Abstract
Pinus radiata (Monterey pine), a tree native to coastal California and Mexico, is widely planted worldwide for timber production. A major threat to Monterey pine plantations is the fungal disease pine pitch canker, caused by Fusarium circinatum (Hypocreales). We present a novel trapping approach using filter paper in combination with a rapid molecular method to detect the presence of inoculum in the air. The assay is also useful for diagnosing the presence of the pathogen on plants. The test is based on the F. circinatum specific primer pair CIRC1A-CIRC4A, which amplifies a 360-bp DNA fragment in the intergenic spacer region of the nuclear ribosomal operon. Real-time PCR was used to calculate the number of fungal spores present in each reaction mixture by comparing the threshold cycle (Ct) of unknown spore samples to the Ct values of standards with known amounts of F. circinatum spores. The filter paper method allows prolonged and more sensitive spore sampling in the field compared to traditional traps using petri dishes filled with selective medium. A field test at two sites in coastal California infested with pine pitch canker was carried out during the summer and fall of 2002. Spore counts were in the range of ca. 1 × 103 to ca. 7 × 105/m2, with the highest spore counts in the fall, suggesting a seasonal fluctuation.
Monterey pine (Pinus radiata D. Don) is found in three native populations in the coastal range of California and in two insular populations in northwestern Mexico. This species is one of the most popular ornamental trees in the United States and is used widely for timber and pulp production in New Zealand, Australia, Chile, South Africa, and Spain (21).
The narrow genetic base of Monterey pine used in exotic and vast monoculture plantations is the cause of concern for the potential rise of destructive epidemics caused by either pests or pathogens (25). Pine pitch canker, caused by the ascomycete Fusarium circinatum Nirenberg and O'Donnell (= F. subglutinans [Wollenweb and Reinking] Nelson, Toussoun, and Marasas f. sp. pini [teleomorph: Gibberella circinata Nirenberg and O'Donnell]), is a serious disease of Monterey pine, with symptoms ranging from brown flagging of twigs, stem cankers associated with a strong ooze, dieback, and often tree death. Pine pitch canker was first detected in the southeastern United States (13) and was hypothesized to be endemic there and in Mexico (10, 12). More recently, the disease has been introduced into California (5, 10), South Africa (25), Japan (2), and Chile (26). Long-range spread of the disease may be connected to the movement of infected plant material for commercial use, including infected seeds.
Medium- and short-range dispersal of the pathogen is easily linked to the presence of abundant airborne inoculum (5, 8) and may be enhanced by insects either by vectoring or by the creation of optimal infection courts through their feeding activity. Pathogen phoresy has been shown for insects in the families Scolitydae, Anobiidae, and Cercopidae (9, 11, 14, 20), but true vectoring still awaits experimental confirmation.
Both G. circinata mating types, MAT-1 and MAT-2, are known to occur in nature, and their nucleotide sequences have been reported (7, 17, 23), but the importance of the sexual stage is poorly understood. Recent introductions of a pathogen into a new area are often characterized by the presence or at least dominance of a single mating type (e.g., in California, MAT-1) (24). As in other exotic pathosystems (P. infestans, etc.), subsequent introduction of the missing mating type is extremely likely.
Several molecular methods have been developed recently to differentiate F. circinatum from other species of the G. fujikuroi species complex. These include PCR-restriction fragment length polymorphisms (RFLPs) of the histone H3 gene (16) and mitochondrial RFLP (6). RFLP methods, however, are poorly suited for quantification of target DNA copy numbers, their sensitivity is likely to be too low for a reliable diagnosis directly from infected plant tissue, and this method can be problematic when plants have been infected by more than one Fusarium species. Molecular methods can be used to identify a Fusarium culture, but their utility can be better appreciated when identifying the pathogen directly from infected plants. Another important application of DNA-based diagnostics relies on its ability to be used directly on trapped spores, without the need for spores to be germinated. Inoculum pressure (e.g., spore load in the air) is of paramount importance in developing epidemiological models for most airborne diseases. All of the spore trapping methods used to date (e.g., bark washes, air sampling with petri dishes) include a step in which colony growth on a selective medium is observed and quantified. The use of medium, which is prone to desiccation, effectively limits the trapping period to 1 to 3 days. The quantification step, involving colony counts, also presents some limitations, as plates can easily become overcrowded or overgrown, making precise counting impossible. Trapping methods suitable for long-term measurements in the field, matched with an alternative quantification approach, are highly desirable for epidemiological studies in which inoculum pressure needs to be determined.
The objectives of our study were (i) to establish a fast and reliable diagnostic test for the presence of F. circinatum independently of the presence of disease symptoms, (ii) to develop a new spore trapping assay that can be used for extended trapping periods and, (iii) to quantify airborne spores at infested sites with a real-time PCR assay.
MATERIALS AND METHODS
Fungal strains.
The strains used in this study are listed in Table 1. The geographic origins and hosts or substrates of the 31 strains selected for sequencing of the nuclear ribosomal intergenic spacer (IGS) region have already been described (15), except for NRRL 26431 and NRRL 26432, which were isolated from Pinus luchuensis (Ryukyu Island pine) in Kyushu and Okinawa, Japan, respectively. All strains are stored in the Agricultural Research Service Culture Collection (Northern Regional Research Laboratory), National Center for Agricultural Utilization Research, Peoria, Ill.
TABLE 1.
List of Fusarium strains for which the nuclear ribosomal IGS region was sequenceda
| Organism | Agricultural Research Service culture collection no. | GenBank accession no. |
|---|---|---|
| F. anthophilumb | NRRL 13602 | AY249392 |
| F. bactridioidesb | NRRL 20476 | AY249389 |
| F. begoniaeb | NRRL 25300 | AY249393 |
| F. bulbicolab | NRRL 13618 | AY249395 |
| F. circinatumb | NRRL 25331 | AY249397 |
| F. circinatumb | NRRL 25332 | AY249400 |
| F. circinatumb | NRRL 25333 | AY249401 |
| F. circinatumb | NRRL 25621 | AY249403 |
| F. circinatumb | NRRL 25707 | AY249402 |
| F. circinatumb | NRRL 26431 | AY249398 |
| F. circinatumb | NRRL 26432 | AY249399 |
| F. concentricum | NRRL 25303 | AY249385 |
| F. fractiflexum | NRRL 28852 | AY249386 |
| F. fujikuroi | NRRL 13556 | AY249382 |
| F. globosum | NRRL 26131 | AY249384 |
| F. oxysporum | NRRL 22902 | AY249409 |
| F. proliferatum | NRRL 22944 | AY249383 |
| F. pseudoanthophilum | NRRL 25206 | AY249380 |
| F. subglutinans | NRRL 22016 | AY249387 |
| F. succisae | NRRL 13613 | AY249391 |
| Fusarium sp.b | NRRL 25195 | AY249394 |
| Fusarium sp.b | NRRL 25204 | AY249405 |
| Fusarium sp.b | NRRL 25346 | AY249404 |
| Fusarium sp.b | NRRL 25622 | AY249390 |
| Fusarium sp.b | NRRL 25623 | AY249408 |
| Fusarium sp.b | NRRL 25807 | AY249396 |
| Fusarium sp.b | NRRL 26756 | AY249406 |
| Fusarium sp. | NRRL 26757 | AY249407 |
| Fusarium sp.b | NRRL 29123 | AY249388 |
| F. thapsinum | NRRL 22045 | AY249381 |
| F. verticillioides | NRRL 22172 | AY249379 |
The specificity of the primer pair CIRC1A-CIRC4A was tested by using the strains belonging to G. fujikuroi clade 2.
Member of G. fujikuroi clade 2.
Medium.
Fusarium selective (FS) medium consists of the following per liter: 15 g of Bacto Peptone, 20 g of agar, 1 g of KH2PO4, 0.5 g of MgSO4 · 7H2O, 0.3 g of streptomycin sulfate, 0.1 g of ampicillin, 0.2 g of pentachloronitrobenzene (Aldrich, Milwaukee, Wis.), and 39 g of potato dextrose agar (Difco, Detroit, Mich.).
DNA isolation, amplification, and sequencing of fungal isolates.
Strains were cultured and DNA was isolated as described by O'Donnell et al. (15). The concentrations of DNA from mycelium used for calculating the calibration curves were determined by UV spectrophotometry at 260 nm. Primers NL11 and CNS1 (Table 2) were used to amplify the entire IGS region with Platinum Taq DNA polymerase Hi-Fi (Invitrogen Life Technologies, Carlsbad, Calif.) in accordance with the supplier's recommendations, in a 9700 thermocycler (Applied Biosystems, Foster City, Calif.), with the following cycling parameters: 94°C for 90 s, 40 cycles of 94°C for 30 s, 52°C for 30 s, and 68°C for 3 min, followed by 68°C for 5 min and a 4°C soak. Amplicons were purified with Multiscreen plates (Millipore Corp., Bedford, Mass.) and then cycle sequenced with Applied Biosystems BigDye chemistry version 2 in a 9700 cycler as follows: 96°C for 15 s, 40 cycles of 96°C for 15 s, 50°C for 10 s, and 60°C for 4 min, ending in a 4°C soak. The primers used for sequencing are listed in Table 2. Sequencing reaction mixtures were purified via ethanol precipitation and then run on a Prism 3100 genetic analyzer (Applied Biosystems-Hitachi, Tokyo, Japan). DNA sequences were edited and aligned with Sequencher 4.1.2 (Gene Codes, Ann Arbor, Mich.), and alignments were improved manually.
TABLE 2.
Primers used for specific detection of F. circinatum, determination of mating type, and sequencing of the nuclear ribosomal IGS region
| Primer | Sequence | Target sequence |
|---|---|---|
| F. circinatum detection | ||
| CIRC1A | 5′ CTTGGCTCGAGAAGGG | IGS forward |
| CIRC4A | 5′ ACCTACCCTACACCTCTCACT | IGS reverse |
| Mating type determination | ||
| GcHMG1 | 5′ CTTTACCGTAAGGAGCGTCACCAT | MAT-2 |
| GcHMG2 | 5′ TGATCCGCCATCTGCTTGTAGAGT | MAT-2 |
| MAT1p2 | 5′ AGAAACTGACTGATACATCAAGGGG | MAT-1 |
| MAT1p3 | 5′ TCATAAGAAGTGTTGAAGGAATCACAG | MAT-1 |
| IGS sequencing | ||
| NL11 | 5′ CTGAACGCCTCTAAGTCAG | American, Asian, and African clades of F. oxysporum |
| GCNS3 | 5′ CTGCAAAGCTGTACAGAGGG | American, Asian, and African clades of F. oxysporum |
| GCNS5d | 5′ CACCTACCCTACACCACC | F. oxysporum |
| GCNS7f | 5′ GACACCGCGCCTCTTAACAAC | American, Asian, and African clades |
| GCNS7r | 5′ GTTGTTAAGAGGCGCGGTGTC | American, Asian, and African clades |
| G5-2G | 5′ TGTGACKACCTACCCTACACC | American clade |
| G5-4C | 5′ TGTGAYGACCTACCCTATACC | Asian clade |
| G5-A5 | 5′ TGTGACTRCCTACCCTATACC | African clade |
| CNS1 | 5′ GAGACAAGCATATGACTAC | American, Asian, and African clades of F. oxysporum |
Phylogenetic analysis.
The heuristic search option of PAUP* V4.0b4a (22) was used with 1,000 random addition sequences to conduct a maximum-parsimony analysis of the aligned IGS sequences, after excluding 37 ambiguously aligned nucleotide positions. Of the 2,593 included characters, 382 and 485 were autapomorphic and synapomorphic, respectively. On the basis of the results of a previous phylogenetic analysis (15), a sequence of F. oxysporum NRRL 22902 was used to root the parsimony tree by the outgroup method. Clade stability was assessed via 1,000 parsimony bootstrap replications with PAUP* (22).
DNA extraction from spores.
Spore suspensions (100 μl) were mechanically disrupted by shaking with glass beads (0.5 mm; Biospec Products, Inc., Bartlesville, Okla.) for 30 s. After addition of 0.3 ml of cetyltrimethylammonium bromide extraction buffer (15), the samples were subjected to three freeze-thaw cycles (2 min on dry ice, 2 min at 75°C, repeated three times with the last thaw extended to 30 min). DNA was extracted with 0.35 ml of phenol-chloroform-isoamyl alcohol, followed by purification with the GeneClean kit (Bio 101, Inc., Carlsbad, Calif.), in accordance with the instructions supplied by the manufacturer. DNA from fungal colonies grown on FS medium was extracted by a “hyphal tipping” method in which mycelium was suspended in 100 μl of distilled water, frozen on dry ice for 3 min, thawed at 75°C, and vortexed for 2 min. Freezing and thawing were repeated three times, with the last thaw extended to 15 min. Samples were centrifuged for 5 min at 15,700 × g, and 6.25 μl of the supernatant was used for PCRs.
Primers for specific detection of F. circinatum.
Specific primers for F. circinatum (CIRC1A and CIRC4A; Table 2) were designed with primer3 software (www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi) on the basis of the alignment of the nuclear ribosomal IGS region of the 31 Fusarium strains and synthesized by Qiagen-Operon (Alameda, Calif.). The amplicon size is 360 bp. The specificity of the primers used to amplify F. circinatum DNA was tested with a PCR comparison of F. circinatum strains with all strains belonging to the same clade (clade 2) of the G. fujikuroi complex on the basis of the IGS tree (Fig. 1). The PCR mixture contained 12 μl of H2O, 2.5 μl of PCR buffer, 1.0 μl of 50 mM MgCl2, 2.5 μl of deoxynucleoside triphosphate, 0.25 μl of 50 mM primer CIRC1A, 0.25 μl of 50 mM primer CIRC4A, 0.25 μl of Platinum Taq polymerase, and 6.25 μl of template DNA. The annealing temperature for the primers was tested in the range of 58.0 to 70.0°C. The optimal cycling profile was denaturation at 94.0°C for 180 s, followed by 45 cycles of 94°C for 35 s, 66°C for 55 s, and 72°C for 50 s and a final extension of 72°C for 12 min.
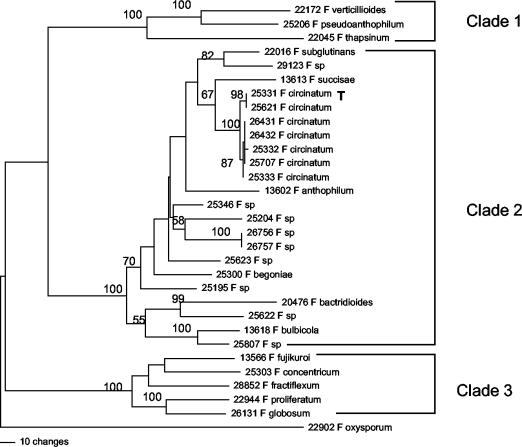
FIG. 1.
Phylogenetic tree of 31 Fusarium strains based on parsimony analyses (heuristic search) of 2,593 bp of the nuclear ribosomal IGS region. The number above each internode represents the bootstrap interval from 1,000 replications. Strain numbers are those of the Agricultural Research Service Culture Collection (Northern Regional Research Laboratory). T = type strain.
Determination of the mating type.
The mating type was determined in a multiplex PCR with the primers listed in Table 2 as described by Covert et al. (7) and Wallace and Covert (23). DNA standards of strain NRRL 25331 (MAT-1) in the range of 101 to 104 pg were used to measure the detection limit for this single-copy gene.
Environmental samples may consist of a mixture of strains of both mating types in different concentrations. In order to simulate this condition, DNA templates consisting of mixtures of strains NRRL 25331 (MAT-1) and NRRL 25333 (MAT-2) at relative concentrations of 1:1, 1:10, 1:100, and 1:1,000 were used in a multiplex reaction.
Real-time PCR.
Real-time PCRs with both CIRC1A-CIRC4A and the mating type primers sets were carried out with the iCycler from Bio-Rad (Hercules, Calif.). The SYBR-Green method used SYBR-Green I (Applied Biosystems) as a fluorescent dye, which intercalates specifically with double-stranded DNA during the extension phase of the PCR. For quantification of the starting amount of template DNA, the threshold cycle (Ct) for each sample was calculated. The Ct value, which is the cycle number when the fluorescence of SYBR-Green measured is significantly different from the background level for the first time, was calculated for baseline cycles 2 to 10. The Ct value is proportional to the logarithm of the initial DNA concentration. The data analysis window was set at 10.00% of a cycle and centered at the end of the cycle. The real-time PCR conditions were as follows: 8.0 μl of H2O, 2.5 μl of PCR buffer, 2.5 μl of 50 mM MgCl2, 2.5 μl of deoxynucleoside triphosphate, 0.25 μl of 50 mM forward primer, 0.25 μl of 50 mM reverse primer, 1.25 μl of SYBR-Green, 1.25 μl of fluorescein (Bio-Rad), 0.25 μl of Platinum Taq polymerase, and 6.25 μl of template DNA. Cycling parameters were as follows: 94.0°C for 180 s, 45 cycles of 94°C for 35 s, 66°C for 55 s, and 72°C for 50 s, followed by a final extension of 72°C for 12 min. Standard curves were calculated for DNAs from F. circinatum NRRL 25331 (type strain, MAT-1) and NRRL 25333 (MAT-2) obtained from mycelium at a concentration of 101 to 104 pg per reaction. For direct comparisons with environmental samples, additional standard curves were calculated with spore suspensions at concentrations of 105, 104, 103, and 102 spores per 100 μl. Three independent amplifications were conducted for each DNA concentration and strain. Melting curves were measured for 110 cycles of 10 s each, starting at 60°C and increasing by 0.3°C each cycle. The melting curve is displayed as the negative first derivative of the fluorescence-versus-temperature plot [−d(RFU)/dT] over temperature. The iCycler software identifies the so-called “melting peaks” and assigns melting temperatures from this plot (1). PCR products were subjected to gel electrophoreses by loading 5 μl of each PCR product onto a 1.5% agarose gel, running it for 60 min at 20 V/cm, and staining it with ethidium bromide prior to visualization over UV.
Environmental test.
Spore traps were placed on two sites. Site I was Presidio National Park, San Francisco, Calif., and site II was San Bruno, Calif. Both sites are nonnative stands of P. radiata. Trees at site I showed light symptoms of pine pitch canker (<25% of the trees), while trees at site II were heavily infected, with numerous dead or dying trees (ca. 90% of the trees were infected). Three platforms (height, 1 m) were placed on each site at a distance of ca. 25 m from each other, each holding a petri dish with FS medium and a filter paper (Whatman no. 1). To increase its trapping capacity, each filter paper was wetted with 4× TE buffer (40 mM Tris HCl, 4 mM EDTA; Sigma Chemical Co., St. Louis, Mo.). Samples were collected from the field after 2 weeks in June, July, and October of 2002 and processed in the laboratory by three different methods.
Method 1 (growth on FS medium).
Petri dishes containing FS medium were cultured for 2 weeks at room temperature, at which time fungal colonies were examined morphologically and counted. The number of F. circinatum colonies growing on each petri dish (surface area, 56.34 cm2) was transformed to the number per square meter.
Method 2 (growth on filter paper).
Twenty percent of the surface of the filter paper was cut into pieces of ca. 5 mm2 each and laid over FS medium. After 2 weeks of incubation at room temperature, the number of F. circinatum colonies was transformed to the number per square meter (area of Whatman no. 1 filter paper, 0.0095 m2; 20%, i.e., 0.0019 m2, was used for culture growth). To test the cospecificity of the cultures with F. circinatum, two representative cultures per plate (methods 1 and 2) were analyzed by PCR with the primer pair CIRC1A-CIRC4A.
Method 3 (real-time PCR).
Spores from the remaining filter paper (80% of the surface) were washed with 20 ml of hot (65°C) 4× TE buffer and resuspended by vortexing (maximum speed) for 5 min. The suspensions were centrifuged at 1,000 × g for 90 min to concentrate the spores. After the supernatant was removed, DNA was extracted from the pellet (100 μl). To prove the efficacy of the spore harvest, 0.3 ml of the supernatant and the filter paper were plated on FS medium, incubated at room temperature for 2 weeks, and inspected for fungal growth. DNA extracts of the spores were amplified by real-time PCR. The resulting Ct values were used for quantification of the starting copy number of F. circinatum DNA in each reaction mixture by interpolation of the standard curve of known DNA and spore concentrations. The value obtained was multiplied by 131.6 to calculate the concentration of F. circinatum per square meter (the surface size of the filter was 0.0076 m2).
Statistical analyses.
Analysis of variance was performed with JMP 5.0.1 software (SAS Institute Inc., Cary, N.C.). Multiple comparisons were performed with the Tukey-Kramer honestly significant difference test. Pairwise comparisons were done with Student's t test.
Nucleotide sequence accession numbers.
The nucleotide sequences determined in this study have been deposited in the GenBank database under accession numbers AY249379 to AY249409.
RESULTS
DNA sequencing.
The nuclear ribosomal IGS region from 30 Fusarium strains in the G. fujikuroi species complex and the outgroup F. oxysporum were sequenced (the accession numbers are in Table 1). The lengths of the fragments ranged from ca. 2,600 to 2,650 bp. Maximum-parsimony analysis of the aligned IGS sequences yielded a single most-parsimonious tree with a length of 1,695 steps (consistency index = 0.65, retention index = 0.73) in which three clades of the G. fujikuroi complex were strongly supported by bootstrapping (Fig. 1).
F. circinatum was deeply nested within clade 2. Two strongly supported subclades were resolved among the exclusive group of F. circinatum strains (100% bootstrap): (i) NRRL 25331 ex P. radiata (Monterey pine) from Georgia and NRRL 25621 ex P. patula (Mexican yellow pine) from South Africa (98% bootstrap) and (ii) NRRL 26431 and NRRL 26432 ex P. luchuensis from Kyushu and Okinawa, Japan, NRRL 25332 ex P. taeda (Loblolly pine) from Georgia, NRRL 25707 ex P. caribaea (Caribbean pine) from North Carolina, and NRRL 25333 ex P. patula from South Africa (87% bootstrap).
Primer design.
A primer pair specific for F. circinatum (CIRC1A-CIRC4A) was designed with the primer3 software on the basis of an alignment of the IGS regions of the 31 Fusarium strains (Table 2). The 360-bp amplicon predicted from the sequence data size was confirmed by gel electrophoreses (Fig. 2).
FIG. 2.
Specificity of the primer pair CIRC1A-CIRC4A for the amplification of a 360-bp fragment in the nuclear ribosomal IGS region of F. circinatum. An ethidium bromide-stained agarose gel of PCR products amplified from stains belonging to the American clade of the G. fujikuroi complex and two environmental samples from spore traps is shown. Lanes: 1, 100-bp DNA ladder (Promega); 2, water negative control; 3, F. circinatum NRRL 25331; 4, F. circinatum NRRL 25332; 5, F. circinatum NRRL 25333; 6, F. circinatum NRRL 25621; 7, F. circinatum NRRL 26431; 8, F. circinatum NRRL 26432; 9, F. circinatum NRRL 25707; 10, environmental sample from San Francisco, Calif. (site I); 11, environmental sample from San Bruno, Calif. (site II); 12, F. subglutinans NRRL 22016; 13, Fusarium sp. strain NRRL 25204; 14, Fusarium sp. strain NRRL 25622; 15, Fusarium sp. strain NRRL 26756; 16, Fusarium sp. strain NRRL 26757; 17, Fusarium sp. strain NRRL 25346; 18, Fusarium sp. strain NRRL 25623; 19, Fusarium sp. strain NRRL 25195; 20, Fusarium sp. strain NRRL 25807; 21, Fusarium sp. strain NRRL 29123; 22, F. bulbicola NRRL 13618; 23, F. bactridioides NRRL 20476; 24, F. succisae NRRL 13613; 25, F. anthophilum NRRL 13602; 26, F. begoniae NRRL 25300; 27, water negative control; 28, DNA ladder.
Specificity of the primers.
The specificity of the primers was shown by PCR analyses of F. circinatum strains belonging to both subclades and other fusaria belonging to clade 2 of the G. fujikuroi complex (Fig. 2). With an annealing temperature of 66°C or higher, only DNA from F. circinatum (both subclades) was amplified.
Determination of linear range of quantification for real-time PCR.
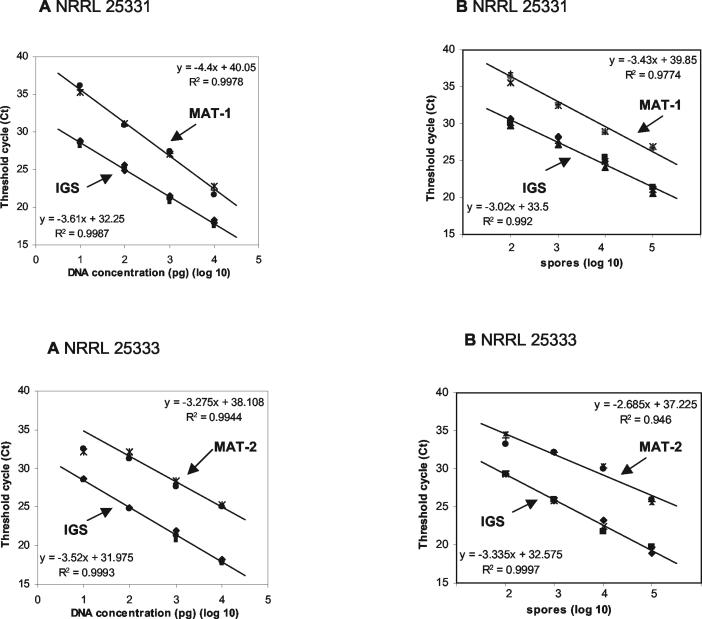
Standard curves were calculated for both the multicopy IGS and the single-copy mating type loci MAT-1 and MAT-2 (Fig. 3). Curves were determined independently in triplicate for DNA extracted from mycelium and from spores of isolates bearing different mating types. In order to obtain comparable data sets for quantification of DNA extracted from spores, an initial spore suspension of 100 μl was used for calibration and for environmental samples. The correlation between Ct and known DNA content in a 10-fold dilution series was high for both mycelium (strain NRRL 25331 R2 = 0.9987, strain NRRL 25333 R2 = 0.9993) and spores (strain NRRL 25331 R2 = 0.9920, strain NRRL 25333 R2 = 0.9997) with amplification of the multicopy locus IGS. With this primer set, the correlation between Ct and DNA concentration was linear from 101 to 104 pg and from 102 to 105 spores/100 μl, and the lower reliable detection threshold was 10 pg or 102 spores/100 μl. With the single-copy mating type genes, 10 pg of DNA was still detectable on an agarose gel (data not shown), but detection by real-time PCR was unreliable because of low melting peaks that were hardly distinguishable from the background. With this method, detection became reliable at a DNA concentration of 102 copies per reaction. Furthermore, the Ct values of the mating type genes at the lowest DNA concentration (101 pg) did not always fit in a linear inverse regression. Ct values of the single-copy genes were higher than those obtained for corresponding concentrations with the multicopy set. The Ct difference (ΔCt) between the two sets averaged 6.1 ± 1.2 (standard deviations [SD]) in the range linear for both methods (102 to 104 pg and 103 to 105 spores/100 μl).
FIG. 3.
Standard curves quantifying DNAs of F. circinatum strains NRRL 25331 (MAT-1) and NRRL 25333 (MAT-2) obtained by real-time PCR with SYBR-Green as the fluorescent dye. Ct values, corresponding to the increase in template DNA above the background level, were plotted against the log of genomic DNA standards of known concentrations (n = 3). DNA amplification was done with the primer combination CIRC1A-CIRC4A for the multicopy ribosomal IGS gene and the primer pairs for the single-copy mating type genes MAT-1 and MAT-2, respectively. (A) Standard curves obtained with DNA extracted from mycelium in the range of 101 to 104 pg per reaction. (B) Standard curves obtained with DNA extracted from a spore suspension in the range of 102 to 105 spores/100 μl.
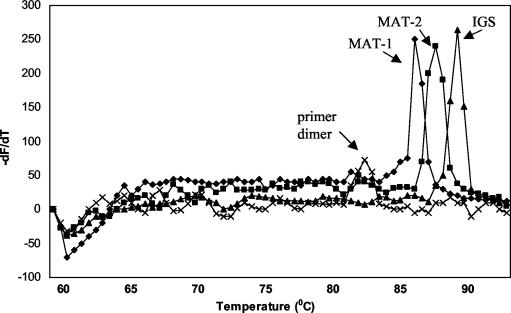
Melting curve analyses.
Specificity of PCR amplicons was measured by calculating the melting curves of double-stranded DNA (Fig. 4). The negative first derivative of the fluorescence-versus-temperature (−dF/dT) plot over temperature displays a melting peak, which is a function of fragment length and base composition, i.e., the relative concentration of pyrimidines. The IGS fragment amplified with the primer pair CIRC1A-CIRC4A gives a melting peak at ca. 88.0 ± 0.5°C. The fragments of the mating type genes MAT-1 and MAT-2 have melting peaks at 85.5 ± 0.5 and 87.0 ± 0.5°C, respectively. Primer dimers (if present) show a melting peak at ca. 82.0 ± 2.0°C.
FIG. 4.
Melting curves of F. circinatum DNA amplicons as measured by real-time PCR. Displayed is the negative first derivative of the fluorescence-versus-temperature plot over temperature (−dF/dT versus temperature). Fragments of the IGS region and the two mating type genes (MAT-1 and MAT-2) can be distinguished by their respective melting temperatures, which are a function of percent G+C content and fragment length. Primer dimers (if present) show a melting peak at ca. 82°C.
Environmental spore sampling.
F. circinatum spores from both sites were detected and quantified by three different methods. For a summary of the numbers of spores from three independent experiments per site and method, see Fig. 6.
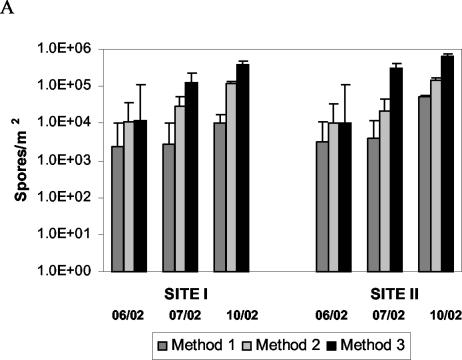
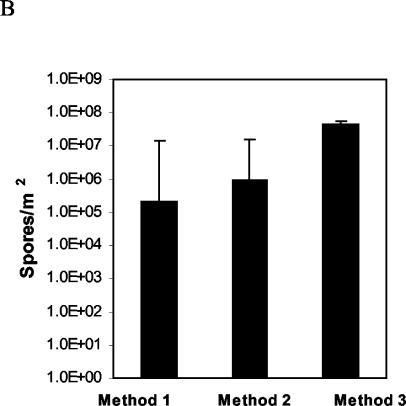
FIG. 6.
Quantification of F. circinatum from spore traps collected after 2 weeks in the field at San Francisco (site I) and San Bruno (site II), Calif. Three spore collection methods were used: 1, trapping with FS medium; 2, trapping with filter paper and subsequent transfer of the paper to FS medium; 3, trapping on filter paper and subsequent real-time PCR with the primer pair CIRC1A-CIRC4A. (A) Numbers of spores were calculated individually for each site, sampling point, and sampling method. (B) Cumulative numbers of spores for each sampling method combining the numbers from all sites and sampling points (vertical bars represent standard errors). Spore counts obtained by method 3 are significantly different from spore counts obtained by methods 1 and 2 (multiple-comparison analysis of variance with the Tukey-Kramer honestly significant difference test, P = 0.05).
Method 1: FS medium.
FS medium remained stable at both field sites for only 1 to 3 days. When plates were collected after 1 or 2 weeks, the medium had dried out and fungal colonies were barely visible. The average numbers of F. circinatum spores from three spore traps collected in June 2002 after 2 weeks in the field were (2.40 ± 0.4) × 103/m2 at site I and (3.20 ± 0.6) × 103 at site II. The corresponding values for spore traps collected in July 2002 were (2.70 ± 0.5) × 103/m2 at site I and (4.00 ± 0.4) × 103/m2 at site II. In October 2002, slightly more were counted, i.e., (0.99 ± 0.07) × 104/m2 at site I and (5.03 ± 0.4) × 104/m2 at site II. To ensure that the counted white colonies really were F. circinatum, DNA was extracted from two representative samples per plate by the hyphal tipping method and the IGS region was amplified with the selective primers CIRC1A and CIRC4A.
Method 2: traps using filter paper.
Filter paper (Whatman no. 1) was stable for 2 weeks in the field without showing any physical degradation. No visible fungal growth was observed on the paper when collected from the field, but after plating of small paper particles (20% of the paper surface) on FS medium, abundant fungal growth was observed after incubation at room temperature for 1 to 2 weeks. Primers CIRC1A and CIRC4A were used as described above to ensue that growing colonies were F. circinatum. The average numbers of F. circinatum spores collected with three spore traps in June 2002 were (11.45 ± 0.2) × 103/m2 at site I and (10.42 ± 1.1) × 103/m2 at site II. The corresponding numbers for July 2002 were (29.00 ± 2.4) × 103/m2 at site I and (21.90 ± 5.4) × 103/m2 at site II. The numbers of spores counted in October 2002 were (1.15 ± 0.03) × 105/m2 at site I and (1.43 ± 0.20) × 105/m2 at site II.
Method 3: real-time PCR quantification of spores on filter paper.
The remaining 80% of the filter paper surface was used for molecular quantification. F. circinatum DNA was extracted from filter paper collected at both sites I (Presidio) and II (San Bruno) and used as the template for real-time PCR amplification (Fig. 5).
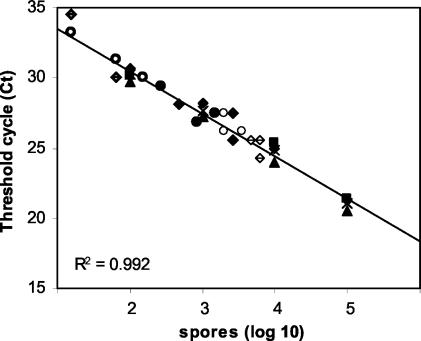
FIG. 5.
Ct values of DNA extracted from airborne conidia of F. circinatum were estimated by real-time PCR of a fragment of the IGS region. Environmental samples were collected from spore traps in San Francisco (site I; June 2002, boldface open circle; July 2002, filled circle; October 2002, lightface open circle) and San Bruno (site II; June 2002, dotted diamond; July 2002, filled diamond; October 2002, bisected diamond), Calif. Standardization was done with DNA extracted from spores of F. circinatum NRRL 25331.
For samples collected in June 2002, the average Ct value from three spore traps was 31.4 ± 2.5 at site I. At site II, one spore sample failed to give an amplification product; the average Ct of the two remaining spore samples was 32.5 ± 2.5. The corresponding numbers of spores, calculated by plotting against the calibration curve of spores (Fig. 3B), were 12.0 × 103/m2 for site I and 9.9 × 103/m2 for site II. For samples collected in July 2002, the average Ct values from three spore traps were 27.7 ± 0.9 at site I and 26.9 ± 1.3 at site II; the corresponding numbers of spores were 1.25 × 105/m2 and 3.00 × 105/m2, respectively. For samples collected in October 2002, the average Ct values from three spore traps were 26.6 ± 0.1 at site I and 25.8 ± 0.2 at site II; the corresponding numbers of spores were 3.95 × 105/m2 and 6.38 × 105/m2, respectively. The spore counts obtained by the three methods are summarized in Fig. 6B. Cumulatively for all sampling times and sites combined, method 1 yielded 2.18 × 105 spores/m2 (n = 18, SD = 0.18 × 105), method 2 yielded 9.92 × 105 spores/m2 (n = 18, SD = 0.55 × 105), and method 3 yielded 44.30 × 105 spores/m2 (n = 18, SD = 2.31 × 105). No significant difference was detected between methods 1 and 2, but spore counts obtained by sampling method 3 were significantly higher (multiple-comparison analysis of variance with the Tukey-Kramer honestly significant difference test, P = 10−4).
No significant difference (Student's t test, P = 0.1173) was observed when comparing the numbers of spores obtained at sites I and II by method 3. Site I had a cumulative number of 15.96 × 105 spores/m2 for all three sampling times (n = 9, SD = 1.70 × 105), and site II had a cumulative number of 28.33 × 105 spores/m2 (n = 9, SD = 2.64 × 105). The lack of difference in spore counts between the two sites was also observed when treating each sampling time independently.
To study the effect of seasonality, the cumulative numbers of spores for both sites obtained by method 3 were compared. In June 2002, 0.56 × 105 spores/m2 (n = 6, SD = 0.01 × 105) were counted, in July, 12.75 × 105 spores/m2 (n = 6, SD = 0.96 × 105) were counted, and in October, 30.99 × 105 spores/m2 (n = 6, SD = 1.33 × 105) were counted. Differences among the three sampling times were significant (Tukey-Kramer honestly significant difference test, P = 10−4).
Mating type analyses.
The PCR-based mating type assay for F. circinatum developed by Wallace and Covert (23) was used to amplify the idiomorphs of the mating type genes in a multiplex PCR with four primers. Amplicons were distinguished by their sizes: approximately 380 bp for MAT-1 versus approximately 190 bp for MAT-2. Four F. circinatum strains from different geographic origins were used as a positive control. F. circinatum strains NRRL 25331 (Georgia) and NRRL 26431 (Japan) gave a positive signal for MAT-1, whereas F. circinatum strains NRRL 25332 (Georgia) and NRRL 25333 (South Africa) possess the MAT-2 idiomorph. We tested the mating type of the environmental F. circinatum samples with pooled DNA from the filter paper traps, which had been identified with the primer pair CIRC1A-CIRC4A. DNA from representative colonies growing on FS medium (at least two per plate) was extracted by the hyphal tipping method and tested for mating type. All environmental samples from our two study sites analyzed so far belong to the MAT-1 mating type (data not shown).
Multiplex PCR with DNA mixtures of both mating types resulted in two distinct fragments on the agarose gel even when the initial relative concentrations differed in a range of 2 orders of magnitude (data not shown). DNA from F. circinatum strain 25331 (MAT-1) was mixed with DNA from strain 25333 (MAT-2) at relative concentrations of 1:1, 1:100, and 1:1,000. The PCR resulted in the amplification of two fragments of the expected size for the mating type genes (MAT-1, 380 bp; MAT-2, 190 bp) with initial relative template concentrations of 1:1, 1:10, and 1:100. When one of the two mating type genes was present at a relative concentration of 0.1% (1:1,000), no fragment of the corresponding size was amplified.
DISCUSSION
In this paper we describe a new method for the detection and quantification of F. circinatum, the causal agent of pine pitch canker. The test is based on the use of the species-specific primer pair CIRC1A-CIRC4A, which amplifies a 360-bp fragment of the nuclear ribosomal IGS region. F. circinatum clusters within the so-called “American clade” sensu O'Donnell (15) of the G. fujikuroi complex (called clade 2 in our analyses) and is the only known pathogen of conifers in this complex. The primer pair CIRC1A-CIRC4A appears to be specific for F. circinatum (both subclades) since it does not amplify the IGS from closely related species in clade 2. New species belonging to the G. fujikuroi complex were discovered recently (2, 27), and cryptic speciation within the complex might occur (18), making it very probable that the full diversity of the complex has not yet been detected. With the exception of new species bearing an IGS sequence identical to that of F. circinatum, sequencing of the CIRC1A-CIRC4A amplicon can be used to confirm the identity of the tested samples. In the course of this study, amplicons were routinely sequenced to ensure that only the target species was being amplified.
One of the main objectives of this study was to establish a method for quantifying F. circinatum spores from air samples. Calibration curves based on DNA extracted from known amounts of spores were the most appropriate standard. A 10-fold dilution series ranging from 102 to 105 spores/100 μl was in linear inverse regression when plotted against Ct values of both the multicopy and single-copy primer sets. A comparison of multicopy versus single-copy regressions indicated that (i) quantification was more reliable at lower DNA concentrations for primers based on the multicopy IGS locus (102 spores/100 μl) than for primers based on the single-copy mating alleles (103 spores/100 μl) and (ii) at any spore concentration, the Ct value obtained with the multicopy primer set will be lower than that obtained with the single-copy set (average ΔCt = 6.1 ± 1.2) (Fig. 3). The exact copy number of the rRNA gene region of F. circinatum or any other Fusarium species has not been measured yet to our knowledge. Our analysis of the comparative sensitivities of the two assays is in agreement with a ca. 100-fold higher copy number of the ribosomal operon compared to the mating type genes.
The assay has been successfully used to detect the presence of the pathogen directly from infected plant tissue, sometimes also from asymptomatic plants (data not shown). Several species within the G. fujikuroi complex are known to be endophytic in their respective hosts. It has been hypothesized that F. circinatum may also remain dormant within the host without causing symptoms of disease for an extended period of time (19). Furthermore, the pathogen was also isolated from pine cones of symptomless trees in Mexico (3).
Measurements of airborne inoculum over an extended period of time are essential for epidemiological studies. We compared airborne inocula at three different times at two sites in California with differential incidences of pine pitch canker. In three parallel experiments, we compared the number of spores obtained from a traditional spore trap with FS medium (method 1) with the number of spores trapped on filter paper subsequently plated on FS medium (method 2) and with the number of spores obtained with filter paper and subsequent DNA extraction and real-time PCR (method 3). The field results indicate comparable concentrations of airborne F. circinatum spores in both P. radiata stands. In all experiments, the numbers of spores obtained with filter paper (methods 2 and 3) were higher than the number of spores obtained with FS medium (method 1), although the difference was significant only for method 3 (P = 10−4). This fact can be explained by the rapid degradation and drying of the medium after 2 to 3 days in the field. Method 1 is therefore not suited for long-term spore sampling. On the other hand, filter paper showed no sign of physical degradation after 2 weeks in the field. The higher counts obtained by real-time PCR (method 3) compared to culturing (method 2) can be explained in part by the slow and retarded growth of some fungal colonies on filter paper because faster-growing colonies had occupied most of the available resources. In addition, several non-Fusarium fungi were able to colonize the medium, competing with F. circinatum colonies for resources. In this study, we could not determine the spore type of the trapped F. circinatum spores. Therefore, the relative frequency of multicelled macroconidia in relation to one-celled microconidia or the number of spores germinating on the filter paper was not quantified and might also be responsible for some of the observed differences.
A clear effect of season on spore production was observed in this study. Samples obtained in June 2002 at both sites reached a total of 0.56 × 105 spores/m2 (n = 6, SD = 0.01 × 105). One out of three samples from site II did not contain an F. circinatum-specific DNA fragment. The numbers of spores from samples collected in July and October of 2002 reached total levels of 12.75 × 105/m2 (n = 6, SD = 0.96 × 105) and 30.99 × 105/m2 (n = 6, SD = 1.33 × 105), respectively. Airborne inoculum of F. circinatum at various levels throughout the year at an infested site near Santa Cruz, Calif., was also observed by Correll et al. (5). Temperature, moisture, and availability of nutrients most probably play a crucial role in spore production.
The relatively high number of spores from site I, which shows a low level of disease, might be explained by the fact that the traps at site I were relatively protected by the shade of neighboring trees. Traps at site II were much more exposed to the sun and UV radiation. Consequently, plates from site II were always drier than those from site I. In addition, site I was subjected to more fog on a regular basis than site II. It is also probable that the dead trees at site II, although providing a striking visual diagnostic trait of an infested site, may not contribute to inoculum production. F. circinatum isolation success is in fact higher when slightly symptomatic living plant tissue is used rather than dead plant material.
F. circinatum has been shown to be heterothallic on the basis of vegetative compatibility tests and mating type segregation (4). In order to assess the mating type of the field isolates of F. circinatum, a multiplex PCR was performed with primer sets previously developed (7, 23). The primer sets are not specific for F. circinatum but amplify the mating type loci of a number of closely related Fusarium species. When both mating types are detected at a site, it is critical to ensure that the results are not due to cross-reactivity with other Fusarium species. In this study, we have determined ΔCt values with primers for the multicopy IGS region and the single-copy mating type genes. This difference remains constant for DNA concentrations ranging from 102 to 104 pg and from 102 to 105 spores/100 μl. At DNA concentrations lower than 102 pg and 103 spores/100 μl, the signal from the single-copy primers becomes unreliable and the linearity of the corresponding standard curve is lost. A ΔCt value smaller than that reported in this study indicates that the mating type primers are also amplifying a number of non-circinatum spores, and the results should be verified by an alternative method if both mating types are detected.
Multiplex PCRs with both primer sets on mixtures of DNAs from isolates with different mating types indicated that it is possible to detect both types even at very uneven ratios (1:100). In our study, only MAT-1 was amplified at both sites; thus, no further validation of the data was necessary. The prevalence of MAT-1 is in agreement with results from previous studies and is consistent with the hypothesis that F. circinatum arrived relatively recently in California (10, 24).
The results presented here demonstrate that an improved trapping method for aerial spores of F. circinatum with filter paper combined with species-specific real-time PCR amplification can improve spore detection significantly compared to more traditional approaches. The real-time PCR method offers the following advantages: (i) it is a diagnostic technique with increased sensitivity and higher selectivity that is independent of the presence of symptoms, (ii) it has the ability to quantify airborne conidia continuously at low cost for long periods of time, and (iii) DNA extracts used for identification and quantification can also be used for mating type or other genetic analyses of bulk collections of spores, thus enhancing our ability to detect the arrival of new genotypes, strains, or mating types.
Acknowledgments
We thank the Presidio Trust and the State of California for the use of the field sites. For technical assistance we thank D. Schmidt and M. A. Smith.
This research was supported by the State of California Department of Forestry and Fire Protection (grant 8CA99067).
REFERENCES
- 1.Anonymous. 2001. The iCycler iQ real-time detection system resource guide, p. 118. Bio-Rad, Hercules, Calif.
- 2.Aoki, T., K. O'Donnell, and K. Ichikawa. 2001. Fusarium fractiflexum sp. nov. and two other species within the Gibberella fujikuroi species complex recently discovered in Japan that form aerial conidia in false heads. Mycoscience 42:461-478. [Google Scholar]
- 3.Britz, H., T. A. Coutinho, T. R. Gordon, and M. J. Wingfield. 2001. Characterisation of the pitch canker fungus, Fusarium circinatum, from Mexico. S. Afr. J. Bot. 67:609-614. [Google Scholar]
- 4.Britz, H., T. A. Coutinho, M. J. Wingfield, and W. F. O. Marasas. 2002. Validation of the description of Gibberella circinata and morphological differentiation of the anamorph Fusarium circinatum. Sydowia 54:9-22. [Google Scholar]
- 5.Correll, J. C., T. R. Gordon, A. H. McCain, J. W. Fox, C. S. Koehler, D. L. Wood, and M. E. Schultz. 1991. Pitch canker disease in California: pathogenicity, distribution and canker development on Monterey pine (Pinus radiata). Plant Dis. 75:676-682. [Google Scholar]
- 6.Correll, J. C., T. R. Gordon, A. H. McCain, and D. J. Jacobsen. 1992. Examination of genetic diversity in populations of the pitch canker pathogen Fusarium subglutinans. Phytopathology 82:415-420. [Google Scholar]
- 7.Covert, S. F., A. Briley, M. M. Wallace, and V. T. McKinney. 1999. Partial MAT-2 gene structure and the influence of temperature on mating success in Gibberella circinata. Fungal Genet. Biol. 28:43-45. [DOI] [PubMed] [Google Scholar]
- 8.Dwinell, L. D., J. B. Barrows-Broaddus, and E. G. Kuhlman. 1985. Pitch canker: a disease complex of southern pines. Plant Dis. 69:270-276. [Google Scholar]
- 9.Fox, J. W., D. L. Wood, C. S. Koehler, and S. T. O'Keefe. 1991. Engraver beetles (Scolytidae: Ips species) as vectors of the pitch canker fungus, Fusarium subglutinans. Can. Entomol. 123:1355-1367. [Google Scholar]
- 10.Gordon, T. R., A. J. Storer, and D. Okamoto. 1996. Population structure of the pitch canker pathogen Fusarium subglutinans f. sp. pini in California. Mycol. Res. 100:850-854. [Google Scholar]
- 11.Gordon, T. R., A. J. Storer, and D. L. Wood. 2001. The pitch canker epidemic in California. Plant Dis. 85:1128-1139. [DOI] [PubMed] [Google Scholar]
- 12.Guerra-Santos, J. 1998. Pitch canker on Monterey pine in Mexico, p. 58-61. In M. Devey, C. Matheson, and T. R. Gordon (ed.), Current and potential impacts of pitch canker in radiata pine. Proceedings of the IMPACT Monterey Workshop, Monterey, Calif., 30 Nov. to 3 Dec. 1998. CSIRO, Collingwood, Victoria, Australia.
- 13.Hepting, G. H., and E. R. Roth. 1946. Pitch canker, a new disease of some southern pines. J. For. 44:742-744. [Google Scholar]
- 14.Hoover, K., D. L. Wood, J. W. Fox, and W. E. Bros. 1995. Quantitative and seasonal association of the pitch canker fungus, Fusarium subglutinans f. sp. pini, with Conophthorus radiatae (Coleoptera:Scolytidae) and Ernobius punctulatus (Coleoptera:Anobiidae) which infest Pinus radiata. Can. Entomol. 127:79-91. [Google Scholar]
- 15.O'Donnell, K., E. Cigelnik, and H. I. Nirenberg. 1998. Molecular systematics and phylogeography of the Gibberella fujikuroi species complex. Mycologia 90:465-493. [Google Scholar]
- 16.Steenkamp, E. T., B. D. Wingfield, T. A. Coutinho, M. J. Wingfield, and W. F. O. Marasas. 1999. Differentiation of Fusarium subglutinans f. sp. pini by histone gene sequence data. Appl. Environ. Microbiol. 65:3401-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steenkamp, E. T., B. D. Wingfield, T. A. Coutinho, K. A. Zeller, M. J. Wingfield, W. F. O. Marasas, and J. F. Leslie. 2000. PCR-based identification of MAT-1 and MAT-2 in the Gibberella fujikuroi species complex. Appl. Environ. Microbiol. 66:4378-4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steenkamp, E. T., B. D. Wingfield, A. E. Desjardins, W. F. O. Marasas, and M. J. Wingfield. 2002. Cryptic speciation in Fusarium subglutinans. Mycologia 94:1032-1043. [PubMed] [Google Scholar]
- 19.Storer, A. J., T. R. Gordon, and S. L. Clark. 1998. Association of the pitch canker fungus, Fusarium subglutinans f. sp. pini, with Monterey pine seeds and seedlings in California. Plant Pathol. 47:649-656. [Google Scholar]
- 20.Storer, A. J., D. L. Wood, K. R. Wikler, and T. R. Gordon. 1998. Association between a native spittlebug (Homoptera:Cercopidae) on Monterey pine and an introduced tree pathogen which causes pitch canker disease. Can. Entomol. 130:783-792. [Google Scholar]
- 21.Sutton, W. R. J. 1997. Radiata pine and the global opportunity for plantation forestry. International forestry reports. Evergreen Forests Limited, Auckland, New Zealand.
- 22.Swofford, D. L. 1998. PAUP phylogenetic analysis using parsimony version 4.0b1. Sinauer Associates, Sunderland, Mass.
- 23.Wallace, M. M., and S. F. Covert. 2000. Molecular mating type assay for Fusarium circinatum. Appl. Environ. Microbiol. 66:5506-5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wikler, K., T. R. Gordon, S. L. Clark, M. J. Wingfield, and H. Britz. 2000. Potential for outcrossing in an apparently asexual population of Fusarium circinatum, the causal agent of pitch canker disease. Mycologia 92:1085-1090. [Google Scholar]
- 25.Wingfield, M. J., B. D. Wingfield, T. A. Coutinho, A. Viljoen, H. Britz, and E. Steenkamp. 1998. Pitch canker: a South African perspective, p. 62-69. In M. E. Devey, A. C. Matheson, and T. R. Gordon, (ed.), Current and potential impacts of pitch canker in radiata pine. Proceedings of the IMPACT Monterey Workshop, Monterey, Calif., 30 Nov. to 3 Dec. 1998. CSIRO, Collingwood, Victoria, Australia.
- 26.Wingfield, M. J., A. Jacobs, T. A. Coutinho, R. Ahumada, and B. D. Wingfield. 2002. First report of the pitch canker fungus, Fusarium circinatum, on pines in Chile. New Dis. Rep., vol. 4, August 2001-January 2002. [Online.] http://www.bspp.org.uk/ndr/jan2002/2001-53.htm.
- 27.Zeller, K. A., B. A. Summerell, S. Bullock, and J. F. Leslie. 2003. Gibberella konza (Fusarium konzum) sp. nov. from prairie grasses, a new species in the Gibberella fujikuroi species complex. Mycologia 95:943-954. [PubMed] [Google Scholar]