Abstract
Examination of supernatant fractions from broth cultures of Lactobacillus fermentum BR11 revealed the presence of a number of proteins, including a 27-kDa protein termed Sep. The amino-terminal sequence of Sep was determined, and the gene encoding it was cloned and sequenced. Sep is a 205-amino-acid protein and contains a 30-amino-acid secretion signal and has overall homology (between 39 and 92% identity) with similarly sized proteins of Lactobacillus reuteri, Enterococcus faecium, Streptococcus pneumoniae, Streptococcus agalactiae, and Lactobacillus plantarum. The carboxy-terminal 81 amino acids of Sep also have strong homology (86% identity) to the carboxy termini of the aggregation-promoting factor (APF) surface proteins of Lactobacillus gasseri and Lactobacillus johnsonii. The mature amino terminus of Sep contains a putative peptidoglycan-binding LysM domain, thereby making it distinct from APF proteins. We have identified a common motif within LysM domains that is shared with carbohydrate binding YG motifs which are found in streptococcal glucan-binding proteins and glucosyltransferases. Sep was investigated as a heterologous peptide expression vector in L. fermentum, Lactobacillus rhamnosus GG and Lactococcus lactis MG1363. Modified Sep containing an amino-terminal six-histidine epitope was found associated with the cells but was largely present in the supernatant in the L. fermentum, L. rhamnosus, and L. lactis hosts. Sep as well as the previously described surface protein BspA were used to express and secrete in L. fermentum or L. rhamnosus a fragment of human E-cadherin, which contains the receptor region for Listeria monocytogenes. This study demonstrates that Sep has potential for heterologous protein expression and export in lactic acid bacteria.
Lactobacilli have been engineered to express a variety of heterologous proteins of interest, including antigens, enzymes, and more recently therapeutic single-chain antibodies and pathogen receptors (5, 8, 12, 30). Certain features of lactobacilli make them attractive as recombinant protein delivery vehicles. These include their generally regarded as safe (GRAS) status; their presence in dairy, meat, and vegetable food fermentation processes; and their ability to reside naturally and persist on mucosal surfaces where they can exert certain beneficial effects to the host.
A number of protein expression and targeting systems in lactobacilli have been developed which allow accumulation of the protein either intracellularly, anchored to the cell surface, or in the extracellular environment. Protein secretion and surface anchoring systems rely on the utilization of functional elements of extracellular proteins to efficiently secrete and/or anchor the heterologous protein to the cell surface. The majority of extracellular proteins in gram-positive bacteria are at some stage anchored to the cell wall either via covalent or noncovalent cell wall binding domains. Covalent attachment to the cell wall or cytoplasmic membrane occurs via the carboxyterminal LPXTG-type or amino-terminal LXXC sorting signals, respectively (18). Noncovalent attachment to the cell wall or cell wall components occurs either via specific repetitive LysM, YG, or choline-binding GW or S-layer homology domains (3, 7, 18) or via nonspecific cationic domains (2, 31). Varying amounts of both covalent and noncovalent surface anchored proteins may be released into the environment due to cell wall turnover, cell lysis, or proteolytic events (4, 21, 23, 24).
Our research is geared towards the development of heterologous protein expression systems using the targeting regions of native lactobacilli surface proteins. Our model organism is Lactobacillus fermentum BR11, which is a vaginal tract isolate of a guinea pig that has been shown to be amenable to genetic manipulation (25). Members of the L. fermentum-Lactobacillus reuteri group are commonly found inhabitants of mucosal surfaces of mammals and are one of the predominant Lactobacillus species found in the intestines of humans. To date, we have characterized a number of different surface proteins from L. fermentum BR11 and tested them as heterologous protein fusion partners. These include the noncovalently anchored cystine binding surface protein, BspA, and the covalently anchored LPXTG-containing proteins Mlp and Rlp (10, 29, 30, 32). Here we report the identification and characterization of a novel abundant small exported protein (Sep) from L. fermentum BR11. This protein was found in the supernatant as well as being associated with cell surface of L. fermentum BR11. Sep was shown to be useful as a heterologous peptide fusion partner in L. fermentum BR11, Lactobacillus rhamnosus GG, and Lactococcus lactis MG1363.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
L. fermentum BR11 and L. rhamnosus GG (ATCC 53103) were grown on solid MRS medium (Oxoid, Basingstoke, United Kingdom) anaerobically or in standing liquid culture tubes. L. lactis MG1363 was grown at 30°C in M17 medium (Oxoid, Basingstoke, United Kingdom) supplemented with 0.5% (wt/vol) glucose (GM17). Escherichia coli JM109 was used in molecular cloning experiments. Ampicillin was used at a concentration of 100 or 200 μg per ml for E. coli, while erythromycin was used at concentrations of 500 to 750 μg per ml for E. coli, 10 μg per ml for L. fermentum and L. rhamnosus, and 5 μg per ml for L. lactis. Expression cassettes were introduced into L. fermentum using pJRS233 (20) or into L. rhamnosus and L. lactis using pGh9:ISS1 (14). Plasmids pUC18, pBluescript II (KS) and pGEM3zf were used for routine cloning.
N-terminal sequencing of Sep.
Proteins from the supernatant fraction of L. fermentum BR11 broth culture were concentrated using ice-cold 5% trichloroacetic acid and were separated using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). They were then electroblotted to an Immobilon-PSQ membrane (Millipore) similarly to that previously described (15), and the band corresponding to Sep was sequenced with a 470a protein sequencer (Applied Biosystems).
PCR amplification, cloning, and DNA sequencing of sep.
A degenerate oligonucleotide based on the N-terminal sequence of Sep was designed (Bam-N-term) (Table 1). This oligonucleotide was used in combination with an oligonucleotide specific for the pUC18 multiple cloning site (pUC-Bam) in a PCR using a plasmid preparation of the L. fermentum BR11 pUC18 genomic library as a template similarly to that described previously (30). Expand long-template polymerase was used according to manufacturer's instructions (Roche). The amplified products were cloned into pBluescript II KS (Stratagene) and sequenced. The sequence of a clone containing DNA encoding amino acids matching all of the Sep N-terminal sequence was used to design an oligonucleotide to obtain the sequence of the sep 5′ end and upstream region (AcmA-N-term). This oligonucleotide was used with pUC-Bam to amplify the L. fermentum BR11 pUC18 library and clone DNA upstream of sep. Finally the 1,463-bp DNA sequence of the sep locus was obtained by sequencing from a PCR product amplified using genomic DNA isolated from L. fermentum BR11 using oligonucleotides (SepUS-PCR and SepDS-PCR). To identify domains in Sep, its sequence was compared with sequences in the NCBI Conserved Domain Database (version 1.62) (http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml), which contains a collection of functional and/or structural domains derived from two sources, Smart and Pfam.
TABLE 1.
Oligonucleotides used in the present study
| Oligonucleotide | Nucleotide sequence (5′ to 3′)a | Amplified product |
|---|---|---|
| Bam-N-term | AAGGATCCGAYACNATHTAYACNGTNCA | sep 3′ and downstream |
| pUC-Bam | CTTGGATCCCTGCAGGTCGACTCTAG | sep 3′ or sep 5′ regions |
| AcmA-N-term | CAGGATCCTTGATCATACTGTTGTCTTTAGC | sep 5′ and upstream |
| SepUS-PCR | AATTCGCGCGAGCATCTC | Entire sep locus |
| SepDS-PCR | TGCGTTTGAATTATTGTTTGC | Entire sep locus |
| Nterm-US-Xba | ATATCTAGAAACCTTCCTGCTGACCT | sep 5′ end and upstream |
| Nterm-Pst-US | AAACTGCAGAGTGATGATGGTGATGATGATCGGTGTAGATAGTGTCAGCA | sep 5′ end and upstream |
| SepDS-PstXho | AAACTGCAGCAGGTTCTCGAGACACTATCTACACCGTACA | sep 3′ end and terminator |
| SepDS-ApaSal | CAGGGGCCCGTCGACCTATACCTGTCGAATCCA | sep 3′ end and terminator |
| E-cad-PstI | AGACCTGCAGGAGACTGGGTTATTCCTCCCA | E-cadherin-encoding region |
| E-cad-XhoI | AGACTCGAGGTTAATCGTTGGTGTCAGTGACTGT | E-cadherin-encoding region |
Underlining indicates restriction endonuclease recognition sites. Single-letter code: Y = C or T; H = A, C, or T; N = A, G, C, or T.
Transformation of L. fermentum, L. rhamnosus, and L. lactis with plasmids.
Transformation of L. fermentum and L. rhamnosus was done using penicillin as a cell wall-weakening agent at concentrations of 1.25 or 10 μg per ml, respectively, as described previously (16, 25). L. lactis was transformed using 1% glycine as a cell wall-weakening agent as described previously (9), except that transformants were selected directly on GM17 plates containing erythromycin at 5 μg per ml. L. lactis transformants (containing pGh9:ISS1 derivatives) were grown at 30°C while L. rhamnosus transformants (containing pGh9:ISS1 derivatives) were grown on plates at 30°C and in liquid at 30 or 37°C. pJRS233 plasmid derivatives integrated into the chromosome of L. fermentum were selected for by incubation at 40°C in the presence of erythromycin.
Construction of Sep expression cassettes.
The first construct (Sep-6xHis-Sep) consists of DNA upstream of sep and the sep 5′ region encoding the secretion signal and a six-histidine (His6) epitope (amplified and cloned using Nterm-US-Xba and Nterm-Pst-US) and DNA encoding the mature Sep protein and the putative sep transcription terminator (amplified and cloned using SepDS-PstXho and SepDS-ApaSal). The second construct (BspA-6xHis-Sep) consists of DNA encoding the mature Sep protein and putative sep transcription terminator as described above but instead contains upstream DNA encoding a full-length BspA protein followed by DNA encoding the BspA secretion signal and a His6 epitope as described previously (30). The full-length bspA gene is required to allow integration of the expression cassette downstream of the bspA promoter (30). These expression cassettes were constructed in pBluescript II and then cloned into the XbaI- and SalI-digested pJRS233. The Sep-6xHis-Sep construct in pBluescript II was also digested with SalI and cloned into XhoI-digested pGh9:ISS1. Note that all the modified pGh9:ISS1 plasmids used in this study contain both the expression construct as well as the full pBluescript II plasmid thereby allowing it to replicate in E. coli at 37°C.
Construction of E-cadherin expression cassettes.
The region encoding the amino-terminal 1 to 216 amino acids of the mature E-cadherin protein was amplified by PCR from cDNA template prepared from cultured mammalian T47D and LNCap cells using oligonucleotides E-cad-PstI and E-cad-XhoI. This fragment was cloned in frame downstream of DNA encoding either the Sep or BspA secretion signals to generate constructs Sep-6xHis-Ecad and BspA-6xHis-Ecad, respectively. The sequence of the cloned E-cadherin DNA fragment which contained an introduced stop codon after codon 216 was checked by DNA sequencing. The putative bspA transcription terminator was amplified using oligonucleotides Term-Xho and Term-Hind (30) and cloned downstream of the E-cadherin encoding DNA.
Cell fractionation, protein extraction, Western blot analysis, and cell surface display assay.
Cell extracts were prepared from late-exponential- or early-stationary-phase cultures, while supernatants were taken from late-exponential-phase cultures. Two different whole-cell protein extraction methods which involved either boiling cells in 2× SDS-PAGE loading buffer (26) or sonication were used as described previously (30). LiCl (5 M) extractions of cells and supernatant fractions were also obtained as described previously (30). Prior to loading the gels for SDS-PAGE, all samples were boiled for 5 min. Proteins were transferred to nitrocellulose, blocked, and then probed with an anti-His5 monoclonal antibody (Qiagen, Hilden, Germany) at 1 in 1,000 dilution. Following washes, the membrane was incubated with rabbit anti-mouse-horseradish peroxidase conjugate (Dako, Glostrup, Denmark). The bound antibodies were detected using the HRP chemiluminescence kit (Roche, Mannheim, Germany). To estimate levels of His6 proteins in extracts, various amounts of His6-labeled protein markers (Qiagen, Glostrup, Denmark) were included alongside the samples. These markers have known quantities of His6-containing proteins in each band, allowing densitometry to be done on films using the TotalLab package (version 1.11; Phoretix, Newcastle upon Tyne, United Kingdom). For detection of E-cadherin a mouse monoclonal anti-human E-cadherin antibody (from clone HECD-1; Zymed Laboratories Inc.) was used at a concentration of 1 in 750. The accessibility of the His6 epitope on whole cells was done the same as that described previously (30).
Nucleotide sequence accession number.
The 1,463-bp nucleotide sequence of the sep locus has been deposited to GenBank under accession number AY486145.
RESULTS
Identification and characterization of an abundant protein from the L. fermentum BR11 culture supernatant.
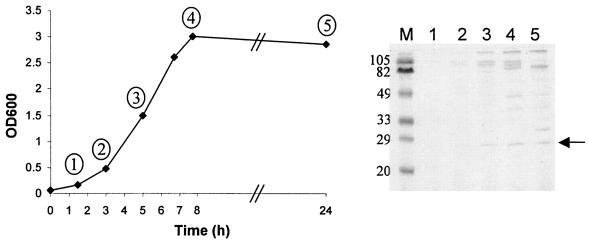
L. fermentum BR11 was grown in standing MRS broth at 37°C for 24 h, and fractions were taken at five time points. SDS-PAGE analysis revealed a number of proteins which accumulated in the supernatant during growth (Fig. 1). The smallest visible protein larger than 20 kDa was still abundant in late stationary phase and was called Sep for small exported protein. To further characterize Sep we identified the N-terminal sequence which was found to be DTIYTVQSGDTLSGI. Using PCR, the sep gene and surrounding regions were amplified and sequenced (see Materials and Methods).
FIG. 1.
Analysis of proteins found in the culture supernatant of L. fermentum BR11 grown in MRS broth. Growth of L. fermentum BR11 was monitored over 24 h by optical density measurements at 600 nm. At various time points, indicated by a number in a circle, aliquots were taken and centrifuged, and the supernatant was filtered and precipitated with 5% trichloroacetic acid. The equivalent of 225 μl of culture supernatant was analyzed by SDS-PAGE followed by staining with Coomassie brilliant blue G-250. The arrow indicates Sep.
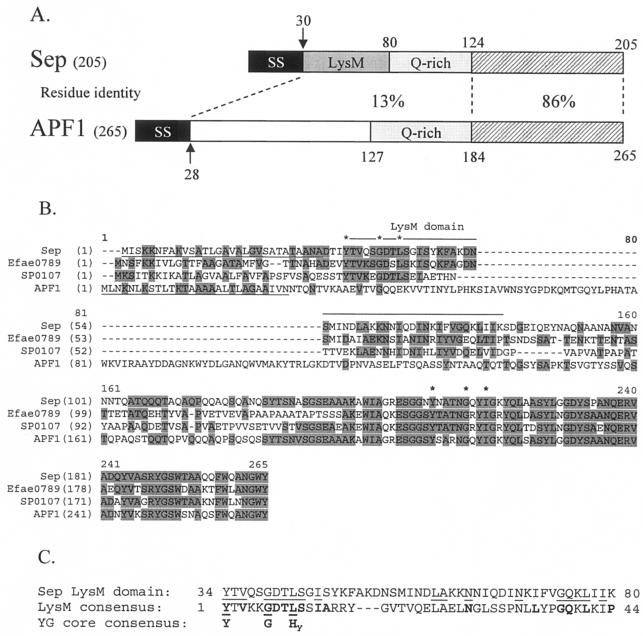
Sep is a 205-amino-acid protein with a 30-amino-acid amino terminal secretion signal giving rise to a predicted 19-kDa mature protein with an isoelectric point of 5.3 (Fig. 2A). Sep has greatest overall similarity to proteins from L. reuteri (Lre0018 [35] is 203 amino acids and 92% identical), Enterococcus faecium (Efae0789 is 202 amino acids long and 50% identical), and Streptococcus pneumoniae (SP0107 is 195 amino acids long and 46% identical) (Fig. 2B). It also has similarity to similarly sized proteins from Lactobacillus plantarum (lp_0304 is 212 amino acids long and 46% identical), Streptococcus agalactiae (gbs2107 is 179 amino acids long and 39% identical), Leuconostoc mesenteroides (Lmes0554 is 212 amino acids long and 36% identical), and Oenococcus oeni (Ooen0238 is 200 amino acids long and 31% identical). All of these proteins contain typical secretion signals and are therefore predicted to be extracellular. Interestingly, there is also localized high similarity between the carboxy termini of Sep and its homologs and the carboxy termini of the recently characterized aggregation-promoting factor (APF) surface proteins (33) from Lactobacillus johnsonii and Lactobacillus gasseri (86% identical over the carboxy-terminal 81 amino acids) (Fig. 2A and B).
FIG. 2.
(A) Amino acid identity between different regions of Sep from L. fermentum BR11 and APF1 from L. johnsonii ATCC 11506. SS indicates a secretion signal, LysM indicates a LysM domain, Q-rich indicates regions rich in glutamine amino acids, and the hatched boxes indicate the highly homologous carboxy-terminal regions. The arrows indicate cleavage sites of the secretion signals. Note: APF1 does not contain a LysM domain. (B) Multiple alignment of Sep, Efae0789 from E. faecium, SP0107 from S. pneumoniae, and APF1 from L. johnsonii ATCC 11506. Shaded residues indicate identity with the consensus sequence determined using the AlignX program in Vector NTI Suite 6.0. The secretion signals of Sep and APF1 are underlined. The amino-terminal LysM domains found in Sep, Efae0789, and SP0107, but not in APF1, are indicated by a line above the sequence (note: there is one LysM domain for these proteins which extends over the alignment gap). The core residues found in the YG motif in the amino-terminal LysM domains and near the carboxy termini are indicated by asterisks. (C) Alignment of the amino-terminal LysM domain of Sep with the consensus LysM domain pfam01476 (see text) and the core amino acids found in YG domains. Letters in boldface type indicate the consensus sequence amino acids for the LysM and YG core domains. Hy stands for a hydrophobic amino acid. Lines indicate amino acid matches.
A search of the NCBI conserved domain database revealed a match with the amino-terminal 34 to 80 amino acids of Sep (amino acids 4 to 50 of mature Sep) and the pfam01476 LysM domain (E = 3 × 10−7) (Fig. 2A and C). LysM or lysin motif domains are close to 40 amino acids in length, are found in a number of peptidoglycan-degrading enzymes, and are thought to function as peptidoglycan binding domains (3). The amino-terminal LysM domain is present in the proteins likely to be true homologs of Sep but is not present in the related but distinct APF proteins (Fig. 2B).
In the course of this study, we recognized that the conserved features of LysM domains display a close similarity to the conserved features of the YG repeat units found in the carbohydrate or cell wall binding domains of streptococcal glucosyltransferases and glucan binding proteins, Clostridium difficile toxins and the S. pneumoniae Psp surface proteins (7, 34, 36). YG repeats are 20 to 21 residues in length and conform to the consensus sequence (N/D/G×4)(AroAro)YXXXXGXXHydXSpX(HydHydHyd) where X represents a poorly conserved residue; (N/D/G×4) represents four residues enriched for N, D, and G; (AroAro) represents two residues of which at least one is aromatic; Hyd represents a hydrophobic residue; and (HydHydHyd) represents three residues of which at least one but more usually two are hydrophobic. The YXXXXGXXHyd motif is normally very highly conserved although in the case of C. difficile toxins, the conserved G is less conserved than in the other sequences and is often replaced by the polar residue S, N, or T. The LysM consensus sequence possesses the YXXXXGXXHyd motif (Fig. 2C), and it appears that the LysM domain is a variant or subgroup of the broader family of YG repeats. Sep and its homologs are unusual in that they contain a LysM domain at their amino termini and a YG sequence that is not specifically related to the LysM domain at their carboxy termini (Fig. 2B). This suggests that these molecules have two different binding functions. In contrast, the APF proteins possess only the carboxy-terminal YG repeat.
Upstream of sep and in the same orientation is an open reading frame which encodes a protein similar to predicted hydrolases of the HAD superfamily, including lp_2759 from L. plantarum (45% identical over 118 amino acids). There is no putative transcription terminator between this open reading frame and sep, indicating that these genes may be cotranscribed. These genes are separated by a reasonably large intergenic region (310 bp) which may contain as yet unknown expression elements. Eleven nucleotides upstream of the sep start codon is a putative ribosome binding site (AGGATGG), and immediately downstream of sep is a stem-loop resembling a rho-independent transcription terminator.
Determination of the subcellular location of a His6-Sep fusion protein in L. fermentum BR11, L. rhamnosus GG, and L. lactis MG1363.
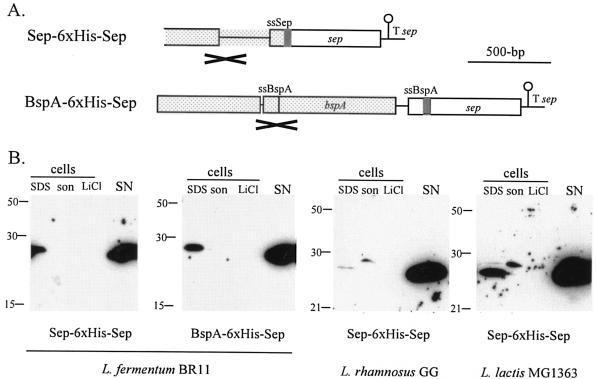
To facilitate the detection of Sep in culture supernatants and subcellular fractions, chimeric genes were constructed that encoded Sep derivatives in which a His6 epitope had been inserted between the secretion signal and the amino terminus of the mature protein (Fig. 3A). Both the Sep and BspA secretion signals were used; the extra amino acids added onto the mature N termini of Sep in the Sep-6xHis-Sep construct were DTIYTDHHHHHHSAAGSR, and those in the BspA-6xHis-Sep construct were ASDDVHHHHHHSAAGSR.
FIG. 3.
Expression and subcellular location of a His6-Sep fusion protein in L. fermentum BR11, L. rhamnosus GG, and L. lactis MG1363. (A) Arrangement of the constructs which were introduced into L. fermentum, L. rhamnosus, or L. lactis. The sep terminator (T sep) and DNA encoding the Sep secretion signal (ssSep), BspA secretion signal (ssBspA), and His6 (grey box) are indicated. The DNA region which is the site of single-crossover homologous recombination into either the sep or bspA loci of L. fermentum BR11 is spotted and is marked with a cross below. (B) Western blot detection of fusion proteins in cell extracts and in the supernatant using an anti-His5 antibody. The strains and constructs are indicated at the bottom of each blot. Sizes of molecular mass markers are indicated in kilodaltons on the left. The lanes containing cell extracts prepared by boiling in 2× SDS-loading dye (SDS), by sonication (son), and with 5 M LiCl (LiCl) and the precipitated supernatant fractions (SN) are indicated. The amount of cells or medium loaded in each lane is the equivalent to 500 μl (SDS), 50 μl (son), 160 μl (LiCl), and 675 μl (SN) of culture.
Protein extracts from culture supernatant or cell fractions from strains containing the Sep-6xHis-Sep and the BspA-6xHis-Sep constructs were analyzed by Western blot using an anti-His5 antibody (Fig. 3B). The predicted molecular mass of the mature His6-Sep fusion proteins is 21 kDa, although the bands in Western blots correspond to proteins 28 kDa in size (Fig. 3B). From Fig. 1 it can be seen that the 19-kDa Sep resolves aberrantly more slowly in SDS-PAGE at 27 kDa. In L. fermentum containing the Sep-6xHis-Sep and the BspA-6xHis-Sep constructs, the Sep fusion protein was found predominantly in the supernatant at levels of ∼2 mg per liter of culture in both cases. Levels of the Sep fusion protein in the SDS cell extracts for L. fermentum containing the Sep-6xHis-Sep and BspA-6xHis-Sep constructs were ∼9 and ∼13% of that found in the supernatants, respectively. No Sep fusion protein was detected in sonicate or 5 M LiCl extracts. When SDS cell and supernatant extracts were run in neighboring lanes, the Sep fusion protein bands migrated identically on SDS-PAGE (data not shown), which suggests that the Sep fusion protein associated with cells is the mature form and therefore does not contain a signal sequence and is located outside the cytoplasmic membrane.
In L. rhamnosus and L. lactis, levels of Sep fusion protein expressed from the Sep-6xHis-Sep construct were found at concentrations of ∼200 and ∼300 μg per liter of culture in the supernatant, respectively (Fig. 3B). Levels of the Sep fusion protein in the SDS cell extracts for L. rhamnosus and L. lactis were <1 and ∼10% of that found in the supernatants, respectively. Interestingly a slightly higher molecular weight His6 reactive protein was observed in the sonicate cell extracts of both L. rhamnosus and L. lactis and in the SDS cell extract of L. lactis. This band most likely corresponds to Sep fusion protein still containing its secretion signal. Like L. fermentum, no His6-Sep was detected in the 5 M LiCl extracts of either L. rhamnosus or L. lactis containing the Sep-6xHis-Sep construct.
To examine if Sep is exposed on the cell surface of L. fermentum, a whole-cell enzyme-linked immunosorbent assay for the His6 epitope was performed as described previously (30). It was found that the A405 signal per optical density unit (optical density at 600 nm) of cells obtained for L. fermentum cells containing the BspA-6xHis-Sep construct (0.0302 ± 0.0003) was significantly greater (1.8-fold) than that for L. fermentum BR11 cells (0.0168 ± 0.0007). This result suggests that at least some cell-associated Sep is located in an exposed form in the cell envelope of L. fermentum.
Expression and secretion of an E-cadherin peptide by L. fermentum and L. rhamnosus using Sep or BspA.
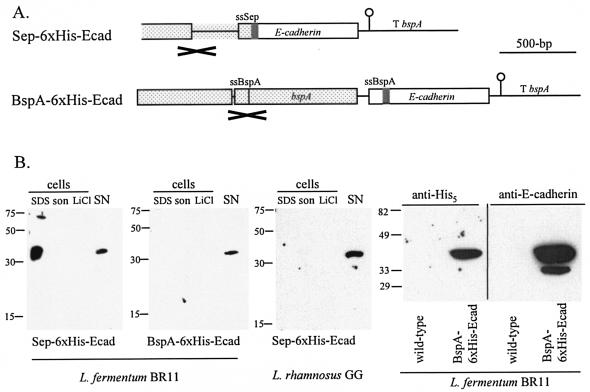
DNA encoding a human E-cadherin (Ecad) fragment was cloned in frame downstream of DNA encoding either the Sep or BspA secretion signals (Fig. 4A). The distribution and levels of His6-tagged E-cadherin protein were examined by Western blotting (Fig. 4B). For the L. fermentum strain containing the Sep-6xHis-Ecad construct, E-cadherin fusion protein was detected in the SDS cell extract (∼370 μg per liter of culture) and supernatant (∼30 μg per liter of culture) from cells grown at 30°C. No E-cadherin fusion protein was detected, however, in extracts from this strain when it was grown at 40°C. For the L. fermentum strain containing the BspA-6xHis-Ecad construct, E-cadherin fusion protein was detected in the supernatant (∼30 μg per liter of culture) from cells grown at 40°C. For the L. rhamnosus strain containing the Sep-6xHis-Ecad construct, E-cadherin fusion protein was detected in supernatant (∼16 μg per liter of culture). Buffering of the growth medium has been shown to stabilize heterologous proteins expressed in lactic acid bacteria (19, 27). An experiment was performed where L. fermentum containing the BspA-6xHis-Ecad construct was grown to exponential phase in either buffered MRS medium (containing 0.2 M potassium phosphate, pH 6.8) or unbuffered MRS medium, and the level of E-cadherin fusion protein in the supernatant was determined. Surprisingly, no E-cadherin fusion protein was detected in the supernatant of the buffered medium, but the protein was detected at normal levels as mentioned previously in the supernatant of the unbuffered medium (data not shown).
FIG. 4.
Expression and secretion of a human E-cadherin fusion protein by L. fermentum BR11 and L. rhamnosus GG. (A) Arrangement of the constructs which were introduced into L. fermentum or L. rhamnosus. The bspA terminator (T bspA) and DNA encoding the Sep secretion signal (ssSep), BspA secretion signal (ssBspA), and His6 (grey box) are indicated. The DNA region which is the site of single-crossover homologous recombination into either the sep or bspA loci of L. fermentum BR11 is spotted and is marked with a cross below. (B) Western blots of fusion proteins in cell extracts and in the supernatant using an anti-His5 antibody, except for the rightmost blot for which an anti-E-cadherin antibody was used. The strains and constructs are indicated at the bottom of each blot. The sizes of the molecular mass markers are indicated on the left. The lanes containing cell extracts prepared by boiling in 2× SDS-loading dye (SDS), by sonication (son), and with 5 M LiCl (LiCl) and the precipitated supernatant fractions (SN) are indicated. The amounts of cells or medium loaded in each lane are the equivalent to 500 μl (SDS), 50 μl (son), 160 μl (LiCl), and 675 μl (SN) of culture. For the two rightmost blots, the equivalent of 1.2 ml of culture supernatant from L. fermentum (wild-type) or L. fermentum containing BspA-6xHis-Ecad (BspA-6xHis-Ecad) was loaded in each lane.
The predicted sizes of the two E-cadherin fusion proteins are 25 kDa; however, the protein recognized by the anti-His5 antibody in the Western blots resolved at ∼38 kDa. To confirm that this protein is indeed E-cadherin, a mouse monoclonal anti-human E-cadherin antibody was used as the primary antibody in a Western blot. Figure 4B (rightmost blot) shows that the anti-E-cadherin antibody recognized a protein the same size as the protein recognized using the anti-His5 antibody. The anti-E-cadherin antibody does not recognize proteins found in the supernatant of the parent L. fermentum BR11 strain. Interestingly there is a smaller protein recognized by the anti-E-cadherin antibody but not by the anti-His5 antibody suggesting that it may be a proteolytically degraded E-cadherin product which does not contain the amino-terminal His6 epitope.
DISCUSSION
This report describes the identification and characterization of the extracellular protein Sep from L. fermentum BR11. Sep was found predominantly in the supernatant but was also associated with the cell surface of L. fermentum BR11. Examination of the sequence of Sep for cell surface anchoring domains revealed that it does not contain any typical covalent anchoring signals such as cell wall-anchoring LPXTG or lipoprotein LXXC signals but instead does contain a single LysM domain at its amino terminus. LysM domains are typically found repeated a number of times in noncovalently anchored surface proteins such as in the autolysins AcmA (contains three LysM domains) and p60 (contains two LysM domains) of L. lactis and Listeria monocytogenes, respectively (3, 4). Recently it has been shown that the three carboxyterminal LysM repeats of the AcmA protein bind to peptidoglycan from a variety of gram-positive bacteria and that this binding is hindered by other cell wall constituents, possibly lipoteichoic acid (28). We identify here that the conserved features of LysM domains display a close similarity to the conserved features of YG units (7, 34, 36). This is probably not overly surprising since LysM domains and YG repeats are both functionally similar in that they both recognize carbohydrates as ligands.
Thus far, the proteins that have been identified as having the greatest similarity to Sep and sharing the same domain structure were mostly from genome sequences and therefore have yet to be fully studied. Comparisons can, however, be drawn with the recently characterized APF surface proteins of L. johnsonii and L. gasseri (11, 33). Sep and the APF proteins have very similar carboxy termini (86% over 81 amino acids) but very different amino termini, and unlike Sep, APF does not have an amino-terminal LysM domain. APF was identified originally as one of the most abundant proteins found in the culture supernatant of L. gasseri 4B2 (formally L. plantarum 4B2) and was hypothesized as being involved in cell aggregation (22). Recent work has since shown that APF proteins are also associated with the cell surface and that overproduction or down-regulation of their synthesis can cause dramatic changes in the bacterial cell shape (11). We have found that L. fermentum cells containing the BspA-6xHis-Sep construct and L. rhamnosus and L. lactis cells containing the Sep-6xHis-Sep construct did not appear different from the parent strains as examined by light microscopy (data not shown). This suggests that Sep does not have a significant influence on cell shape and also that the different amino termini of Sep and APF probably impart different functions to these two proteins.
The question of how Sep and the APF surface proteins are anchored to the cell surface is as yet unresolved. Interestingly, it was found using Western blot analysis that cell extracts from L. gasseri 4B2 contain a 10-kDa fragment of APF2 that is recognized by carboxy-terminus-specific anti-APF antibodies (11). This means that a fragment consisting of the carboxy-terminal one-third of APF2, which has high similarity to the carboxy terminus of Sep, is associated with the cell and probably mediates the cell surface anchoring. Upon examination of the carboxy terminus of APF proteins, Sep, and Sep homologs, we identified the presence of a YG sequence that is not specifically related to LysM domains. This suggests that the carboxy termini of Sep and its homologs are involved, at least in part, in cell surface anchoring and therefore the amino-terminal LysM domain may function other than to anchor these proteins to the cell surface. Physical properties, including isoelectric point, may affect nonspecific cell surface anchoring properties and extractability of the Sep and APF surface proteins. The APF proteins (pI ∼9) are much more positively charged than Sep (pI 5.3) and therefore may be able to interact electrostatically with the negatively charged cell envelope teichoic acid. Consistent with this, cell-bound APF proteins can be extracted from whole cells using 5 M LiCl (33), while cell- bound Sep cannot be extracted in detectable levels from whole cells using 5 M LiCl. Other Lactobacillus surface proteins which have high isoelectric points and can be selectively extracted using 5 M LiCl include S-layer subunits (pI > 9) and BspA (pI 10.6).
Sep was tested as a potential heterologous peptide fusion partner in the native host L. fermentum as well as foreign hosts, L. rhamnosus and L. lactis. The Sep fusion protein was found predominantly in the supernatant in all three hosts. It is possible that cell-associated Sep fusion protein was not efficiently removed using the extraction methods or that the His6 epitope in the Sep fusion protein was more prone to proteolytic degradation when closely associated with the cell envelope. Expression of His6-Sep from the Sep-6xHis-Sep construct in L. rhamnosus and L. lactis suggests that an active promoter is located in the 625-bp DNA upstream of sep. This result also shows that the Sep secretion signal is efficiently recognized in lactobacilli and Lactococcus. To our knowledge this is the first report of the expression and secretion of a heterologous protein in the probiotic strain L. rhamnosus GG. This strain has been shown to have health-promoting properties and is currently marketed as an active ingredient in a number of commercial foodstuffs (1). The results obtained here describe tools which can be used to drive expression and secretion of heterologous proteins in this organism and therefore have potential for the development of orally delivered recombinant proteins of interest.
Sep or BspA was shown to direct the expression and secretion of the human E-cadherin amino-terminal region (amino acids 1 to 216 of the mature protein) in L. fermentum or L. rhamnosus. The amino terminus of this protein has been shown to be the major intestinal cell receptor for the food-borne disease causing pathogen L. monocytogenes (13, 17) and therefore may have potential as an intestinal L. monocytogenes attachment-inhibiting therapeutic. Recently a recombinant E-cadherin peptide (amino acids 1 to 192) expressed and purified from E. coli was shown to inhibit the uptake of L. monocytogenes into Caco-2 cells (6). We calculated that L. fermentum secreted significantly lower levels of E-cadherin fusion protein than Sep fusion protein. The reason for the low level of E-cadherin detected in the supernatant of L. fermentum is not known; however, the presence of multiple bands detected by the anti-E-cadherin antibody suggests that this protein is susceptible to proteolytic degradation. Indeed we have found that this E-cadherin fragment is proteolytically degraded when fused to the carboxy terminus of BspA (data not shown). In the future, experiments to stabilize this protein in culture supernatants could be investigated.
Acknowledgments
We thank Steve Myers for preparing the cDNA from mammalian cells, Tara Veveris-Lowe and Judith Clements for the anti-E-cadherin antibody, Wade Louwrens for preparing the genomic DNA from L. fermentum, Chris Wood for N-terminal sequencing, Scott Chandry for supplying L. lactis, and Alexandra Gruss for supplying pGh9:ISS1.
This work was supported by Dairy Australia (grant QUT10723), a QUT Encouragement Award, and QUT Faculty of Science and School of Life Sciences Support.
REFERENCES
- 1.Alvarez-Olmos, M. I., and R. A. Oberhelman. 2001. Probiotic agents and infectious diseases: a modern perspective on a traditional therapy. Clin. Infect. Dis. 32:1567-1576. [DOI] [PubMed] [Google Scholar]
- 2.Antikainen, J., L. Anton, J. Sillanpaa, and T. K. Korhonen. 2002. Domains in the S-layer protein CbsA of Lactobacillus crispatus involved in adherence to collagens, laminin and lipoteichoic acids and in self-assembly. Mol. Microbiol. 46:381-394. [DOI] [PubMed] [Google Scholar]
- 3.Bateman, A., and M. Bycroft. 2000. The structure of a LysM domain from E. coli membrane-bound lytic murein transglycosylase D (MltD). J. Mol. Biol. 299:1113-1119. [DOI] [PubMed] [Google Scholar]
- 4.Buist, G., J. Kok, K. J. Leenhouts, M. Dabrowska, G. Venema, and A. J. Haandrikman. 1995. Molecular cloning and nucleotide sequence of the gene encoding the major peptidoglycan hydrolase of Lactococcus lactis, a muramidase needed for cell separation. J. Bacteriol. 177:1554-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, T. L., C. H. Chang, D. A. Simpson, Q. Xu, P. K. Martin, L. A. Lagenaur, G. K. Schoolnik, D. D. Ho, S. L. Hillier, M. Holodniy, J. A. Lewicki, and P. P. Lee. 2003. Inhibition of HIV infectivity by a natural human isolate of Lactobacillus jensenii engineered to express functional two-domain CD4. Proc. Natl. Acad. Sci. USA 100:11672-11677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.da Silva Tatley, F., F. E. Aldwell, A. K. Dunbier, and P. J. Guilford. 2003. N-terminal E-cadherin peptides act as decoy receptors for Listeria monocytogenes. Infect. Immun. 71:1580-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giffard, P. M., and N. A. Jacques. 1994. Definition of a fundamental repeating unit in streptococcal glucosyltransferase glucan-binding regions and related sequences. J. Dent. Res. 73:1133-1141. [DOI] [PubMed] [Google Scholar]
- 8.Havenith, C. E. G., J. F. M. L. Seegers, and P. H. Pouwels. 2002. Gut-associated lactobacilli for oral immunisation. Food Res. Int. 35:151-163. [Google Scholar]
- 9.Holo, H., and I. F. Nes. 1989. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl. Environ. Microbiol. 55:3119-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hung, J., C. Rathsam, N. A. Jacques, and P. M. Giffard. 2002. Expression of a streptococcal glucosyltransferase as a fusion to a solute-binding protein in Lactobacillus fermentum BR11. FEMS Microbiol. Lett. 211:71-75. [DOI] [PubMed] [Google Scholar]
- 11.Jankovic, I., M. Ventura, V. Meylan, M. Rouvet, M. Elli, and R. Zink. 2003. Contribution of aggregation-promoting factor to maintenance of cell shape in Lactobacillus gasseri 4B2. J. Bacteriol. 185:3288-3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kruger, C., Y. Hu, Q. Pan, H. Marcotte, A. Hultberg, D. Delwar, P. J. van Dalen, P. H. Pouwels, R. J. Leer, C. G. Kelly, C. van Dollenweerd, J. K. Ma, and L. Hammarstrom. 2002. In situ delivery of passive immunity by lactobacilli producing single-chain antibodies. Nat. Biotechnol. 20:702-706. [DOI] [PubMed] [Google Scholar]
- 13.Lecuit, M., S. Dramsi, C. Gottardi, M. Fedor-Chaiken, B. Gumbiner, and P. Cossart. 1999. A single amino acid in E-cadherin responsible for host specificity towards the human pathogen Listeria monocytogenes. EMBO J. 18:3956-3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maguin, E., H. Prevost, S. D. Ehrlich, and A. Gruss. 1996. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J. Bacteriol. 178:931-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsudaira, P. T. 1989. A practical guide to protein and peptide purification for microsequencing. Academic Press, New York, N.Y.
- 16.McCracken, A., M. S. Turner, P. Giffard, L. M. Hafner, and P. Timms. 2000. Analysis of promoter sequences from Lactobacillus and Lactococcus and their activity in several Lactobacillus species. Arch. Microbiol. 173:383-389. [DOI] [PubMed] [Google Scholar]
- 17.Mengaud, J., H. Ohayon, P. Gounon, R.-M. Mege, and P. Cossart. 1996. E-cadherin is the receptor for internalin, a surface protein required for entry of L. monocytogenes into epithelial cells. Cell 84:923-932. [DOI] [PubMed] [Google Scholar]
- 18.Navarre, W. W., and O. Schneewind. 1999. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol. Mol. Biol. Rev. 63:174-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perez-Arellano, I., and G. Perez-Martinez. 2003. Optimization of the green fluorescent protein (GFP) expression from a lactose-inducible promoter in Lactobacillus casei. FEMS Microbiol. Lett. 222:123-127. [DOI] [PubMed] [Google Scholar]
- 20.Perez-Casal, J., J. A. Price, E. Maguin, and J. R. Scott. 1993. An M protein with a single C repeat prevents phagocytosis of Streptococcus pyogenes: use of a temperature-sensitive shuttle vector to deliver homologous sequences to the chromosome of S. pyogenes. Mol. Microbiol. 8:809-819. [DOI] [PubMed] [Google Scholar]
- 21.Piard, J. C., I. Hautefort, V. A. Fischetti, S. D. Ehrlich, M. Fons, and A. Gruss. 1997. Cell wall anchoring of the Streptococcus pyogenes M6 protein in various lactic acid bacteria. J. Bacteriol. 179:3068-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reniero, R., P. Cocconcelli, V. Bottazzi, and L. Morelli. 1992. High frequency of conjugation in Lactobacillus mediated by an aggregation-promoting factor. J. Gen. Microbiol. 138:763-768. [Google Scholar]
- 23.Rojas, M., F. Ascencio, and P. L. Conway. 2002. Purification and characterization of a surface protein from Lactobacillus fermentum 104R that binds to porcine small intestinal mucus and gastric mucin. Appl. Environ. Microbiol. 68:2330-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roos, S., and H. Jonsson. 2002. A high-molecular-mass cell-surface protein from Lactobacillus reuteri 1063 adheres to mucus components. Microbiology 148:433-442. [DOI] [PubMed] [Google Scholar]
- 25.Rush, C. M., L. M. Hafner, and P. Timms. 1994. Genetic modification of a vaginal strain of Lactobacillus fermentum and its maintenance within the reproductive tract after intravaginal administration. J. Med. Microbiol. 41:272-278. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Schotte, L., L. Steidler, J. Vandekerckhove, and E. Remaut. 2000. Secretion of biologically active murine interleukin-10 by Lactococcus lactis. Enzyme Microb. Technol. 27:761-765. [DOI] [PubMed] [Google Scholar]
- 28.Steen, A., G. Buist, K. J. Leenhouts, M. El Khattabi, F. Grijpstra, A. L. Zomer, G. Venema, O. P. Kuipers, and J. Kok. 2003. Cell wall attachment of a widely distributed peptidoglycan binding domain is hindered by cell wall constituents. J. Biol. Chem. 278:23874-23881. [DOI] [PubMed] [Google Scholar]
- 29.Turner, M. S., and P. M. Giffard. 1999. Expression of Chlamydia psittaci- and human immunodeficiency virus-derived antigens on the cell surface of Lactobacillus fermentum BR11 as fusions to BspA. Infect. Immun. 67:5486-5489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turner, M. S., L. M. Hafner, T. Walsh, and P. M. Giffard. 2003. Peptide surface display and secretion using two LPXTG-containing surface proteins from Lactobacillus fermentum BR11. Appl. Environ. Microbiol. 69:5855-5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turner, M. S., P. Timms, L. M. Hafner, and P. M. Giffard. 1997. Identification and characterization of a basic cell-surface-located protein from Lactobacillus fermentum BR11. J. Bacteriol. 179:3310-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turner, M. S., T. Woodberry, L. M. Hafner, and P. M. Giffard. 1999. The bspA locus of Lactobacillus fermentum BR11 encodes an l-cystine uptake system. J. Bacteriol. 181:2192-2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ventura, M., I. Jankovic, D. C. Walker, R. D. Pridmore, and R. Zink. 2002. Identification and characterization of novel surface proteins in Lactobacillus johnsonii and Lactobacillus gasseri. Appl. Environ. Microbiol. 68:6172-6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.von Eichel-Streiber, C., C. Saurborn, and H. K. Kuramitsu. 1992. Evidence for a modular structure of the homologous repetitive C-terminal carbohydrate-binding sites of Clostridium difficile toxins and Streptococcus mutans glucosyltransferases. J. Bacteriol. 174:6707-6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wall, T., S. Roos, K. Jacobsson, A. Rosander, and H. Jonsson. 2003. Phage display reveals 52 novel extracellular and transmembrane proteins from Lactobacillus reuteri DSM 20016T. Microbiology 149:3493-3505. [DOI] [PubMed] [Google Scholar]
- 36.Wren, B. W. 1991. A family of clostridial and streptococcal ligand-binding proteins with conserved C-terminal repeat sequences. Mol. Microbiol. 5:797-803. [DOI] [PubMed] [Google Scholar]