Abstract
Simkania negevensis is a recently discovered chlamydia-like intracellular microorganism which has been associated with bronchiolitis in infants and with community-acquired pneumonia in adults; a high seroprevalence of antibodies to the microorganism has been found in various population groups. S. negevensis can be grown in various cell lines as well as in free-living amoebae such as Acanthamoeba polyphaga. In this study, evidence for the existence of Simkania or Simkania-like microorganisms in drinking water and in reclaimed wastewater is presented for the first time. Detection of the microorganism was made possible by the development of a specific and sensitive filter membrane immunoassay and was confirmed by PCR detection of microbial DNA in the water samples. The common presence of S. negevensis in water sources together with the high seroprevalence of antibodies to it and early age of acquisition of infection may implicate water as a source of infection. The possible significance of this finding for public health and for municipal water testing and treatment needs to be further examined.
Simkania negevensis is an intracellular bacterium which has been shown to be similar to chlamydiae in some of its characteristics (13, 14, 16). Taxonomic studies have placed S. negevensis in a new family, Simkaniaceae, in the order Chlamydiales (2). This bacterium has been associated with several types of respiratory tract disease including bronchiolitis in infants (by PCR, culture, and serology) (15) and community-acquired pneumonia and acute exacerbation of chronic obstructive pulmonary disease in adults (by serology) (20, 21, 23). Natural infection with S. negevensis seems to be highly prevalent. In the Negev region of Israel, for example, between 55 and 80% of several groups of healthy adults had serum antibodies against the organism (6). However, infection with S. negevensis is not limited to southern Israel, and it now appears that infection with this organism (or very closely related ones) is rather common in many parts of the world (5).
The seroprevalence of S. negevensis has been studied in groups of people of different ages, such as the entire population of a Negev kibbutz (n = 290) and healthy Bedouins of various ages (n = 198). The distribution of seropositivity by age indicated an early exposure to the infectious agent (5) and raised the question of the source of infection and mode of transmission. In the past, early age of acquisition of infection with Helicobacter pylori was associated with its presence in drinking water (18).
Since some newly described members of the Chlamydiales were in fact discovered as endosymbionts (endocytobionts) of free-living amoebae, the ability of S. negevensis to replicate in amoebae in the laboratory was also examined. It was found that not only were the organisms able to replicate successfully in Acanthamoeba polyphaga but they were also able to survive over long periods of time in amoebic cysts (12). In addition, other chlamydia-like microorganisms, such as members of the family Parachlamydiaceae, were shown to be able to grow successfully as endocytobionts of free-living amoebae (9, 11); some other Chlamydiales members, such as Chlamydophila (Chlamydia) pneumoniae, may infect these organisms but grow only slowly (1). Since free-living amoebae such as acanthamoebae may be found in many water sources, as well as in desert sand (22), it may be that the natural source of transmission of S. negevensis to humans is via such amoebae. Similar transmission of legionellae has been described previously (4), and indeed, amoebae are thought to play an important role in the natural history of infections with legionellae and possibly other intracellular microorganisms (10).
In this study, we examined the question of whether S. negevensis can be found in drinking water or in wastewater in the Negev. The development of a simple assay system for detection of the organism in such water samples is described, and evidence for the presence of S. negevensis in various water sources is presented.
MATERIALS AND METHODS
Water sources.
Wastewater in the Negev (southern Israel) is treated intensively (activated sludge) or extensively (oxidation ponds) and is routinely evaluated for quality by the regional public health laboratory. The parameters measured include the biological oxygen demand, the chemical oxygen demand, and total suspended solids. Forty samples of reclaimed wastewater were obtained from the regional public health laboratory after their routine testing was completed but without our knowledge of the results of this testing until our experiments were complete. These were samples of water that had not received tertiary treatment (which would have included sand filtration and chlorine treatment, followed by testing for coliforms).
Samples of tap water were taken from several neighborhoods in Beer Sheva, which obtains its water supply from both the national water carrier and from deep wells, that is, underground water reserves (aquifers) located in the area. The supply alternates between these two sources but not in a set pattern. Drinking water samples were also obtained from three different satellite communities near Beer Sheva, which receive their water from local deep wells. Drinking water brought to the area by the National Water Carrier normally has a turbidity measurement of <1 while the turbidity measurement of deep well water is usually <0.3. Drinking water is chlorinated to 0.1 to 0.5 ppm (or milligrams/liter) of free chlorine. Drinking water in the city is tested according to a municipal plan. In 2003, 684 tests were carried out. In one, 5 coliforms were detected, but upon retesting the source, none were found. All other samples were negative for coliforms.
Chlorination of water and determination of the survival of S. negevensis in chlorinated water.
Chlorine levels in water were determined by a pocket calorimeter (HACH Europe, S. A./N.V., Floriffoux, Belgium) used for routine field testing of chlorine levels. Sterile deionized water (containing 0.07 mg of free chlorine/liter) was exposed for 20 min to chlorination (0.5 mg of free chlorine/liter), generated with free chlorine reagent powder (cat. no. 21055-69; HACH) which was later neutralized with sodium thiosulfate (1 M). S. negevensis particles or S. negevensis in persistently infected amoebae precipitated in a microcentrifuge were suspended in chlorinated water immediately after its chlorination treatment and incubated at room temperature (RT). Control tubes received the same water without chlorine treatment. At the beginning of the incubation, the chlorine levels ranged from 0.48 to 0.52 mg of free chlorine/liter in the different experiments, and at the end of the 20-min incubation, they were 0.22 mg/liter. After neutralization of the chlorine, the contents of the experimental and control tubes were diluted in infection medium for titration on Vero cells, which was carried out by the plate immunoperoxidase assay as previously described (15). In parallel, samples were tested by membrane enzyme immunoassay (MEIA) (see below) to determine the effect of chlorination on antigen detection.
MEIA.
Cellulose nitrate filter membranes (0.45-μm pore size [Schleicher & Schuell] or 0.8-μm pore size [Pall Supor 800]) were used for filtration of control or tested water samples. Samples were mixed, and any coarse particles, if present, were removed by gravity drainage through several layers of viscose polypropylene material (this is a light gauze-like [but nonwoven] fabric composed of viscose and polypropylene fibers and used in clinics as disposable bed covers). Then a sample volume of 100 to 500 μl of reclaimed wastewater or 500 ml of drinking water was filtered (using a 96-place manifold system [S&S] with the 0.45-μm-pore-size membrane sheet or the 47-mm-diameter Pall Supor filter, respectively).
For each test, S. negevensis-infected Vero cell lysates or lysates of A. polyphaga persistently infected with S. negevensis (prepared as described below) served as controls. Twofold serial dilutions of these controls in distilled water (200-μl volumes) were filtered in duplicate through the manifold as calibration standards (Fig. 1B). Filters were processed as follows. They were frozen for 10 min at −70°C and incubated for 30 min at RT in 0.1% saponin in distilled water with vigorous shaking. The solution was removed, and the filter was frozen again at −70°C. The filter was then fixed for 15 min in a solution consisting of 30% methanol and 5% hydrogen peroxide, then fixed for 5 min in 70% ethanol, and fixed for 5 min in 96% ethanol. When the filter was almost dry, it was incubated for 45 min in a blocking solution containing 0.3% polyvinylpyrrolidone dissolved in TBS-Tween buffer, containing 10 mM Tris (pH 7.6), 150 mM NaCl, and 0.05% Tween-20, to which normal swine immunoglobulins (X-0906; DAKO), diluted 1:5,000, were added. The addition of swine immunoglobulins to the blocking buffer reduced the background due to nonspecific binding of rabbit or mouse immunoglobulins or of labeled secondary antibodies to biologic material on the filters.
FIG. 1.
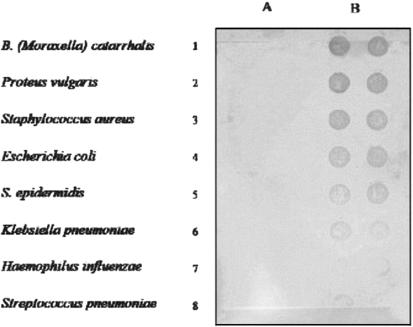
Specificity of MEIA with respect to other bacteria commonly found in the respiratory tract. The MEIA was performed as described in Materials and Methods. Column A, 100 μl of various bacteria filtered in duplicate; column B, 100 μl of S. negevensis suspension in twofold dilutions.
For detection of the organisms trapped on the filters in the MEIA, the filters prepared as described above were incubated for 45 min at RT in rabbit anti-S. negevensis polyclonal hyperimmune serum diluted 1:20,000 in blocking solution. After washing three times for 5 min with TBS-Tween, the filters were incubated with affinity-purified horseradish peroxidase (HRP)-conjugated goat anti-rabbit antibodies (111-035-003; Jackson ImmunoResearch Laboratories, Inc.) diluted at 1:20,000 in blocking solution. After further washing, they were incubated in diaminobenzidine substrate solution. In each assay a duplicate control filter was prepared in parallel and processed as described above but omitting the serum incubation step. Only signals stronger than those obtained on the control filter were considered positive.
The same procedure was employed for detection of amoebic antigens with mouse polyclonal anti-A. polyphaga (at 1:5,000) and HRP-conjugated, affinity-purified anti-mouse antibodies (115-035-003; Jackson), diluted 1:2,000.
Preparation of purified S. negevensis, Chlamydia trachomatis, and C. pneumoniae.
S. negevensis, C. trachomatis (L2/434/Bu), and C. pneumoniae (TW-183) were grown in Vero cells and purified by sucrose density gradient centrifugation as described previously (13, 17), except that the latter two organisms were grown in the absence of penicillin.
Preparation of positive controls for water studies.
Vero cells infected with S. negevensis were scraped off the growth surface with glass beads at 3 to 5 days postinfection; aliquots were frozen at −80°C in the presence of 50% fetal calf serum. Infectivity titers were determined by titration on Vero cells in the plate immunoperoxidase assay as previously described (15). A. polyphaga organisms persistently infected with S. negevensis were grown in PYG medium containing antibiotics as detailed previously (12). A. polyphaga organisms were counted, and their S. negevensis infectivity was determined by titration on Vero cells, as described previously (12). The infected amoebae served as positive controls in MEIA in those assays in which the presence of both S. negevensis and amoebic antigens were determined.
Hyperimmune sera used in assays.
Hyperimmune rabbit sera were prepared against S. negevensis, C. trachomatis, and C. pneumoniae as described previously for C. trachomatis and C. pneumoniae (7). Hyperimmune murine sera against A. polyphaga were produced by immunizing BALB/c mice with lysates of A . polyphaga grown in PYG.
List of bacterial strains and their American Type Culture Collection numbers.
A panel of eight standard bacterial strains obtained from the American Type Culture Collection were used in developing the present protocol of the MEIA. They were as follows: Branhamella (Moraxella) catarrhalis, ATCC 25238; Proteus vulgaris, ATCC 33420; Staphylococcus aureus, ATCC 25923; Escherichia coli, ATCC 25922; Staphylococcus epidermidis, ATCC 12228; Klebsiella pneumoniae, ATCC 13883; Haemophilus influenzae, ATCC 19418; Streptococcus pneumoniae, ATCC 49619. A loop of each of the bacteria in the panel was suspended in water (concentration, about 107/ml) and stored frozen at −70°C.
PCR assays.
DNA from drinking water samples was prepared as follows. A 500-ml volume of water sample was drained (by gravity) through a cellulose acetate filter (5-μm pore size, catalog no. 12342-47-N; Sartorius). The filter was incubated with Page's saline buffer (24) for 1 h. The solution containing the biological material was centrifuged for 20 min at 800 × g to sediment amoebae, and the precipitate was used to prepare DNA with the QIAamp kit (Qiagen). Assays for the detection of the S. negevensis genome by PCR were performed by using two sets of nested primers.
The first set consisted of primers ccF (CTT CGG GTT GTA AAG CAC TTT CGC) and ccR (CCC CGT CAA TTC TTT TGA GTT T), recognizing conserved chlamydial 16S ribosomal DNA (rDNA) sequences (433 to 457 and 933 to 945, respectively) and amplifying a 512-bp fragment, followed by nested ZpF and ZpR primers specific for S. negevensis Z (as previously described) (15), amplifying a 405-bp fragment.
A second set of nested primers consisted of AF and BR, amplifying a 1,099-bp fragment within the 23S rDNA of S. negevensis containing the large subunit intron (3), and IntD and IntR nested primers, amplifying a 338-bp fragment within this intron. If the DNA tested does not contain the intron, a 441-bp fragment is obtained with the AF and BR primers and no sequence is amplified by the IntF-IntR primer pair.
The use of the nested primers allowed increased sensitivity of the PCR assay when it was necessary; however, when DNA was abundant, primer set AF-BR, IntF-IntR, or ZpF-ZpR could also be used alone. PCR assays for the presence of amoebic sequences were carried out with the primers AmP1 and Amp2 described by Lai et al. (19) by using the cycling program described below for S. negevensis.
PCR amplifications were performed with Taq DNA polymerase (Takara Shuzo Co., Ltd., Kyoto, Japan). Amplification conditions were 1 cycle for 6 min at 94°C; 30 cycles of 60 s at 94°C, 60 s at 53°C, and 60 s at 73°C; and then 1 cycle of 10 min at 73°C (3).
When amplification was with the ccF-ccR primers, the same amplification program was used, except that annealing was performed at 58°C instead of 53°C. When nested primers were used, the material amplified with the first pair of primers was diluted 1:100, of which 1 μl was used in the second reaction mixture. The high dilution was needed to ensure the specificity of the assay.
RESULTS
MEIA sensitivity and specificity.
The sensitivity of the MEIA protocol was evaluated by determination of the number of S. negevensis inclusion-forming units (IFU) or number of amoebae present in the highest dilution of control material giving a positive signal. Using anti-S. negevensis serum diluted 1:20,000, 121 IFU of S. negevensis could be detected in the lysate and 468 infected amoebae could be detected. Uninfected A. polyphaga amoebae served as a negative control in the latter case. No signal was detected in the absence of anti-S. negevensis serum in the assay. When the same sample volumes were used for detection of amoebic antigen, the MEIA was able to detect 1,000 amoebae. S. negevensis-containing cell culture lysates and a filter used in the assay without the first antibody served as negative controls for the amoebic assay.
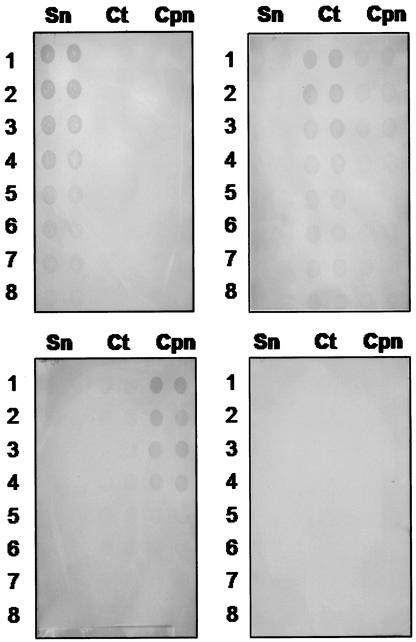
Figures 1 and 2 demonstrate the specificity of the MEIA for Simkania with respect to a number of common bacterial respiratory pathogens (Fig. 1) and members of the Chlamydiaceae (Fig. 2). A volume of 100 μl was used for the assay. None of the panel of standard bacteria gave a positive signal with antibodies to S. negevensis and affinity-purified goat anti-rabbit HRP conjugates (Fig. 1), nor did C. trachomatis or C. pneumoniae antigens under the same conditions (Fig. 2).
FIG. 2.
Specificity of MEIA with respect to members of the Chlamydiaceae. Duplicate samples of 100-μl volumes of twofold dilutions of bacterial suspensions were applied to each of the membranes, as indicated. Sn, S. negevensis; Ct, C. trachomatis L2; Cpn, C. pneumoniae. Starting concentrations were as follows: S. negevensis, 4 × 104 organisms per dot; C. trachomatis L2, 2.5 × 104 organisms per dot; C. pneumoniae, 2.5 × 104 organisms per dot. Upper left panel, rabbit anti-S. negevensis serum used at 1:20,000; upper right panel, rabbit anti-C. trachomatis serum used at 1:20,000; lower left panel, rabbit anti-C. pneumoniae serum used at 1:20,000; lower right panel, serum diluent with secondary antibody only.
The possibility of demonstrating the presence of S. negevensis in water sources depends on the sensitivity and specificity of the detection assay; however, the significance of such detection depends on the continuous presence of the bacteria in the water source, which may be affected by various additional factors. Therefore, a number of experiments were performed to evaluate S. negevensis survival in the laboratory under simulated natural conditions.
Effect of chlorination on S. negevensis particles and on S. negevensis in persistently infected amoebae.
In three separate experiments, the chlorinated suspension of Simkania particles had the same amount of infectivity as the control suspension that did not undergo chlorination. Control infectivity levels for the three experiments ranged from 1.2 × 104 to 2.8 × 105 IFU/ml. Survival values for S. negevensis particles ranged from 90 to 104% of the control untreated samples, with a mean of 96.8%. The variation obtained was within the experimental variation of titration. Similarly, S. negevensis in trophozoites of A. polyphaga were not sensitive to the treatment. In addition, chlorination treatment had no effect on antigen detection by MEIA (data not shown).
Survival in water of S. negevensis compared with C. trachomatis.
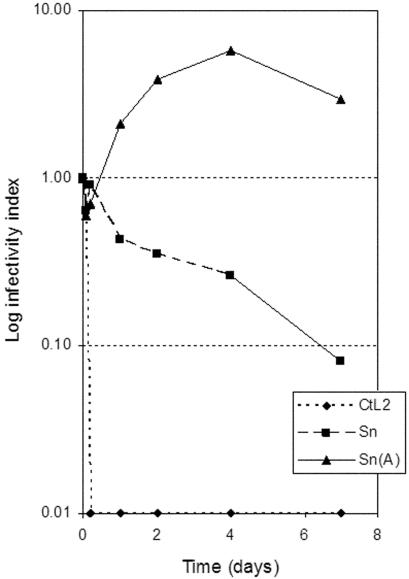
S. negevensis particles, S. negevensis-infected amoebae, and C. trachomatis L2 particles were exposed for various periods of time to sterile distilled water at RT, and both infectivity and antigen detection were monitored (Fig. 3). S. negevensis particle infectivity was remarkably preserved even after 7 days of incubation (ranging from 8 to 15% in various experiments). S. negevensis titers in infected amoebae even increased significantly under the same conditions. The infectivity of C. trachomatis L2 particles was reduced to 65% within 1 h and was completely destroyed by 5 h of exposure to distilled water. However, antigens were detected by MEIA in all samples tested at all time points. Even when infectivity was no longer present, no significant change in the level of antigen detection was observed (data not shown).
FIG. 3.
Residual infectivity of S. negevensis and C. trachomatis after incubation for various times in sterile distilled water at RT. Sn, S. negevensis; CtL2, C. trachomatis L2; Sn(A), S. negevensis in persistently infected A. castellani. The infectivity index is the ratio of infectivity at a given time point to the infectivity at time zero.
Detection of S. negevensis in water samples by MEIA.
The MEIA was used to detect the possible existence of S. negevensis in drinking water or in reclaimed wastewater. On three different occasions, at intervals of 1 month and then 2 months, drinking water samples were collected from houses located in various parts of Beer Sheva, with each location being visited on three different occasions. The results are shown in Table 1 (one sample was disqualified for inclusion in the table). As shown in Table 1, both S. negevensis and amoebic antigens were detected in most of the locations. In addition, 9 samples of drinking water from 3 satellite communities of Beer Sheva that obtain their water from aquifers were also examined on two different occasions, 10 days apart. In the majority of samples (7 of 9) both S. negevensis and amoebic antigens were detected by MEIA. In contrast, drinking water from Eilat, which is obtained by desalination (by reverse osmosis) had no traces of either S. negevensis or amoebic antigens (data not shown).
TABLE 1.
Detection of S. negevensis and Acanthamoeba antigens in samples of drinking watera
| Neighborhood no. | No. of positive samples/no. of samples tested for:
|
|
|---|---|---|
| S. negevensis antigen | Acanthamoeba antigen | |
| 1 | 12/12 | 12/12 |
| 2 | 3/3 | 3/3 |
| 3 | 5/5 | 5/5 |
| 4 | 1/3 | 3/3 |
| 5 | 0/3 | 1/3 |
Samples (500 ml) were collected from a number of houses located in Beer Sheva on three different occasions: 30 January 2003, 8 April 2003, and 6 May 2003) (days 1, 67, and 95, respectively).
Reclaimed wastewater is routinely examined by Israeli Ministry of Health laboratories before being approved for use in agriculture. Forty samples of such water obtained during a 1.5-month interval from various locations in the Negev were tested for the presence of S. negevensis and amoebic antigens by MEIA (Table 2). The majority of samples (39 of 40, or 97.5%) were shown to contain S. negevensis antigens. No samples were found to contain antigens of S. negevensis in the absence of amoebic antigens.
TABLE 2.
Presence of Simkania and amoebic antigens (as detected by MEIA) in samples of reclaimed wastewater and quality characteristics of the watera
| Treatment type/water qualityb | No. of positive samples/no. of samples tested for:
|
BOD range | COD range | (T)SS range | |
|---|---|---|---|---|---|
| S. Simkania antigen | Amoebic antigen | ||||
| Intensive/good | 7/7 | 6/6 | 9.5-26.1 | 28-62 | 5.5-26 |
| Intensive/poor | 2/2 | 2/2 | 104-107 | 170-181 | 33-144 |
| Extensive/good | 25/25 | 17/17 | 12.2-77.4 | 12.8-250 | 30-190 |
| Extensive/poor | 5/6 | 4/4 | 86.5-208.1 | 220-610 | 63-297 |
BOD, biological oxygen demand; COD, chemical oxygen demand; (T)SS, total suspended solids.
Intensive, activated sludge; extensive, oxidation ponds.
Detection of S. negevensis and Acanthamoeba DNA sequences in water samples by PCR.
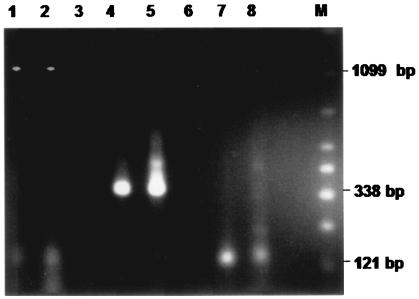
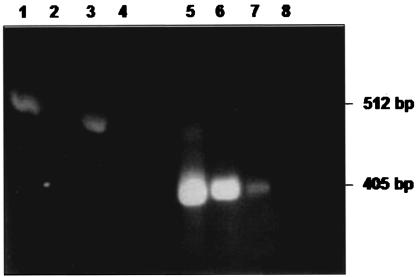
To confirm the finding of the S. negevensis antigen in drinking water, DNA was prepared from natural water samples by using the same volume of water as for the MEIA. The procedure for DNA preparation from water samples was optimized by using A. polyphaga persistently infected with S. negevensis and mixed with sterile water. All samples containing Simkania or Simkania-like antigens detected by the MEIA that were tested for the presence of S. negevensis DNA contained Simkania DNA as detected by PCR. The results from a representative experiment are shown in Fig. 4. Figure 4 also demonstrates the presence of amoebic DNA in both control and test samples (121-bp amplicon). Figure 5 shows the amplification of S. negevensis rDNA sequences with the ccF-ccR and ZpF-ZpR nested primers for a number of DNA samples obtained from water. It should be noted that the nested PCR proved to be more sensitive for the detection of S. negevensis DNA in water samples than the standard PCR previously described (2).
FIG. 4.
Intron and amoebic amplicons detected by PCR assay of DNA prepared from water samples (lanes 2, 5, and 8) compared with DNA from purified cell culture-grown S. negevensis (lanes 1, 4, and 7), with the following primer pairs: AF-BR, lanes 1 and 2; intF-intR, lanes 4 and 5; amp1-amp2, lanes 7 and 8. M, marker (50- to 2,000-bp ladder; Bio-Rad).
FIG. 5.
Nested PCR for detection of 16S rDNA S. negevensis sequences in DNA prepared from water samples. The primers used were as follows: lanes 1 to 4, ccF-ccR; lanes 5 to 8, ZpF-ZpR (nested primers). Lanes 1 and 5: positive control; lanes 2 to 3 and 6 to 7: DNA prepared from two water samples; lanes 4 and 8, negative controls (no DNA).
DISCUSSION
S. negevensis was initially isolated as a contaminant of cell cultures, and its natural host or hosts were unknown. In this study, evidence for the presence of S. negevensis or Simkania-like bacteria in drinking water or in reclaimed wastewater is presented for the first time. A MEIA was developed, and it was shown that this MEIA could be used for detection of Simkania antigens. Under the conditions used, there was no cross-reaction with members of the Chlamydiaceae. The method is simple and does not require sophisticated equipment or manipulations. PCR assays that require only one set of specific primers may not be sufficient to detect various unknown strains of the microorganism, more than one set may be needed, and in general, PCR assays are more expensive than the MEIA. Since the MEIA employs polyclonal antisera recognizing a wide range of Simkania antigens, it may be a more appropriate method for detection of various Simkania strains, the pathogenicity of which needs to be further investigated.
Chlamydia-like organisms other than Simkania, members of the family Parachlamydiaceae, have been shown to be present in the environment and to be able to grow and survive in protozoa, such as Acanthamoeba castellani and Hartmanella (9, 11), and lately, to replicate in human monocyte-derived macrophages (8).
Because we are able to grow S. negevensis in human and simian cells lines, we are able to isolate it from human clinical samples. We have also succeeded in isolating it from natural water samples (unpublished data). However, since the methods described in this study detect bacterial antigens or genomes only and are not capable of determining whether the organisms are viable, further studies are needed to understand the significance of the detection of S. negevensis or S. negevensis-like microorganisms in water. Aspects of such studies could include determination of the possible correlation between a high seroprevalence of antibodies to S. negevensis in healthy population samples and the presence of the organisms in the drinking water or agricultural irrigation water used in the same area. Sera obtained from a number of groups of healthy pregnant women (n = 192) living in the same areas from which water samples were obtained for this study showed an average prevalence of antibodies to S. negevensis of 75% (range, 50 to 92% for the different groups) (unpublished data), as assayed by enzyme-linked immunosorbent assay (6). The serum samples were from Jewish and Bedouin women living in rural and urban locations. The seropositivity ranges are consistent with published data (5, 6).
If the organisms found in drinking water are indeed viable, it is possible that their detection is of relevance to infection of the respiratory tract, in that drinking water is also generally used for oral hygiene, bathing, and showering, which can result in the formation of inhalable aerosols. Also, organisms found in drinking water may infect the oropharyngeal epithelium, thus gaining access to the respiratory and gastrointestinal tracts. In an ongoing study in our laboratory, S. negevensis strains isolated from children with respiratory disease are being compared to strains that may be isolated from their households.
Although laboratory experiments indicate that S. negevensis is more resilient than other similar bacteria, such as Chlamydia, we are well aware that under environmental conditions, bacteria and their antigens may have a shorter survival time due to the presence of microorganisms and factors such as various chemicals in the water. Clearly, it would be advantageous to have simple surrogate markers for viability of environmental chlamydia-like bacteria, but these are not presently available. The possibility of transmission of potential pathogens in drinking water is clear, but reclaimed wastewater may be used for irrigation of crops and its pathogenic potential is also of concern. The significance of the ability of S. negevensis or S. negevensis-like microorganisms to survive in various water sources needs to be further studied to establish public health guidelines taking this into account. Future research will enable conclusions about the environmental, epidemiological, and health care implications of the very common existence of simkaniae in various water sources.
Acknowledgments
We thank I. Belmaker for interest and encouragement and M. Grabarnik for assistance in processing samples for testing.
This study was supported by grant no. TA-Mou-99-C19-033 funded by the United States-Israel Cooperative Development Research Program, Bureau for Economic Growth, Agriculture, and Trade, U.S. Agency for International Development; by grant no. 4672/0 from the Office of the Chief Scientist of the Israel Ministry of Health, via the Keren Kayemet LeIsrael; and by a grant from the “Pinchas Sapir” fund of Mifal Hapayis.
REFERENCES
- 1.Essig, A., M. Heinemann, U. Simnacher, and R. Marre. 1997. Infection of Acanthamoeba castellani by Chlamydia pneumoniae. Appl. Environ. Microbiol. 63:1396-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Everett, K. D. E., R. M. Bush, and A. A. Andersen. 1999. Emended description of the order Chlamydiales, proposal of Parachlamydiaceae fam. nov. and Simkaniaceae fam. nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiaceae including a new genus and five new species, and standards for the identification of organisms. Int. J. Syst. Bacteriol. 49:415-440. [DOI] [PubMed] [Google Scholar]
- 3.Everett, K. D. E., S. Kahane, R. Bush, and M. G. Friedman. 1999. An unspliced group I intron in 23S rRNA links Chlamydiales, chloroplasts, and mitochondria. J. Bacteriol. 181:4734-4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fallon, R. J., and T. J. Rowbotham. 1990. Microbiological investigations into an outbreak of Pontiac fever due to Legionella micdadei associated with use of a whirlpool. J. Clin. Pathol. 43:479-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Friedman, M. G., B. Dvoskin, and S. Kahane. 2003. Infections with the chlamydia-like microorganism Simkania negevensis, a possible emerging pathogen. Microbes Infect. 5:1013-1021. [DOI] [PubMed] [Google Scholar]
- 6.Friedman, M. G., A. Galil, S. Greenberg, and S. Kahane. 1999. Seroprevalence of IgG antibodies to the Chlamydia-like microorganism “Simkania Z” by ELISA. Epidemiol. Infect. 122:117-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gonen, R., Y. Shemer-Avni, P. A. Csango, B. Sarov, and M. G. Friedman. 1993. Serum reactivity to Chlamydia trachomatis and C. pneumoniae antigens in patients with documented infection and in healthy children by microimmunofluorescence and immunoblotting techniques. APMIS 101:719-726. [PubMed] [Google Scholar]
- 8.Greub, G., J.-L. Mege, and D. Raoult. 2003. Parachlamydia acanthamoeba enters and multiplies within human macrophages and induces their apoptosis. Infect. Immun. 71:5979-5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greub, G., and D. Raoult. 2002. Parachlamydiaceae, potential emerging pathogens. Emerg. Infect. Dis. 8:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harb, O. S., L. Y. Gao, and Y. Abu Kwaik. 2000. From protozoa to mammalian cells: a new paradigm in the life cycle of intracellular bacterial pathogens. Environ. Microbiol. 2:251-265. [DOI] [PubMed] [Google Scholar]
- 11.Horn, M., M. Wagner, K.-D. Müller, E. N. Schmid, T. R. Fritsche, K.-H. Schliefer, and R. Michel. 2000. Neochlamydia hartmannellae gen. nov., sp. nov. (Parachlamydiaceae), an endoparasite of the amoeba Hartmannella vermiformis. Microbiology 146:1231-1239. [DOI] [PubMed] [Google Scholar]
- 12.Kahane, S., B. Dvoskin, M. Mathias, and M. G. Friedman. 2001. Infection of Acanthamoeba polyphaga with Simkania negevensis and S. negevensis survival within amoebal cysts. Appl. Environ. Microbiol. 67:4789-4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kahane, S., K. D. E. Everett, N. Kimmel, and M. G. Friedman. 1999. Simkania negevensis strain ZT: growth, antigenic and genome characteristics. Int. J. Syst. Bacteriol. 49:815-820. [DOI] [PubMed] [Google Scholar]
- 14.Kahane, S., R. Gonen, C. Sayada, J. Elion, and M. G. Friedman. 1993. Description and partial characterization of a new chlamydia-like microorganism. FEMS Microbiol. Lett. 109:329-334. [DOI] [PubMed] [Google Scholar]
- 15.Kahane, S., D. Greenberg, M. G. Friedman, H. Haikin, and R. Dagan. 1998. High prevalence of “Simkania Z,” a novel Chlamydia-like bacterium, in infants with acute bronchiolitis. J. Infect. Dis. 177:1425-1429. (Erratum, 178:1553.) [DOI] [PubMed] [Google Scholar]
- 16.Kahane, S., E. Metzer, and M. G. Friedman. 1995. Evidence that the novel microorganism “Z” may belong to a new genus in the family Chlamydiaceae. FEMS Microbiol. Lett. 126:203-208. [DOI] [PubMed] [Google Scholar]
- 17.Kahane, S., N. Kimmel, and M. G. Friedman. 2002. The growth cycle of Simkania negevensis. Microbiology 148:735-742. [DOI] [PubMed] [Google Scholar]
- 18.Klein, P. D., D. Y. Graham, A. Gaillou, A. R. Opekun, and E. O. Smith. 1991. Water source as risk factor for Helicobacter pylori infection in Peruvian children. Gastrointestinal Physiology Working Group. Lancet 337:1503-1506. [DOI] [PubMed] [Google Scholar]
- 19.Lai, S., M. Asgari, and H. R. Henney, Jr. 1994. Non-radioactive DNA probe and polymerase chain reaction procedures for the specific detection of Acanthamoeba. Mol. Cell. Probes 8:81-89. [DOI] [PubMed] [Google Scholar]
- 20.Lieberman, D., S. Kahane, D. Lieberman, and M. G. Friedman. 1997. Pneumonia with serological evidence of acute infection with the chlamydia-like microorganism “Z.” Am. J. Respir. Crit. Care Med. 156:578-582. [DOI] [PubMed] [Google Scholar]
- 21.Lieberman, D., B. Dvoskin, D. V. Lieberman, S. Kahane, and M. G. Friedman. 2002. Serological evidence of acute infection with the Chlamydia-like microorganism Simkania negevensis (Z) in acute exacerbation of chronic obstructive pulmonary disease. Eur. J. Clin. Microbiol. Infect. Dis. 21:307-309. [DOI] [PubMed] [Google Scholar]
- 22.Marshall, M. M., D. Naumovitz, Y. Ortega, and C. R. Sterling. 1997. Waterborne protozoan pathogens. Clin. Microbiol. Rev. 10:67-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petrich, A., M. Smieja, K. Luinstra, T. Sinha, S. Chong, R. Leigh, M. Loeb, and J. Mahony. 2002. Development of a PCR assay to determine prevalence of Simkania negevensis DNA in specimens from patients with respiratory disease, p. 471-474. In J. Schachter et al. (ed.), Chlamydial infections: proceedings of the Tenth International Symposium on Human Chlamydial Infections. International Chlamydia Symposium, San Francisco, Calif.
- 24.Visvesvara, G. S. 1992. Parasite culture: Acanthamoeba and Naeglaria spp., p. 7.9.2.1-7.9.2.8. In H. D. Isenberg (ed.), Clinical microbiology procedures handbook. American Society for Microbiology, Washington, D.C.