The actinomycete genus Gordonia has attracted much interest in recent years for a variety of reasons. Most species were isolated due to their abilities to degrade xenobiotics, environmental pollutants, or otherwise slowly biodegradable natural polymers as well as to transform or synthesize possibly useful compounds. The variety of chemical compounds being transformed, biodegraded, and synthesized by gordoniae makes these bacteria potentially useful for environmental and industrial biotechnology. However, because some gordoniae are opportunistic pathogens, their application in the environment may be restricted in some cases. Valuable phenotypic features of Gordonia species can be genetically transferred to other microorganisms, as has been demonstrated by Park et al. (76) and Bröker et al. (12). Recent studies have revealed improved cloning vectors allowing the transfer of genes between different Gordonia species or the transfer of foreign genes from Escherichia coli to Gordonia spp. and the expression of these genes (3, 12). Current research also focuses on several species that are known to cause infections, especially in humans. Gordonia species have been isolated from various native biotopes such as soil or mangrove rhizosphere, from extensively industrially influenced habitats such as oil-producing wells or hydrocarbon-contaminated soil, from artificial sources such as wastewater treatment bioreactors or biofilters, and from diseased humans. The genus Gordonia belongs phylogenetically to the suborder Corynebacterineae, the mycolic acid group within the order Actinomycetales (86), and its classification has changed drastically in recent years, with several species being reclassified and many novel species being described.
This review provides an overview of recent research on Gordonia species and summarizes the importance of this emerging genus in industrial and environmental biotechnology and its role as a pathogen of humans. Since the introduction of the correct name in 1997 (see below), the number of publications per annum listed by the Science Citation Index if “Gordonia” is entered as the key word and only studies on prokaryotes are considered has increased steadily: from 3 in 1998 to 7 in 1999, 10 in 2000, 15 in 2001, 13 in 2002, and 18 in 2003 (until November). In addition, as of November 2003, more than 40 patents relating to Gordonia species have been registered, indicating their increasingly recognized commercial potential.
TAXONOMY OF THE GENUS GORDONIA
In 1971, Tsukamura proposed Gordona as a new genus for coryneform bacteria isolated from sputa of patients with pulmonary disease or from soil (91). The name for this novel genus was chosen to pay tribute to the American bacteriologist Ruth E. Gordon. Members of this genus are distinguished from fast-growing mycobacteria by their slight acid fastness and the absence of arylsulfatase, and from the genus Nocardia by their ability to reduce nitrate and the absence of a mycelium. Only six years later, Goodfellow and Alderson discarded this taxon and transferred Mycobacterium rhodochrous and the “rhodochrous complex,” including representatives of the genus Gordona, to the genus Rhodococcus as “a home for the rhodochrous complex” (34). However, studies of the mycolic acid (high-molecular-weight α-branched 3-hydroxy fatty acid) and menaquinone composition revealed heterogeneous variations within the genus Rhodococcus (33, 90): while Rhodococcus spp. usually contain mycolic acids with 34 to 52 carbon atoms and menaquinones with 8 isoprene units and 2 hydrogen atoms added to the isoprene side chain double bonds [MK-8(H2)] as the major form of menaquinone, three Rhodococcus species originally assigned to the genus Gordona (Gordona bronchialis, Gordona rubripertincta [formerly Gordona rubra], and Gordona terrae) were found to contain mycolic acids of 48 to 66 carbon atoms and dihydrogenated menaquinones with 9 isoprene units [MK-9(H2)] as the predominant menaquinone. Additional analysis of 16S rRNA similarities led to a revival of the genus Gordona by Stackebrandt et al. (87) with the three species mentioned above and, additionally, Rhodococcus sputi (92) as Gordona sputi. After reclassification of Rhodococcus aichiensis (93) and Nocardia amarae (59) to the genus Gordona as Gordona aichiensis and Gordona amarae (50), respectively, this taxon became a well-defined genus within the order Actinomycetales. In 1997, the etymologically correct name Gordonia instead of Gordona was proposed by Stackebrandt et al. (86). According to their newly proposed hiercharchic classification system for the actinomycete line, Gordonia is the type genus of Gordoniaceae (the Gordonia family) within the suborder Corynebacterineae. This suborder includes also the families Corynebacteriaceae, Dietziaceae, Mycobacteriaceae, Nocardiaceae, Tsukamurellaceae, and Williamsiaceae. Since the report of Gordonia hydrophobica as a new species (7), the number of validly described members of this genus has increased considerably. At present, the genus Gordonia comprises 19 validly published species, and at least 2 further species are in the process of classification (Table 1). Meanwhile, gordoniae have been shown to be ubiquitously distributed in nature.
TABLE 1.
Overview of described Gordonia species and their relevant characteristics
| Type strain | Characteristic or source | Reference for type strain |
|---|---|---|
| G. aichiensis DSM 43978T | Clinical isolate from sputum | 93 |
| G. alkanivorans DSM 44369T | Alkane-degrading bacterium | 55 |
| G. amarae DSM 43392T | Foam-forming bacterium | 59 |
| G. amicalis DSM 44461T | Dibenzothiophene-desulfurizing bacterium | 49 |
| G. bronchialis DSM 43247T | Isolated from sputum of a woman with cavitary pulmonary tuberculosis | 91 |
| G. desulfuricans DSM 44462T | Benzothiophene-desulfurizing bacterium | 48 |
| G. hirsuta DSM 44140T | Isolated from a biofilter of an animal-rendering plant | 51 |
| G. hydrophobica DSM 44015T | Isolated from a biofilter of an animal-rendering plant | 7 |
| G. jacobaea MV-1a | Canthaxanthin-containing bacterium | 22 |
| G. namibiensis DSM 44568T | Nitrile-metabolizing bacterium | 11 |
| G. nitida DSM 44499T | 3-Ethyl- and 3-methylpyridine-degrading bacterium | 95 |
| G. paraffinivorans DSM 44604T | Hydrocarbon-degrading bacterium | 94 |
| G. polyisoprenivorans DSM 44302T | cis-1,4-Polyisoprene-degrading bacterium | 62 |
| G. rhizosphera DSM 44383T | Isolated from mangrove rhizosphere | 89 |
| G. rubripertincta DSM 43197T | Mycobactin-producing bacterium | 91 |
| G. sihwensis DSM 44576T | Nitrate-reducing bacterium | 47 |
| G. sinesedis DSM 44455T | Isolated from soil | 28, 66 |
| G. spumae T4a | Isolated from activated sludge foam | Sukhoom et al., unpublished data |
| G. sputi DSM 43896T | Isolated from sputum of a patient with pulmonary disease | 92 |
| G. terrae DSM 43249T | Isolated from soil | 91 |
| G. westfalica DSM 44215T | cis-1,4-Polyisoprene-degrading bacterium | 64 |
Not a validly described species.
Interestingly, the taxon Gordonia also exists in the plant kingdom. There, the genus Gordonia comprises evergreen trees, and it belongs to the family Theaceae (27). Because the bacterial taxon Gordonia was introduced before the revised principle 2 of the “Code of Nomenclature of Prokaryotes” was published (24), this name can continue to be used for these bacteria.
MORPHOLOGICAL AND CHEMOTAXONOMIC PROPERTIES
Morphological and structural aspects.
Members of the genus Gordonia are aerobic, catalase-positive, gram-positive to gram-variable, slightly acid-fast, nonmotile, nocardioform actinomycetes. They have an oxidative carbohydrate metabolism and are arylsulfatase negative; susceptibility to lysozyme has been shown. The term “nocardioform” is morphologically descriptive and refers to mycelial growth with fragmentation into rod-shaped or coccoid elements (58). Gordoniae do not generate spores. The colony morphology of Gordonia species varies from slimy, smooth, and glossy to irregular and rough; it may even differ within one species depending on the medium used for growth (64). Some strains, such as Gordonia alkanivorans DSM 44369T and Gordonia westfalica DSM 44215T, are also able to form both smooth and rough colonies (55, 64). Usually, cultures with smooth phenotypes can generate rough colonies; this alteration seems to be irreversible and could be due to mutations in genes encoding methyltransferases involved in glycopeptidolipid synthesis, as described for Mycobacterium smegmatis (78). The importance of glycosylated peptidolipids for colony morphology has also been demonstrated for G. hydrophobica (69). The colors of the colonies cover a broad range, including white, yellow, tannish, orange, red, and pink (94). The cell wall belongs to wall chemotype IV sensu Lechevalier and Lechevalier; the peptidoglycan is of the A1 γ type, containing meso-diaminopimelic acid as the only diamino acid and muramic acid with N-glycolyl residues (2). The major cell wall sugars are arabinose and galactose. Characteristically, mycolic acids with 44 to 66 carbon atoms and major portions of straight-chain, saturated, monounsaturated, and 10-methyl (tuberculostearic) cellular fatty acids are formed (8). Polar lipids are typically composed of diphosphatidylglycerol, phosphatidylethanolamine, phosphatidylinositol, and phosphatidylinositol mannosides. Cells contain MK-9(H2) as the predominant isoprenologue. Only in some species can traces of MK-8(H2) be found. The cell wall composition characteristics and cellular chemotaxonomic properties mentioned above separate Gordonia clearly from related genera such as Mycobacterium, Rhodococcus, and Skermania (Table 2).
TABLE 2.
Chemotaxonomic markers of genera of the suborder Corynebacterineae
| Family | Genus | Acyl typea | Major menaquinone | PEb | Fatty acid compositionsc | Mycolate size (no. of carbons) | Pyrolysis esters of mycolatesd (no. of carbons) | G+C content (mol%) |
|---|---|---|---|---|---|---|---|---|
| Gordoniaceae | Gordonia | G | MK-9(H2) | + | S, U, T | 48-66 | 16-18 | 63-69 |
| Skermania | G | MK-8(H4 ω-cyc) | + | S, U, T | 58-64 | 16-20 | 68 | |
| Corynebacteriaceae | Corynebacterium | A | MK-8(H2) | −e | S, Uf | 22-36 | 8-18 | 51-67 |
| Turicella | ND | MK-10, MK-11 | ND | S, U, T | -h | -h | 65-72 | |
| Dietziaceae | Dietzia | A | MK-8(H2) | − | S, U, T | 34-38 | ND | 73 |
| Mycobacteriaceae | Mycobacterium | G | MK-9(H2) | + | S, U, T | 70-90 | 22-26 | 70-72 |
| Nocardiaceae | Nocardia | G | MK-8(H4 ω-cyc) | + | S, U, T | 50-62 | 12-18 | 64-72 |
| Rhodococcus | G | MK-8(H2) | + | S, U, T | 34-54 | 12-16 | 63-73 | |
| Tsukamurellaceae | Tsukamurella | G | MK-9 | + | S, U, T | 64-78 | 20:1g | 67-68 |
| Williamsiaceae | Williamsia | G | MK-9(H2) | + | S, U, T | 50-56 | ND | 64-65 |
G, glycolated muramic acid; A, acetylated muramic acid; ND, not determined.
PE, phosphatidylethanolamine; +, present; −, absent.
S, saturated; U, unsaturated; T, tuberculostearic acid.
Fatty acid methyl esters released by pyrolysis of mycolic acid methyl esters.
Present in Corynebacterium bovis and Corynebacterium urealyticum.
Tuberculostearic acid present in Corynebacterium anmoniagenes, C. bovis, C. minutissimum, C. urealyticum, and C. variabile.
20:1, eicosanoic acid.
Mycolic acid free.
Taxonomic aspects related to DNA and RNA sequence data.
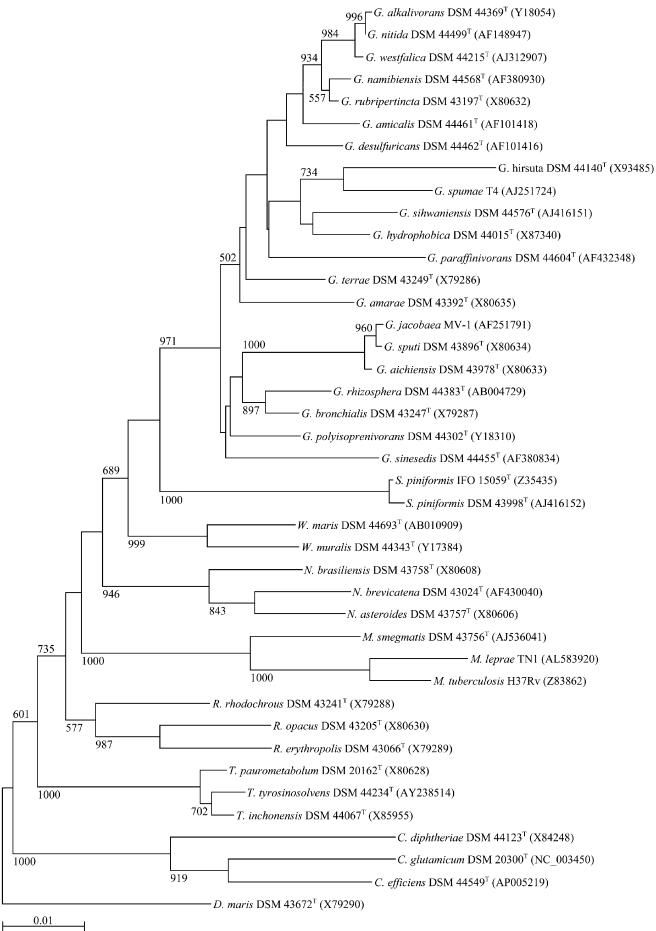
The G+C content of Gordonia DNA ranges from 63 to 69 mol% (Table 2), and intrageneric DNA-DNA relatednesses ranges from 3.7% (shared by Gordonia nitida and G. amarae) (95) to 52% (shared by G. alkanivorans and G. rubripertincta) (55). The 16S ribosomal DNA (rDNA) sequences of Gordonia species reveal similarities ranging from 94.8% (between G. amarae and Gordonia sihwensis) to 99.9% (between G. alkanivorans and G. nitida). Phylogenetic trees based on sequence alignments with species of other genera of the suborder Corynebacterineae show the distinctly defined inter- and intrageneric phylogenetic positions of Gordonia species (Fig. 1). The deviations in the 16S rDNA sequences of different Gordonia species occur mostly in two hypervariable regions between nucleotide positions 136 and 229 and between nucleotides 996 and 1028 (according to the numbering of the E. coli K-12 16S rDNA sequence) (Fig. 2). All species differ from each other in these regions except G. alkanivorans and G. nitida. The latter were published individually as novel species at the same time. DNA-DNA-hybridization experiments must still be carried out to confirm the phylogenetic relationship between these species.
FIG. 1.
Neighbor-joining tree (82) based on nearly complete 16S rDNA sequences of gordoniae and representatives of other mycolic acid-containing actinomycetes, including members of the genera Corynebacterium, Dietzia, Mycobacterium, Nocardia, Rhodococcus, Skermania, Tsukamurella, and Williamsia. Numbers at nodes indicate the level of bootstrap support based on a neighbor-joining analysis of 1,000 resampled data sets; only values above 50% are given. Bar, 0.01 substitution per nucleotide position.
FIG. 2.
Comparison of the two major hypervariable regions of the 16S rDNA sequences of the type strains of all Gordonia species by multiple alignment. Highlighted nucleotide positions have at least 70% identical nucleotides. Nucleotide positions are given according to the numbering of the Escherichia coli K-12 16S rDNA sequence.
PATHOGENIC GORDONIA SPECIES
Most Gordonia species have been isolated from environmental sources; however, a few are also sporadically associated with human infections (Table 3). In almost all cases, patients were immunosuppressed after underlying diseases, and infections by Gordonia species occurred only secondarily. Altogether, approximately 20 case reports of gordonial infections can be found in the literature; most of these infections were caused by G. bronchialis and were associated with sternal wounds resulting from surgery (Table 3). A few infections also occurred after coronary artery bypass surgery and were associated with heart-lung machines or applications of central venous or Hickman catheters (Table 3). Two additional infections with Gordonia spp. have been reported in the literature; however, these isolates were not identified to species level (61). Interestingly, one of these reports described an infection by a Gordonia species taxonomically closely related to G. sputi. The pattern of its p-bromophenacyl esters of mycolic acids was similar to that of G. sputi DSM 43896T (also classified as ATCC 29627), and several biochemical tests revealed characteristics of Gordonia spp., especially those of G. bronchialis. The first 450 bp at the 5′ end of the 16S rDNA totally matched those of G. sputi. Surprisingly, DNA-DNA relatedness to G. sputi, as analyzed by the stringent nuclease S1 method, did not meet the genetic criterion of more than 70% similarity to belong to the same species. Consequently, one must suggest that this strain could belong to a novel gordonial species. So far, reports of human gordonial infections are rather rare in comparison to reports on infections caused by other opportunistically pathogenic bacteria belonging to taxonomically related genera such as Rhodococcus and Nocardia.
TABLE 3.
Infections of humans with gordoniae and underlying conditions
| Gordonia species | Type of infection | No. of cases | Underlying condition(s) | Reference |
|---|---|---|---|---|
| G. bronchialis | Sternal wound | 7 | Surgery | 80 |
| G. polyisoprenivorans | Bacteremia due to Hickman catheter | 1 | Bone marrow transplantation | 45 |
| G. rubripertincta | Lung infection | 1 | Tuberculosis | 37 |
| G. sputi | Mediastinitis | 1 | Surgery | 57 |
| Bacteremia due to cutaneous lesions | 1 | Metastatic melanoma; interleukin-2 treatment | 81 | |
| G. terrae | Brain abscess | 1 | None | 25 |
| 1 | Cerebral tumor | 26 | ||
| Skin infection | 1 | None | 67 | |
| Bacteremia due to central venous catheter | 1 | Chronic intestinal pseudo-obstruction syndrome | 13 | |
| Gordonia sp. | Bacteremia due to central venous catheter | 1 | Breast and ovarian cancer | 13 |
| 1 | Multiple | 61 |
CAPABILITIES FOR BIODEGRADATION AND BIOREMEDIATION
Gordoniae are probably important in natural environments and are powerful candidates for bioremediation processes because of their capacity to degrade substituted and nonsubstituted hydrocarbons, widespread toxic environmental pollutants, other xenobiotics, and natural compounds that are not readily biodegradable. Gordonia species may play an important role during wastewater treatment and in biofilters (9). Several Gordonia species were isolated due to their capabilities to degrade or modify aliphatic and aromatic hydrocarbons, halogenated aromatic compounds, benzothiophene, nitrile, polyisoprene, xylene, and so forth. Incorporation of the long aliphatic chains of the mycolic acids into the cell wall is associated with hydrophobicity and surface adhesion (8) and may play a role in the degradation of hydrophobic pollutants. Examples of this ability are the adhesive growth of several Gordonia strains during the biodegradation of rubber materials (2, 63, 65) and the utilization of hydrophobic hydrocarbons by many species of this genus (55, 94).
Phthalic acid esters.
Chatterjee and Dutta (15) described Gordonia sp. strain MTCC 4818, isolated from creosote-contaminated soil, which was able to metabolize several phthalic acid esters as sole sources of carbon and energy. Phthalic acid esters exhibit hormonal action due to their binding activity toward the estrogen receptor (71) and can cause proliferation of MCF-7 cells (85). Obviously, this strain was able to hydrolyze both ester bonds of butyl benzyl phthalate and utilized the released benzyl alcohol and butanol for growth. The esterase activity was shown to be induced by butyl benzyl phthalate and/or monobenzyl phthalate, indicating a specificity of the enzyme for this xenobiotic. Surprisingly, Gordonia sp. strain MTCC 4818 was unable to metabolize phthalic acid, which therefore accumulated as a dead-end product in the culture broth. Under aerobic conditions, phthalic acid is usually further degraded via protochatechuate (88).
s-Triazine and alkylpyridines.
Further examples of xenobiotic pollutants degraded or transformed by members of the gordoniae are heterocyclic compounds such as s-triazines and alkylpyridines. The relative persistence of s-triazines such as atrazine and simazine has led to increasing concern about environmental contamination by these compounds. So far, only a very few microbial isolates capable of transforming substituted s-triazines have been identified. Mulbry purified and characterized a unique inducible s-triazine hydrolase from extracts of G. rubripertincta DSM 10347 (formerly Rhodococcus corallinus) (70); this enzyme was able to dechlorinate atrazine and deethylsimazine as well as to deaminate melamine and related compounds (17, 18). The ability of the triazine hydrolase to dechlorinate s-triazine compounds makes this unique enzyme potentially useful for environmental remediation. Alkylpyridines are also toxic environmental pollutants, and despite their occurrence and toxicity, only a few studies have focused on the biodegradation of these compounds. Detailed studies of the degradation pathway were previously performed with Pseudomonas sp. strain KM3; however, recent research revealed a novel degradation pathway of 3-ethylpyridine and 3-methylpyridine in the newly isolated and described species G. nitida (60, 95).
DBT and biodesulfurization of fuels.
The modification of heterocyclic compounds can also be used in the processing of important industrial products such as fossil fuels. Because regulations concerning the sulfur content of motor fuels have become increasingly stringent in an effort to reduce SOX emissions, sulfur removal by biocatalytic means is often considered a potential alternative to the conventional deep hydrodesulfurization processes used in refineries. The development of convenient and economically feasible microbial processes for the removal of organic sulfur compounds from fossil fuels led to the isolation of Rhodococcus rhodochrous ATCC 53968 (44). Dibenzothiophene (DBT) is widely regarded as a model compound that is representative of the aromatic organosulfur fraction of coal and crude oil. DBT is usually transformed to 2-hydroxybiphenyl and inorganic sulfite via the 4S pathway (46, 74). Several Gordonia species or strains have been characterized for their ability to desulfurize DBT or benzothiophene (1, 32, 48, 49). The first representative of the genus Gordonia described to be capable of DBT desulfurization was Gordonia sp. strain CYKS1 (79). The desulfurization genes (dszABC) of this strain and the closely related strain G. nitida LSSEJ-1 have been cloned (GenBank accession no. AY396519 and AY293884) and functionally expressed in E. coli. The resulting recombinant strains were applied for deep desulfurization of diesel oil (76). Chang et al. (14) described in detail the production of Gordonia sp. strain CYKS1 as a desulfurization biocatalyst by a two-stage process and its application for treatment of model and diesel oils. In addition, the phase separation in diesel oil biodesulfurization was optimized by the addition of ethanol as a de-emulsifier to a three-phase (diesel oil-biocatalyst-aqueous phase) emulsion in order to recover the biocatalyst (Gordonia sp. strain CYKS1) and the desulfurized oil (16). The enhanced processes allowed a decrease in the sulfur content of diesel oil from 250 to approximately 60 mg liter−1 and should enable the production of ultra-low-sulfur fuel oils by biotechnological means.
Other xenobiotics.
Several other poorly accessible carbon sources, such as t-butyl ether, methyl t-butyl ether, t-amyl methyl ether, cyclic alkanes, and polycyclic aromatic hydrocarbons, can be degraded by species of the genus Gordonia (38, 42, 43, 52). These catabolic abilities are probably closely related to the production of biosurfactants by microbial cells (41).
Natural and synthetic rubber.
The ability to biodegrade natural and synthetic isoprene rubber (cis-1,4-polyisoprene) is not widely distributed among bacteria. As described previously, degradation of this polymer can follow one of at least two strategies, and the most potent rubber-degrading bacteria belong to the genus Gordonia (2, 65). Until recently, all rubber-degrading bacteria had in common the production of translucent halos on solid medium containing cis-1,4-polyisoprene in the form of latex. The first gordoniae capable of utilizing cis-1,4-polyisoprene as a source for carbon and energy were described by Linos and Steinbüchel (63). Surprisingly, these isolates did not produce clear zones on latex-containing solid medium but nevertheless degraded the substrate more effectively. Presumably, this group of rubber-degrading bacteria was not recognized earlier due to the method used for enrichment and characterization. Later studies resulted in the description of the novel species Gordonia polyisoprenivorans (62) and G. westfalica (64), which are characterized by adhesive growth during rubber degradation. The molecular basis of rubber degradation is still unknown, but one can presume that the hydrophobicity of the cell surfaces of gordoniae affected by the presence of mycolic acids is significantly involved in this process. Probably, the occurrence of mycolic acids and the production of biosurfactants (see below) are important for the formation of biofilms enabling direct contact with cis-1,4-polyisoprene in solid rubber materials, which is required for rubber degradation by these strains.
ANABOLIC CAPABILITIES
Gordonia species also produce a variety of compounds which are useful for various applications. The number of publications describing novel compounds produced by Gordonia strains has increased steadily. These compounds include l-lysine and some of its analogues (56) and also complex molecules such as gordonan. Some of these are synthesized exclusively by Gordonia strains; in other cases, Gordonia strains may represent an alternative source.
Biosurfactants.
The capability for biodegradation of water-insoluble and hydrophobic compounds is often associated with the production of biogenic surface-active, amphiphilic compounds (biosurfactants) by a bacterium (6). The relevance of biosurfactants to the biodegradation of pollutants is threefold: (i) cellular biosurfactants such as mycolic acids cause adherence of the microbial cells to hydrophobic phases in two-phase systems (73); (ii) surfactants promote the access of hydrophobic compounds to microbial cells by decreasing the interfacial tension between the phases (29); and (iii) surfactants disperse hydrophobic compounds, leading to an increased surface area accessible for microbial attack (30). On the other hand, biosurfactants may cause undesired mobilization and movement of hydrophobic pollutants in natural environments, resulting in, e.g., the release of the pollutants into groundwater.
In general, biosurfactants can be subdivided into low-molecular-weight compounds such as glycolipids and lipopeptides and high-molecular-weight polymeric compounds such as polysaccharides, lipoproteins, and lipopolysaccharides. Production of surface-active compounds has been reported for several Gordonia strains. In this regard G. amarae is the most extensively studied species of the genus. By morphological and physiological means, it has been shown that G. amarae is strongly associated with foaming activated sludge in wastewater treatment plants, as also confirmed by phylogenetic hybridization-based experiments and comparative rRNA sequence analysis (20, 21). Pagilla et al. (75) confirmed that foaming problems in activated sludge are due to the production of biosurfactants by G. amarae when hydrophobic substrates such as hexadecane are present. They found that the lowest surface tension was 55 dynes/cm in the presence of acetate and less than 40 dynes/cm when hexadecane was provided as the sole source of carbon. It was also demonstrated that foam formation depended on the presence of both bacterial cells and the culture supernatant (39). The authors identified a high-molecular-weight fraction (Mw, >10,000) of the supernatant that was able to emulsify n-hexadecane in addition to exhibiting foaming activity, indicating the presence of surface-active compounds (39). They also suggested that the hydrophobicity of the cell surface was involved in the attachment of cells to air bubbles, generating stable foam due to the high affinity of the microbial cells for hexadecane. Nazina et al. (72) characterized the formation of biosurfactants by Gordonia sp. strain 321, along with that by several strains of different genera. Culture broth of cells grown on a mixture of liquid paraffins significantly decreased the surface tension and interfacial tension, while increasing emulsifying activity, in two-phase systems consisting of an aqueous phase (culture broth) and oil. Gordonia sp. strain 321 produced exopolysaccharides, which are known to exhibit surface activity and may be responsible for the results obtained. Another exopolysaccharide-producing Gordonia strain, isolated from polluted subtropical groundwater, was identified as G. polyisoprenivorans (31). These examples demonstrate that biosynthesis of biosurfactants is important for microbial cells and influences access to hydrophobic substrates.
Carotenoids.
The abilities of several gram-positive bacteria, including species of the genera Corynebacterium, Flavobacterium, Micrococcus, and Mycobacterium, to synthesize different carotenoids are known (19, 40, 54, 77). Many Gordonia species also produce colonies with reddish pigmentation, indicating their capacity to synthesize significant amounts of carotenoids (Fig. 3; see also below). Gordonia jacobaea MV-1, which is not yet validly described, was isolated because of its property of producing large amounts of carotenoids (22) and was the first Gordonia strain characterized with regard to carotenoid biosynthesis. G. jacobaea MV-1 was found to contain approximately 200 μg of the ketocarotenoid all-trans-canthaxanthin (4,4′-diketo-β-carotene)/g (dry weight) and smaller amounts ofall-trans-astaxanthin (3,3′-dihydroxy-4,4′-diketo-β-carotene)as a product of later steps in carotenoid synthesis (4). Both compounds are currently produced by Hoffmann-La Roche, Ltd., and are approved by the Food and Drug Administration as food additives in poultry and fish feeding. Carotenoid-overproducing strains were obtained after ethyl methanesulfonate treatment, yielding hyperpigmented mutants with a carotenoid content 10-fold-higher than that of the wild type (23). Because canthaxanthin, unlike other carotenoids, is not converted to vitamin A, and because it leads to crystalline deposition at the retina, its content in animal feed has been limited to 25 mg/kg of body weight by the European Union. In contrast, astaxanthin is still categorized as harmless. Engineering of carotenoid-overproducing strains of Gordonia species could provide a new biological source for these economically important substances.
FIG. 3.
Tn5096-induced mutants of G. polyisoprenivorans DSM 44266 with modified carotenoid biosynthesis. Mutants 44-07 and 49-05 were defective in carotenoid synthesis; mutants 33-45 and 38-33 exhibited a coloration slightly different from that of the wild type; and mutant 47-46 obviously overproduced a red carotenoid.
Imidazol-2-yl amino acids.
Substituted imidazoles exhibit manifold biological activities and have been extensively used as pharmaceuticals. They can act as antihistamine drugs, defined as H1, H2, and H3 receptor antagonists with structures related to histamine (10, 83), and they are often used as pharmaceuticals against allergic diseases and insomnia because of their distinctive sedating properties. Some substituted imidazoles exhibit antifungal properties or act as anticonvulsants. Among 61 alkane-oxidizing bacterial strains tested for their abilities to oxidize N-(2-hexylamino-4-phenylimidazol-1-yl)-acetamide to imidazol-2-yl amino acids applicable for pharmaceutical purposes, 5 strains producing high yields of the desired products were identified (68). Two of these strains belonged to the genus Gordonia and were taxonomically characterized as G. rubropertincta and G. terrae, respectively. Each strain was able to transform N-(2-hexylamino-4-phenylimidazol-1-yl)-acetamide to 1-amino-4-phenylimidazol-2-yl-4-aminobutanoic acid. Two different pathways comprising several enzymatic steps were revealed.
Gordonan, an acidic, cell aggregation-inducing polysaccharide.
In general, many bacterial species are able to produce extracellular polysaccharides in response to specific environmental circumstances. Well-known examples are Staphylococcus aureus, Staphylococcus epidermidis, and Pseudomonas aeruginosa, producing acidic polysaccharides including alginate as a main component to form sessile communities such as biofilms (53). The formation of these biofilms is considered essential for the pathogenicity of these bacteria. During screening for bioactive metabolites produced by microorganisms affecting the BM-N4 insect cell line in a study of insect cell growth and hormone signaling, Gordonia sp. strain Y-102, synthesizing a novel acidic polysaccharide referred to as gordonan, was isolated (53). BM-N4 is an insect cell line established from an ovarian tissue of the domestic silkworm Bombyx mori, and characteristic morphological changes of this cell line are induced by some insect hormones such as ecdysteroids and bombyxin-II. Gordonan, with an average molecular weight of 5 × 106, is composed of the trisaccharide repeating unit →3)-4-O-(1-carboxyethyl)-β-d-Manp-(1→4)-β-d-GlcAp-(1→4)-β-d-Glcp-(1→. This polymer induced aggregation of BM-N4 cells at concentrations higher than 4 μg/ml. The aggregated cells were alive and proliferated to become confluent upon further incubation. Control experiments with dextran sulfate and hyaluronic acid indicated that the bioactivity of gordonan was not due to its polyanionic structure. Since the genus Gordonia is closely related to the genera Nocardia and Mycobacterium, members of which cause tuberculosis or leprosy (5), and since members of the genus Gordonia are also known to be involved in bronchopulmonary disorders (35) or metastatic melanoma (81), gordonan might support the process of infection or the fixation of Gordonia spp. to a host, as do biofilms produced by P. aeruginosa (53). Whether gordonan also has effects on mammalian cells is still unclear. However, the isolation of Gordonia sp. strain Y-102, producing the novel polysaccharide gordonan, demonstrates that gordoniae are potential sources for very specific compounds. Further highly bioactive substances, which could be of great interest to the pharmaceutical industry, may well be detected in Gordonia strains in the future.
Gordonin and other glycosylated peptidolipids.
During an investigation of cell wall components responsible for the difference between the rough and smooth colony morphologies of G. hydrophobica strains isolated from biofilters for waste gas treatment (9), compounds were identified in the fraction of the most-polar lipids of the smooth variant that were lacking in the rough variant, whereas the less-polar lipids were found to be identical (69). The differing cell wall components were identified as (mono-) glycosylated peptidolipids consisting of a tridecapeptide interlinked by a β-hydroxylated fatty acid to form a cyclic lactone ring structure. The main fraction of glycosylated peptidolipids, for which the name gordonin was proposed, was identified as 3-hydroxyeicosanoyl-l-seryl-l-phenylalanyl-l-seryl-l-seryl-d-alanyl-l-(O-β-d-glucopyranosyl)-threonyl-glycyl-d-leucyl-l-valyl-l-seryl-l-phenylalanyl-glycyl-l-valyl lactone (69). Gordonin differed from the other glycosylated peptidolipids in the nature of the β-hydroxylated fatty acid [20:0 (3-OH) versus 22:1 (3-OH)] as well as in one amino acid (d-leucine versus d-phenylalanine). The amphiphilic character of many peptidolipids gives rise to several biological functions of these molecules such as hemolytic properties, antibiotic activities, or enzyme-inhibiting properties. The amphiphilic nature of gordonin is due to the fact that 20:0 (3-OH) fatty acid and several hydrophobic amino acids are clustered at one side of the proposed structure, while several l-serine residues and glucose are located on the opposite side. Therefore, the authors suggested that gordonin is able to confer a hydrophilic cell surface by intercalating with the hydrophobic tail into the very hydrophobic mycolic acid layer, exposing the hydrophilic head of the molecule to the exterior of the cell. As indicated by the different colony morphologies of rough and smooth variants, gordonin and the other peptidolipids may contribute significantly to the physicochemical character of the cell surface and may enable smooth variants of G. hydrophobica to grow in suspension in liquid culture. The authors also reported that gordonin could act as a surfactant, dissolving hydrophobic substrates, if it was released to the environment during growth. However, gordonin was not shown to reduce the interfacial tension between air and water or between water and organic solvents.
GENETIC MANIPULATION OF GORDONIA STRAINS
Investigation of the interesting metabolic pathways existing in gordoniae and of the genes encoding the respective enzymes was hampered until recently by the unavailability of suitable genetic tools. The first vectors appropriate for gene transfer to Gordonia species have been described only recently, by Arenskötter et al. (3); these are based on an origin of replication (oriV) of a native Rhodococcus plasmid, and genes can be transferred either by electrotransformation or by conjugational transfer. Also recently, Bröker et al. (12) discovered and sequenced a native 101-kbp megaplasmid, pKB1, occurring in G. westfalica DSM 44215T. Evidence was obtained that pKB1 encodes genes essential for cadmium resistance and rubber biodegradation. Based on the oriV region of this megaplasmid, E. coli-Gordonia shuttle vectors suitable for gene cloning and expression in several Gordonia species and members of related taxa were constructed (12). However, vectors appropriate for transposon mutagenesis or for generation of Gordonia knockout mutants are still lacking. Unpublished studies document that the temperature-sensitive mycobacterial transposon vector pCG79 (36) also transposed in G. polyisoprenivorans DSM 44266, but the mutants obtained were unstable and frequently reverted to a wild-type phenotype (M. Arenskötter et al., unpublished data). Suicide transposon vectors harboring Tn5096 (84) look promising for stable transposon-induced mutagenesis. Auxotrophic mutants with defective amino acid-synthesizing pathways and mutants in which carotenoid biosynthesis is affected (Fig. 3) have been isolated by using these vectors and were subsequently characterized at the molecular level (Arenskötter et al., unpublished).
CONCLUDING REMARKS
As clearly indicated by the increasing number and range of publications, patents, and gene sequence depositions relating to the genus Gordonia in recent years, this genus is of significant interest in a wide range of fields. The polyphasic approach for taxonomic classification of gordoniae and closely related genera such as Rhodococcus, Mycobacterium, and Nocardia have established the genus Gordonia as a relatively stable entity within the mycolic-acid-containing actinomycetes. However, one may expect that additional new species will be isolated and assigned to the genus Gordonia and also that several species already isolated are probably mislabeled and will be reclassified as members of this genus in the future.
The biotechnological interest in members of the genus Gordonia is mainly due to the diverse range of enzymatic reactions catalyzed by these bacteria that occur seldom or not at all in other organisms. Many potential applications, such as bioremediation, biodegradation, biotransformation, and production of biosurfactants, seem to be associated with the presence of hydrophobic, mycolic-acid-containing cell envelope structures on gordoniae, but so also do pathogenicity and problems such as foam formation in activated sludge. The potential of microbial processes for the bioremediation and biodegradation of toxic and/or xenobiotic compounds is still high, and despite the high costs and economic infeasibility of many processes, there is continuing industrial interest, as indicated by the increasing number of patents relating to the genus Gordonia. Beyond doubt, the metabolic capabilities of Gordonia species will continue to attract the interest of the chemical biotechnological industry, and novel applications will probably be identified and explored. One must, however, consider that some members of the genus Gordonia are opportunistic pathogens, which will restrict the use of these bacteria.
Acknowledgments
The principal investigator's studies on gordoniae and rubber biodegradation were financially supported by the Fonds der Chemischen Industrie (Frankfurt, Germany), the Max-Buchner Forschungsstiftung of DECHEMA e.V. (Frankfurt), and the Deutsche Bundesstiftung Umwelt (Osnabrück, Germany) in the context of an ICBIO project (AZ. 13072).
REFERENCES
- 1.Abbad-Andaloussi, S., M. Warzywoda, and F. Monot. 2003. Microbial desulfurization of diesel oils by selected bacterial strains. Oil Gas Sci. Technol. 58:505-513. [Google Scholar]
- 2.Arenskötter, M., D. Baumeister, M. M. Berekaa, G. Pötter, R. M. Kroppenstedt, A. Linos, and A. Steinbüchel. 2001. Taxonomic characterization of two rubber degrading bacteria belonging to the species Gordonia polyisoprenivorans and analysis of hypervariable regions of 16S rDNA sequences. FEMS Microbiol. Lett. 205:277-282. [DOI] [PubMed] [Google Scholar]
- 3.Arenskötter, M., D. Baumeister, R. Kalscheuer, and A. Steinbüchel. 2003. Identification and application of plasmids suitable for transfer of foreign DNA to members of the genus Gordonia. Appl. Environ. Microbiol. 69:4971-4974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Armstrong, G. A. 1994. Eubacteria show their true colors: genetics of carotenoid pigment biosynthesis from microbes to plants. J. Bacteriol. 176:4795-4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aspinall, G. O., D. Chatterjee, and P. J. Brennan. 1995. The variable surface of mycobacteria: structures, synthesis of epitopes, and biological properties. Adv. Carbohydr. Chem. Biochem. 51:169-242. [DOI] [PubMed] [Google Scholar]
- 6.Banat, I. M., R. S. Makkar, and S. S. Cameotra. 2000. Potential commercial applications of microbial surfactants. Appl. Microbiol. Biotechnol. 53:495-508. [DOI] [PubMed] [Google Scholar]
- 7.Bendinger, B., F. A. Rainey, R. M. Kroppenstedt, M. Moormann, and S. Klatte. 1995. Gordona hydrophobica sp. nov., isolated from biofilters for waste gas treatment. Int. J. Syst. Bacteriol. 45:544-548. [Google Scholar]
- 8.Bendinger, B., H. H. M. Rijnaarts, K. Altendorf, and A. J. B. Zehnder. 1993. Physicochemical cell surface and adhesive properties of coryneform bacteria related to the presence and chain length of mycolic acids. Appl. Environ. Microbiol. 59:3973-3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bendinger, B., R. M. Kroppenstedt, S. Klatte, and K. Altendorf. 1992. Chemotaxonomic differentiation of coryneform bacteria isolated from biofilters. Int. J. Syst. Bacteriol. 42:474-486. [DOI] [PubMed] [Google Scholar]
- 10.Black, J. W., W. A. M. Duncan, G. J. Durant, J. C. Ganellin, and M. E. Parsons. 1972. Definition and antagonism of histamine H2-receptors. Nature (London) 236:385-390. [DOI] [PubMed] [Google Scholar]
- 11.Brãndao, P. F. B., L. A. Maldonado, A. C. Ward, A. T. Bull, and M. Goodfellow. 2001. Gordonia namibiensis sp. nov., a novel nitrile metabolising actinomycete recovered from an African sand. Syst. Appl. Microbiol. 24:510-515. [DOI] [PubMed] [Google Scholar]
- 12.Bröker, D., M. Arenskötter, A. Legatzki, D. H. Nies, and A. Steinbüchel. 2004. Characterization of the 101-kilobase-pair megaplasmid pKB1, isolated from the rubber-degrading bacterium Gordonia westfalica Kb1. J. Bacteriol. 186:212-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buchman, A. L., M. M. McNeil, J. M. Brown, B. A. Lasker, and M. E. Ament. 1992. Central venous catheter sepsis caused by unusual Gordona (Rhodococcus) species: identification with a digoxigenin-labeled rDNA probe. Clin. Infect. Dis. 15:694-697. [DOI] [PubMed] [Google Scholar]
- 14.Chang, J. H., Y. J. Kim, B. H. Lee, K.-S. Cho, H. W. Rye, Y. K. Chang, and H. N. Chang. 2001. Production of a desulfurization biocatalyst by two-stage fermentation and its application for the treatment of model and diesel oils. Biotechnol. Prog. 17:876-880. [DOI] [PubMed] [Google Scholar]
- 15.Chatterjee, S., and T. K. Dutta. 2003. Metabolism of butyl benzyl phthalate by Gordonia sp. strain MTCC 4818. Biochem. Biophys. Res. Commun. 309:36-43. [DOI] [PubMed] [Google Scholar]
- 16.Choi, O. K., K. S. Cho, H. W. Ryu, and Y. K. Chang. 2003. Enhancement of phase separation by the addition of de-emulsifiers to three-phase (diesel oil/biocatalyst/aqueous phase) emulsion in diesel biodesulfurization. Biotechnol. Lett. 25:73-77. [DOI] [PubMed] [Google Scholar]
- 17.Cook, A. M., and R. Hütter. 1984. Deethylsimazine: bacterial dechlorination, deamination, and complete degradation. J. Agric. Food Chem. 32:581-585. [Google Scholar]
- 18.Cook, A. M., and R. Hütter. 1986. Ring dechlorination of deethylsimazine by hydrolases from Rhodococcus corallinus. FEMS Microbiol. Lett. 34:335-338. [Google Scholar]
- 19.David, H. L. 1974. Carotenoid pigments of Mycobacterium kansasii. Appl. Microbiol. 28:696-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De los Reyes, M. F., F. L. de los Reyes III, M. Hernandez, and L. Raskin. 1998. Quantification of Gordonia amarae strains in foaming activated sludge and anaerobic digester systems with oligonucleotide hybridization probes. Appl. Environ. Microbiol. 64:2503-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De los Reyes, M. F., F. L. de los Reyes, M. Hernandez, and L. Raskin. 1998. Identification and quantification of Gordona amarae strains in activated sludge systems using comparative rRNA sequence analysis and phylogenetic hybridization probes. Water Sci. Technol. 37:521-525. [Google Scholar]
- 22.De Miguel, T., C. Sieiro, M. Poza, and T. G. Villa. 2000. Isolation and taxonomic study of a new canthaxanthin-containing bacterium, Gordonia jacobaea MV-1 sp. nov. Int. Microbiol. 3:107-111. [PubMed] [Google Scholar]
- 23.De Miguel, T., C. Sieiro, M. Poza, and T. G. Villa. 2001. Analysis of canthaxanthin and related pigments from Gordonia jacobaea mutants. J. Agric. Food. Chem. 49:1200-1202. [DOI] [PubMed] [Google Scholar]
- 24.De Vos, P., and H. G. Trüper. 2000. Judicial Commission of the International Committee on Systematic Bacteriology. IXth International (IUMS) Congress of Bacteriology and Applied Microbiology. Minutes of the meetings, 14, 15 and 18 August 1999, Sydney, Australia. Int. J. Syst. Evol. Microbiol. 50:2239-2244. [Google Scholar]
- 25.Drancourt, M., J. Pelletier, A. A. Cherif, and D. Rault. 1997. Gordonia terrae central nervous system infection in immunocompetent patient. J. Clin. Microbiol. 35:379-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Drancourt, M., M. M. McNeil, J. M. Brown, B. A. Lasker, M. Mauron, M. Choux, and D. Raoult. 1994. Brain-abscess due to Gordona terrae in an immunocompromised child: case report and review of infections caused by G. terrae. Clin. Infect. Dis. 19:258-262. [DOI] [PubMed] [Google Scholar]
- 27.Ellis, J. 1770. A copy of a letter from John Ellis, Esq; F. R. S. to Dr. Linnæus, F. R. S. &c. with the figures and characters of the elegant American evergreen-tree, called by the Gardiners the Loblolly-Bay, taken from Blossoms blown near London, and shewing that it is not an Hibiscus, as Mr. Miller calls it; nor an Hypericum, as Dr. Linnæus supposes it; but an intire new genus, to which Mr. Ellis gives the name of Gordonia. Philos. Trans. 60:518-523. [Google Scholar]
- 28.Euzéby, J. 2003. Valid publication of new names and new combinations previously effectively published outside the IJSEM. Int. J. Syst. Evol. Microbiol. 53:1219-1220. [Google Scholar]
- 29.Fiechter, A. 1992. Biosurfactants: moving towards industrial application. Trends Biotechnol. 10:208-217. [DOI] [PubMed] [Google Scholar]
- 30.Finnerty, W. M. 1994. Biosurfactants in environmental biotechnology. Curr. Opin. Biotechnol. 5:291-295. [Google Scholar]
- 31.Fusconi, R., and M. J. L. Godinho. 2002. Screening for exopolysaccharide-producing bacteria from sub-tropical polluted groundwater. Braz. J. Biol. 62:363-369. [DOI] [PubMed] [Google Scholar]
- 32.Gilbert, S. C., J. Morton, S. Buchanan, C. Oldfield, and A. McRoberts. 1998. Isolation of a unique benzothiophene-desulphurizing bacterium, Gordona sp. 213E (NCIMB 40816), and characterization of the desulphurization pathway. Microbiology 144:2545-2553. [DOI] [PubMed] [Google Scholar]
- 33.Goodfellow, M. 1986. Genus Rhodococcus Zopf 4891, p. 1472-1481. In P. H. A. Sneath, N. S. Mair, M. E. Sharpe, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 2. The Williams & Wilkins Co., Baltimore, Md. [Google Scholar]
- 34.Goodfellow, M., and G. Alderson. 1977. The actinomycete genus Rhodococcus: a home for the rhodochrous complex. J. Gen. Microbiol. 100:99-122. [DOI] [PubMed] [Google Scholar]
- 35.Gugnani, H. C., I. C. Unaogu, F. Provost, and P. Boiron. 1998. Pulmonary infections due to Nocardiopsis dassonvillei, Gordonia sputi, Rhodococcus rhodochrous and Micromonospora sp. in Nigeria and literature review. J. Mycol. Med. 8:21-25. [Google Scholar]
- 36.Guilhot, C., I. Otal, I. van Rompaey, C. Martín, and B. Gicquel. 1994. Efficient transposition in mycobacteria: construction of Mycobacterium smegmatis insertional mutant libraries. J. Bacteriol. 176:535-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hart, D. H. L., M. M. Peel, J. H. Andrew, and J. G. W. Burdon. 1988. Lung infection caused by Rhodococcus. Aust. N. Z. J. Med. 18:790-791. [DOI] [PubMed] [Google Scholar]
- 38.Hernandez-Perez, G., F. Fayolle, and J.-P. Vandecasteele. 2001. Biodegradation of ethyl t-butyl ether (ETBE), methyl t-butyl ether (MTBE) and t-amyl methyl ether (TAME) by Gordonia terrae. Appl. Microbiol. Biotechnol. 55:117-121. [DOI] [PubMed] [Google Scholar]
- 39.Iwahori, K., T. Tokutomi, N. Miyata, and M. Fujita. 2001. Formation of stable foam by cells and culture supernatant of Gordonia (Nocardia) amarae. J. Biosci. Bioeng. 92:77-79. [DOI] [PubMed] [Google Scholar]
- 40.Jagannadham, M. V., M. K. Chattopadhyay, and S. Shivaji. 1996. The major carontenoid pigment of a psychrotrophic Micrococcus roseus strain: fluorescence properties of the pigment and its binding membranes. Biochem. Biophys. Res. Commun. 220:724-728. [DOI] [PubMed] [Google Scholar]
- 41.Johnsen, A. R., and U. Karlson. 2003. Evaluation of bacterial strategies to promote the bioavailability of polycyclic aromatic hydrocarbons. Appl. Microbiol. Biotechnol. 63:452-459. (First published 5 March 2003; 10.1007/s00253-003-1265-z.) [DOI] [PubMed] [Google Scholar]
- 42.Kästner, M., M. Breuer-Jammali, and B. Mahro. 1994. Enumeration and characterization of the soil microflora from hydrocarbon-contaminated soil sites able to mineralize polycyclic aromatic hydrocarbons (PAH). Appl. Environ. Biotechnol. 41:267-273. [Google Scholar]
- 43.Kästner, M., M. Breuer-Jammali, and B. Mahro. 1998. Impact of inoculation protocols, salinity, and pH on the degradation of polycyclic aromatic hydrocarbons (PAHs) and survival of PAH-degrading bacteria introduced into soil. Appl. Environ. Microbiol. 64:359-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kayser, K. J., B. A. Bielaga-Jones, K. Jackowski, O. Odusan, and J. J. Kilbane. 1993. Utilization of organosulphur compounds by axenic and mixed cultures of Rhodococcus rhodochrous strain IGTS8. J. Gen. Microbiol. 139:3123-3129. [Google Scholar]
- 45.Kempf, V. A. J., M. Schmalzing, A. Y. Yassin, K. P. Schaal, D. Baumeister, M. Arenskötter, A. Steinbüchel, and I. B. Autenrieth. 2004. Gordonia polyisoprenivorans septicemia in a bone marrow transplant patient. Eur. J. Clin. Microbiol. Infect. Dis. 23:226-228. (First published 4 February 2004; 10.1007/s10096-003-1087-z.) [DOI] [PubMed] [Google Scholar]
- 46.Kertesz, M. A., and C. Wietek. 2001. Desulfurization and desulfonation: applications of sulfur-controlled gene expression in bacteria. Appl. Microbiol. Biotechnol. 57:460-466. [DOI] [PubMed] [Google Scholar]
- 47.Kim, K. K., C. S. Lee, R. M. Kroppenstedt, E. Stackebrandt, and S. T. Lee. 2003. Gordonia sihwensis sp. nov., a novel nitrate-reducing bacterium isolated from a wastewater treatment bioreactor. Int. J. Syst. Evol. Micriobiol. 53:1427-1433. [DOI] [PubMed] [Google Scholar]
- 48.Kim, S. B., R. Brown, C. Oldfield, S. C. Gilbert, and M. Goodfellow. 1999. Gordonia desulfuricans sp. nov., a benzothiophene-desulfurizing actinomycete. Int. J. Syst. Bacteriol. 49:1845-1851. [DOI] [PubMed] [Google Scholar]
- 49.Kim, S. B., R. Brown, C. Oldfield, S. C. Gilbert, S. Iliarionov, and M. Goodfellow. 2000. Gordonia amicalis sp. nov., a dibenzothiophene-desulfurizing actinomycete. Int. J. Syst. Bacteriol. 50:2031-2036. [DOI] [PubMed] [Google Scholar]
- 50.Klatte, S., F. A. Rainey, and R. M. Kroppenstedt. 1994. Transfer of Rhodococcus aichiensis Tsukamura 1982 and Nocardia amarae Lechevalier and Lechevalier 1974 to the genus Gordona as Gordona aichiensis comb. nov. and Gordona amarae comb. nov. Int. J. Syst. Bacteriol. 44:769-773. [DOI] [PubMed] [Google Scholar]
- 51.Klatte, S., R. M. Kroppenstedt, P. Schumann, K. H. Altendorf, and F. A. Rainey. 1996. Gordona hirsuta sp. nov. Int. J. Syst. Bacteriol. 46:876-880. [DOI] [PubMed] [Google Scholar]
- 52.Koma, D., Y. Sakashita, K. Kubota, Y. Fujii, F. Hasumi, S.-Y. Chung, and M. Kubo. 2003. Degradation of car engine base oil by Rhodococcus sp. NDKK48 and Gordonia sp. NDKY76A. Biosci. Biotechnol. Biochem. 67:1590-1593. [DOI] [PubMed] [Google Scholar]
- 53.Kondo, T., D. Yamamoto, A. Yokota, A. Suzuki, H. Nagasawa, and S. Sakuda. 2000. Gordonan, an acidic polysaccharide with cell aggregation-inducing activity in insect BM-N4 cells, produced by Gordonia sp. Biosci. Biotechnol. Biochem. 64:2388-2394. [DOI] [PubMed] [Google Scholar]
- 54.Krubasik, P., M. Kobayashi, and G. Sandmann. 2001. Expression and functional analysis of a gene cluster involved in synthesis of decaprenoxanthin reveals the mechanisms for C50 carotenoid formation. Eur. J. Biochem. 268:3702-3708. [DOI] [PubMed] [Google Scholar]
- 55.Kummer, C., P. Schumann, and E. Stackebrandt. 1999. Gordonia alkanivorans sp. nov., isolated from tar-contaminated soil. Int. J. Syst. Bacteriol. 49:1513-1522. [DOI] [PubMed] [Google Scholar]
- 56.Kurimura, Y., Y. Furutani, N. Makiguchi, and K. Souda. September 1975. Method of producing l-lysine by fermentation. U.S. patent 3,905,867.
- 57.Kuwabara, M., T. Onitsuka, K. Nakamura, M. Shimada, S. Ohtaki, and Y. Mikami. 1999. Mediastinitis due to Gordonia sputi after CABG. J. Cardiovasc. Surg. 40:675-677. [PubMed] [Google Scholar]
- 58.Lechevalier, H. A. 1989. Nocardioform actinomycetes, p. 2348-2404. In S. T. Williams, M. E. Sharpe, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 4. The Williams & Wilkins Co., Baltimore, Md.
- 59.Lechevalier, M. P., and H. A. Lechevalier. 1974. Nocardia amarae sp. nov., an actinomycete common in foaming activated sludge. Int. J. Syst. Bacteriol. 24:278-288. [Google Scholar]
- 60.Lee, J. J., S.-K. Rhee, and S.-T. Lee. 2001. Degradation of 3-methylpyridine and 3-ethylpyridine by Gordonia nitida LE31. Appl. Environ. Microbiol. 67:4342-4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lesens, O., Y. Hansmann, P. Riegel, R. Heller, M. Benaissa-Djellouli, M. Martinot, H. Petit, and D. Christmann. 2000. Bacteremia and endocarditis caused by a Gordonia species in a patient with a central venous catheter. Emerg. Infect. Dis. 6:382-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Linos, A., A. Steinbüchel, C. Spröer, and R. M. Kroppenstedt. 1999. Gordonia polyisoprenivorans sp. nov., a rubber-degrading actinomycete isolated from an automobile tire. Int. J. Syst. Bacteriol. 49:1785-1791. [DOI] [PubMed] [Google Scholar]
- 63.Linos, A., and A. Steinbüchel. 1998. Microbial degradation of natural and synthetic rubbers by novel bacteria belonging to the genus Gordona. Kautsch. Gummi. Kunstst. 51:496-499. [Google Scholar]
- 64.Linos, A., M. M. Berekaa, A. Steinbüchel, K. K. Kim, C. Spöer, and R. M. Kroppenstedt. 2002. Gordonia westfalica sp. nov., a novel rubber-degrading actinomycete. Int. J. Syst. Evol. Microbiol. 52:1133-1139. [DOI] [PubMed] [Google Scholar]
- 65.Linos, A., M. M. Berekaa, R. Reichelt, U. Keller, J. Schmitt, H. C. Flemming, R. M. Kroppenstedt, and A. Steinbüchel. 2000. Biodegradation of cis-1,4-polyisoprene rubbers by distinct actinomycetes: microbial strategies and detailed surface analysis. Appl. Environ. Microbiol. 66:1639-1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Maldonado, L. A., F. M. Stainsby, A. C. Ward, and M. Goodfellow. 2003. Gordonia sinesedis sp. nov., a novel soil isolate. Antonie Leeuwenhoek 83:75-80. [DOI] [PubMed] [Google Scholar]
- 67.Martin, T., D. J. Hogan, F. Murphy, I. Natyshak, and E. P. Ewan. 1991. Rhodococcus infections of the skin with lymphadenitis in a nonimmunocompromised girl. J. Am. Acad. Dermatol. 24:328-332. [DOI] [PubMed] [Google Scholar]
- 68.Mikolasch, A., E. Hammer, and F. Schauer. 2003. Synthesis of imidazol-2-yl amino acids by using cells from alkane-oxidizing bacteria. Appl. Environ. Microbiol. 69:1670-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Moormann, M., U. Zähringer, H. Moll, R. Kaufmann, R. Schmid, and K. Altendorf. 1997. A new glycosylated lipopeptide incorporated into the cell wall of a smooth variant of Gordonia hydrophobica. J. Biol. Chem. 272:10729-10738. [DOI] [PubMed] [Google Scholar]
- 70.Mulbry, W. W. 1994. Purification and characterization of an inducible s-triazine hydrolase from Rhodococcus corallinus NRRL B-15444R. Appl. Environ. Microbiol. 60:613-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nakai, M., Y. Tabira, D. Asai, Y. Yakabe, T. Shimyozu, M. Noguchi, M. Takatsuki, and Y. Shimohigashi. 1999. Binding characteristics of dialkyl phthalates for the estrogen receptor. Biochem. Biophys. Res. Commun. 254:311-314. [DOI] [PubMed] [Google Scholar]
- 72.Nazina, T. N., D. S. Sokolova, A. A. Grigor'yan, Y. F. Xue, S. S. Belyaev, and M. V. Ivanov. 2003. Production of oil-releasing compounds by microorganisms from the Daqing oil field, China. Microbiology 72:206-211. [PubMed] [Google Scholar]
- 73.Neu, T. R. 1996. Significance of bacterial surface-active compounds in interaction of bacteria with interfaces. Microbiol. Rev. 60:151-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Oldfield, C., O. Pogrebinsky, J. Simmonds, E. Olson, and C. F. Kulpa. 1997. Elucidation of the metabolic pathway for dibenzothiophene desulphurization by Rhodococcus sp. strain IGTS8 (ATCC 53968). Microbiology 143:2961-2973. [DOI] [PubMed] [Google Scholar]
- 75.Pagilla, K. R., A. Sood, and H. Kim. 2002. Gordonia (Nocardia) amarae foaming due to biosurfactant production. Water Sci. Technol. 46:519-524. [PubMed] [Google Scholar]
- 76.Park, S. J., I. S. Lee, Y. K. Chang, and S. Y. Lee. 2003. Desulfurization of dibenzothiophene and diesel oil by metabolically engineered Escherichia coli. J. Microbiol. Biotechnol. 13:578-583. [Google Scholar]
- 77.Pasamontes, L., D. Hug, M. Tessier, H.-P. Hohmann, J. Schierle, and A. P. G. M. van Loon. 1997. Isolation and characterization of the carotenoid biosynthesis genes of Flavobacterium sp. strain R1534. Gene 185:35-41. [DOI] [PubMed] [Google Scholar]
- 78.Patterson, J. H., M. J. McConville, R. E. Haites, R. L. Coppel, and H. Bilman-Jacobe. 2000. Identification of a methyltransferase from Mycobacterium smegmatis involved in glycopeptidolipid synthesis. J. Biol. Chem. 275:24900-24906. [DOI] [PubMed] [Google Scholar]
- 79.Rhee, S. K., J. H. Chang, and H. N. Chang. 1998. Desulfurization of dibenzothiophene and diesel oils by a newly isolated Gordona strain, CYKS1. Appl. Environ. Microbiol. 64:2327-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Richet, H. M., P. C. Craven, J. M. Brown, B. A. Lasker, C. D. Cox, M. M. McNeil, A. D. Tice, W. R. Jarvis, and O. C. Tablan. 1991. A cluster of Rhodococcus (Gordona) bronchialis-sternal wound infections after coronary-artery bypass-surgery. N. Engl. J. Med. 324:104-109. [DOI] [PubMed] [Google Scholar]
- 81.Riegel, P., R. Ruimy, D. de Briel, F. Eichler, J.-P. Bergerat, R. Christen, and H. Monteil. 1996. Bacteremia due to Gordona sputi in an immunocompromised patient. J. Clin. Microbiol. 34:2045-2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 83.Schwartz, J. C., J. M. Arrang, M. Garbarg, H. Pollard, and M. Ruat. 1991. Histaminergic transmission in the mammalian brain. Physiol. Rev. 71:1-51. [DOI] [PubMed] [Google Scholar]
- 84.Solenberg, P. J., and R. H. Baltz. 1991. Transposition of Tn5096 and other IS493 derivatives in Streptomyces griseofuscus. J. Bacteriol. 173:1096-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Soto, A. M., C. Sonnenschein, K. L. Chung, M. F. Fernandez, N. Olea, and F. O. Serrano. 1995. The E-SCREEN assay as a tool to identify estrogens: an update on estrogenic environmental pollutants. Environ. Health Perspect. 103:113-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stackebrandt, E., F. A. Rainey, and N. L. Ward-Rainey. 1997. Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int. J. Syst. Bacteriol. 47:479-491. [Google Scholar]
- 87.Stackebrandt, E., J. Smida, and M. D. Collins. 1988. Evidence of phylogenetic heterogeneity within the genus Rhodococcus: revival of the genus Gordona (Tsukamura). J. Gen. Appl. Microbiol. 34:341-348. [Google Scholar]
- 88.Staples, C. A., D. R. Peterson, T. F. Parkerton, and W. J. Adams. 1997. The environmental fate of phthalate esters: a literature review. Chemosphere 35:667-749. [Google Scholar]
- 89.Takeuchi, M., and K. Hatano. 1998. Gordonia rhizosphera sp. nov. isolated from the mangrove rhizosphere. Int. J. Syst. Bacteriol. 48:907-912. [DOI] [PubMed] [Google Scholar]
- 90.Tomiyasu, I., and I. Yano. 1986. The mycolic acids and the chemotaxonomy of the genera Nocardia and Rhodococcus. J. Jpn. Soc. Actinomycetes 48:1-20. [Google Scholar]
- 91.Tsukamura, M. 1971. Proposal of a new genus, Gordona, for slightly acid-fast organisms occurring in sputa of patients with pulmonary disease and in soil. J. Gen. Microbiol. 68:15-26. [DOI] [PubMed] [Google Scholar]
- 92.Tsukamura, M. 1978. Numerical classification of Rhodococcus (formerly Gordona) organisms recently isolated from sputa of patients: description of Rhodococcus sputi Tsukamura sp. nov. Int. J. Syst. Bacteriol. 28:169-181. [Google Scholar]
- 93.Tsukamura, M. 1982. Numerical analysis of the taxonomy of norcardiae and rhodococci. Division of Nocardia asteroides sensu stricto into two species and description of Nocardia paratuberculosis sp. nov. Tsukamura (formerly the Kyoto-I group of Tsukamura), Nocardia nova sp. nov. Tsukamura, Rhodococcus aichiensis sp. nov. Tsukamura, Rhodococcus chubuensis sp. nov. Tsukamura, and Rhodococcus obuensis sp. nov. Tsukamura. Microbiol. Immunol. 26:1101-1119. [DOI] [PubMed] [Google Scholar]
- 94.Xue, Y., X. Xuesong, P. Zhou, R. Liu, F. Liang, and Y. Ma. 2003. Gordonia paraffinivorans sp. nov., a hydrocarbon-degrading actinomycete isolated from an oil-producing well. Int. J. Syst. Evol. Micriobiol. 53:1643-1646. [DOI] [PubMed] [Google Scholar]
- 95.Yoon, J.-H., J. J. Lee, S. S. Kang, M. Takeuchi, Y. K. Shin, S. T. Lee, K. H. Kang, and Y. H. Park. 2000. Gordonia nitida sp. nov., a bacterium that degrades 3-ethylpyridine and 3-methylpyridine. Int. J. Syst. Evol. Microbiol. 50:1203-1210. [DOI] [PubMed] [Google Scholar]