Abstract
All biological processes take place in highly crowded cellular environments. However, the effect that molecular crowding agents have on the folding and catalytic properties of RNA molecules remains largely unknown. Here, we have combined single-molecule fluorescence resonance energy transfer (smFRET) and bulk cleavage assays to determine the effect of a molecular crowding agents on the folding and catalysis of a model RNA enzyme, the hairpin ribozyme. Our single-molecule data reveal that PEG favors the formation of the docked (active) structure by increasing the docking rate constant with increasing PEG concentrations. Furthermore, Mg2+ ion-induced folding in the presence of PEG occurs at concentrations ∼7-fold lower than in the absence of PEG, near the physiological range (∼1 mM). Lastly, bulk cleavage assays in the presence of the crowding agent show that the ribozyme’s activity increases while the heterogeneity decreases. Our data is consistent with the idea that molecular crowding plays an important role in the stabilization of ribozyme active conformations in vivo.
Ribonucleic acid (RNA) folding is required for biological activity and occurs hierarchically, such that tertiary structure follows secondary structure formation.1 RNA folding into the native state can be challenged by local folding traps on rugged potential energy surfaces.2−7 The minimal hairpin ribozyme is a well-characterized model system to study the folding dynamics of small RNA enzymes.4,8−10 Derived from the negative strand of the tobacco ringspot virus satellite RNA, it is involved in processing the product of rolling circle replication through backbone cleavage and ligation.10−12 The ribozyme adopts a docked (active) and an undocked (inactive) conformation in the presence of divalent metal ions to generate the cleavage products. Numerous in vitro experiments have shown that the ribozyme requires higher than physiological metal ion concentrations (≥10 mM Mg2+) to effectively form the active state, and it cleaves its substrate with slow biphasic kinetics.3,8,10,13,14 Furthermore, the ribozyme exhibits nonergodic, heterogeneous folding kinetics.4,5,9
In living cells, enzymatic reactions take place in crowded environments, containing high concentration (∼40%) of macromolecules such as proteins, DNA, and RNA.15−17 Experiments conducted in vitro are typically more than 100 times more dilute and may not accurately mimic cellular conditions.18 Molecular crowding can alter the thermodynamic and kinetic properties of biopolymers, including folding dynamics and catalysis of RNA.19−29 For example, in the presence of molecular crowding agents the catalytic rate of the hammerhead ribozyme increases 2- to 6-fold,22 and the active conformation of group I intron ribozyme is stabilized and its activity increased.23,27,30 Several methods such as UV and CD spectroscopy and FRET have been used to determine the effect of crowding agents on the folding of RNA.31−33 However, such bulk studies cannot avoid ensemble averaging, which can mask the true molecular behavior. One prior single molecule study has investigated the role of crowding agents in the folding of a noncatalytic RNA.29 Here, we directly visualize the effect of crowding agents on the folding of a ribozyme by combining a single-molecule approach with bulk cleavage assays. The single-molecule data show that molecular crowding agents increase the stability and enhance the formation the ribozyme’s most compact conformation (the active state) by specifically accelerating the docking rate constant and reducing folding heterogeneity. Moreover, the required magnesium concentration for efficient folding decreases to near the physiological range. Lastly, the cleavage rate increases at higher percentages of crowding agents, consistent with the stabilization of the native state and the reduction in folding heterogeneity. These results support the idea that crowded environments may play an important role in the activity of RNA enzymes.
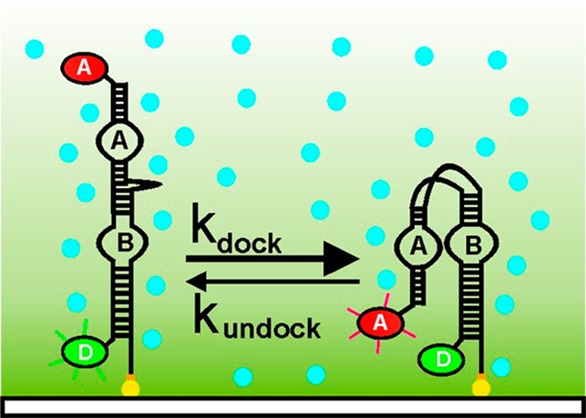
To visualize the effect of molecular crowding on the hairpin ribozyme folding, we labeled the RNA with a FRET donor and acceptor (Cy3 and Cy5, respectively, Figure 1a). The labeled ribozyme was immobilized onto the surface of a quartz microscope slide using biotin and streptavidin. Single ribozyme molecules were imaged by exciting the donor fluorophore with a 532 nm laser and their filtered fluorescence collected using a CCD. As expected, the resulting single molecule trajectories (Figure 1b) show that, in the absence of molecular crowding agents, the ribozyme adopts either a docked, compact active state (0.8 FRET) or an undocked, extended inactive state (0.2 FRET). Time binning 115 time trajectories into a FRET histogram shows that, under standard conditions (50 mM Tris-HCl, pH 7.5, 10 mM MgCl2), the ribozyme samples both dynamic conformations with almost equal probability (Figure 1b,c).
Figure 1.

Molecular crowding stabilizes the docked conformation of the hairpin ribozyme. (a) Schematic diagram of single molecule FRET experiments. Fluorophore labeled ribozyme is surface immobilized on a quartz slide via a biotin–streptavidin linkage. The donor fluorophore (D) is excited in a prism-based total internal reflection microscope. Arrow indicates cleavage site. (b) Characteristic single molecule FRET time trajectory shows the ribozyme dynamically switching between the docked (0.8 FRET) and the undocked (0.2 FRET) conformations. (c) Time binned FRET histograms reveal the distribution in the docked and undocked states. The docked fraction increases with increasing amounts of PEG. (d) Fraction of docked molecules as a function of %PEG and fit to a binding isotherm (solid line). (e) Change in docking Gibbs free energy change (ΔΔG°) as a function of %PEG. PEG stabilizes the docked conformation. (f) Docking rate constant increases with %PEG. (g) A scatter plot showing the folding heterogeneity in the presence (red) and absence (blue) of crowding agents. In the presence of 25% PEG, the distribution narrows and its average increases (dashed lines).
We use polyethylene glycol (PEG, MW 8000) to mimic crowded environments in vitro because it is neutral, has low background fluorescence, and does not interact directly with RNA.28 In the presence of PEG (Figure 1c), the FRET histograms show that the docked state becomes more favored relative to the undocked state. To quantify this effect, we vary the fraction of PEG in solution (5–25 wt %/vol). The resulting titration (Figure 1d) shows that the docked state fraction increases gradually with increasing PEG concentration (PEG50 = 9 ± 2%) and saturates near 0.83. A calculation of the docking Gibbs free energy change (ΔΔG) from the integrated area of each conformation in the FRET histograms shows that the docked state is stabilized by −1.2 kcal/mol in 25% PEG (Figure 1e). A similar increase in native state stability was recently reported for the group I intron ribozyme in the presence of PEG as a crowding agent.23 The increased stability of the native state may result from an entropic effect, whereby the crowded environment restricts the available space, thus favoring the most compact (docked) conformation. Indeed, previous temperature-dependent crowding studies have demonstrated that PEG derived effects are mostly entropic with almost negligible enthalpic contributions.29,34 This relative docked state stabilization can arise from either an acceleration of the docking rate constant or a deceleration of the undocking rate constant. To distinguish between these two scenarios, we measured the docking and undocking rate constants from the distribution of dwell times in the undocked and docked states (Figure S1, Supporting Information (SI)). The resulting docking rate constant increases with increasing PEG (Figure 1f), whereas the undocking rate constants remain approximately constant (Figure S2, SI). This result indicates that both the folding transition state and the docked state are similarly stabilized by molecular crowding relative to the undocked state. This is consistent with the observation that the folding transition state and the docked state have similar structural compactness.9 The same kinetic behavior was previously demonstrated for a noncatalytic RNA.29 A similar rate enhancement effect was observed for the human telomere G-quadruplex formation over duplex formation,35 highlighting the importance of crowded environments in cellular conditions, which may lead to significant increases in the stability of compact active states and their activity in vivo.
To confirm that the observed effects arise from molecular crowding and not from the specific interactions between the PEG and the RNA, we performed control experiments in the presence of PEG monomer (ethylene glycol, EG), which should not have a crowding effect, and another crowding agent, dextran (a 10 kDa polysaccharide).30 As expected, the presence of EG monomers does not change the FRET distribution between the docked and undocked states (Figure S3a, SI). Conversely, the presence of dextran significantly increases the fraction of the docked state as observed with PEG results (Figure S3b, SI). A smaller crowding agent (PEG 3350) results in a lesser effect, confirming that molecular crowding effects depend on the size of the agent (Figure S4a, SI). To show that the effect is also present with natural crowding agents, we tested a non-RNA binding protein (bovine serum albumin, BSA), an RNA binding protein (polypyrimidine tract binding protein, PTB),36 and cellular extract (Figure S4b–d, SI). The data show that all of these favor the most compact conformation to various degrees, consistent with the idea that the crowded environment of a cell can play an important role in ribozyme folding.
It has been previously shown that the hairpin ribozyme adopts heterogeneous folding kinetics with a strong memory effect where each molecule rarely switches between different docked populations.3,9 To assess the effect of molecular crowding on folding heterogeneity, we determined docking and undocking rate constants from individual trajectories in the presence or absence of PEG, as described for the c-di-GMP riboswitch.37 The resulting scatter plot showing the molecular docking and undocking rate constants reveals the folding heterogeneity (Figure 1g).37 In the absence of PEG, the observed docking rate constants span a range between 0.01 and 1 s–1, while the observed undocking rate constants span a range between 0.01 and 10 s–1, consistent with previously reported data.38 In the presence of 20% PEG, the distribution narrows and its average shifts to higher kdock values on the y-axis, consistent with the idea that molecular crowding accelerates the docking rate constant (Figures 1d and S2, SI).
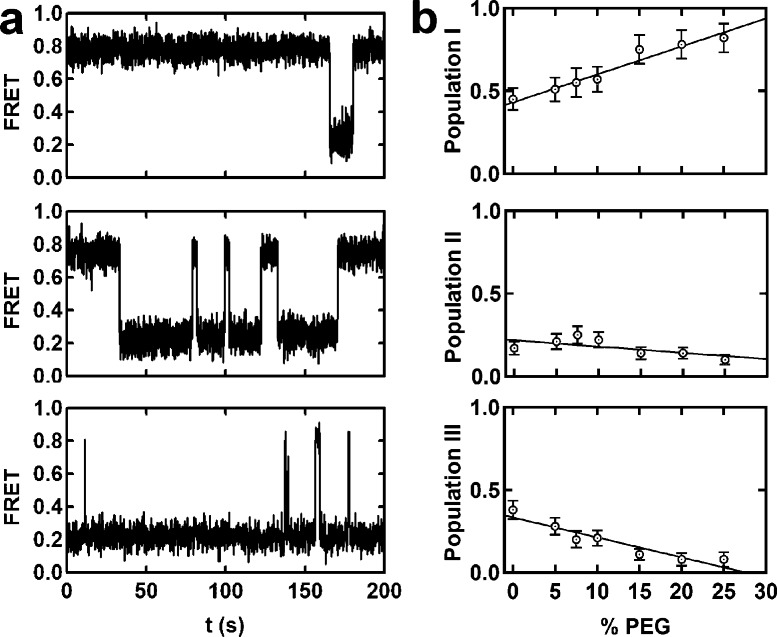
To further investigate the influence of crowding agents on the ribozyme’s folding heterogeneity, we categorized the observed trajectories into three populations based on their dynamics. This categorization is based on previous more-in-depth analyses of the hairpin ribozyme’s kinetic behavior.3,4,9,10 Population I comprises molecules residing mostly in the docked state (Figure 2, top), population II comprises molecules distributed equally among both states (Figure 2a, middle), and population III those residing mostly in the undocked state (Figure 2a, bottom). It appears that the fraction of molecules in population I increases with increasing PEG concentration, at the expense of the other two populations (Figure 2b). This result suggests that the folding heterogeneity observed in vitro may be reduced or eliminated in the crowded environment of a living cell.
Figure 2.
Molecular crowding reduces folding heterogeneity by favoring the most compact folding population. (a) The hairpin ribozymes exhibit at least three folding subpopulations with a strong memory effect. Population I resides mostly in the docked state (0.8 FRET), population II resides equally in both states, and population III remains mostly in the undocked state (0.2 FRET). (b) Fraction of molecules in population I increases with %PEG at the expense of the other two populations.
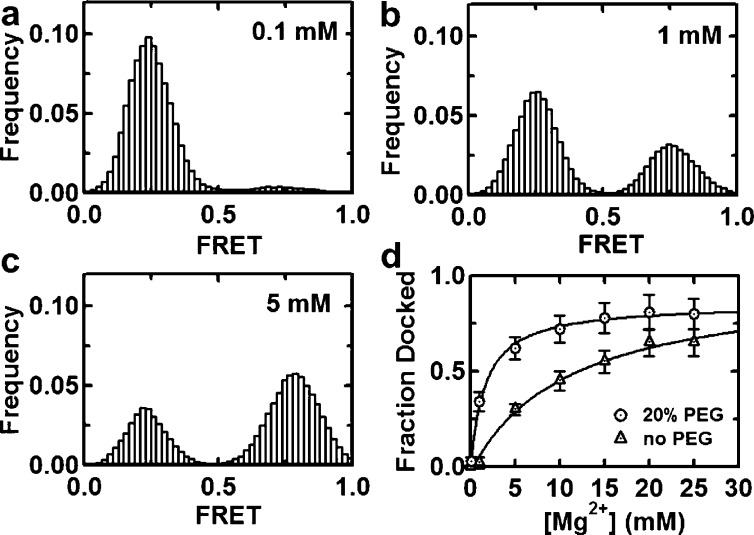
Next, we investigated the effect of crowding agents on the requirement of metal ions for ribozyme folding in vitro. To this aim, we carried out experiments under physiological Mg2+ ion concentrations (1 mM), in the absence and presence of 20% PEG (Figure S5, SI). The resulting FRET histograms show that, in the absence of PEG, the ribozyme remains in the docked state only for a small fraction of time (4%). In the presence of PEG, however, the molecules remain in the docked state 40% of the time. This result shows that, under near-physiological conditions, molecular crowding can increase the active state population by ∼10-fold, indicating that crowding agents can significantly reduce the magnesium requirement, consistent with similar bulk observations for the hammerhead and group I intron ribozymes.22,23 To quantify this effect, we titrated magnesium ions in the absence and presence of saturating PEG (20%, Figure 3). The FRET histograms show that Mg2+ ions stabilize the docked state with 7-fold lower binding constant (11 to 1.6 mM), near the physiological range.
Figure 3.
Molecular crowding helps the ribozyme fold in near-physiological conditions. (a–c) Representative FRET histograms with increasing [Mg2+] in the presence of 20% PEG. Each histogram comprises ≥97 single-molecule trajectories. The docked state fraction increases with [Mg2+]. (d) Magnesium titration in the absence and presence of PEG. The resulting Mg binding constant decreases from 11 to 1.6 mM, near physiological concentrations.
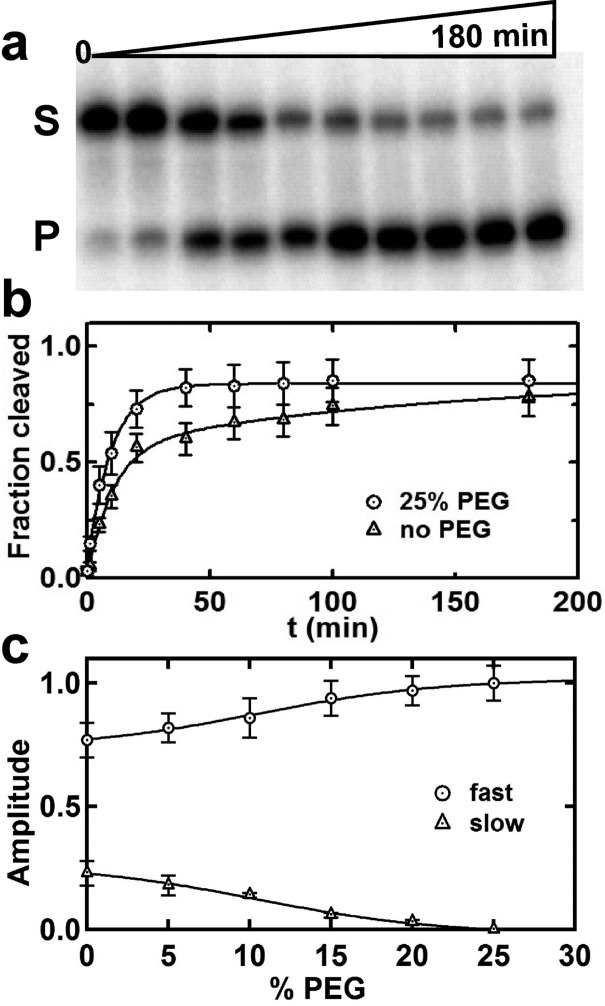
The increased stability of the native state and the decreased heterogeneity in folding suggest that molecular crowded environments may enhance the activity of the ribozyme. To test this hypothesis, we compared the ribozyme’s activity in the absence and presence of PEG. The hairpin ribozyme cleaves its own substrate at a specific backbone site in loop A (Figure 1a).4,8,10 To measure the cleavage activity, we performed radiolabeled cleavage assays, as described8 (Figure 4a). In the absence of PEG, the fraction of cleaved product increases over time, showing that the ribozyme actively cleaves its substrate. A quantification of the fraction cleaved as a function of time shows that the ribozyme exhibits biphasic kinetics (Figures 4b and S6, SI). In the presence of 25% PEG, however, the slow phase disappears, yielding a higher cleaved fraction at all-time points. A PEG titration shows that the slow fraction decreases with increasing PEG concentration (Figure 4c), consistent with the increased stability of the native state and decreased folding heterogeneity in the presence of PEG (Figure 2b). Similar rate enhancements have been reported for the self-cleavage of the hammerhead and the HDV-like CPEB3 ribozyme.26,39 These data show that the presence of crowding agents increases ribozyme activity. Next, we wanted to test whether the presence of crowding agents can promote ribozyme activity under near-physiological conditions (∼1 mM Mg2+). In the absence of PEG the ribozyme exhibits very low activity levels (Figure S7a, SI). In 20% PEG and 1 mM Mg2+, however, the activity of the ribozyme increases significantly (Figure S7b,c, SI). A Mg2+ titration shows that the presence of 20% PEG increases the Mg2+ binding affinity by ∼4-fold into the physiological range (from 8 to 2 mM; Figure S7d, SI).
Figure 4.
Crowding agents accelerate ribozyme cleavage. (a) Fraction of intact substrate (S) and cleaved product (P) as a function of time (0–180 min) after separation by 20% denaturing polyacrylamide gel electrophoresis. The fraction of P increases with time. (b) Fraction of cleaved product as a function of time in the presence and absence of 25% PEG. The presence of PEG increases the fraction of cleaved product. (c) Amplitude of the fast and slow cleavage phases as a function of %PEG. The slow cleaving fraction decreases with increasing PEG concentration.
In summary, we have used the hairpin ribozyme as a model system to investigate the influence of crowding agents in the folding and catalysis of small ribozymes. Our single-molecule data show that molecular crowing agents such as PEG and dextran, proteins, and cellular extract increase the stability of the most compact conformation, the docked state, which in the case of the hairpin ribozyme corresponds to the active state. This effect is achieved almost exclusively by acceleration of the docking rate constant, which suggests that both the folding transition state and the docked state are similarly stabilized (Figure 5b). We also observe a decrease in folding heterogeneity, whereby the most compact population is favored over the others. Furthermore, the amount of magnesium ions required to form the active, docked state decreases to near-physiological concentrations (∼1.6 mM). Together these effects result in less heterogeneous, faster catalysis. Although PEG is not normally present in living cells, our results are consistent with the idea that, in vivo, RNA enzymes have evolved to function optimally within the crowded environment of the cell.
Figure 5.
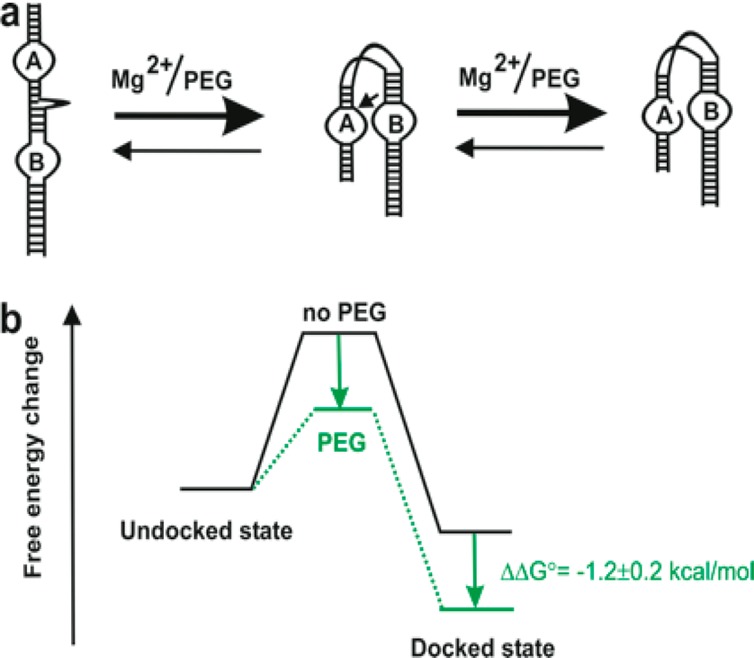
Proposed model of ribozyme folding and stability in crowding environments. (a) Cartoon represents the acceleration of the docking and catalysis of the ribozyme in the presence of crowding agents and divalent metal ions. (b) The free energy change diagram shows the docked state is stabilized by −1.2 kcal/mol of energy in the presence of PEG.
Acknowledgments
We thank Dr. S. Woodson, Dr. K. Chaurasiya, and Dr. J. Hardin for reading the manuscript. We thank Dr. Amit Gautam for kindly providing cell extract for our experiments. This work was supported by the National Science Foundation (NSF CAREER award 0747285), the NIH (GM085116), the Medical Research Council (UK, MC_A658_5TY10), and Imperial College London startup funds.
Supporting Information Available
Single-molecule experimental results of ribozyme folding and dynamics. Methods and protocols. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Doudna J. A.; Cech T. R. Nature 2002, 418, 222. [DOI] [PubMed] [Google Scholar]
- Treiber D. K.; Williamson J. R. Curr. Opin. Struct. Biol. 2001, 11, 309. [DOI] [PubMed] [Google Scholar]
- Zhuang X.; Kim H.; Pereira M. J.; Babcock H. P.; Walter N. G.; Chu S. Science 2002, 296, 1473. [DOI] [PubMed] [Google Scholar]
- Ditzler M. A.; Rueda D.; Mo J.; Hakansson K.; Walter N. G. Nucleic Acids Res. 2008, 36, 7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R.; Rueda D. In Biophysics in RNA Folding; Russel R., Ed.; Springer: New York, 2013; Vol. 3, p 236. [Google Scholar]
- Woodson S. A. Annu. Rev. Biophys. 2010, 39, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R.; Rueda D. Methods 2009, 49, 112. [DOI] [PubMed] [Google Scholar]
- Walter N. G.; Burke J. M. Curr. Opin. Chem. Biol. 1998, 2, 24. [DOI] [PubMed] [Google Scholar]
- Bokinsky G.; Rueda D.; Misra V. K.; Rhodes M. M.; Gordus A.; Babcock H. P.; Walter N. G.; Zhuang X. Proc. Natl. Acad. Sci. U.S.A. 2003, 100, 9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueda D.; Bokinsky G.; Rhodes M. M.; Rust M. J.; Zhuang X.; Walter N. G. Proc. Natl. Acad. Sci. U.S.A. 2004, 101, 10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzayan J. M.; Gerlach W. L.; Bruening G. Proc. Natl. Acad. Sci. U.S.A. 1986, 83, 8859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter N. G.; Burke J. M. Curr. Opin. Chem. Biol. 1998, 2, 303. [DOI] [PubMed] [Google Scholar]
- Murray J. B.; Seyhan A. A.; Walter N. G.; Burke J. M.; Scott W. G. Chem. Biol. 1998, 5, 587. [DOI] [PubMed] [Google Scholar]
- Wilson T. J.; Lilley D. M. RNA 2002, 8, 587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minton A. P.; Wilf J. Biochemistry 1981, 20, 4821. [DOI] [PubMed] [Google Scholar]
- Ellis R. J. Trends Biochem. Sci. 2001, 26, 597. [DOI] [PubMed] [Google Scholar]
- Miyoshi D.; Sugimoto N. Biochimie 2008, 90, 1040. [DOI] [PubMed] [Google Scholar]
- Luby-Phelps K. Int. Rev. Cytol. 2000, 192, 189. [DOI] [PubMed] [Google Scholar]
- Zimmerman S. B.; Minton A. P. Annu. Rev. Biophys. Biomol. Struct. 1993, 22, 27. [DOI] [PubMed] [Google Scholar]
- Minton A. P. Curr. Opin. Biotechnol. 1997, 8, 65. [DOI] [PubMed] [Google Scholar]
- Minton A. P. Methods Enzymol. 1998, 295, 127. [DOI] [PubMed] [Google Scholar]
- Nakano S.; Karimata H. T.; Kitagawa Y.; Sugimoto N. J. Am. Chem. Soc. 2009, 131, 16881. [DOI] [PubMed] [Google Scholar]
- Kilburn D.; Roh J. H.; Guo L.; Briber R. M.; Woodson S. A. J. Am. Chem. Soc. 2010, 132, 8690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Z. J.; Chen S. J. Biophys. J. 2012, 103, 827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutsioubas A.; Lairez D.; Combet S.; Fadda G. C.; Longeville S.; Zalczer G. J. Chem. Phys. 2012, 136, 215101. [DOI] [PubMed] [Google Scholar]
- Strulson C. A.; Yennawar N. H.; Rambo R. P.; Bevilacqua P. C. Biochemistry 2013, 52, 8187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilburn D.; Roh J. H.; Behrouzi R.; Briber R. M.; Woodson S. A. J. Am. Chem. Soc. 2013, 135, 10055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano S.; Miyoshi D.; Sugimoto N. Chem. Rev. 2014, 114, 2733. [DOI] [PubMed] [Google Scholar]
- Dupuis N. F.; Holmstrom E. D.; Nesbitt D. J. Proc. Natl. Acad. Sci. U.S.A. 2014, 111, 8464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai R.; Kilburn D.; Lee H. T.; Woodson S. A. J. Biol. Chem. 2014, 289, 2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuriki N.; Kinjo M.; Negi S.; Hoshino M.; Goto Y.; Urabe I.; Yomo T. Protein Sci. 2004, 13, 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi T.; Galiacy S. D.; Briscoe G.; Erickson H. P. Protein Sci. 2007, 16, 1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L. N.; Wu B.; Kong D. M. Nucleic Acids Res. 2013, 41, 4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung M. S.; Klimov D.; Thirumalai D. Proc. Natl. Acad. Sci. U.S.A. 2005, 102, 4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J.; Wei C.; Jia G.; Wang X.; Feng Z.; Li C. Biochimie 2009, 91, 1104. [DOI] [PubMed] [Google Scholar]
- Lamichhane R.; Daubner G. M.; Thomas-Crusells J.; Auweter S. D.; Manatschal C.; Austin K. S.; Valniuk O.; Allain F. H.; Rueda D. Proc. Natl. Acad. Sci. U.S.A. 2010, 107, 4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood S.; Ferre-D’Amare A. R.; Rueda D. ACS Chem. Biol. 2012, 7, 920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan E.; Wilson T. J.; Nahas M. K.; Clegg R. M.; Lilley D. M.; Ha T. Proc. Natl. Acad. Sci. U.S.A. 2003, 100, 9308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strulson C. A.; Molden R. C.; Keating C. D.; Bevilacqua P. C. Nat. Chem. 2012, 4, 941. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.