst. Evol. Microbiol. 53:1155-1163. [DOI] [PubMed] [Google Scholar]
- 57.Zheng, D., E. W. Alm, D. A. Stahl, and L. Raskin. 1996. Characterization of universal small-subunit rRNA hybridization probes for quantitative molecular microbial ecology studies. Appl. Environ. Microbiol. 62:4504-4513. [DOI] [PMC free article] [PubMed] [Google Scholar]