Abstract
Pep5 is a cationic pore-forming lantibiotic produced by Staphylococcus epidermidis strain 5. The producer strain protects itself from the lethal action of its own bacteriocin through the 69-amino-acid immunity peptide PepI. The N-terminal segment of PepI contains a 20-amino-acid stretch of apolar residues, whereas the C terminus is very hydrophilic, with a net positive charge. We used green fluorescent protein (GFP)-PepI fusions to obtain information on its localization in vivo. PepI was found to occur outside the cytoplasm and to accumulate at the membrane-cell wall interface. The extracellular localization appeared essential for conferring immunity. We analyzed the functional role of the specific segments by constructing various mutant peptides, which were also fused to GFP. When the hydrophobic N-terminal segment of PepI was disrupted by introducing charged amino acids, the export of PepI was blocked and clones expressing such mutant peptides were Pep5 sensitive. When PepI was successively shortened at the C terminus, in contrast, its export properties remained unchanged whereas its ability to confer immunity was gradually reduced. The results show that the N-terminal part is required for the transport of PepI and that the C-terminal part is important for conferring the immunity phenotype. A concept based on target shielding is proposed for the PepI immunity mechanism.
Lantibiotics, a subgroup of bacteriocins from gram-positive bacteria, are polycyclic peptides containing modified amino acids: in particular, the thioether amino acids lanthionine and 3-methyllanthionine and the dehydroamino acids 2,3-didehydroalanine and 2,3-didehydrobutyrine. Lantibiotics are ribosomally synthesized as precursor peptides, consisting of a leader sequence and a propeptide part (31). The precursor peptides are converted into the mature peptide by posttranslational modification followed by processing and export from the producing cell (for a review, see reference 27). The cationic lantibiotic Pep5 is produced by Staphylococcus epidermidis strain 5 (28) and is classified along with nisin, subtilin, and epidermin as a pore-forming type-A lantibiotic (27). The biosynthetic gene cluster of Pep5 contains the structural gene pepA as well as the genes for posttranslational modification (pepB and pepC), processing (pepP), transport (pepT), and immunity (pepI) (18).
Bacteriocin production generally requires a self-protection mechanism for the producer strain (1, 29). Various immunity concepts have been elaborated by the different groups of bacteria; e.g., the immunity proteins of the channel-forming colicins (e.g., colicin A, E1, and B) reside in the cytoplasmic membrane and protect the producer by forming a stoichiometric complex with the respective colicin (10, 32, 36). For the lantibiotic bacteriocins of gram-positive bacteria generally two different mechanisms, which in some cases may even complement each other, are found: the small immunity peptides collectively defined as LanI and the ABC (ATP-binding cassette) transporters LanFEG. The latter have been identified in the biosynthetic gene clusters of the nisin (34), subtilin (15), epidermin (21), lacticin 481 (26, 26), and mersacidin (12) producer strains. They share sequence homology to the type-B ABC transporter family and consist typically of three separate proteins, which are most likely located in the membrane and act by expelling the active lantibiotic from the cytoplasmic membrane (12, 19, 21). In contrast, the molecular function of immunity peptides and proteins remains enigmatic. The 165-amino-acid NisI and the 245-amino-acid SpaI immunity proteins confer immunity to nisin and subtilin, respectively. These immunity proteins are characterized by the presence of a typical N-terminal lipoprotein signal sequence, suggesting that these proteins are attached to the outside of the cytoplasmic membrane (15, 16, 23). However, NisI and SpaI do not share sequence similarity to other known proteins and do not provide cross-immunity. The 116-amino-acid protein LtnI, which confers immunity to the two-peptide lantibiotic lacticin 3147, is also unrelated to any known protein and is predicted to be localized within the cytoplasmic membrane (17). PepI, the 69-amino-acid immunity peptide of the Pep5 producer strain, is characterized by a hydrophobic N-terminal segment and a strongly hydrophilic C-terminal part. PepI displays a high degree (74.2%) of sequence similarity to EciI, the immunity protein of the epicidin 280 producer S. epidermidis BN 280 (13). Epicidin 280 is related to Pep5, and their producer strains show cross-immunity, which indicates a similar self-protection mechanism for both lantibiotics. PepI and EciI seem to be representatives of a unique class of immunity peptides, since in the gene clusters of the structurally unrelated lantibiotic lactocin S (35) and of the nonlantibiotic divergicin A (38) similar genes which code for peptides of comparable size, charge distribution, and significant sequence similarity have been identified (13). The structural similarity of these immunity peptides and the absence of any obvious structural similarity of the corresponding bacteriocins could indicate that the immunity mechanisms are related. In contrast to the channel-forming colicins, however, their mechanism may not be based on direct stoichiometric interaction of the bacteriocin and the immunity peptide. In a first attempt to elaborate a concept on how such immunity peptides may antagonize pore formation, we used green fluorescent protein (GFP) fusion and site-specific mutagenesis to localize PepI in the cell to correlate structural and functional features.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used are listed in Table 1 and Table 2. All staphylococcal strains were maintained on blood agar or tryptone soy agar (TSA) (Merck, Darmstadt, Germany) supplemented with the appropriate antibiotic (25 μg of tetracycline ml−1 or 40 μg of ampicillin ml−1) (Sigma-Aldrich, Deisenhofen, Germany). Staphylococcus carnosus TM300 (30) was used for heterologous expression, and S. epidermidis strain 25 was used for homologous expression. Escherichia coli TB1 was used as the host strain for pEGFP-1 (BD Biosciences, Erembodegem, Belgium) and was maintained on Luria-Bertani agar containing 25 μg of kanamycin ml−1 (Sigma-Aldrich).
TABLE 1.
Bacterial strains or plasmids
| Bacterial strain or plasmid | Characteristics | Reference or source |
|---|---|---|
| Strains | ||
| Staphylococcus epidermidis strain 5 | Pep5+ Imm+, wild-type producer of Pep5, harbors pED503 (20 kb) | 28 |
| Staphylococcus epidermidis strain 25 | Pep5− Imm−, S. epidermidis 5, pED503 removed | 9 |
| Staphylococcus carnosus TM300 | Cloning host, sensitive to Pep5 | 30 |
| Escherichia coli TB1 | araΔ(lac-proAB), rpsL(φ80, lacZΔM15), hsdR | 14 |
| Plasmids | ||
| pCU1 | Ampr Cmr; 4.95-kb shuttle vector, contains pC194 and pUC19 sequences and multiple cloning site from pUC19 | 4 |
| pAG1/1 | Ampr Cmr, contains a mutated pepI gene in which Ile17 has been exchanged for Arg cloned in pCU1 | 20 |
| pAG4/1 | Ampr Cmr, contains a mutated pepI gene in which Phe13 and Ile17 have been exchanged for Asp and Arg, respectively, cloned in pCU1 | 20 |
| pAG4/2 | Ampr Cmr, contains a mutated pepI gene in which Phe13 and Ile17, and Lys65 have been exchanged for Asp, Arg and Stop, respectively, cloned in pCU1 | This study |
| pTX15 | Tetr, 7.2-kb staphylococcal expression vector | 22 |
| pUP10 | Tetr, contains 0.244-kb PCR-pepI and 0.024-kb six-His tag cloned in pTX15 (Δlip) | This study |
| pAH3 | Tetr, contains 0.772-kb PCR-eciO and 0.024-kb His tag cloned in pTX15 (Δlip) | This study |
| pEGFP-1 | Kanr, Neor, 4.2-kb promotorless expression vector contains a 0.720-kb gfp gene | BD Biosciences |
| pAH-PepI1-68 | Tetr, contains 0.244-kb PCR-pepI and 0.732-kb PCR-gfp cloned in pTX15 (Δlip) | This study |
| pAH-EciO | Tetr, contains 0.772-kb PCR-eciO and 0.732-kb PCR-gfp cloned in pTX15 (Δlip) | This study |
TABLE 2.
Plasmids generated from pTX15 used for mutated PepI-Gfp expression and designation of mutated peptides
| Plasmida | Designation and size of the mutated PepI peptides |
|---|---|
| pTS(I17R)-PepI1-68 | I17R-PepI1-68-Gfp |
| pTS(I17R;K59T)-PepI1-68 | I17R;K59T-PepI1-68-Gfp |
| pTS(F13D;I17R)-PepI1-68 | F13D;I17R-PepI1-68-Gfp |
| pTS(F13D;I17R)-PepI1-63 | F13D;I17R-PepI1-63-Gfp |
| pTS-PepI1-63 | PepI1-63-Gfp |
| pTS-PepI1-57 | PepI1-57-Gfp |
| pTS-PepI1-53 | PepI1-53-Gfp |
The source for all plasmids was this study.
DNA isolation and cloning.
Plasmid DNAs of E. coli TB1 and staphylococcal strains were isolated with QIAprep spin columns (QIAGEN, Hilden, Germany). Lysis of staphylococcal cells was achieved by adding 200 μg of lysostaphin ml−1 to buffer P1 (QIAGEN) and incubating the cells for 30 to 60 min at 37°C.
S. carnosus TM300 protoplasts were transformed as described by Götz and Schumacher (11). E. coli TB1 and S. epidermidis strain 25 were transformed by electroporation (3). DNA sequencing was performed by Sequiserve. Restriction enzymes and T4 DNA ligase were obtained from Roche (Mannheim, Germany).
PCR amplification.
Plasmid DNAs of pUP10, pEGFP-1, pAH3, pAG1/1, pAG4/1, pAG4/2, and pTS-PepI1-63 served as templates for PCR amplification of pepI, mutated pepI, and gfp genes. Oligonucleotides used as primers were purchased from Metabion (Planegg-Martinsried, Germany). Primer sequences and restriction sites are shown in Table 3. The Pwo DNA polymerase and deoxynucleoside triphosphates were obtained from Hybaid-AGS (Heidelberg, Germany) and Roche, respectively.
TABLE 3.
Primers used in this study
| Primer | Sequence (5′-3′)a | Restric- tion site |
|---|---|---|
| GFP5′ | GTCGCCGTGCACGTGAGCAAGGGCGAGGA GCTG | Alw44I |
| GFP3′ | CTAGAGACGCGTCCGCTTTACTTGTACAGC | MluI |
| pepIMut 5′ | TAAGGGATCCATAAGATTATCTAAATATAT TTAA AAAG | BamHI |
| ptxpepI 3′ | ACATAGTGCACTTTATCTTTTTTGTTGCTATT TA | Alw44I |
| pepIMut 5′ | TAAGGGATCCATAAGATTATCTAAATATATT TAAAAAG | BamHI |
| 3′pepI63b | GTTTATCGTGCACGTTGCTATTTATTCTTTTT TC | Alw44I |
| 3′pepI57b | ATTTGTGCACACTTAATTTCGTTGAATAATCG | Alw44I |
| 3′pepI53b | TTCTGTGCACATATTTTTTATCACTTAATTTCG TTG | Alw44I |
Restriction sites in the primer sequence are underlined. Mutations in the primer sequence are marked in boldface.
The number to the right of “pepI” indicates the amino acid number of PepI.
Plasmid construction and mutagenesis.
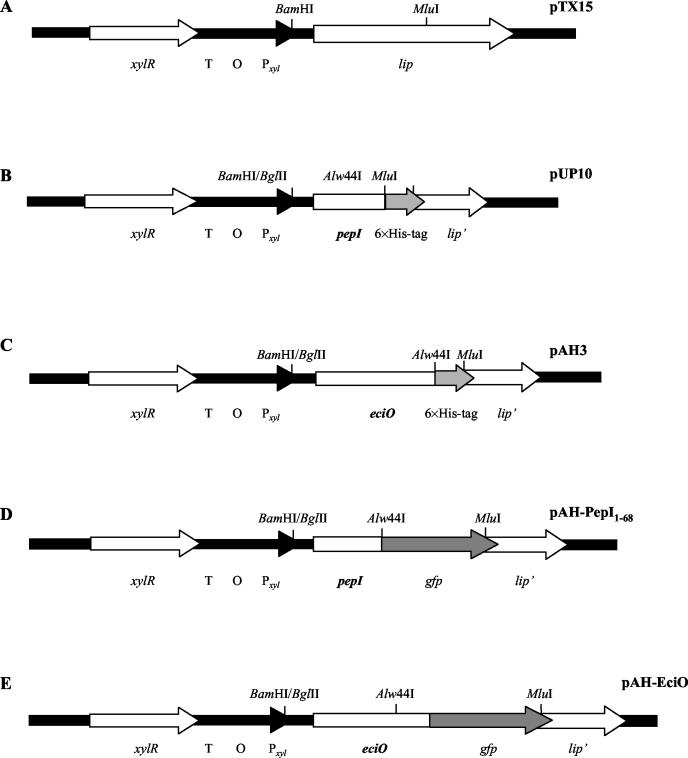
The plasmid pAH-PepI (Fig. 1) containing the 0.98-kb pepI-gfp fusion gene was constructed by cloning the gfp gene into pUP10 (Fig. 1). For amplification of gfp, the primer pair GFP5′-GFP3′ (Table 3) and the template pEGFP-1 (BD Biosciences) were used. The six-His tag fragment was excised from pUP10 and exchanged for the 0.73-kb gfp gene. For construction of pAH-EciO, the six-His tag in pAH3 was exchanged for gfp, resulting in the 1.5-kb fusion gene eciO-gfp (Fig. 1).
FIG. 1.
Construction of plasmids for xylose-inducible expression of pepI-six-His tag, pepI-gfp, and eciO-gfp fusion genes in pTX15. (A) Staphylococcal expression vector pTX15 (22), containing the xylR repressor gene, the terminator (T), the operator XylR (O), the xylA promoter (Pxyl), and the lipase gene (lip). (B) Plasmid pUP10, containing pepI-six-His tag under the control of the xylA promoter, was constructed by replacing the BamHI/MluI lip gene fragment of pTX15 with pepI-six-His tag. PUP10 was the origin for the construction of the plasmids pAH-PepI1-68 for PepI-GFP expression and pTS(I17R)-PepI1-68, pTS(I17R;K59T)-PepI1-68, pTS(F13D;I17R)-PepI1-68, pTS(F13D;I17R)-PepI1-63, pTS-PepI1-63, pTS-PepI1-57, and pTS-PepI1-53 for expression of mutated PepI-GFP fusion proteins. (C) Plasmid pAH3, containing eciO-six-His tag under the control of the xylA promoter, was constructed by ligating eciO-six-His tag to the BamHI/MluI-digested pTX15. (D) Plasmid pAH-PepI1-68 was used for expression of PepI-GFP. The six-His tag of pUP10 was replaced by the Alw44I/MluI gfp fragment. (E) The plasmid pAH-EciO, which was used for EciO-GFP expression, was constructed by replacing the six-His tag of pAH3 with the Alw44I/MluI-restricted gfp gene.
Mutated pepI genes, which contain amino acid exchanges in the N-terminal segment, were amplified with the primer pair pepImut5′-pTXpepI3′ (Table 3), with the plasmids pAG1/1, pAG4/1 (20), and pAG4/2 (unpublished results) as templates. Following amplification, mutated pepI fragments were digested with the restriction enzymes BamHI and Alw44I, ligated to the Alw44I/MluI-restricted gfp genes, and then cloned into the BamHI/MluI-digested pTX15. Shortened pepI genes were obtained by amplification of intact pepI in pUP10 with the primer pairs pepImut5′-3′pepI63, pepImut5′-3′pepI57, and pepImut5′-3′pepI53 (Table 3). Following BamHI/Alw44I digestion, the mutated pepI fragments were ligated to the Alw44I/MluI-restricted gfp genes and cloned into the BamHI/MluI-restricted pTX15. The resulting plasmids and PepI peptides containing mutations are listed in Table 2, and the respective peptide structures are indicated in Fig. 2.
FIG. 2.
Primary structure of PepI and PepI mutants.
By the use of the Alw44I restriction site for cloning pepI in frame to gfp, the leucine residue at position 69 was exchanged for a valine residue followed by gfp. Further use of this cloning site for construction of the other gfp fusions resulted in a carryover of the valine residue in front of the gfp gene. Therefore, we chose the designations PepI1-68-GFP, PepI1-63-GFP, PepI1-57-GFP, and PepI1-53-GFP to consistently indicate the unchanged segment of PepI.
Expression of pepI-gfp, eciO-gfp, and mutated pepI-gfp fusion genes.
Recombinant staphylococcal strains were grown in B-Broth (10 g of casein hydrolysate, 5 g of yeast extract, 5 g of NaCl, 20 mM lactate, 1 g of K2HPO4, pH 7.3). Expression of fusion genes and mutated PepI-GFP was induced by adding 0.5% xylose. At an optical density at 600 nm (OD600) of 1, intact cells were separated from the culture supernatant by centrifugation of 2 ml of culture at 4,500 × g for 10 min. Cells were lysed in 100 μl of buffer P1 (50 mM Tris-HCl, 10 mM EDTA, 100 μg of RNAse A ml−1) containing 300 μg of lysostaphin ml−1 for 30 to 60 min at 37°C. The cell suspension was mixed with 20 μl of fivefold sodium dodecyl sulfate (SDS) sample buffer and boiled for 10 min. Proteins in the culture supernatant were precipitated by addition of trichloroacetic acid (20%) and incubation at 4°C for 1 to 2 h. Following centrifugation at 20,000 × g at 4°C for 15 min, the precipitated proteins were dissolved in 2 μl of 1 M Tris and 30 μl of SDS sample buffer.
Proteins were analyzed by SDS-polyacrylamide gel electrophoresis and immunoblotting with a polyclonal anti-GFP horseradish peroxidase conjugate (BD Biosciences). A recombinant GFP (QBIOgene, Heidelberg, Germany) and soluble cell fraction of S. epidermidis strain 25 producing PepI-GFP were used as a positive control and size standard.
For growth curves and subsequent Western blot analysis of cell pellets and culture supernatants, culture aliquots of S. epidermidis strain 25 pAH-PepI1-68 and pAH-EciO were harvested at defined ODs ranging from 0.4 to 2.0 and treated as described above.
Protein preparation of Staphylococcus cell fractions.
The preparation of soluble cytoplasmic and membrane fractions of S. epidermidis strain 25 strains was done by disruption of cells with glass beads and differential centrifugation, as previously described (24).
MIC determination.
The MIC determination for Pep5 was performed in a microtiter plate assay with half-concentrated tryptone soy broth as previously described (5). For induction of fusion gene expression 0.5% xylose was added to the medium. To reduce binding of positively charged Pep5 to the surface, wells were coated with bovine serum albumin (BSA) by incubation of each well with 200 μl of 1% BSA in phosphate-buffered saline buffer for 30 min at 37°C and subsequent washing with phosphate-buffered saline buffer.
Gradient agar sensitivity test.
Inoculum plates were filled with 40 ml of half-concentrated TSA containing 0.5% xylose and lifted on one side to create a slope ending exactly in one corner of the plate. Then 40 ml of half-concentrated TSA supplemented with 0.5% xylose and 100 nM Pep5 was filled onto the horizontally placed plate. The two-layer bedding of the agar resulted in a Pep5 concentration gradient from 0 to 100 nM. Diluted bacterial culture (0.5 McFarland units) was applied with a sterile cotton swab along the Pep5 gradient and incubated overnight at 37°C.
Fluorescence microscopy.
An overnight culture was diluted 50-fold in half-concentrated tryptone soy broth supplemented with 25 μg of tetracycline ml−1 and 0.5% xylose and shaken at 37°C for 24 h. Cells of a 100-μl volume were harvested by centrifugation at 4,500 × g for 5 min. The pellet was dissolved in 25 μl of 0.9% NaCl solution and mixed with 25 μl of 2% agarose solution. Then 7 μl of the solution was applied to a microscope slide and fixed with a cover glass. Cells were viewed with a Axioplan microscope (Zeiss, Jena, Germany) equipped with an oil-immersion plan neofluar objective (100×; numeric aperture = 1.3) and a HVC20A charge-coupled-device video camera (Hitachi). For fluorescence imaging a fluorescein isothiocyanate filter (Filter set BP 450-490, FT 510, and LP 515; Zeiss) was used. Digital images were obtained by using DISKUS version 4.20.34 software (Hilgers, Königswinter, Germany).
RESULTS
In previous cell fractionation studies, PepI was found in the soluble as well as in the membrane fraction (24) and it was suggested that PepI might be loosely associated with the cytoplasmic membrane, where it can antagonize Pep5 activity through a mechanism similar to the that of the complex formation of channel-forming colicins with their respective immunity proteins. To study functional aspects of PepI, it was important to obtain precise information on its cellular localization. We chose to fuse the C terminus of PepI to the GFP, which was expected to allow in vivo detection and Western blot analysis using GFP antibodies. To control the localization of PepI, we used EciO, a cytoplasmic oxidoreductase which is putatively involved in posttranslational modification of the N terminus of epicidin 280, a lantibiotic related to Pep5 (13).
GFP fusion constructs were expressed in the staphylococcal expression vector pTX15 (22) in which the respective genes are under control of the xylose-inducible xylA promoter (Fig. 1). The resulting plasmids (Table 1 and Table 2) were introduced into the Pep5-sensitive strains S. carnosus TM300 for heterologous expression and S. epidermidis strain 25 for homologous expression. S. epidermidis strain 25, a variant of the Pep5 producer S. epidermidis strain 5, has been cured from the plasmid pED503 harboring the Pep5 biosynthetic gene cluster including pepI (9).
PepI-GFP fusion proteins confer Pep5 immunity.
To test the impact of GFP fusions on the antagonistic activity of PepI, we compared the susceptibility of cloning hosts expressing various PepI peptides with that of the wild-type producer (Table 4). The Pep5 MIC for control strain S. epidermidis strain 5 (Pep5 producer, wild-type immunity level) was 1,470 nM, that for S. epidermidis strain 25 (Pep5−, Imm−; S. epidermidis strain 5 cured from pED503) was 3 nM, and that for S. carnosus TM300 (cloning host) was 0.1 nM.
TABLE 4.
Pep5 susceptibility of strains producing PepI-Gfp and six-His-tagged PepI1-68
| Plasmid | MIC (nM) for expression host:
|
|
|---|---|---|
| S. epidermidis strain 25 | S. carnosus TM300 | |
| pTX15 (cloning vector) | 3 | 0.1 |
| pAH-EciO (EciO-Gfp, negative control) | 3 | 0.1 |
| pAH-PepI (PepI1-68-Gfp) | 103 | 1.5 |
| pUP10 (six-His-tagged PepI1-68) | 735 | 5 |
As a standard assay, we used MIC determination in twofold broth dilution; however, it was important to include BSA-precoated microtiter plates, since Pep5 adsorbs significantly to plastic, which strongly reduces its bioavailability and increases the MICs. This was of particular importance at low Pep5 concentrations, e.g., in the nanomolar range, at which the Pep5 MIC for S. carnosus TM300 shifted from 0.09 nM in coated plates to 2.7 nM in uncoated plates. At micromolar concentrations, the amount of Pep5 adsorbed was too small to reduce the MIC when determined in twofold dilution series. Additional information on intermediate susceptibilities was obtained from a Pep5 gradient agar plate.
The wild-type Pep5 producer S. epidermidis strain 5 was found to be 500-fold less susceptible than the cured variant S. epidermidis strain 25 when tested under such conditions (Table 4). Introduction of the empty cloning vector pTX15 and expression of the control protein EciO did not change the susceptibility of the respective strains. When six-His-tagged PepI was expressed from pTX15, the variant was 250 times less susceptible, almost reaching wild-type levels of immunity. Six-His-tagged PepI had been previously shown to have the same potency for antagonization of Pep5 as wild-type PepI (U. Pag, unpublished results). Expression of a C-terminal fusion of GFP to PepI increased the MIC from 3 to 103 nM; i.e., the expression host was approximately 10-fold more susceptible than wild-type or His-tagged PepI-producing strains. Considering that the 69-amino-acid peptide is turned into a 35-kDa fusion protein in which sterical hindrance of PepI activity may be expected, however, the residual potency of PepI-GFP is still remarkable. When the PepI plasmids were heterologously expressed in S. carnosus, we observed a similar trend. His-tagged PepI reduced the susceptibility of the cloning host by a factor of 50 and of PepI-GFP by a factor of 15.
PepI-GFP is exported outside the cells.
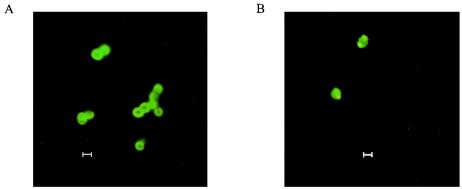
We further inspected growing PepI-GFP-producing clones by fluorescence microscopy (Fig. 3); the results for both host strains were similar. Cells expressing the PepI fusion protein mostly showed bright fluorescence, with a clear ring surrounding the entire cell, including the newly synthesized septal area of the cell. The average diameter of such cells was in the range of 1.4 to 1.6 μm. In contrast, cells expressing GFP fused to the cytoplasmic protein EciO showed homogenous fluorescence, occasionally with irregular bright spots, presumably representing inclusion bodies. Such cells lacked the fluorescent ring, and the fluorescent area was considerably smaller (1 to 1.3 μM). We conclude from these observations that while EciO remains inside the cells, PepI is largely exported but remains trapped at the membrane and within the cell wall compartment.
FIG. 3.
Localization of PepI-GFP (A) and EciO-GFP (B) fusion proteins by fluorescence microscopy. S. carnosus TM300 pAH-PepI1-68 and pAH-EciO, respectively, were grown in cultures in B-broth in the presence of 0.5% xylose, which induces the expression of the fusion proteins PepI-GFP and EciO-GFP. Cells were harvested and embedded in 1% agarose for fluorescence microscopy. Bars, 1 μm.
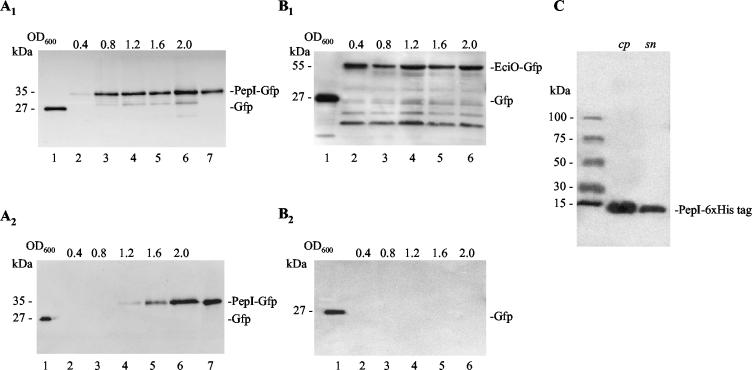
To verify the microscopic observations, the producer strains, S. epidermidis strain 25 pAH-PepI1-68 and S. epidermidis pAH-EciO, of both fusion proteins were analyzed to determine the occurrence of the fusion protein in the cell and the supernatant fractions (Fig. 4A and B). PepI-GFP (35 kDa) was detected for the first time in cells at an OD600 of 0.4 and subsequently appeared in the culture supernatant at an OD600 of 1.2. Detection of PepI-GFP in the supernatant at this early stage during the exponential growth phase indicated active transport of PepI-GFP to the outside. In contrast, the intracellularly located EciO-GFP (55 kDa) was only detected in the cell pellet, making it unlikely that the appearance of PepI outside the cells resulted from cell lysis. Similar results were obtained when both fusion proteins were expressed in S. carnosus TM300 (data not shown). Furthermore, the six-His-tagged PepI was similarly detected at first in the cell pellet and subsequently in the culture supernatant (Fig. 4C). These results demonstrated that PepI-GFP and His-tagged PepI are transported outside the cells whereas EciO-GFP accumulated in the cytoplasm. Surprisingly, transport of PepI was not inhibited by the fusion to GFP.
FIG. 4.
Western blot analysis of the cellular localization of PepI-GFP and EciO-GFP in the course of a growth curve. Western blots show cellular protein (A1 and B1) and excreted protein (A2 and B2) of S. epidermidis strain 25 pAH-PepI1-68 (A) and S. epidermidis strain 25 pAH-EciO (B), respectively. Equivalent amounts of cells were harvested at optical densities of 0.4, 0.8, 1.2, 1.6, and 2, and the proteins were extracted and analyzed as described in Materials and Methods. Recombinant GFP (QBIOgene) and PepI-GFP were used as positive controls and size standards. Lane 1, 0.01 μg of recombinant GFP (27 kDa); lanes 2 to 6, lysed cells (A1 and B1) or culture supernatant (A2 and B2) obtained at an OD600 of 0.4, 0.8, 1.2, 1.6, and 2.0; lane 7, PepI-GFP. Partial degradation was observed when mutations in PepI caused intracellular accumulation of the respective fusion protein (see also Fig. 6). (C) Western blot of cellular and excreted proteins of S. carnosus TM300 pUP10 expressing six-His-tagged PepI.
Mutations in the N-terminal segment of PepI affect transport.
PepI is characterized by a striking charge distribution. The positively charged C terminus is strongly hydrophilic, whereas the N-terminal segment contains a 20-amino-acid stretch of apolar residues. To analyze the functional role of the N-terminal segment of PepI, mutant peptides were constructed in which the hydrophobic stretch was disrupted by replacing uncharged with charged amino acids.
The exchange of isoleucine at position 17 for arginine (I17R-PepI1-68-GFP) resulted in a considerable decrease of the immunity level in S. carnosus TM300 and S. epidermidis strain 25 compared to the results seen with strains expressing native PepI-GFP (Table 5). When in addition Phe13 was replaced by a negatively charged amino acid (F13D;I17R-PepI1-68-GFP), interestingly, the level of immunity was slightly increased (Table 5 and Fig. 5A).
TABLE 5.
Pep5 susceptibility of strains producing PepI and mutant PepI-Gfp
| Plasmid and characteristic | MIC (nM) for expression host
|
|
|---|---|---|
| S. epidermidis strain 25 | S. carnosus TM300 | |
| No plasmid | 3 | 0.1 |
| pAH-PepI1-68 | 103 | 1.5 |
| N-terminal mutations in PepI | ||
| pTS(I17R)-PepI1-68 | 9 | 0.3 |
| pTS(F13D;I17R)-PepI1-68 | 15 | 0.35 |
| C-terminal shortening of PepI | ||
| pTS-PepI1-63 | 103 | 1.5 |
| pTS-PepI1-57 | 20 | 0.3 |
| pTS-PepI1-53 | 7.5 | 0.15 |
| N- and C-terminal mutations in PepI | ||
| pTS(I17R;K59T)-PepI1-68 | 9 | 0.3 |
| pTS(F13D;I17R)-PepI1-63 | 15 | 0.35 |
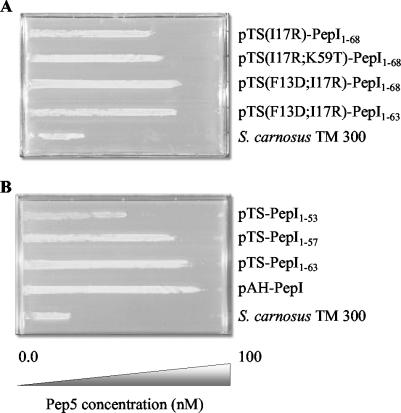
FIG. 5.
Pep5 susceptibility of PepI-producing variants. Cultures of S. carnosus TM300 strains producing PepI-GFP and mutated PepI-GFP were diluted up to 0.5 McFarland units with 0.9% NaCl solution, inoculated onto a Pep5 gradient plate, and incubated overnight at 37°C. S. carnosus TM300 containing the staphylococcal expression vector pTX15 served as a control.
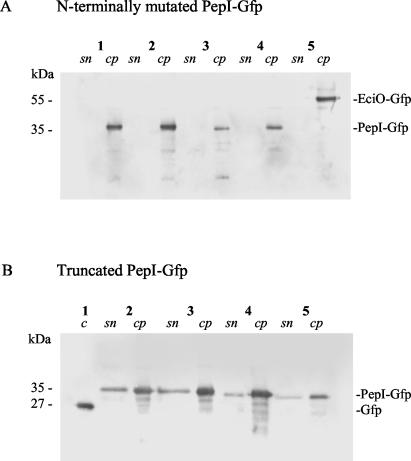
Western blot analysis demonstrated that both N-terminally mutated PepI-GFP proteins, just like the EciO control, were localized in the cell pellet but not in the culture supernatant (Fig. 6, lanes 1 and 4). Moreover, fluorescence microscopy showed that cells producing the PepI-GFP mutant peptides exhibited homogenously green fluorescence (data not shown) without the characteristic ring around the cell (Fig. 3B). This strongly suggests that disruption of the hydrophilic stretch between Val7 and Ile24 interferes with transport of PepI to the outside. Partial restoration of immunity (and transport compatibility) in the Asp13/Lys17 double mutant may be explained by the high-level helix propensity of this PepI segment (25). A helical conformation should allow Asp13 and Lys17 to form a salt bridge, which would neutralize the charges, thus reducing their impact on the hydrophobic segment.
FIG. 6.
Cellular localization of mutant PepI-GFP by Western blot analysis. A total of 30 μl of each culture supernatant (sn) and a total of 10 μl of each cell pellet (cp) obtained at an OD600 of 1 were used. Recombinant GFP (QBIOgene) (0.1 μg) was used as a positive control (c) and size standard. (A) N-terminally mutated PepI-GFP. Lanes: 1, S. carnosus TM300 pTS(F13D;I17R)-PepI1-68; 2, S. carnosus TM300 pTS(F13D;I17R)-PepI1-63; 3, S. carnosus TM300 pTS(I17R;K59T)-PepI1-68; 4, S. carnosus TM300 pTS(I17R)-PepI1-68; 5, S. carnosus TM300 pAH-EciO. (B) Truncated PepI-GFP. Lanes: 1, 0.01 μg of recombinant GFP (27 kDa); 2, S. carnosus TM300 pAH-PepI1-68; 3, S. carnosus TM300 pTS-PepI1-63; 4, S. carnosus TM300 pTS-PepI1-57; 5, S. carnosus TM300 pTS-PepI1-53.
The C-terminal segment of PepI is essential for immunity.
We successively shortened PepI and constructed PepI1-63-GFP, PepI1-57-GFP, and PepI1-53-GFP. Each segment removed contains a regular pattern of two positively charged residues next to one negatively charged side chain (Fig. 2). Removal of the first segment (PepI1-63-GFP) did not result in a visible loss of immunity upon MIC determination. However, when tested on the gradient plate (Fig. 5B), the reduction of Pep5-antagonizing activity was detectable. Further truncation to PepI1-57-GFP reduced the bioactivity of PepI by a factor of 5 in both expression hosts. The activity of PepI1-53-GFP was further decreased and almost reached the level of the empty vector upon MIC testing (Table 5), whereas the gradient plate still indicated a significant degree of activity (Fig. 5B).
Western blotting confirmed that the shortened PepI peptides were excreted into the supernatant (Fig. 6B). In addition, the fluorescent microscopy images of the mutants strongly resembled the wild-type PepI-GFP picture, with the characteristic ring surrounding each individual cell. Therefore, we concluded that the mutants properly reach the site of action and that the C-terminal segment with its characteristic charge distribution provides Pep5 immunity. This interpretation is supported by results obtained with two spontaneous mutants detected after PCR amplification of the respective pepI mutant gene. In the first clone, in addition to the desired N-terminal mutation one Lys residue of the C-terminal segment had been replaced by a Thr residue [pTS(I17R;K59T)-PepI1-68] (Fig. 5A); in the second clone a C-terminal deletion had been created [pTS(F13D;I17R)-PepI1-63]. In particular, in pTS(I17R)-PepI1-68 the Lys57Thr exchange reduced PepI activity, although the effect was only visible on the gradient plate (Table 5; Fig. 5A).
DISCUSSION
Pep5 is a well-studied member of the type-A lantibiotics for which nisin is the prototype peptide. This group of lantibiotics comprises elongated amphiphilic cationic peptides which kill gram-positive bacteria by formation of pores in the cytoplasmic membrane. Recently, we were able to show that nisin pore formation is 103-fold enhanced when it uses the cell wall precursor lipid II for docking onto the membrane and subsequent pore formation (6, 7, 37). Although the possibility can be excluded that lipid II serves a similar function for Pep5, there are significant indications leading to the assumption that Pep5 similarly uses a defined membrane target, particularly in strains such as Staphylococcus simulans 22 and S. carnosus TM300 which are susceptible to Pep5 in the nanomolar range of concentrations (7).
Nisin-producing strains protect themselves primarily by means of a dedicated ABC transporter (34). Homologs of the nisin transporter have been identified in the gene clusters of the lantibiotics epidermin and mersacidin (2, 21) and have been shown to repel the peptides from the cytoplasmic membrane (12, 19). In contrast, Pep5 producer immunity is solely dependent on the production of the 69-amino-acid peptide PepI. Pag et al. recently demonstrated that the half-life of the pepI mRNA strongly depends on the presence of an inverted loop within pepI-containing transcripts and that the level of immunity was directly correlated to the amount of PepI produced (20). While this suggested that in analogy to channel-forming colicins PepI may antagonize Pep5 by forming a stoichiometric complex with the lantibiotic, the overall physicochemical similarity of the peptides (both are strongly cationic) argues against such a molecular model for immunity.
Here, we demonstrate that the ability of PepI to provide immunity depends on a C-terminal stretch of approximately 20 amino acids with eight positively and three negatively charged residues arranged in a regular pattern (Fig. 2). Moreover, PepI appears to be transported out of the cell and to accumulate and to act at the membrane-cell wall interface. Based on these data a new working hypothesis emerges on how PepI and related immunity peptides from various bacteriocin systems may work. Provided that Pep5 uses a defined integral membrane target for specific docking onto the membrane and subsequent pore formation, PepI may bind the target molecule itself, thus shielding it from the lantibiotic. In such a scenario, it is not unlikely that the putative Pep5 target would be an anionic compound; in analogy to the nisin-lipid II binding, the target might be a membrane-bound precursor for the biosynthesis of anionic cell wall polymers such as teichoic or lipoteichoic acids. Such a target seems ideal for binding the 20-amino-acid segment of PepI containing the eight positively charged residues as well as the 34-amino-acid lantibiotic Pep5, which also contains eight positive charges regularly distributed along the peptide chain. Moreover, when emerging from the membrane such a target would provide an excellent docking site for subsequent pore formation. When bound to such a negatively charged target, PepI may slowly be moved in the course of a few cell cycles from the membrane to the cell surface and be shed into the supernatant as a result of cell wall turnover. This interpretation would explain the time lag of approximately two to three generation times which was observed between the first appearance of PepI in the cells and that in the cell-free supernatant (Fig. 4A).
The data presented here leave little doubt that PepI is translocated across the cytoplasmic membrane and yet raise a question respecting how transport may be accomplished. Frequently, small peptides such as PepI are transported by dedicated ABC transporters. However, such a transporter is not encoded in the Pep5 biosynthesis gene cluster or in any of the other clusters. Furthermore, a dedicated transporter presumably would be unable to handle a bulky GFP fusion protein; efficient heterologous synthesis without coexpression of the transporter, as achieved here, seems hardly possible. Another option would be cotranslocation of PepI with its hypothetical target, e.g., a growing teichoic acid chain. Such a mechanism would provide optimal producer safety, since the target is shielded as soon as it emerges from the membrane. However, this again would raise questions as to interference of comparatively large GFP fusions with the transport process. A third possibility may be based on the overall similarity of the N-terminal segment of PepI with leader peptides of the sec pathway. Within the first five N-terminal residues of PepI there are one Asn residue and one Lys residue, which are then followed by an uninterrupted stretch of 20 apolar amino acids. Insertion of charges into this segment, as shown here, strongly interfered with export. Clearly, a signal peptidase motif is missing, allowing PepI or any PepI fusion protein to escape from secP-mediated cleavage. Moreover, the sec machinery appears to be able to translocate GFP fusion proteins to the bacterial surface, as has been demonstrated, e.g., for OmpA-GFP (33) and ChoD-GFP (8) fusion proteins. Certainly, more experimental work is needed for full understanding of the PepI-mediated bacteriocin immunity; however, the data presented here provide an excellent basis for further studies of this unique phenomenon.
Acknowledgments
We gratefully acknowledge the Deutsche Forschungsgemeinschaft for financial support of the project (Sa 292/8-3). Additional support was provided by the BONFOR program of the Medical Faculty, University of Bonn.
We thank G. Bierbaum for valuable discussions, M. Josten and M. Oedenkoven for providing Pep5, J. Gebel, Bonn, Germany, for making the fluorescence microscope available to us, and A. Rechenburg for helping with digital images. We also thank the group of F. Götz, Tübingen, Germany, for providing the staphylococcal expression vector pTX15.
REFERENCES
- 1.Abee, T. 1995. Pore-forming bacteriocins of gram-positive bacteria and self-protection mechanisms of producer organisms. FEMS Microbiol. Lett. 129:1-10. [DOI] [PubMed] [Google Scholar]
- 2.Altena, K., A. Guder, C. Cramer, and G. Bierbaum. 2000. Biosynthesis of the lantibiotic mersacidin: organization of a type B lantibiotic gene cluster. Appl. Environ. Microbiol. 66:2565-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Augustin, J., and F. Götz. 1990. Transformation of Staphylococcus epidermidis and other staphylococcal species with plasmid DNA by electroporation. FEMS Microbiol. Lett. 66:203-208. [DOI] [PubMed] [Google Scholar]
- 4.Augustin, J., R. Rosenstein, B. Wieland, U. Schneider, N. Schnell, G. Engelke, K.-D. Entian, and F. Götz. 1992. Genetic analysis of epidermin biosynthetic genes and epidermin-negative mutants of Staphylococcus epidermidis. Eur. J. Biochem. 204:1149-1154. [DOI] [PubMed] [Google Scholar]
- 5.Bierbaum, G., M. Reis, C. Szekat, and H.-G. Sahl. 1994. Construction of an expression system for engineering of the lantibiotic Pep5. Appl. Environ. Microbiol. 60:4332-4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breukink, E., I. Wiedemann, C. van Kraaij, O. P. Kuipers, H.-G. Sahl, and B. de Kruijff. 1999. Use of the cell wall precursor lipid II by a pore-forming peptide antibiotic. Science 286:2361-2364. [DOI] [PubMed] [Google Scholar]
- 7.Brötz, H., G. Bierbaum, K. Leopold, P. E. Reynolds, and H.-G. Sahl. 1998. The lantibiotic mersacidin inhibits peptidoglycan synthesis by targeting lipid II. Antimicrob. Agents Chemother. 42:154-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cowley, S. C., and Y. Av-Gay. 2001. Monitoring promoter activity and protein localization in Mycobacterium spp. using green fluorescent protein. Gene 264:225-231. [DOI] [PubMed] [Google Scholar]
- 9.Ersfeld-Dressen, H., H.-G. Sahl, and H. Brandis. 1984. Plasmid involvement in production of and immunity to the staphylococcin-like peptide Pep5. J. Gen. Microbiol. 130:3029-3035. [DOI] [PubMed] [Google Scholar]
- 10.Geli, V., D. Baty, F. Pattus, and C. Lazdunski. 1989. Topology and function of the integral membrane protein conferring immunity to colicin A. Mol. Microbiol. 3:679-687. [DOI] [PubMed] [Google Scholar]
- 11.Götz, F., and B. Schumacher. 1987. Improvements of protoplast transformation in Staphylococcus carnosus. FEMS Microbiol. Lett. 40:285-288. [Google Scholar]
- 12.Guder, A., T. Schmitter, I. Wiedemann, H. G. Sahl, and G. Bierbaum. 2002. Role of the single regulator MrsR1 and the two-component system MrsR2/K2 in the regulation of mersacidin production and immunity. Appl. Environ. Microbiol. 68:106-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heidrich, C., U. Pag, M. Josten, J. W. Metzger, R. W. Jack, G. Bierbaum, G. Jung, and H.-G. Sahl. 1998. Isolation, characterization, and heterologous expression of the novel lantibiotic epicidin 280 and analysis of its biosynthetic gene cluster. Appl. Environ. Microbiol. 64:3140-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnston, T. C., R. B. Thompson, and T. O. Baldwin. 1986. Nucleotide sequence of the luxB gene of Vibrio harveyi and the complete amino acid sequence of the beta subunit of bacterial luciferase. J. Biol. Chem. 261:4805-4811. [PubMed] [Google Scholar]
- 15.Klein, C., and K.-D. Entian. 1994. Genes involved in self-protection against the lantibiotic subtilin produced by Bacillus subtilis ATCC 6633. Appl. Environ. Microbiol. 60:2793-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuipers, O. P., M. M. Beerthuyzen, R. J. Siezen, and W. M. de Vos. 1993. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis Requirement of expression of the nisA and nisI genes for development of immunity. Eur. J. Biochem. 216:281-291. [DOI] [PubMed] [Google Scholar]
- 17.McAuliffe, O., C. Hill, and R. P. Ross. 2000. Identification and overexpression of ltnl, a novel gene which confers immunity to the two-component lantibiotic lacticin 3147. Microbiology 146:129-138. [DOI] [PubMed] [Google Scholar]
- 18.Meyer, C., G. Bierbaum, C. Heidrich, M. Reis, J. Süling, M. I. Iglesias-Wind, C. Kempter, E. Molitor, and H.-G. Sahl. 1995. Nucleotide sequence of the lantibiotic Pep5 biosynthesis gene cluster and functional analysis of PepP and PepC. Eur. J. Biochem. 232:478-489. [DOI] [PubMed] [Google Scholar]
- 19.Otto, M., A. Peschel, and F. Götz. 1998. Producer self-protection against the lantibiotic epidermin by the ABC transporter EpiFEG of Staphylococcus epidermidis Tü3298. FEMS Microbiol. Lett. 166:203-211. [DOI] [PubMed] [Google Scholar]
- 20.Pag, U., C. Heidrich, G. Bierbaum, and H.-G. Sahl. 1999. Molecular analysis of expression of the lantibiotic Pep5 immunity phenotype. Appl. Environ. Microbiol. 65:591-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peschel, A., and F. Götz. 1996. Analysis of the Staphylococcus epidermidis genes epiF, -E, and - G involved in epidermin immunity. J. Bacteriol. 178:531-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peschel, A., B. Ottenwälder, and F. Götz. 1996. Inducible production and cellular location of the epidermin biosynthetic enzyme EpiB using an improved staphylococcal expression system. FEMS Microbiol. Lett. 137:279-284. [DOI] [PubMed] [Google Scholar]
- 23.Qiao, M., T. Immonen, O. Koponen, and P. E. J. Saris. 1995. The cellular location and effect on nisin immunity of the NisI protein from Lactococcus lactis N8 expressed in Escherichia coli and L. lactis. FEMS Microbiol. Lett. 131:75-80. [DOI] [PubMed] [Google Scholar]
- 24.Reis, M., M. Eschbach-Bludau, M. I. Iglesias-Wind, T. Kupke, and H.-G. Sahl. 1994. Producer immunity towards the lantibiotic Pep5: identification of the immunity gene pepI and localization and functional analysis of its gene product. Appl. Environ. Microbiol. 60:2876-2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reis, M., and H.-G. Sahl. 1991. Genetic analysis of the producer self-protection mechanism (“immunity”) against Pep5, p. 320-331. In G. Jung and H.-G. Sahl (ed.), Nisin and novel lantibiotics. Escom, Leiden, The Netherlands.
- 26.Rincé, A., A. Dufour, P. Uguen, J. P. Le Pennec, and D. Haras. 1997. Characterization of the lacticin 481 operon: the Lactococcus lactis genes lctF, lctE, and lctG encode a putative ABC transporter involved in bacteriocin immunity. Appl. Environ. Microbiol. 63:4252-4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sahl, H.-G., and G. Bierbaum. 1998. Lantibiotics: biosynthesis and biological activities of uniquely modified peptides from Gram-positive bacteria. Annu. Rev. Microbiol. 52:41-79. [DOI] [PubMed] [Google Scholar]
- 28.Sahl, H.-G., and H. Brandis. 1981. Production, purification and chemical properties of an antistaphylococcal agent produced by Staphylococcus epidermidis. J. Gen. Microbiol. 127:377-384. [DOI] [PubMed] [Google Scholar]
- 29.Saris, P. E. J., T. Immonen, M. Reis, and H.-G. Sahl. 1996. Immunity to lantibiotics. Antonie Leeuwenhoek 69:151-159. [DOI] [PubMed] [Google Scholar]
- 30.Schleifer, K. H., and U. Fischer. 1982. Description of a new species of the genus Staphylococcus: Staphylococcus carnosus. Int. J. Syst. Bacteriol. 32:153-156. [Google Scholar]
- 31.Schnell, N., K.-D. Entian, U. Schneider, F. Götz, H. Zähner, R. Kellner, and G. Jung. 1988. Prepeptide sequence of epidermin, a ribosomally synthesized antibiotic with four sulphide-rings. Nature 333:276-278. [DOI] [PubMed] [Google Scholar]
- 32.Schramm, E., T. Ölschläger, W. Troger, and V. Braun. 1988. Sequence, expression and localization of the immunity protein for colicin B. Mol. Gen. Genet. 211:176-182. [DOI] [PubMed] [Google Scholar]
- 33.Shi, H., and S. W. Wen. 2001. Display of green fluorescent protein on Escherichia coli cell surface. Enzyme Microb. Technol. 28:25-34. [DOI] [PubMed] [Google Scholar]
- 34.Siegers, K., and K.-D. Entian. 1995. Genes involved in immunity to the lantibiotic nisin produced by Lactococcus lactis 6F3. Appl. Environ. Microbiol. 61:1082-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skaugen, M., C. I. M. Abildgaard, and I. F. Nes. 1997. Organization and expression of a gene cluster involved in the biosynthesis of the lantibiotic lactocin S. Mol. Gen. Genet. 253:674-686. [DOI] [PubMed] [Google Scholar]
- 36.Song, H. Y., and W. A. Cramer. 1991. Membrane topography of ColE1 gene products: the immunity protein. J. Bacteriol. 173:2935-2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wiedemann, I., E. Breukink, C. van Kraaij, O. P. Kuipers, G. Bierbaum, B. de Kruijff, and H. G. Sahl. 2001. Specific binding of nisin to the peptidoglycan precursor lipid II combines pore formation and inhibition of cell wall biosynthesis for potent antibiotic activity. J. Biol. Chem. 276:1772-1779. [DOI] [PubMed] [Google Scholar]
- 38.Worobo, R. W., M. J. van Belkum, M. Sailer, K. L. Roy, J. C. Vederas, and M. E. Stiles. 1995. A signal peptide secretion-dependent bacteriocin from Carnobacterium divergens. J. Bacteriol. 177:3143-3149. [DOI] [PMC free article] [PubMed] [Google Scholar]