Abstract
Enormous potential of cell-based therapeutics is hindered by the lack of effective means to control genetically engineered cells in mammalian tissues. Here, we describe a synthetic module for remote photocontrol of engineered cells that can be adapted for such applications. The module involves photoactivated synthesis of cyclic dimeric GMP (c-di-GMP), a stable small molecule that is not produced by higher eukaryotes and therefore is suitable for orthogonal regulation. The key component of the photocontrol module is an engineered bacteriophytochrome diguanylate cyclase, which synthesizes c-di-GMP from GTP in a light-dependent manner. Bacteriophytochromes are particularly attractive photoreceptors because they respond to light in the near-infrared window of the spectrum, where absorption by mammalian tissues is minimal, and also because their chromophore, biliverdin IXα, is naturally available in mammalian cells. The second component of the photocontrol module, a c-di-GMP phosphodiesterase, maintains near-zero background levels of c-di-GMP in the absence of light, which enhances the photodynamic range of c-di-GMP concentrations. In the E. coli model used in this study, the intracellular c-di-GMP levels could be upregulated by light by >50-fold. Various c-di-GMP-responsive proteins and riboswitches identified in bacteria can be linked downstream of the c-di-GMP-mediated photocontrol module for orthogonal regulation of biological activities in mammals as well as in other organisms lacking c-di-GMP signaling. Here, we linked the photocontrol module to a gene expression output via a c-di-GMP-responsive transcription factor and achieved a 40-fold photoactivation of gene expression.
Keywords: bacteriophytochrome, photoreceptor, optogenetics, protein engineering, second messenger, cell-based therapeutics
Therapies based on engineered mammalian or microbial cells are poised to revolutionize biomedicine.1,2 One of the major obstacles in applying engineered cells is the lack of tools to control their behavior in live animals. Chemicals (drugs) used for this purpose offer very limited spatial and temporal resolution and often generate undesired side effects. Light, on the other hand, is a noninvasive stimulus that offers higher spatiotemporal resolution for regulating biological activities than the resolution achievable by chemicals.3 Protein photoreceptors belonging to several major classes have been engineered to control biological activities in a light-dependent manner. These include UV-light receptors, blue-light sensing LOV, cryptochrome and BLUF receptors, red- and green-light sensing phytochromes, and rhodopsins that can sense visible light in a broad range of the spectrum.4,5 Several regulatory modalities have been successfully placed under light control, including protein–protein interactions,6,7 protein degradation,8 recombination,9 gene expression,10,11,5 and synthesis of second messengers.12−15
Second messengers, particularly cyclic mono- and dinucleotides such as cAMP, cGMP, c-di-AMP, and c-di-GMP have remained underexplored as optogenetic tools. These second messengers are attractive for several reasons. (i) Cyclic mono- (cNMP) and di- (c-di-NMP) nucleotide second messengers regulate diverse biochemical processes, including ion transport across membranes, transcription, translation, protein activities, and protein–protein interactions.16−19 Therefore, cNMP/c-di-NMP sensory modules can be adapted to regulate various kinds of outputs. (ii) No known organism possesses signaling pathways involving all cyclic mono- and dinucleotide second messengers; therefore, orthogonal regulation that does not interfere (or only minimally interferes) with the host signaling pathways can be developed for various animal, plant, or microbial organisms. (iii) The substrates for nucleotide cyclases (ATP or GTP) are omnipresent and available in millimolar concentrations, in all cell types. (iv) Synthesis of cNMP or c-di-NMP involves a single enzymatic step that can be readily controlled by external signals (e.g., by light). cNMP/c-di-NMP degradation also involves single dedicated phosphodiesterases that produce benign, nontoxic products (AMP or GMP).
cNMP/c-di-NMP synthases with high photodynamic ranges (i.e., light-to-dark activity ratios) are particularly useful for controlling cNMP/c-di-NMP levels with light. At present, natural blue light-activated adenylate cyclases with high photodynamic ranges have been identified and characterized,13−15 and a blue light-activated guanylate cyclase has been engineered.13 The utility of these cyclases for regulating cNMP-dependent signaling processes in animals has been proven.20,21 However, cNMPs are inappropriate for orthogonal regulation in mammals where native cAMP- and cGMP-dependent signaling pathways are prevalent. Another major obstacle in using these light-activated cNMP synthases is poor penetration of blue light through tissues of red-blooded animals, which limits their use to cell cultures, small transparent organisms and in mammalian organs accessible to light.
Most photons in the UV/visible spectrum penetrate mammalian tissues to the maximal depths of only one to several millimeters due high absorption by hemoglobin and melanin. However, light in the so-called near-infrared window (NIRW) of the spectrum, ∼680–880 nm,22 penetrates much deeper. NIRW light absorption by mammalian tissues is several fold to several orders of magnitude lower than absorption of light below ∼680 nm. Even deep-red light (650–670 nm) is significantly inferior, as demonstrated by measurements of light penetration depths through human tissues.23,24 Optimal penetration makes NIRW light a promising tool for remote photocontrol of biological activities in live mammals.25
Among known protein photoreceptors, only bacteriophytochromes sense NIRW light. Not surprisingly, fluorescent bacteriophytochrome derivatives engineered in recent years have readily outperformed fluorescent proteins of other classes in whole animal imaging involving murine models.26−29 Another advantage of bacteriophytochromes for application in animals, compared, for example, to the related classes of plant and cyanobacterial phytochromes is that the bacteriophytochrome chromophore, biliverdin IXα, is naturally made in animal tissues as the first intermediate in the heme degradation pathway, which obviates the need to supply the chromophore.25 Bacteriophytochromes bind biliverdin covalently via intrinsic biliverdin ligase activity. However, in cells with limited biliverdin levels, biliverdin can be generated by a single enzyme, heme oxygenase, that can be supplied along with a bacteriophytochrome gene.30,31
In this study, we describe a NIRW-light activated c-di-GMP synthesis/degradation module. It combines advantages of NIRW light for deep mammalian tissue penetration with second messenger-mediated regulation that can be adapted to control diverse output targets. Cyclic di-GMP is absent from higher eukaryotes and therefore can be used for orthogonal regulation. The synthetic module engineered here comprises (i) a bacteriophytochrome c-di-GMP synthase (diguanylate cyclase, DGC) originating from the Rhodobacter sphaeroides BphG1 protein described by us earlier,32 and (ii) a constitutive c-di-GMP-specific phosphodiesterase (PDE), YhjH from E. coli.33 These two enzymes keep intracellular c-di-GMP levels minimal in the absence of light and allow c-di-GMP accumulation of >50-fold following exposure of E. coli to NIRW light. To demonstrate system performance, we engineered a c-di-GMP-responsive gene expression output and observed high, ∼40-fold, change in gene expression in response to NIRW light. Because numerous natural protein and RNA modules involved in c-di-GMP sensing have been discovered,16 various output modules can be engineered to regulate diverse biological activities by NIRW light.
Results and Discussion
Engineering a Potent Chimeric NIRW Light-activated DGC
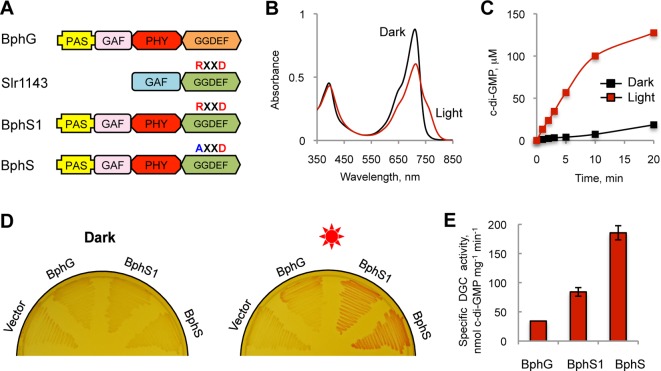
Bacteriophytochromes possess a common photoreceptor module, PAS-GAF-PHY, usually linked to a histidine kinase output.31 Earlier, we described an unorthodox bacteriophytochrome, BphG1, from Rhodobacter sphaeroides that contains a C-terminal GGDEF-EAL domain tandem as an output.32 The full-length BphG1 has constitutive c-di-GMP PDE activity and no DGC activity. However, a truncated derivative of BphG1 lacking the C-terminal EAL domain (PAS-GAF-PHY-GGDEF) possesses low-level DGC activity. We hereby designate this derivative BphG (Figure 1A). As is typical of bacteriophytochromes, BphG exists in a Pr form (712 nm, absorption maximum) in the dark and is converted to the Pfr form (756 nm, absorption maximum) upon irradiation with red light (Figure 1B).
Figure 1.
Engineering a potent photoactivated DGC. (A) Domain architectures of the proteins used for engineering BphS, a high-potency photoactivated DGC. The RXXD motif of the I-site is indicated above the GGDEF domain. (B) UV–vis absorption spectroscopy of BphG. The BphG protein undergoes reversible photoconversion between the Pr (712 nm, absorption maximum) and Pfr (756 nm, absorption maximum) forms. (C) Kinetics of the DGC activity of BphG showing an ∼11-fold higher specific activity in the light versus dark. (D) Congo red test for c-di-GMP-dependent curli fimbriae production. E. coli cells expressing a light-activated DGC (BphG, BphS1 or BphS) together with the heme oxygenase BphO were incubated on plates supplemented with Congo red dye (50 μg/mL) in the absence or presence of light. pET23a(+) was used as a control. Red sun, irradiation with red light. (E) Maximal specific activity of the original, BphG, and engineered photoactivated DGCs, BphS1 and BphS, measured in the light.
The photoactivation ratio of DGC activity of BphG (activity in the light versus activity in the dark) observed earlier was ∼4-fold.32 Upon careful re-examination of this parameter using freshly prepared BphG (Figure 1B), we determined that the ratio is actually higher, ∼11-fold (Figure 1C). To our knowledge, this is the highest photoactivation ratio described for any bacteriophytochrome for which such a ratio has been quantified. This makes BphG particularly attractive for optogenetic applications.
To evaluate the potential of BphG in regulating c-di-GMP levels in a light-dependent manner, we overexpressed it in E. coli BL21[DE3], a bacterial host with well-characterized c-di-GMP signaling pathways.34 Since E. coli does not synthesize biliverdin, we cloned the R. sphaeroides heme oxygenase gene, bphO (RSP_4190), immediately downstream of bphG as the second gene in the artificial bphG-bphO operon. To test for light-dependent c-di-GMP synthesis by the bphG-bphO operon, we grew E. coli, in the dark or under red light, on agar medium containing Congo red and monitored production of curly fimbriae, which bind this dye.35 The bphG-bphO operon produced barely observable red pigmentation in BL21[DE3] in the light (Figure 1D) indicating poor accumulation of c-di-GMP, insufficient for developing a versatile c-di-GMP-dependent regulatory system. We extracted intracellular nucleotides from the biomass of BL21[DE3] expressing the bphG-bphO operon grown in liquid culture under constant irradiation. Cyclic di-GMP was separated and quantified by LC-MS-MS. The maximal intracellular c-di-GMP concentration achieved was ∼90 nM. This low level is consistent with the poor c-di-GMP synthesis by BphG assessed by the Congo red colony staining. Since the majority of c-di-GMP receptors have dissociation constants, Kd, for c-di-GMP in the submicromolar-to-low micromolar range,16 BphG is clearly not sufficiently potent for activating c-di-GMP-dependent processes.
To increase the DGC activity, we attempted to replace the low activity GGDEF domain of BphG with the GGDEF domain from a more active DGC, Synechocystis sp. Slr1143, characterized by us earlier36 (Figure 1A). The site for domain swapping was chosen based on protein domain alignments (Supporting Information (SI) Figure S1). The chimeric protein, BphS1, composed of the N-terminal photosensory module of BphG (1–511 aa) and the C-terminal GGDEF domain of Slr1143 (175–343 aa) showed more intense Congo red staining in the curli fimbriae test. Upon irradiation, BphS1 showed ∼2.5-fold higher DGC activity than that of BphG (Figure 1E). To enhance BphS1 potency further, we mutated the conserved RXXD sequence motif located in the GGDEF domain of Slr1143 (Figure 1A). This motif is part of the c-di-GMP binding I-site involved in the feedback inhibition of DGC activity.35 We introduced the R587A mutation in the RXXD motif to abolish feedback inhibition. The constructed mutant protein, designated BphS (SI Figure S2), produced more intense pigmentation on Congo red plates than BphS1 (Figure 1D). Its DGC activity in the light was ∼2.2-fold higher compared to BphS1 (Figure 1E). Therefore, removal of feedback inhibition further increased the range of c-di-GMP concentrations achievable by BphS. Overall, BphS had ∼5.5-fold higher specific activity compared to BphG (in the dark or light), but importantly, it retained the same photoactivation ratio as BphG (not shown). Biliverdin produced by BphO is essential for BphS activity (SI Figure S3). The photochemical properties of BphS were not different from the properties of BphG.32
Improving the Photodynamic Range of a Light-activated Synthetic c-di-GMP Module
While the ∼11-fold ratio is high among bacteriophytochromes, the higher DGC activity of BphS resulted in an undesirable increase in c-di-GMP synthesis in the dark, compared to BphG. To decrease the level of c-di-GMP accumulated in the dark and thereby to increase the range of the c-di-GMP concentrations that can be manipulated by light, we added a c-di-GMP PDE as a second component. Ideally, this PDE would be inactivated by NIRW light; however, no such enzyme has been found in nature or engineered. Therefore, we added an enzyme with constitutive activity, E. coli YhjH, which contains a single EAL domain responsible for PDE activity.33 The yhjH gene was placed downstream of bphS-bphO as part of the three-gene operon (Figure 2A).
Figure 2.
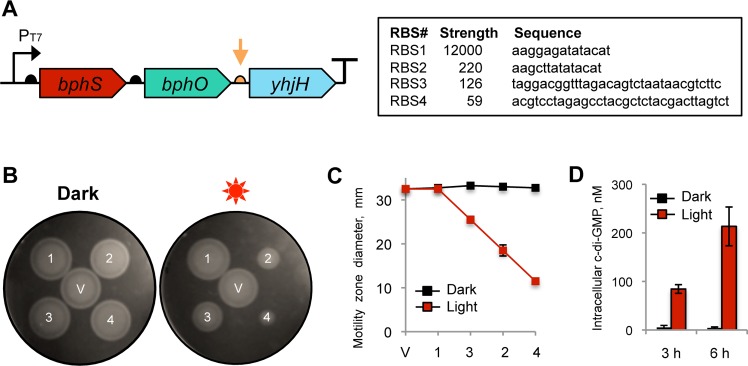
Synthetic operon for light-activated c-di-GMP synthesis. (A) Structure of the synthetic operon for light-activated c-di-GMP synthesis. The genes encoding light-activated DGC (bphS), heme oxygenase (bphO), and c-di-GMP PDE (yhjH) are assembled in a single operon, bphS-bphO-yhjH. A semicircle in front of each gene indicates a RBS; a T-sign at the end of the operon indicates a transcription terminator. The expression levels of yhjH were altered by using RBS sequences (orange arrow) of varying strengths shown in the box. (B) Adjustment of the yhjH RBS strength using semisolid agar motility assays in E. coli MG1655[DE3]. V, pMQ56 (empty vector); 1, RBS1; 2, RBS2; 3, RBS3; 4, RBS4. Increased intracellular c-di-GMP levels decrease the size of a motility zone. (C) Diameters of the swimming zones from panel B. (D) Intracellular c-di-GMP levels measured in liquid-grown cultures of MG1655[DE3] expressing bphS-bphO-yhjH with RBS3 upstream of yhjH.
The purpose of YhjH is to maintain near-zero levels of intracellular c-di-GMP by degrading c-di-GMP produced by the dark DGC activity of BphS. However, the PDE activity of YhjH must not interfere or interfere only minimally with c-di-GMP synthesis in the light. To adjust YhjH expression to the desired levels, we optimized the strength of a ribosome-binding site (RBS) driving yhjH mRNA translation. To monitor relative strengths of DGC and PDE activities, we needed a system responsive to changes in c-di-GMP levels at the low end of c-di-GMP concentrations, unlike the curli fimbriae system that responds to relatively higher intracellular c-di-GMP. E. coli flagellar motility in semisolid agar presented a suitable system. The c-di-GMP receptor YcgR37,33 is one of the most sensitive c-di-GMP receptors described thus far.38 YcgR works as a backstop motility break that binds to a flagellum rotor and, in the presence of c-di-GMP, decreases its ability to reverse rotation direction. This, in turn, impairs the ability of cells to change the direction of their movement and gets them stuck in the blind alleys of semisolid agar.39,40 Sizes of motility zones in semisolid agar can therefore serve as proxies for intracellular c-di-GMP levels.33 Since strain BL21[DE3] used in Congo red staining tests is nonmotile, we constructed an alternative host derived from the highly motile E. coli strain MG1655.41 To enable bphS-bphO-yhjH operon expression in MG1655, we inserted into its chromosome a recombinant λDE3 phage carrying an IPTG-inducible T7 RNA polymerase gene.
When cells expressing the originally constructed bphS-bphO-yhjH operon were tested, they showed equally good motility irrespective of irradiation, which suggested that PDE activity of YhjH overpowered the DGC activity of BphS not only in the dark (as desired) but also in the light (Figure 2B). To decrease YhjH expression, we designed a series of ribosome-binding sites (RBS) with predicted lower translation initiation efficiencies42 and used them to replace the original RBS (RBS1) upstream of yhjH (Figure 2A). We tested three RBS (RBS2–4) whose strengths span a 200-fold range below the strength of RBS1 (Figure 2A). The RBS strength was predicted by a RBS calculator.43 Constructs with RBS with the predicted lower strengths generally corresponded to the smaller motility zones in the light than the RBS1 construct (Figure 2B and C); however, the calculated RBS values did not always correlate with the expected phenotypes (e.g., reverse order in motility zone sizes in RBS3 and RBS2). None of the tested constructs with lower RBS strengths showed motility inhibition in the dark (Figure 2B and C), which indicated that YhjH expression in the dark in all constructs was sufficient to overcome the background activity of BphS.
Kinetics of the NIRW Light-Induced c-di-GMP Accumulation in E. coli
To determine the kinetics of intracellular c-di-GMP accumulation in response to NIRW light, we used MG1655[DE3] expressing the bphS-bphO-yhjH operon with the RBS3 upstream of yhjH. The data in Figure 2D indicate that intracellular c-di-GMP rose from ∼4 nM in the dark to ∼84 nM after 3 h of irradiation (>20-fold increase) and continued to rise reaching ∼210 nM after additional 6 h of irradiation (>50-fold increase).
It is noteworthy that in this experiment we chose a relatively strong RBS (RBS3) for YhjH to overpower not only the dark DGC activity of BphS but also native E. coli DGCs that may have contributed an additional ∼0.1–1 μM c-di-GMP.44 In organisms lacking native c-di-GMP signaling pathways, where the role of YhjH will be restricted to degrading c-di-GMP produced only by the dark DGC activity of BphS, it may be possible to select a BphS-YhjH module with lower YhjH expression and therefore achieve higher than 50-fold photodynamic range of c-di-GMP concentrations.
NIRW Light-Activated, c-di-GMP-Mediated Gene Expression
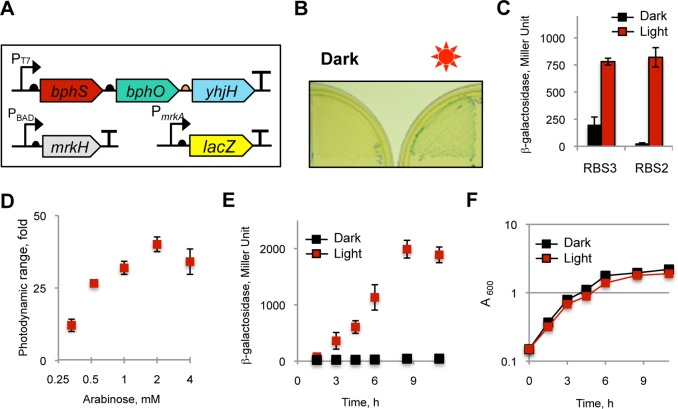
Here, we assessed the performance of the synthetic NIRW light-activated module in photoactivating gene expression. We engineered two c-di-GMP-dependent expression systems and linked them to the BphS-YhjH module. The first system involved the CRP-like protein Clp from Xanthomonas axonopodis,45 where CRP is an E. coli cAMP receptor/activator protein46 (Figure 3A). Clp recognizes the same target sequence as CRP; its affinity to DNA is decreased upon c-di-GMP binding45 (i.e., opposite to the effect of cAMP on CRP). We placed the lacZ reporter gene downstream of the Pclp promoter containing two CRP-binding sites,47 while clp gene expression was under the arabinose-inducible PBAD promoter. The bphS-bphO-yhjH operon expressing YhjH under RBS4, clp and lacZ constructs were expressed in the constructed BL21[DE3] crp null mutant to monitor reduction in LacZ expression in response to light (SI Figure S4). Light-induced c-di-GMP synthesis resulted in the expected decrease in β-galactosidase levels (Figure 3B). The observed fold-reduction, measured in liquid culture, was moderate, that is, ∼3-fold (Figure 3C). We modified the promoter strength, number of Clp binding sites, and relative position of these sites and yet did not succeed in significantly improving this parameter (not shown), which suggests that system limitation may be Clp-specific. We therefore linked a different transcription factor downstream of the BphS-YhjH photocontrol module.
Figure 3.
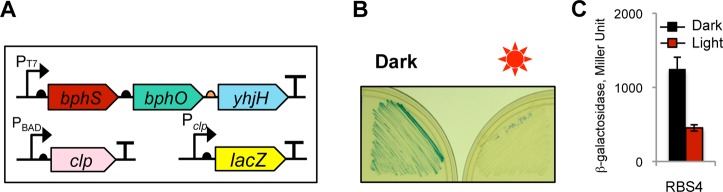
NIRW light-inactivated gene expression module. (A) Structure of a NIRW light-inactivated synthetic module. (B) Light-inactivated β-galactosidase activity in the E. coli T7 Express crp mutant expressing a c-di-GMP-dependent Clp transcription factor. Plates contained 0.1% arabinose, 0.015 mM IPTG, and 40 μg/mL X-gal. (C) β-galactosidase activity in liquid cultures grown in the dark or light to A600, ∼1.0.
Klebsiella pneumoniae MrkH is a transcriptional factor that binds c-di-GMP via a high-affinity c-di-GMP-binding PilZ domain.48,49 Cyclic di-GMP binding enhances MrkH affinity to DNA, that is, opposite to the effect of c-di-GMP on Clp (SI Figure S4). We placed a lacZ reporter under the control of the MrkH-dependent PmrkA promoter, where mrkA is the MrkH-activated gene target.48,49mrkH was expressed from the arabinose-inducible PBAD promoter (Figure 4A). We adjusted the strength of the RBS upstream of lacZ (300 arbitrary units, according to the RBS calculator43) to produce blue colonies on X-gal plates when MrkH is expressed at moderate levels (0.1% arabinose). The mrkH expression was then lowered (0.008% arabinose) to allow for light-dependent c-di-GMP activation of MrkH activity, and β-galactosidase levels were measured. The bphS-bphO-yhjH operon expressing YhjH from RBS3 produced colorless colonies in the dark and blue colonies in the light (Figure 4B). β-galactosidase levels in the liquid cultures in the dark were very low for the RBS3 construct (but not for RBS2) (Figure 4C). The expression level of mrkH was further optimized (0.03%, or 2 mM arabinose) to achieve a photoactivation ratio of ∼40 (Figure 4D). The kinetics of β-galactosidase levels (Figure 4E) shows that an ∼10-fold activation of gene expression was reached after ∼4 h, and the maximal ∼40-fold activation was reached after ∼8.5 h of irradiation. Red or NIRW light has no known targets in E. coli except for BphS; therefore, it was not surprising that this light did not inhibit E. coli growth under constant irradiation (Figure 4F).
Figure 4.
NIRW light-activated gene expression module. (A) Structure of a NIRW light-activated synthetic module. The DNA fragment encoding the c-di-GMP synthetic photocontrol module is ∼3.5 kb; the mrkH module is ∼0.8 kb. (B) Light-activated β-galactosidase activity in the E. coli T7 Express strain expressing a c-di-GMP-dependent MrkH transcription activator. Plates contained 0.008% arabinose, 0.015 mM IPTG, and 40 ug/mL X-gal. (C) β-galactosidase activities in liquid cultures grown in the dark or light to A600, ∼1.5. The medium contained 0.008% arabinose, 0.025 mM IPTG. Higher YhjH expression in the RBS3 construct abolished the undesired basal LacZ expression in the dark, whereas the RBS2 construct produced undesired LacZ expression in the dark. (D) Optimization of the photodynamic range of β-galactosidase levels by adjusting MrkH expression from the arabinose-inducible PBAD promoter. (E) Time-course of β-galactosidase levels. The E. coli T7 Express strain expressing the NIRW light-activated module with RBS3 was grown in LB medium containing 2 mM arabinose and 0.025 mM IPTG. (F) Growth curve of liquid cultures from panel E grown in the dark or light.
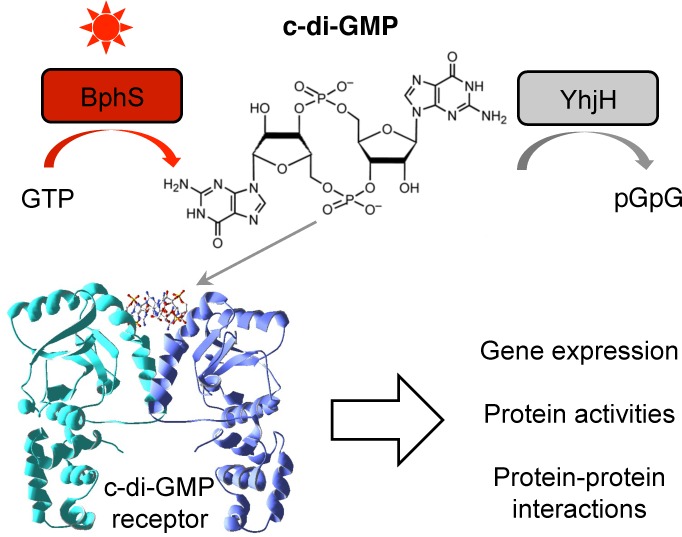
In this study, we combined two concepts whose utility in synthetic biology applications has thus far been underexplored. First, we engineered a module for highly controlled synthesis and degradation of a bacterial second messenger, c-di-GMP. Because c-di-GMP is not made by higher eukaryotes, it can be used for orthogonal regulation of biological activities in these organisms. Second messengers, including c-di-GMP, offer two important advantages over other regulatory modalities. (i) They can amplify the magnitude of the change achieved by primary signals. In the photocontrol module described here, an ∼11-fold activation of the DGC activity by light (primary signal) is converted to a >50-fold increase in intracellular c-di-GMP levels. Importantly, because second messenger levels can be controlled at both synthesis and degradation levels, establishing a near-zero background levels is feasible. In our case, this was accomplished by integrating a c-di-GMP PDE, YhjH, with the engineered DGC, BphS. (ii) Another advantage of second messengers lies in the versatility of the downstream targets that can respond to changes in concentrations of second messengers. In the case of c-di-GMP, several protein types that bind this ligand via diverse modes have been discovered.16 Some of them bind c-di-GMP via well-defined protein domains, such as the PilZ domain of the K. pneumoniae transcription activator MrkH used in this study. Conformational changes occurring in PilZ in response to c-di-GMP are structurally characterized and can be used for protein engineering. Other proteins, that bind c-di-GMP at the homodimeric50 or heterodimeric51 protein interfaces, can be used for engineering homologous or heterologous c-di-GMP-dependent protein–protein interactions. Further, c-di-GMP-specific riboswitches52 and ribozymes53 can be employed to affect mRNA stability and/or transcription and translation efficiencies. Therefore, complex, multilayer signaling cascades involving c-di-GMP can be engineered (Figure 5).
Figure 5.
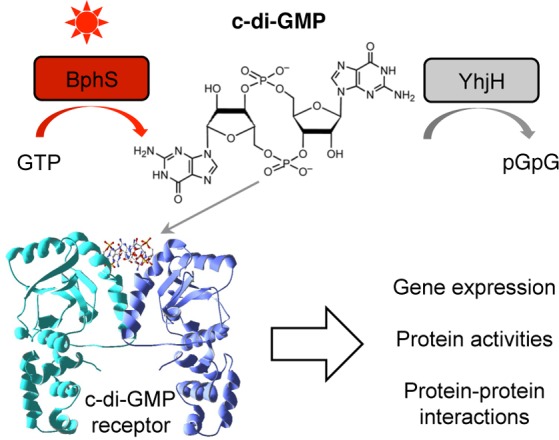
Regulation of diverse biological activities via the NIRW light-activated c-di-GMP module. NIRW light-controlled synthetic c-di-GMP module can be linked to output modules via c-di-GMP receptor proteins and RNA to regulate various biological activities. The structure of a VpsT receptor (PDB ID, 3klo) is shown as an example.
Nucleotide second messengers are particularly attractive for synthetic biology applications. Briefly, they are synthesized in a single step from ubiquitous substrates (GTP or ATP) and degraded into nontoxic products (GMP or AMP). Since no organism possesses signaling pathways involving all known cNMP/c-di-NMP second messengers54,18 orthogonal regulatory systems can be designed for various organisms. Finally, several highly regulated adenylate and guanylate cyclases have been discovered or engineered.13−15 BphS engineered in this study is the first dinucleotide cyclase with a high photoactivation ratio and an activity level suitable for orthogonal applications.
The second concept explored in this study deals with engineering of photoreceptors of the bacteriophytochrome class. Bacteriophytochromes sense light in the farthest region of spectrum compared to other known photoreceptor types. Their absorption falls directly in the NIRW (∼680–880 nm), the optimal spectral region for accessing deep mammalian tissues.22 Bacteriophytochrome absorption is separated from the absorption of related plant- or cyanobacterial type phytochromes by a relatively small (∼30–100 nm) red shift; however, this results in a 2–10-fold gain in the depth of light penetration into mammalian tissues,23,24,55 which may be critical in biomedical applications. An additional advantage of bacteriophytochromes over other phytochromes that have been engineered to control gene and protein activities10,5 is that their chromophore, biliverdin IXα, is naturally produced in mammalian cells as a first intermediate in the heme degradation pathway. Where biliverdin availability is limited, it can be supplied by a single heme oxygenase gene. In the case of plant- and cyanobacterial phytochromes, the chromophore biosynthetic pathway requires an extra enzymatic step.30
While second messenger-mediated NIRW light activated systems are attractive because versatile downstream regulatory modules that can be linked to them, they have inherent pitfalls. For example, they respond to light relatively slowly because of the time required for second messenger accumulation (Figure 4E). Another potential concern in using c-di-GMP- (or c-di-AMP-) mediated systems in mammals is that c-di-NMPs are recognized as pathogen-associated molecular patterns that may stimulate the innate immune system. In mammalian cells expressing the STING receptor, c-di-GMP activates interferon β response via the DDX1-STING pathway.56 However, the Kd for c-di-GMP of DDX1 and STING lies in the 5–15 μM range,56−59 which is significantly higher than the Kd of most bacterial c-di-GMP receptors; for example, 120 nM for MrkH44 or 1 μM for Clp.45 Therefore, the NIRW light-induced c-di-GMP systems can operate below the level of interferon activation. In the models intolerable to interferon induction, animals lacking the STING receptor may be used, for example, Goldenticket mice.60
Finally, while we envision the brightest prospects for the use of the engineered c-di-GMP-mediated photocontrol module in deep mammalian tissues (from rodents to humans), it may of course be used in other animals (flies and worms), as well as in plant or microbial organisms that lack c-di-GMP signaling pathways. For example, major bacterial pathogens (staphylococci, streptococci, enterococci, mycoplasmas, etc.) as well as probiotic bacteria (lactobacilli, bacteroidetes, etc.) do not make c-di-GMP (http://www.ncbi.nlm.nih.gov/Complete_Genomes/c-diGMP.html). The c-di-GMP photocontrol module can be introduced in these bacteria to activate or inactivate genes and proteins in animal tissues in vivo, which may be invaluable for gaining insights into host-microbial interactions. The described BphS-YhjH module can of course also be applied as a tool to regulate c-di-GMP-dependent behavior of bacteria that have c-di-GMP signaling pathways, as shown here using E. coli motility and curli fimbriae formation.
Methods
Bacterial Strains and Growth Conditions
Bacterial strains and plasmids used for this study are listed in SI Table S1. E. coli DH5α and BL21[DE3] were used for cloning and protein overexpression, respectively, and the E. coli BL21 derivative, T7 Express (NEB Biolabs), which contains a deletion in the lacZ gene, was used for light experiments. The T7 Express crp mutant was constructed by a one-step gene inactivation method,61 as described earlier.13
A motile E. coli strain expressing an IPTG-inducible T7 RNA polymerase was constructed as follows. A recombinant λDE3 phage carrying the T7 RNA polymerase gene under a Plac promoter was inserted into the chromosome of MG1655 using the λDE3 lysogenization kit according to the instructions of the manufacturer (Novagen). Unexpectedly, λDE3 insertion abrogated the motility of all phage integrants. Therefore, a spontaneous suppressor that regained swimming ability in semisolid agar was selected.
All E. coli strains were grown at 37 or 30 °C in Luria–Bertani (LB) medium.62 For plate assays, cells were streaked on LB agar supplemented with X-gal (40 μg/mL), 0.008% arabinose, 0.025 mM IPTG, and appropriate antibiotics. Plates were grown at 30 °C with or without irradiation provided by light-emitting diode panels All-red (660 nm) LED Grow Light panel 225 (30.5 × 30.5 cm square, LED Wholesalers, CA) at an irradiance of 2 W/m2. Antibiotics were used at the following concentrations (μg/mL): ampicillin, 100; kanamycin, 25; gentamycin 15; tetracycline 5.
Plasmid and Strain Construction
The plasmids for protein expression and purification were constructed using pET23a(+) (Invitrogen). The engineered bphS1 gene was generated by linking the PAS-GAF-PHY encoding fragment (1–511 aa) of bphG1 with the GGDEF encoding fragment (175–343 aa) of slr1143 via fusion PCR. The fragment was ligated in pET23a(+) digested with XbaI and HindIII to yield pETBphS1::His6. The R587A mutation in the RXXD site of BphS1 was introduced by site-directed mutagenesis using the QuickChange kit (Stratagene) to create BphS. The bphS-bphO-yhjH operon under the T7 promoter was constructed using yeast recombineering based on shuttle plasmid pMQ56.63 The bphO and yhjH gene were amplified by PCR from the genomic DNA of R. sphaeroides and E. coli MG1655, respectively, and assembled as a single operon in yeast by homologous recombination, yielding pMQbSHY. The unique HindIII site was introduced in the RBS (RBS1) of yhjH by site-directed mutagenesis.
The clp gene was cloned into pJN105 carrying the pBBR1MCS origin of replication and gentamycin-resistance marker for arabinose-inducible expression.64 The lacZ gene was placed under the control of the lac promoter carrying two CRP binding sites in yeast shuttle vector pMQ125 containing the p15a origin of replication and gentamycin-resistance marker.63 The mrkH gene was cloned into the modified in-house vector pBAD/Myc-HisB (Invitrogen) that was made tetracycline resistant by the insertion of a tetracycline resistance gene into the ScaI site of the ampicillin resistance gene to generate plasmid pBATmrkH. The LacZ reporter plasmids were constructed in pMQ125. The lacZ gene was placed downstream of the 131-bp mrkA promoter (positions −91...+39 relative to the mrkA transcription initiation site48,49 along with the medium-strength RBS (∼300 au according to the RBS Calculator43).
Protein Overexpression, Purification, and Spectroscopy
The His6-tagged bacteriophytochrome proteins (BphG, BphS1, and BphS) were purified using Ni-NTA affinity chromatography as previously described.32 Protein purification was performed under safe green light. Briefly, 10-ml overnight cultures of E. coli BL21[DE3] containing the pETBph::His6 vector together with the heme oxygenase encoding plasmid pT7-ho1-1 described previously32 were transferred into 1 L of fresh LB and grown to A600 0.7 at room temperature. Protein expression was induced with 0.5 mM IPTG and cultures were incubated with shaking at 250 rpm at 18 °C for an additional 20 h. E. coli cells were harvested by centrifugation at 4000g for 10 min and disrupted in the purification buffer (50 mM sodium phosphate buffer (pH 8.0), 300 mM NaCl) plus 0.2 mM phenylmethylsulfonyl fluoride and 10 mM imidazole using a French pressure cell, and cell debris was removed by centrifugation at 10 000g for 30 min at 4 °C. Crude soluble cell extracts were agitated with 3 mL (bed volume) of Ni-NTA resin equilibrated with the buffer for 1 h at 4 °C. The resin was washed with 100 mL of the buffer plus 20 mM imidazole and eluted with the buffer plus 250 mM imidazole. The proteins were used immediately or stored at −80 °C in 20% glycerol. Protein concentrations were measured using a Bradford protein assay kit (Bio-Rad) with bovine serum albumin as the protein standard. Proteins were analyzed using SDS-PAGE.
For spectral analysis, we used light originated from a halogen lamp (EKE21 V150W, GE) with a flexible light guide to which optical filters were attached. The 12.5-mm diameter interference band filters (Andover Corporation) with center wavelengths of 694.3 nm and a 50% bandwidth of 9.8 ± 0.5 nm were used (approximate fluence 3.5 μmol m–2 s–1).
Motility Assays
Flagellar motility was assayed in semisolid agar (1% tryptone, 0.5% NaCl, 0.25% agar) containing 0.02 mM IPTG as previously described.33 Motility zones were measured after 6 h of growth in the dark or light.
Enzymatic Assays
The DGC assays in vitro were performed by measuring the rate of GTP conversion to c-di-GMP as described earlier.36 The protein was irradiated with NIRW light at an irradiance of 0.2 mW/cm2 (which is saturating) or kept in green light. The reaction was initiated by the addition of 200 μM GTP. Aliquots were withdrawn at different time points and boiled for 5 min. The precipitated protein was removed by centrifugation at 15 000g for 5 min. The supernatant was filtered through a 0.22-μm pore size filter (MicroSolv) and analyzed by reversed-phase HPLC.36
β-galactosidase assays were performed as follows. Overnight cultures grown without a chemical inducer were diluted 1:50 in fresh LB medium containing appropriate antibiotics and 2 mM arabinose, 0.025 mM IPTG for MrkH, or 6 mM arabinose and 0.5 mM IPTG for Clp. The transferred cultures were grown at 30 °C, and shaken at 250–275 rpm. The cultures were illuminated at an irradiance of 0.2 mW/cm2 or kept in the dark (wrapped in foil). β-galactosidase activity was analyzed in the aliquots withdrawn at different time points using o-nitrophenyl-β-galactoside as described previously.65 Absorbances were recorded with a UV-1601 PC UV–visible spectrophotometer (Shimadzu). Error bars represent mean ± SD of at least three independent experiments.
Intracellular c-di-GMP Measurements
Intracellular nucleotides were extracted with 40% methanol, 40% acetonitrile in 0.1 N formic acid, following the protocol described earlier.66 Cells were grown at the same conditions used for β-galactosidase assays. C-di-GMP concentrations were quantified by liquid chromatography-tandem mass spectrometry (LC-MS-MS) carried out at the Mass Spectrometry Core at Michigan State University. Quantification of extracted c-di-GMP was determined based on c-di-GMP standards. Intracellular c-di-GMP concentrations were estimated based on the total number of cells. One molecule per bacterial cell yields a concentration of ∼10 nM under the assumption that a typical E. coli cell has an internal volume of ∼2 × 10–16 L.67,68
Acknowledgments
This study was supported in part by the National Science Foundation (MCB1052575) and National Cancer Institute (National Institutes of Health, NIH 5R21CA167862). We are grateful to Marina Tarutina for characterization of R. sphaeroides bphO and Kurt W. Miller for manuscript proofreading.
Supporting Information Available
List of stains and plasmids used in this work (Table S1) as well as alignment of the GGDEF domains from BphG and Slr1143 (Figure S1), protein sequence of BphS (Figure S2), role of BphO-produced chromophore (Figure S3), and modes of light-dependent regulation by transcription factors Clp and MrkH (Figure S4). This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
M.H.R. and M.G. conceived the project. M.H.R. performed experiments. M.H.R. and M.G. analyzed the data and wrote the manuscript.
The authors declare no competing financial interest.
Author Present Address
† For M.-H.R.: Department of Biological Engineering, Massachusetts Institute of Technology, Cambridge, MA 02139, USA
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Mason C.; Brindley D. A.; Culme-Seymour E. J.; Davie N. L. (2011) Cell therapy industry: Billion dollar global business with unlimited potential. Regen. Med. 6, 265–272. [DOI] [PubMed] [Google Scholar]
- Fischbach M. A.; Bluestone J. A.; Lim W. A. (2013) Cell-based therapeutics: The next pillar of medicine. Sci. Transl. Med. 5, 179ps7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenno L.; Yizhar O.; Deisseroth K. (2011) The development and application of optogenetics. Annu. Rev. Neurosci. 34, 389–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak G. P.; Vrana J. D.; Tucker C. L. (2013) Optogenetic control of cell function using engineered photoreceptors. Biol. Cell 105, 59–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller K.; Weber W. (2013) Optogenetic tools for mammalian systems. Mol. BioSyst. 9, 596–608. [DOI] [PubMed] [Google Scholar]
- Levskaya A.; Weiner O. D.; Lim W. A.; Voigt C. A. (2009) Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature 461, 997–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazawa M.; Sadaghiani A. M.; Hsueh B.; Dolmetsch R. E. (2009) Induction of protein–protein interactions in live cells using light. Nat. Biotechnol. 27, 941–945. [DOI] [PubMed] [Google Scholar]
- Renicke C.; Schuster D.; Usherenko S.; Essen L.-O.; Taxis C. (2013) A LOV2 domain-based optogenetic tool to control protein degradation and cellular function. Chem. Biol. 20, 619–626. [DOI] [PubMed] [Google Scholar]
- Kennedy M. J.; Hughes R. M.; Peteya L. A.; Schwartz J. W.; Ehlers M. D.; Tucker C. L. (2010) Rapid blue-light-mediated induction of protein interactions in living cells. Nat. Methods 7, 973–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levskaya A.; Chevalier A. A.; Tabor J. J.; Simpson Z. B.; Lavery L. A.; Levy M.; Davidson E. A.; Scouras A.; Ellington A. D.; Marcotte E. M.; Voigt C. A. (2005) Synthetic biology: Engineering Escherichia coli to see light. Nature 438, 441–442. [DOI] [PubMed] [Google Scholar]
- Ohlendorf R.; Vidavski R. R.; Eldar A.; Moffat K.; Möglich A. (2012) From dusk till dawn: One-plasmid systems for light-regulated gene expression. J. Mol. Biol. 416, 534–542. [DOI] [PubMed] [Google Scholar]
- Airan R. D.; Thompson K. R.; Fenno L. E.; Bernstein H.; Deisseroth K. (2009) Temporally precise in vivo control of intracellular signaling. Nature 458, 1025–1029. [DOI] [PubMed] [Google Scholar]
- Ryu M.-H.; Moskvin O. V.; Siltberg-Liberles J.; Gomelsky M. (2010) Natural and engineered photoactivated nucleotidyl cyclases for optogenetic applications. J. Biol. Chem. 285, 41501–41508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stierl M.; Stumpf P.; Udwari D.; Gueta R.; Hagedorn R.; Losi A.; Gärtner W.; Petereit L.; Efetova M.; Schwarzel M.; Oertner T. G.; Nagel G.; Hegemann P. (2011) Light modulation of cellular cAMP by a small bacterial photoactivated adenylyl cyclase, bPAC, of the soil bacterium Beggiatoa. J. Biol. Chem. 286, 1181–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffelberg S.; Wang L.; Gao S.; Losi A.; Gärtner W.; Nagel G. (2013) A LOV-domain-mediated blue-light-activated adenylate (adenylyl) cyclase from the cyanobacterium Microcoleus chthonoplastes PCC 7420. Biochem. J. 455, 359–365. [DOI] [PubMed] [Google Scholar]
- Römling U.; Galperin M. Y.; Gomelsky M. (2013) Cyclic di-GMP: The first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. 77, 1–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaccolo M. (2011) Spatial control of cAMP signalling in health and disease. Curr. Opin. Pharmacol. 11, 649–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalia D.; Merey G.; Nakayama S.; Zheng Y.; Zhou J.; Luo Y.; Guo M.; Roembke B. T.; Sintim H. O. (2013) Nucleotide, c-di-GMP, c-di-AMP, cGMP, cAMP, (p)ppGpp signaling in bacteria and implications in pathogenesis. Chem. Soc. Rev. 42, 305–341. [DOI] [PubMed] [Google Scholar]
- Corrigan R. M.; Gründling A. (2013) Cyclic di-AMP: Another second messenger enters the fray. Nat. Rev. Microbiol. 11, 513–524. [DOI] [PubMed] [Google Scholar]
- Efetova M.; Petereit L.; Rosiewicz K.; Overend G.; Haußig F.; Hovemann B. T.; Cabrero P.; Dow J. A. T.; Schwärzel M. (2013) Separate roles of PKA and EPAC in renal function unraveled by the optogenetic control of cAMP levels in vivo. J. Cell Sci. 126, 778–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann A.; Arroyo-Olarte R. D.; Imkeller K.; Hegemann P.; Lucius R.; Gupta N. (2013) Optogenetic modulation of an adenylate cyclase in Toxoplasma gondii demonstrates a requirement of the parasite cAMP for host-cell invasion and stage differentiation. J. Biol. Chem. 288, 13705–13717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissleder R. (2001) A clearer vision for in vivo imaging. Nat. Biotechnol. 19, 316–317. [DOI] [PubMed] [Google Scholar]
- Wan S.; Parrish J. A.; Anderson R. R.; Madden M. (1981) Transmittance of nonionizing radiation in human tissues. Photochem. Photobiol. 34, 679–681. [DOI] [PubMed] [Google Scholar]
- Petrov G. I.; Doronin A.; Whelan H. T.; Meglinski I.; Yakovlev V. V. (2012) Human tissue color as viewed in high dynamic range optical spectral transmission measurements. Biomed. Opt. Express 3, 2154–2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatkevich K. D.; Subach F. V.; Verkhusha V. V. (2013) Engineering of bacterial phytochromes for near-infrared imaging, sensing, and light-control in mammals. Chem. Soc. Rev. 42, 3441–3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu X.; Royant A.; Lin M. Z.; Aguilera T. A.; Lev-Ram V.; Steinbach P. A.; Tsien R. Y. (2009) Mammalian expression of infrared fluorescent proteins engineered from a bacterial phytochrome. Science 324, 804–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filonov G. S.; Piatkevich K. D.; Ting L.-M.; Zhang J.; Kim K.; Verkhusha V. V. (2011) Bright and stable near-infrared fluorescent protein for in vivo imaging. Nat. Biotechnol. 29, 757–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbakova D. M.; Verkhusha V. V. (2013) Near-infrared fluorescent proteins for multicolor in vivo imaging. Nat. Methods 10, 751–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatkevich K. D.; Subach F. V.; Verkhusha V. V. (2013) Far-red light photoactivatable near-infrared fluorescent proteins engineered from a bacterial phytochrome. Nat. Commun. 4, 2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell N. C.; Su Y.-S.; Lagarias J. C. (2006) Phytochome structure and signaling mechanisms. Annu. Rev. Plant Biol. 57, 837–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auldridge M. E.; Satyshur K. A.; Anstrom D. M.; Forest K. T. (2012) Structure-guided engineering enhances a phytochrome-based infrared fluorescent protein. J. Biol. Chem. 287, 7000–7009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarutina M.; Ryjenkov D. A.; Gomelsky M. (2006) An unorthodox bacteriophytochrome from Rhodobacter sphaeroides involved in turnover of the second messenger c-di-GMP. J. Biol. Chem. 281, 34751–34758. [DOI] [PubMed] [Google Scholar]
- Ryjenkov D. A.; Simm R.; Römling U.; Gomelsky M. (2006) The PilZ domain is a receptor for the second messenger c-di-GMP: the PilZ domain protein YcgR controls motility in enterobacteria. J. Biol. Chem. 281, 30310–30314. [DOI] [PubMed] [Google Scholar]
- Povolotsky T. L.; Hengge R. (2012) Life-style” control networks in Escherichia coli: Signaling by the second messenger c-di-GMP. J. Biotechnol. 160, 10–16. [DOI] [PubMed] [Google Scholar]
- Christen B.; Christen M.; Paul R.; Schmid F.; Folcher M.; Jenoe P.; Meuwly M.; Jenal U. (2006) Allosteric control of cyclic di-GMP signaling. J. Biol. Chem. 281, 32015–32024. [DOI] [PubMed] [Google Scholar]
- Ryjenkov D. A.; Tarutina M.; Moskvin O. V.; Gomelsky M. (2005) Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J. Bacteriol. 187, 1792–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M.; Park C. (2000) Two novel flagellar components and H-NS are involved in the motor function of Escherichia coli. J. Mol. Biol. 303, 371–382. [DOI] [PubMed] [Google Scholar]
- Pultz I. S.; Christen M.; Kulasekara H. D.; Kennard A.; Kulasekara B.; Miller S. I. (2012) The response threshold of Salmonella PilZ domain proteins is determined by their binding affinities for c-di-GMP. Mol. Microbiol. 86, 1424–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul K.; Nieto V.; Carlquist W. C.; Blair D. F.; Harshey R. M. (2010) The c-di-GMP binding protein YcgR controls flagellar motor direction and speed to affect chemotaxis by a “backstop brake” mechanism. Mol. Cell 38, 128–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X.; Gomelsky M. (2010) A post-translational, c-di-GMP-dependent mechanism regulating flagellar motility. Mol. Microbiol. 76, 1295–1305. [DOI] [PubMed] [Google Scholar]
- Girgis H. S.; Liu Y.; Ryu W. S.; Tavazoie S. (2007) A Comprehensive genetic characterization of bacterial motility. PLoS Genet. 3, e154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfleger B. F.; Pitera D. J.; Smolke C. D.; Keasling J. D. (2006) Combinatorial engineering of intergenic regions in operons tunes expression of multiple genes. Nat. Biotechnol. 24, 1027–1032. [DOI] [PubMed] [Google Scholar]
- Salis H. M. (2011) The ribosome binding site calculator. Meth. Enzymol. 498, 19–42. [DOI] [PubMed] [Google Scholar]
- Ho C. L.; Chong K. S. J.; Oppong J. A.; Chuah M. L. C.; Tan S. M.; Liang Z.-X. (2013) Visualizing the perturbation of cellular cyclic di-GMP levels in bacterial cells. J. Am. Chem. Soc. 135, 566–569. [DOI] [PubMed] [Google Scholar]
- Leduc J. L.; Roberts G. P. (2009) Cyclic di-GMP Allosterically Inhibits the CRP-Like Protein (Clp) of Xanthomonas axonopodis pv. Citri. J. Bacteriol. 191, 7121–7122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber I. T.; Steitz T. A. (1987) Structure of a complex of catabolite gene activator protein and cyclic AMP refined at 2.5 Å resolution. J. Mol. Biol. 198, 311–326. [DOI] [PubMed] [Google Scholar]
- Joung J. K.; Le L. U.; Hochschild A. (1993) Synergistic activation of transcription by Escherichia coli cAMP receptor protein. Proc. Nat. Acad. Sci. 90, 3083–3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilksch J. J.; Yang J.; Clements A.; Gabbe J. L.; Short K. R.; Cao H.; Cavaliere R.; James C. E.; Whitchurch C. B.; Schembri M. A.; Chuah M. L. C.; Liang Z.-X.; Wijburg O. L.; Jenney A. W.; Lithgow T.; Strugnell R. A. (2011) MrkH, a novel c-di-GMP-dependent transcriptional activator, controls Klebsiella pneumoniae biofilm formation by regulating type 3 fimbriae expression. PLoS Pathog. 7, e1002204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. G.; Murphy C. N.; Sippy J.; Johnson T. J.; Clegg S. (2011) Type 3 fimbriae and biofilm formation are regulated by the transcriptional regulators MrkHI in Klebsiella pneumoniae. J. Bacteriol. 193, 3453–3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasteva P. V.; Fong J. C. N.; Shikuma N. J.; Beyhan S.; Navarro M. V. A. S.; Yildiz F. H.; Sondermann H. (2010) Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science 327, 866–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner S.; Lori C.; Boehm A.; Jenal U. (2013) Allosteric activation of exopolysaccharide synthesis through cyclic di-GMP-stimulated protein–protein interaction. EMBO J. 32, 354–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsan N.; Lee E. R.; Weinberg Z.; Moy R. H.; Kim J. N.; Link K. H.; Breaker R. R. (2008) Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science 321, 411–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E. R.; Baker J. L.; Weinberg Z.; Sudarsan N.; Breaker R. R. (2010) An allosteric self-splicing ribozyme triggered by a bacterial second messenger. Science 329, 845–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomelsky M. (2011) cAMP, c-di-GMP, c-di-AMP, and now cGMP: Bacteria use them all!. Mol. Microbiol. 79, 562–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques S. L. (2013) Optical properties of biological tissues: A review. Phys. Med. Biol. 58, R37. [DOI] [PubMed] [Google Scholar]
- Parvatiyar K.; Zhang Z.; Teles R. M.; Ouyang S.; Jiang Y.; Iyer S. S.; Zaver S. A.; Schenk M.; Zeng S.; Zhong W.; Liu Z.-J.; Modlin R. L.; Liu Y.; Cheng G. (2012) The helicase DDX41 recognizes the bacterial secondary messengers cyclic di-GMP and cyclic di-AMP to activate a type I interferon immune response. Nat. Immunol. 13, 1155–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdette D. L.; Monroe K. M.; Sotelo-Troha K.; Iwig J. S.; Eckert B.; Hyodo M.; Hayakawa Y.; Vance R. E. (2011) STING is a direct innate immune sensor of cyclic di-GMP. Nature 478, 515–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang G.; Zhu D.; Li N.; Zhang J.; Zhu C.; Lu D.; Liu C.; Yu Q.; Zhao Y.; Xu S.; Gu L. (2012) Crystal structures of STING protein reveal basis for recognition of cyclic di-GMP. Nat. Struct. Mol. Biol. 19, 725–727. [DOI] [PubMed] [Google Scholar]
- Yin Q.; Tian Y.; Kabaleeswaran V.; Jiang X.; Tu D.; Eck M. J.; Chen Z. J.; Wu H. (2012) Cyclic di-GMP Sensing via the innate immune signaling protein STING. Mol. Cell 46, 735–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer J.-D.; Sotelo-Troha K.; von Moltke J.; Monroe K. M.; Rae C. S.; Brubaker S. W.; Hyodo M.; Hayakawa Y.; Woodward J. J.; Portnoy D. A.; Vance R. E. (2011) The N-ethyl-N-nitrosourea-induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infect. Immun. 79, 688–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko K. A.; Wanner B. L. (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. 97, 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E., and Maniatis T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- Shanks R. M. Q.; Kadouri D. E.; MacEachran D. P.; O’Toole G. A. (2009) New yeast recombineering tools for bacteria. Plasmid 62, 88–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman J. R.; Fuqua C. (1999) Broad-host-range expression vectors that carry the l-arabinose-inducible Escherichia coli araBAD promoter and the araC regulator. Gene 227, 197–203. [DOI] [PubMed] [Google Scholar]
- Tai T.-N.; Havelka W. A.; Kaplan S. (1988) A broad-host-range vector system for cloning and translational lacZ fusion analysis. Plasmid 19, 175–188. [DOI] [PubMed] [Google Scholar]
- Bobrov A. G.; Kirillina O.; Ryjenkov D. A.; Waters C. M.; Price P. A.; Fetherston J. D.; Mack D.; Goldman W. E.; Gomelsky M.; Perry R. D. (2011) Systematic analysis of cyclic di-GMP signalling enzymes and their role in biofilm formation and virulence in Yersinia pestis. Mol. Microbiol. 79, 533–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halling P. J. (1989) Do the laws of chemistry apply to living cells?. Trends Biochem. Sci. 14, 317–318. [DOI] [PubMed] [Google Scholar]
- Gu H.; Furukawa K.; Breaker R. R. (2012) Engineered allosteric ribozymes that sense the bacterial second messenger cyclic diguanosyl 5′-monophosphate. Anal. Chem. 84, 4935–4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.