Abstract
Rhizobium etli, as well as some other rhizobia, presents nitrogenase reductase (nifH) gene reiterations. Several R. etli strains studied in this laboratory showed a unique organization and contained two complete nifHDK operons (copies a and b) and a truncated nifHD operon (copy c). Expression analysis of lacZ fusion demonstrated that copies a and b in strain CFN42 are transcribed at lower levels than copy c, although this copy has no discernible role during nitrogen fixation. To increase nitrogenase production, we constructed a chimeric nifHDK operon regulated by the strong nifHc promoter sequence and expressed it in symbiosis with the common bean plant (Phaseolus vulgaris), either cloned on a stably inherited plasmid or incorporated into the symbiotic plasmid (pSym). Compared with the wild-type strain, strains with the nitrogenase overexpression construction assayed in greenhouse experiments had, increased nitrogenase activity (58% on average), increased plant weight (32% on average), increased nitrogen content in plants (15% at 32 days postinoculation), and most importantly, higher seed yield (36% on average), higher nitrogen content (25%), and higher nitrogen yield (72% on average) in seeds. Additionally, expression of the chimeric nifHDK operon in a poly-β-hydroxybutyrate-negative R. etli strain produced an additive effect in enhancing symbiosis. To our knowledge, this is the first report of increased seed yield and nutritional content in the common bean obtained by using only the genetic material already present in Rhizobium.
The common bean (Phaseolus vulgaris L.) is the most important crop in Mexico after maize and represents the main protein source for large sectors of the population. Bean plants tolerate a wide range of environments and are cultivated from tropical to temperate regions covering up to 2-million hectares in Mexico and 22-million hectares in the rest of the world. Their seeds are consumed either fresh or dry (7). Most of the fields used for their cultivation are fertilized with agrochemicals.
Biological nitrogen fixation is an exclusively prokaryotic process in which atmospheric dinitrogen is converted in an easily assimilable metabolite, ammonia. Rhizobium bacteria, and related genera, induce nodules and fix nitrogen in the roots of legumes in a complex regulated process (12). Given the current world food demand, increasing biological nitrogen fixation offers economic, agricultural, and environmental benefits. Improvement of this process can be obtained by the use of genetically manipulated Rhizobium bacteria. Historically, researchers have had limited success in trying to improve the Rhizobium-legume relationship in agronomically important crops. Strategies used to enhance symbiotic nitrogen fixation include: (i) transgenic expression of hydrogenase uptake in Rhizobium strains (1), (ii) construction and expression of a hybrid nodulation regulatory nodD gene (31), (iii) increasing expression of NifA and C4-dicarboxylic acid transport genes (3), and (iv) obtention of an acid-tolerant R. leguminosarum biovar trifolii strain (9). None of these strategies improved nitrogen fixation ability, compared with inoculation with the wild type, more than 20% for any parameter measured.
The common bean is nodulated by different species of Rhizobium; the majority of strains isolated from bean nodules in Mexican agricultural fields belong to Rhizobium etli (29). The R. etli type strain is CFN42. This strain contains three copies of the nifH gene (named a, b, and c) which code for the nitrogenase reductase component, two of them (a and b) are linked to the nifDK genes which code for dinitrogenase (23, 26). Reiteration c is linked to a truncated nifD homolog (nifD*) gene (35). The three nifH copies are actively expressed during symbiosis although the nifHDK operons are expressed at lower levels than the third nifHc copy. The nitrogenase activity is encoded by only the two complete nifHDK operons in a gene dosage-dependent manner (27). All these genes are located on a 371-kb symbiotic plasmid (pSym) (14).
Both operons a and b are preceded by identical RpoN (σ54)-dependent promoters and canonical NifA (nitrogen fixation activator)-binding sites named upstream activator sequences located at 90 bp from the promoter (26). The third copy, nifHc, is preceded by an identical RpoN-dependent promoter and is activated by NifA bound to a nonconsensus-binding site 85 bp upstream (35). The asymetric arrangement of regulatory elements could contribute to the nifH differential expression observed during symbiosis (35).
Poly-β-hydroxybutyrate (PHB) is a poly-β-hydroxyalkanoate accumulated by a wide range of rhizobia as a carbon and reductive power storage polymer in free life (32, 34) and/or in symbiosis (18, 19, 38). R. etli produces PHB not only in free life but also during symbiosis (8, 10). Although the role of PHB in symbiosis is not well understood, mutation of the R. etli phaC gene, the product of which catalyzes the PHB polymerization step, produced a mutant with increased nitrogenase activity and a slight increase in bean seed yield compared to those of the wild-type strain CFN42 (8). Physiological characterization showed that the PHB− strain excreted a huge quantity of metabolites, mainly from the tricarboxylic acid (TCA) cycle as fumarate, malate, and 2-oxoglutarate, suggesting that the mutant is unable to oxidize the carbon source present in the growth medium. The PHB− strain showed a lower NAD+/NADH ratio. The abundance of reduced cofactors is apparently related to the absence of a reductive power sink (PHB) (8). Encarnación et al. (10) proposed that in R. etli, PHB serves as a reductive power sequester, so that the TCA cycle continues functioning under microaerobic conditions. The PHB− strain shows an increased ability to fix nitrogen (at late stages of symbiosis), in contrast to the notion that PHB could help to prolong or sustain symbiotic nitrogen fixation as proposed by Bergersen et al. (2). In the case of R. etli, apparently part of the excess reducing power present in the PHB− strain is channeled to nitrogen fixation.
The main purpose of our work was to significantly improve the symbiotic efficiency in the R. etli-P. vulgaris relationship by an in vitro manipulation approach of the bacterial genetic material, specifically that which encodes nitrogenase enzyme production. In view of the previously mentioned knowledge about nifH transcriptional activation, we intended to improve nitrogen fixation efficiency by modifying the nitrogenase genes transcription rate. To increase this rate and at the same time to conserve NifA-dependent regulation, we constructed a chimeric complete nitrogenase nifHDK operon coupled to the strong nifHc promoter region and expressed it either on a stably inherited plasmid or in the symbiotic plasmid itself. We assessed the effects of such constructions on symbiosis with common bean plants in greenhouse experiments and compared them to those of inoculation with the parent strain. Additionally, the chimeric nitrogenase operon was expressed in a PHB− background to determine if the carbon and reducing power not stored in the polymer could be derived to fuel nitrogen fixation.
The improved symbiotic relationship obtained in this way is the highest reported for R. etli to date and involves the use of only genetic elements already present in the bacterial genome. Greenhouse experiments with the modified strains support their potential application to obtain better crop yields and more nutritive bean seeds.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture media.
Plasmids and strains used in this work are listed in Table 1. Escherichia coli strains were grown at 37°C in Luria-Bertani complex medium (28). R. etli strains were grown, as described elsewhere, in peptone-yeast extract (PY) or minimal medium containing 1.2 mM K2HPO4, 0.8 mM MgSO4, 10 mM succinic acid, 10 mM NH4Cl, 1.5 mM CaCl2, and 0.0005% FeCl3, with the pH adjusted to 6.8 (5). The following antibiotics were added to the indicated final concentrations (in micrograms per milliliter): kanamycin, 30; nalidixic acid, 20; carbenicillin, 100; and tetracycline, 6 or 10. Plasmids were conjugated into either wild-type R. etli CFN42T (or streptomycin-resistant derived strain CE3) or strain SAM100 (phaC) by triparental mating with pRK2013 as a helper plasmid (11).
TABLE 1.
Bacterial strains and plasmids used in this work
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| R. etli | ||
| CFN42 | Wild-type strain, Smr Nalr | 5 |
| DEM153 | CFN42 with a nifHa-lacZ reporter fusion in pSym, Kmr | 35 |
| DEM233 | CFN42 with a nifHc-lacZ reporter fusion in pSym, Kmr | 35 |
| SAM100 | CFN42 derivative, phaC Kmr | 8 |
| HP55 | CFN42 containing pHP55 plasmid, Tcr | This work |
| HP310 | CFN42 derivative containing chimeric operon pr c nifHcDK in pSym | This work |
| HP210 | CFN42 derivative containing pHP210 plasmid (pTR101, nifHc with its own regulatory region), Tcr | This work |
| HP220 | CFN42 derivative containing pHP220 plasmid (pTR101, nifHDKb with its own regulatory region), Tcr | This work |
| HP310 lac | HP310 derivative containing a nifH-lacZ fusion in the chimeric operon pr c nifHcDK in pSym, Kmr | This work |
| E. coli HB101 | F−hsd S20-recA 13 | 4 |
| Plasmids | ||
| pAM341 | pTR101, with a fragment containing the nifHc promoter region cloned into the XbaI site, Tcr | 24 |
| pRK2013 | Helper plasmid, ColE1, mob+ Tra+ Kmr | 11 |
| pCQ12 | pBR328, with a 4.5-kb EcoRI fragment containing the R. etli nifHDKb operon, Tcr | 26 |
| pCQ23 | pBR328, with a 4.2-kb EcoRI fragment containing the R. etli nifHc gene, Tcr | 26 |
| pTR101 | pTR100 (mini-RK2), with a 0.8-kb stability locus, Tcr | 36 |
| pIC20H | Cloning vector, Ampr | 22 |
| pKOK6 | pSUP202, with a lacZ-kan cassette | 20 |
| pWS233 | Mobilizable replicon ColE1, Gmr TcrsacRB | 30 |
| pHP40 | pCQ12, pr c nifHcDK construction in a 4.5-kb EcoRI fragment | This work |
| pHP50 | pIC20H, with a 4.5-kb EcoRI fragment containing the pr c nifHcDK construct | This work |
| pHP55 | pTR101, containing the pr c nifHcDK construct in a 4.5-kb fragment cloned on an XbaI site | This work |
| pHP100 | pWS233, with a 4.5-kb EcoRI fragment containing the pr c nifHcDK construct cloned into the EcoRI site | This work |
| pHP789 | pHP100, with a 1.6-kb PstI-EcoRI fragment from pCQ23, cloned downstream of the pr c nifHcDK construct | This work |
| pHP210 | pTR101, with a 4.5-kb EcoRI fragment containing the nifHc gene with its own regulatory region | This work |
| pHP220 | pTR101, with a 5-kb EcoRI fragment containing nifHDKb operon with its own regulatory region | This work |
DNA manipulations.
DNA manipulations, such as isolation, transformation, restriction analysis, agarose gel electrophoresis, and hybridization, were performed by standard procedures (28). DNA fragments were purified from agarose gels with the use of the GeneClean kit (Bio101 Inc., Buena Vista, Calif.) or Wizard PCR Resin (Promega, Madison, Wis.). The Eckhardt method as modified by Hynes and McGregor (17) was used to determine plasmid profiles.
RNA isolation and dot blot hybridization.
RNA from 18 days postinoculation (dpi) nodules or free-living cells was isolated by phenol extraction (28) and purified with a MicrobExpress kit (Ambion, Austin, Tex.) according to the manufacturer's instructions. For dot blot hybridization, the membrane was loaded with samples and fixed with UV light with a StrataLinker 1800 apparatus (Stratagene, La Jolla, Calif.). The nifH probe was a 300-bp fragment obtained by PCR with nifH forward and nifH reverse oligonucleotides (described below). A 16S rRNA gene probe was obtained by PCR with universal oligonucleotides fd1 and rd1 (37). The labeled probes were prepared with 32P and a MegaPrime kit (Amersham, Little Chalfont, United Kingdom). Membranes were hybridized at high stringency at 65°C, washed three times with 0.1% sodium dodecyl sulfate in 0.1 × SSC (1 × SSC is 0.15 M NaCl plus 0.015 M sodium citrate) at 60°C, and then exposed to standard film (28) or on a Phosphor Imager screen for signal quantification in a Molecular Dynamics (Amersham, United Kingdom) scanner.
Construction of plasmid pHP55.
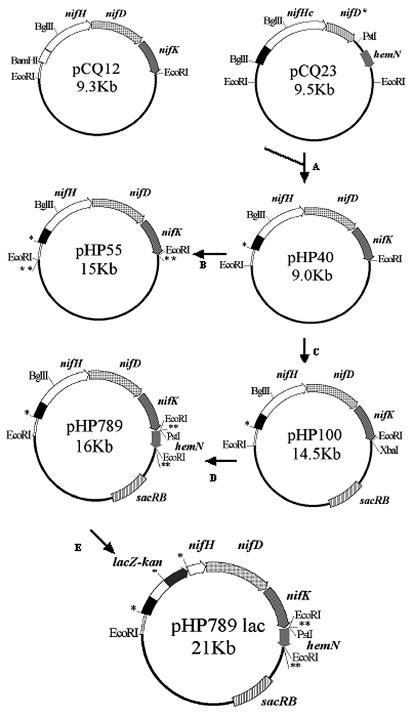
To produce a chimeric nifHDK operon controlled by the nifHc promoter region (hereafter referred as pr c nifHcDK), the promoter region of the nifHDKb operon contained in plasmid pCQ12 (26) was replaced by the nifHc promoter region as follows (see Fig. 1). A 1.5-kb fragment containing the nifHc promoter region and part of the nifHc gene was isolated by digesting pCQ23 (26) with BglII. pCQ12 was digested with BglII and BamHI to eliminate the nifHDKb promoter and part of the nifHb gene, resulting in loss of a 1.8-kb segment. The largest fragment of that digestion was ligated with the 1.5-kb BglII fragment and a plasmid with the correct orientation was chosen and named pHP40. Since the nucleotide sequence of both nifHb and nifHc genes are identical, the nifH gene formed by the joined fragments remains functional.
FIG. 1.
Scheme of plasmid construction. (A) pCQ12 was digested with BamHI and BglII; the largest fragment was ligated to a 1.8-kb BglII fragment from pCQ23. The promoter region of nifHDKb (pr b) is represented by an open box, and the promoter region of nifHc (pr c) is represented by a closed box. (B) pHP40 was digested with EcoRI, and the fragment containing pr c nifHcDK was cloned into the EcoRI site of pIC20H, which was then digested with SpeI, and the fragment of interest was cloned into the XbaI site of pTR101. (C) pHP40 was digested with EcoRI, and the 4.5-kb fragment containing pr c nifHcDK was cloned into the EcoRI site of pWS233, generating pHP100 plasmid. (D) pHP100 was digested with XbaI and ligated into a 1.6-kb SpeI fragment containing part of the hemN gene from pCQ23 digested with PstI and EcoRI, obtaining pHP789 plasmid. (E) pHP789 was digested with BglII and ligated to a 5-kb BamHI fragment containing lacZ-kan genes. *, site formed by BamHI-BglII joining; **, site formed by XbaI/SpeI joining; plasmids are not drawn to scale.
The 4.5-kb EcoRI fragment carrying the pr c nifHcDK construction from pHP40 was cloned into plasmid pIC20H (22) to produce pHP50. The 4.5-kb SpeI fragment containing pr c nifHcDK from plasmid pHP50 was cloned into the XbaI restriction site of the Rhizobium stably inherited vector, pTR101, (36) to produce pHP55.
Strain HP55 was obtained by triparental mating with E. coli HB101/pHP55 as donor, E. coli HB101/pRK2013 (11) as helper, and R. etli CFN42 as recipient. Selection was made on PY plates plus nalidixic acid and tetracycline (10 μg ml−1).
Construction of strain HP310 containing the pr c nifHcDK chimeric operon.
To obtain double-recombinant Rhizobium strains containing the pr c nifHcDK construction, we ligated a suicide vector, pWS233 (30), digested with EcoRI, to the 4.5-kb EcoRI fragment carrying pr c nifHcDK from pHP40. The plasmid obtained was named pHP100. In the vector XbaI site, we cloned a 1.6-kb PstI-EcoRI fragment bordered by SpeI sites and containing a fragment of the hemN gene located downstream of nifHcD* genes (35). The plasmid obtained, pHP789, was conjugated to R. etli CFN42 with pRK2013 as helper by selection on PY plates with nalidixic acid and tetracycline (6 μg ml−1). Selected colonies were cultured overnight in liquid PY and again grown overnight in liquid PY with 10% sucrose, a condition under which cells containing vector sequences were lysed. Surviving cells were plated onto PY plates with nalidixic acid, and a colony was chosen and named HP310.
A nifH-lacZ fusion was obtained by cloning the lacZ-kan cassette from pKOK6 (20), digested with BamHI, into the BglII site of pHP789. A plasmid with the correct orientation was chosen and named pHP789 lac. Incorporation into HP310 was done by triparental conjugation by selection with tetracycline (6 μg ml−1) and kanamycin (30 μg ml−1), and colonies selected by growth in liquid PY with 10% sucrose. Surviving cells were selected on PY plates with nalidixic acid and kanamycin (30 μg ml−1). A colony showing the incorporation of the cassette into the nifHb reiteration by a hybridization assay (data not shown) was chosen and named HP310 lac.
PCR assays and DNA sequencing.
PCR assays were performed with a GeneAmp PCR kit (PerkinElmer Applied Biosystems, Foster City, Calif.) following the manufacturer's instructions. Primers used were nifHc EcoRV forward (5′-GGC CGG ATA TCG CCT GAG A), nifHa forward (5′-CCG TCT GTC GGC TTT GTC TG), intra-nifH1 reverse (5′-GTA AAA TGC GAT TTG ACG C), intra-nifH forward (5′-GAG GAC GTG CTC AAG GCC GGC TAC), end-nifH reverse (5′-CAG CAC GCC GAG CTC AGG AAG ATG), nifD forward (5′-GGC GTG ATG ACG ATC CG), nifD reverse (5′-GCA TTC CGA CTG CAC GC), nifK forward (5′-CCA GGC TCT TCC CAT CG), nifK reverse (5′-GGC CGG GTT CAC GAC C), and 238 reverse (5′-CGT TCC TGG TTG ATA TCG AGC CAA GGT GTC) located downstream to nifK. DNA sequencing of the pr c nifHcDK construct in pSym from strain HP310 was done on a 5-kb product obtained with primers nifHc EcoRV forward and 238 reverse with HP310 total DNA as template and then with all of the mentioned oligonucleotides as primers to obtain the sequence of the product with a PerkinElmer DNA sequencer.
Nodulation test, nitrogenase activity, and nitrogen content determination in bean plants and seeds.
Seeds of P. vulgaris cv. Negro Jamapa were surface sterilized and germinated as previously reported (5). R. etli strains used for inoculation were grown overnight in PY complex medium, washed twice with a 0.85% NaCl solution, and diluted to an A540 of 0.05. Seedlings were planted in groups of five in autoclaved pots containing vermiculite as support material, and then each one was inoculated with 1 ml of bacterial suspension (approximately 106 cells per plant). As controls, experiments included noninoculated plants fertilized with 10 mM KNO3-2 mM NH4NO3 or without added nitrogen. Plant growth and watering were carried out under aseptic conditions in a greenhouse.
Greenhouse conditions included temperature of 22 to 28°C and relative humidity of 50 to 60%. Groups of 10 plants for each experimental condition were harvested at 18, 25, and 32 dpi, and the nodule dry weight, nitrogenase activity, total plant dry weight, and nitrogen content were determined for each plant including the noninoculated (control) plants. Bacteria were isolated from nodules, and their identities verified by their antibiotic resistance patterns. Nitrogenase specific activity (expressed as μmoles of ethylene h−1 g of nodule dry weight−1) was determined by incubating the detached root with 1/80 (vol/vol) acetylene. Ethylene production was estimated with a model 3300 gas chromatograph (Varian, Middelburg, The Netherlands). Plants or seeds were dried in an oven at 60°C for 3 days. Total nitrogen content of samples from dry plants or seeds was determined with a nitrogen analyzer (model ANTEK 9000; Antek Instruments, Inc., Houston, Tex.) and reported as milligrams of nitrogen per gram of dry plant or per gram of powdered seed. Nitrogen yield was calculated by multiplying the nitrogen content in seed times the yield and is expressed as milligrams of N in seed plant−1. Statistical analysis was performed according to the method of Steel and Torrie (33).
β-Galactosidase activity determination in R. etli cultures and plant nodules.
Cultures of R. etli strains were grown overnight in PY medium, collected, and washed with minimal medium as described above. Flasks containing minimal medium were inoculated at an initial A540 value of 0.05. Aliquots (20 ml) were injected into 150-ml bottles sealed with rubber stoppers, flushed with several volumes of 1% oxygen-99% argon mixture (analytical grade; Linde, Mexico City, Mexico), and incubated at 30°C with shaking at 200 rpm. Replicas of the cultures were simultaneously incubated in cotton-stoppered flasks to evaluate aerobic conditions. After 8 h, 1-ml samples were withdrawn, centrifuged at 10,000 × g at 4°C, and resuspended in 1 ml of cold Z buffer for β-galactosidase activity determination as described elsewhere (28). Replica 1-ml samples were pelleted and resuspended in 5% TCA, and their protein content was determined by the method of Lowry et al. (21). Specific activities were reported as nmoles of o-nitrophenol minute−1 milligram of protein−1.
Nodules from single plants were crushed in 1 ml of cold Z buffer (28) and centrifuged at 4°C for 5 min at 8,000 × g in a benchtop centrifuge, and a 0.05-ml aliquot of clear supernatant was transferred to a tube containing 0.95 ml of Z buffer and thoroughly mixed with 2 drops of chloroform. β-Galactosidase activity was measured in a Beckman DU7500 spectrophotometer at 420 nm as recommended by the manufacturer (28). Additional aliquots of the nodule extract (0.05 ml) were precipitated with 0.5 ml of 5% TCA, and the protein content was measured by the method of Lowry et al. (21). Specific activities were reported as nmoles of o-nitrophenol minute−1 milligram of protein−1.
Strain deposition.
The chimeric pr c nifHcDK construct, strains containing it, and other relevant sequences have been submitted for patents. Strain HP310 was deposited under accession no. NRRL B-30606 in the Culture Collection of the USDA Agricultural Research Service, Peoria, Ill.
RESULTS
Construction and transcriptional expression analysis of the chimeric pr c nifHcDK operon.
R. etli contains three copies of nifH that encode nitrogenase reductase (26, 27, 35). One of these, nifHc, is expressed at higher levels than the other two and is induced during nodule development in a NifA-dependent manner (35). Its regulatory region contains an unusual NifA-binding site upstream of the RpoN-dependent promoter, which differs from the canonical NifA-binding site located upstream from the nifHa and nifHb copies (35). To construct a chimeric pr c nifHcDK operon, the nifHDKb genes lacking their promoter were cloned downstream of the nifHc promoter and subcloned into the stable vector plasmid pTR101 or the suicide plasmid pWS233 for conjugation, as described in Materials and Methods (Fig. 1). The construct was cloned into plasmid pTR101, which is stably inherited in R. etli, and the plasmid obtained was named pHP55 and introduced to CFN42 by a triparental mating. To integrate the construct into the pSym of CFN42, a mating was made with E. coli cells containing suicide plasmid pHP789 with the construct. Recombinant cells were grown on PY plates with tetracycline and then grown in PY liquid medium plus sucrose for positive selection of double recombinants, presumably containing no vector sequences. A colony was chosen and named HP310.
To determine the expression of the chimeric pr c nifHcDK construct in R. etli CFN42, a nifH-lacZ fusion was created by inserting a lacZ-kan cassette into the BglII site of pHP789 and then introduced by triparental mating into strain HP310. β-Galactosidase activity of the strain containing this fusion, named HP310 lac, was determined in free-living cultures under a low-oxygen atmosphere (1% oxygen, 99% argon) and in symbiosis. This fusion presented a 4.4-fold induction respect to aerobic conditions. Low oxygen is a well-known physiological condition for the NifA-mediated induction of nifH (35). For comparison, strains DEM153 (nifHa-lacZ in pSym) and DEM233 (nifHc-lacZ in pSym) were used (35), and as described above, the nifHc-lacZ fusion was more highly expressed (21-fold induction) than the nifHa-lacZ fusion (6.5-fold induction) under microaerobic conditions relative to aerobic conditions (Table 2).
TABLE 2.
Transcriptional activity of nifH-lacZ fusions in free-living cultures and nodules
| Strain | Sp act of β-galactosidase (nmol of ONP min−1 mg of protein−1) ina: |
|||
|---|---|---|---|---|
| Free living cultures |
Nodules |
|||
| 1% O2 | 20% O2 | 11 dpi | 18 dpi | |
| CFN42 | 7 ± 2 | 9 ± 2 | NA | 65 ± 15 |
| HP310 lac (nifHc-lacZ) | 363 ± 75 | 82 ± 10 | 492 ± 137 | 929 ± 136 |
| DEM153 (nifHa-lacZ) | 136 ± 25 | 20 ± 3 | 276 ± 57 | 372 ± 190 |
| DEM233 (nifHc-lacZ) | 426 ± 40 | 21 ± 5 | 1,005 ± 143 | 1,158 ± 358 |
Data from free-living cultures are the averages of three different experiments. Nodules from five plants per strain per day were analyzed. Values are means ± standard deviations. NA, not analyzed; ONP, o-nitrophenol.
β-Galactosidase activity from nodules formed by R. etli strains carrying the reporter gene fused to nifH under the transcriptional control of pr c (HP310 lac and DEM233 strains) had the highest values during the early days of symbiosis (11 and 18 dpi). For the latter day, two independent experiments were conducted which gave similar results, and one is shown in Table 2. Activity of the fusion in strain HP310 lac was similar to that of DEM233 at 18 dpi (Table 2).
Construction and genetic characterization of strains containing the pr c nifHcDK chimeric operon.
The chimeric pr c nifHcDK operon contained on pHP55 plasmid was transferred to R. etli CFN42 and SAM100 (phaC) (8) as described in Materials and Methods. Plasmid DNA was extracted from the transconjugants, and the BamHI digestion profile was found to be identical to that of pHP55 (data not shown). An R. etli strain with the chimeric operon incorporated into pSym was made by a triparental mating with E. coli HB101/pHP789 as donor, E. coli HB101/pRK2013 as helper, and R. etli CFN42 as recipient, as described in Materials and Methods.
To confirm genetic exchange, we carried out a PCR assay with an upper oligonucleotide designated nifHc EcoRV forward, which specifically hybridizes with the nifHc promoter region, and a lower oligonucleotide, nifD reverse, corresponding to the 3′ end of nifD. This segment is absent in the wild-type nifHcD* reiteration. Only the pHP789 plasmid (Fig. 2A) and strains derived from the mating mentioned contained the expected 1.8-kb fragment; one of these, designated HP310, was selected (Fig. 2A). In contrast, strain CFN42 did not produce a PCR product with this oligonucleotide combination (Fig. 2A). The 1.8-kb fragment was sequenced on both ends, and adequate priming was confirmed (data not shown). The recombinant nifHDK operon was obtained by PCR with strain HP310 DNA as template with nifHc EcoRV forward and 238 reverse (located downstream of nifK) oligonucleotides, and its nucleotide sequence was obtained. The sequence revealed that the nifHDK operon was coupled to an intact nifHc promoter region (data not shown).
FIG. 2.
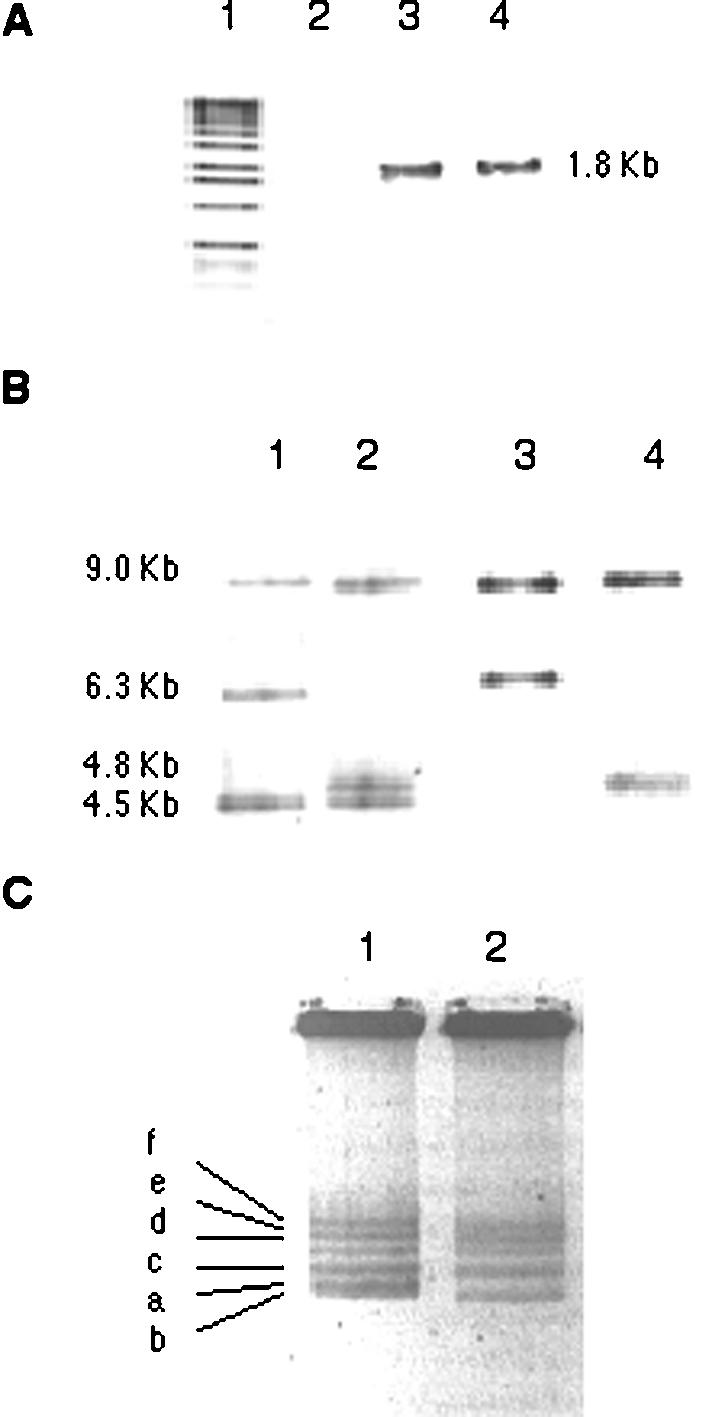
Genetic characterization of strain HP310. (A) PCR with oligonucleotides nifHc EcoRV forward and nifD reverse. Lanes: 1, DNA size marker; 2, CFN42; 3, HP310; and 4, plasmid pHP789. (B) Southern hybridization using as probes an intra-nifH PCR product (lanes 1 and 2) and an intra-nifK PCR product (lanes 3 and 4). Lanes: 1 and 3, CFN42; 2 and 4, HP310. (C) Eckhardt plasmid profile of strains CFN42 (lane 1) and HP310 (lane 2). a to f, plasmids p42a to p42f, respectively.
To determine the genetic modifications in nifHDKb operon caused by the double-recombination process with plasmid pHP789, we hybridized total DNA digested with BamHI from HP310 and CFN42 against nifH and nifK probes. With BamHI digestion, strain CFN42 presents three nifH signals of approximately 9.0, 6.3 and 4.5 kb (26). The first two correspond to nifHDK nitrogenase operons a and b. However, in strain HP310, the nifH hybridization showed that the wild-type nifHDKb band (6.3 kb) was absent and instead the strain contained a 4.8-kb band, very close to the 4.5-kb band corresponding to wild-type nifHc (Fig. 2B). A nifK hybridization demonstrated that the 4.8-kb band was a complete nifHDK operon (Fig. 2B).
Incorporation of the chimeric construct in the nifHDKb reiteration was not unexpected because plasmid pHP789 was constructed based on the nifHDKb reiteration and a 300-bp fragment belonging to region b remained upstream of the pr c nifHcDK construct (Fig. 1). It is possible that this 300-bp segment participated in the recombination process. All isolates obtained by mating with pHP789 and analyzed by hybridization showed the chimeric construct always incorporated into the nifHDKb reiteration (data not shown). Furthermore, we hybridized total DNA digested with BamHI from HP310 and CFN42 against a cosmid collection which covers the entire CFN42 pSym sequence (13). In strain HP310 we observed a band of 4.8-kb instead of the 6.3-kb band in the nifHDKb region, while the rest of the symbiotic plasmid appeared intact (data not shown). By sequencing downstream of the end of nifKb, a single change of one base, which created a BamHI site, was found and was not present in the wild-type sequence (data not shown). The latter explained the reduction in band length.
R. etli CFN42 contains six high-molecular-weight plasmids (with DNA sizes of 150 to 600 kb), named p42a to p42f. The symbiotic plasmid is p42d (371 kb) (14). Plasmids p42b and p42a have similar sizes (150 kb) and appear as a doublet (Fig. 2C). Eckhardt plasmid profile analysis revealed that the symbiotic plasmid of HP310 was similar in size to that of CFN42, but that p42a was absent (Fig. 2C). However, the rest of the plasmids appeared intact. Curing of p42a could be due to additional recombination events originated by reiterated identical sequences shared by both replicons p42a and p42d. It has been previously shown that curing of p42a from the wild-type strain does not alter its symbiotic properties (6). A 7-kb fragment of the symbiotic plasmid, upstream of pr c nifHcDK in strain HP310, was sequenced and found to be identical to that reported for CFN42 pSym (14), except for a 20-bp deletion located close to a transposase (data not shown). This 7-kb sequence is also present in p42a (G. Dávila and V. González, unpublished data).
Symbiotic performance of an R. etli strain overexpressing nitrogenase.
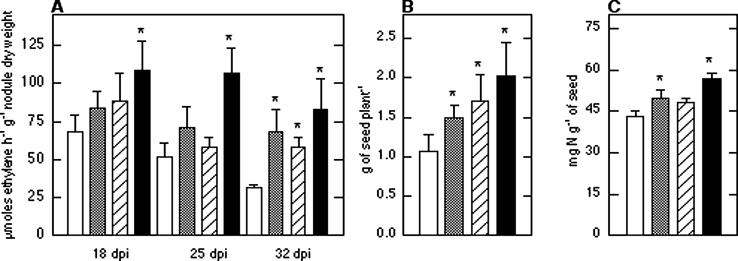
The nitrogenase expression-enhanced pr c nifHcDK operon harbored on plasmid pHP55 was introduced into strain CFN42, and its symbiotic effectiveness was evaluated on bean plants (Fig. 3). A control CFN42 strain harboring plasmid pAM341 (strain AM341 [24]), containing only the nifHc promoter region (pr c) cloned into pTR101, was included in all experiments, and no differences were observed in comparison with CFN42 (data not shown). Three independent assays with each modified strain were performed in the greenhouse. Data presented correspond to a representative experiment. There were no differences between strains with regard to number of nodules and internal (determined by optical microscopy) or external morphology (data not shown). Plants inoculated with HP55 had increases of about 23, 38, and 120% in nitrogenase activity at 18, 25, and 32 dpi, respectively, compared with plants inoculated with the parent strain CFN42, although it was only significantly different at 32 dpi (P < 0.05) (Fig. 3A).
FIG. 3.
Symbiotic performance of R. etli strains with modified nitrogenase expression construct in greenhouse experiments. (A) Specific nitrogenase activity □ Strains. □, CFN42; ░⃞, HP55 (pTR101, pr c nifHcDK); ▨, SAM100 (phaC); ▪, SAM100/pHP55 (phaC, pTR101, pr c nifHcDK). Values are means ± standard deviations of a representative experiment with 10 P. vulgaris plants for each condition and time (n = 30). (B) Seed yield from 10 plants. (C) Nitrogen content in seeds from five plants. Asterisks indicate that the means of the samples are different at (P of <0.05) with respect to CFN42. (D) Growth response of P. vulgaris plants (45 dpi) inoculated with R. etli strains in the greenhouse. Images: 1, Noninoculated nonfertilized; 2, inoculated with CFN42; 3, inoculated with HP55; 4, noninoculated fertilized with 10 mM KNO3-2 mM NH4NO3.
Correlating with the higher nitrogenase activity observed in bean plants inoculated with strain HP55, there was an increase of 25% in plant weight (mean ± standard deviation, 0.56 ± 0.11 and 0.70 ± 0.13 g plant−1 for CFN42 and HP55, respectively) at 32 dpi. For nitrogen content in plants, HP55 had an increase of 15% (24.5 ± 5.9 mg of N plant−1) with respect to CFN42 (21.3 ± 4.7 mg of N plant−1) at 32 dpi. A major difference was observed when seed yields were compared. Plants inoculated with strain HP55 produced a significant increase of 39% (at P < 0.05) in seed yield (1.49 ± 0.16 g of seed plant−1) compared with plants inoculated with the parent strain CFN42 (1.07 ± 0.21 g of seed plant−1) (Fig. 3B). Furthermore, it is noteworthy that not only was seed yield increased by the expression of the recombinant pr c nifHcDK operon but also the nitrogen content of seeds was increased by 16% (from 43 ± 2 to 50 ± 3 mg of N g of seed−1 in plants inoculated with strains CFN42 and HP55, respectively) (Fig. 3C). As a result of the increase in yield and nitrogen content in seeds, an increase of 62% in nitrogen yield was obtained with strain HP55 (74.5 mg of N in seed plant−1) compared to that with CFN42 (46.0 mg of N in seed plant−1). The growth responses of P. vulgaris plants (45 dpi) inoculated with strain HP55 compared with plants inoculated with wild-type strain CFN42 are shown in Fig. 3D. More than 99% of the bacteria isolated from strain HP55-induced nodules retained the pHP55 plasmid after 25 or 32 dpi.
Symbiotic contribution of nifHc or nifHDKb overexpression in R. etli.
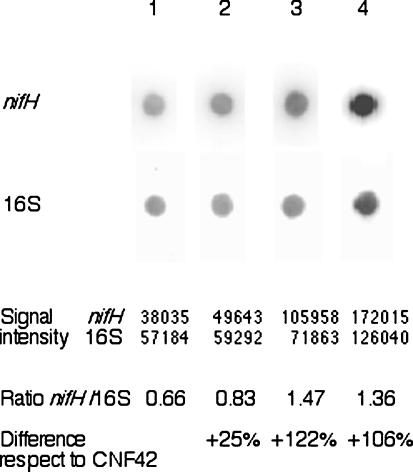
To determine the contribution to symbiotic performance of overexpression of nifHc or nifHDK, and to compare with that produced by pr c nifHcDK, we cloned into plasmid pTR101 the respective reiterations of strain CFN42. The plasmids obtained, pHP210 (pTR101, nifHc) and pHP220 (pTR101, nifHDKb), were incorporated by triparental mating into CFN42 strain and assayed in the greenhouse. The numbers of nodules and the morphology formed by all these strains appeared normal and were similar to those for the wild-type strain CFN42 (data not shown). A dot blot hybridization was made with mRNA extracted from 18-dpi nodules, showing that the nifH transcript was more abundant in nodules obtained for HP210 and HP55 inoculation (122 and 106%, respectively) than those formed by CFN42. HP220 presented 25% more nifH transcript than CFN42 (Fig. 4). The relative intensity signal was calibrated with use of the 16S rRNA gene.
FIG. 4.
Dot blot hybridization of mRNA extracted from P. vulgaris nodules inoculated with R. etli strains at 18 dpi. Lanes: 1, CFN42; 2, HP220; 3, HP210; 4, HP55. Hybridization was done with an intra nifH PCR product or 16S DNA as a probe. Intensity signal (in counts) was obtained by exposure in a PhosphorImager screen.
Nitrogenase activity in bean plants produced by inoculation with strains HP210 and HP220 was slightly increased by 20 and 13%, respectively, while strain HP55 had a significant increase of 68% (at P of <0.05), compared with that for strain CFN42 at 18 dpi (Table 3). In plant weight determination (at 32 dpi), HP210 and HP220 produced increases of 39 and 22%, respectively, compared with CFN42. HP55 produced a significant increase of 50% against CFN42. In regard to seed yield, HP210 and HP220 had increases of 21 and 9%, respectively, compared to CFN42 (Table 3). However, HP55 inoculation produced 2.50 ± 0.23 g plant−1; this is a significant increase of 75% compared to that of CFN42. As expected, nitrogen-fertilized plants produced the highest value (2.60 ± 0.48 g plant−1 [Table 3]). With regard to nitrogen content in seed, HP220 had higher values than HP210. In this parameter, strain HP55 had an increase of 29% compared to CFN42 strain and 21% more than the nitrogen-fertilized plants. With regard to nitrogen yield, strains HP220 and HP210 had increases of 11 and 33% (64.6 and 54.0 mg of N in seed plant−1, respectively) compared with CFN42 (48.5 mg of N in seed plant−1), while HP55 increased 125% (109.0 mg of N in seed plant−1) compared to CFN42 and 16% above nitrogen-fertilized plants (93.9 mg of N in seed plant−1). As observed, symbiotic overexpression of pr c nifHcDK (HP55) in R. etli produced the highest increases in all parameters measured in comparison with results for overexpression of nifHDKb (HP220) or nifHc (HP210).
TABLE 3.
Symbiotic performance of R. etli strains with modified nitrogenase expression constructs in greenhouse experimentsa
| Strain or treatment | Nitrogenase activityb | Plant dry wt (g plant−1)c | Seed yield (g plant−1)d | N content in seed (mg of N g−1)e |
|---|---|---|---|---|
| CFN42 (wild type) | 64.5 ± 9.2A | 0.54 ± 0.13A | 1.43 ± 0.13A | 33.9 ± 2.3A |
| HP220(pTR101, nifHDKb) | 72.7 ± 12.0A | 0.66 ± 0.21AB | 1.56 ± 0.32AB | 41.4 ± 7.5B |
| HP210(pTR101, nifHc) | 77.3 ± 18.5A | 0.75 ± 0.29AB | 1.73 ± 0.24A | 31.2 ± 3.1A |
| HP55(pTR101, pr c nifHcDK) | 108.2 ± 8.9B | 0.81 ± 0.16B | 2.50 ± 0.23B | 43.6 ± 9.1B |
| With added N | 1.22 ± 0.22C | 2.60 ± 0.48B | 36.1 ± 2.3AB |
Values are means ± standard deviations. Different letters represent significant differences (P < 0.05).
Nitrogenase activity is expressed as micromoles of ethylene hour−1 gram of nodule−1.
Dry weight was measured at 18 dpi. Ten plants were dried and weighed at 32 dpi.
Seed yield from 10 plants (80 days old).
Seeds from 5 plants were evaluated.
Symbiotic performance of an R. etli PHB− strain expressing the pr c nifHcDK construction.
A PHB− R. etli strain showed 5- to 21%-higher nitrogenase activity compared with that for wild-type strain CFN42 in late stages of symbiosis with P. vulgaris (8). Additionally, increases in seed yield (8%) and nitrogen content in seed (15%) were observed (8). To determine if an additive effect could be obtained by combining the expression of the chimeric pr c nifHcDK construct and a PHB− background, plasmid pHP55 containing pr c nifHcDK was introduced by conjugation into strain SAM100 (8). Strain SAM100 had increases of 29, 13, and 87% in nitrogenase activity at 18, 25, and 32 dpi (Fig. 3A), 4% in plant weight (0.58 ± 0.11 g plant−1), 34% in nitrogen content per plant (28.6 ± 6.6 mg of N plant−1), 60% in seed yield (1.71 ± 0.34 g plant−1), 12% in nitrogen content in seed (Fig. 3C), and 46% in nitrogen yield (78.7 mg of N in seed plant−1) with respect to its parent strain CFN42. Nitrogenase activities in bean plants inoculated with SAM100/pHP55 were higher than and significantly different (at P of <0.05) from those in plants inoculated with its parent strain SAM100 at 25 and 32 dpi by 82 and 42%, respectively (Fig. 3A). Furthermore, increases of 19% in plant weight (0.69 ± 0.09 g plant−1), 12% in nitrogen content per plant (31.7 ± 5.5 mg of N plant−1), 18% in seed yield (Fig. 3B), 19% in nitrogen content in seed (Fig. 3C), and 32% in nitrogen yield (104.3 mg of N in seed plant−1) were observed for SAM100/pHP55 inoculation compared to SAM100 inoculation.
Symbiotic effect of an R. etli strain with the pr c nifHcDK construct incorporated into pSym.
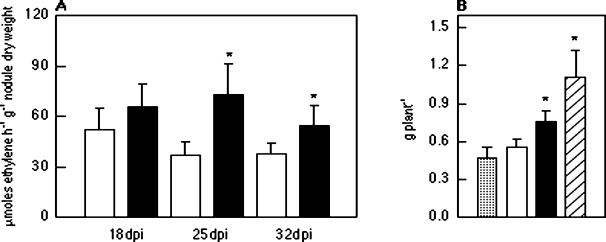
We assessed the symbiotic effect on bean plants inoculated with strain HP310, which contains the pr c nifHcDK construct in pSym. In the greenhouse, plants inoculated with HP310 had increases of 25, 97, and 44% in nitrogenase activity at 18, 25, and 32 dpi, respectively, compared with plants inoculated with parent strain CFN42 (Fig. 5A), with significant differences obtained at 25 and 32 dpi (P < 0.05). HP310 had a significant increase of 38% in plant weight (0.76 ± 0.08 g plant−1) compared with CFN42 (0.55 ± 0.06) (Fig. 5B) at 32 dpi. For comparison, results for noninoculated (0.47 ± 0.08) and fertilized (1.11 ± 0.21 g plant−1) plants are shown in Fig. 5B. Seed yield of plants inoculated with strain HP310 produced a significant increase (P < 0.05) of 33% (1.34 ± 0.3 g of seed plant−1) compared with that in plants inoculated with the parent strain CFN42 (1.01 ± 0.2 g of seed plant−1). In this case, nitrogen content in seed produced by HP310 was significantly increased by 34% compared with that produced by wild-type strain CFN42 (59 ± 3 and 44 ± 2 mg of N g of seed−1, respectively). Nitrogen yield obtained with HP310 was 81% higher than that obtained with CFN42 (79 and 44 mg of N in seed plant−1, respectively). Plants inoculated with HP310 had an appearance similar to those inoculated with HP55 (data not shown).
FIG. 5.
Symbiotic performance of an R. etli strain with a modified nitrogenase expression construct incorporated into pSym. (A) Specific nitrogenase activity of P. vulgaris plants inoculated with CFN42 or HP310; (B) plant weight at 32 dpi. Values are means ± standard deviations for 10 plants for each condition and time (n = 30). Asterisks indicate that the means of the samples are different at P <0.05. □, inoculation with CFN42; ▪, inoculation with HP310; ░⃞, noninoculated nonfertilized; ▨, noninoculated fertilized with 10 mM KNO3-2 mM NH4NO3.
DISCUSSION
Functional analysis of the elements located upstream of the reiterated nifH genes in R. etli CFN42 revealed an asymmetric arrangement of the regulatory regions of the two nifHDK operons (copies a and b) and the third reiterated nifH copy (35). Copies a and b are activated by NifA bound to a canonical binding site, while nifHc is activated by NifA bound to a divergent site. This asymmetric arrangement involves a dissimilar facing of the NifA-binding sites located in these promoter regions, which may imply a particular initiation complex architecture resulting in different transcription levels (35). By sequence alignment, a similar arrangement can be found in the reiterated nifH regulatory regions of R. leguminosarum biovars phaseoli and trifolii and Rhizobium sp. strain NGR234, where the NifA binding site in one copy differs by about a half helical turn in distance to its promoter with respect to another copy(ies) (G. Guerrero and J. Mora, unpublished results).
We have found that all strains of R. etli analyzed to date carry three nifH reiterations, two of them in nifHDK operons and the third reiteration linked to a truncated nifD* gene (35; B. Valderrama, unpublished results). The third nifH gene has been analyzed in two strains closely related to R. etli isolated from bean nodules, and the corresponding upstream region sequence highly resembles that from CFN42 (data not shown). It is important that polar insertions in nifHc have no obvious symbiotic phenotype (35).
Rhizobium bacteria undergo a complex differentiation process once they infect legume roots. Bacteroids present a particular structural and physiological adaptation to the nodule environment. One of these physiological changes is nitrogenase induction, mediated by the regulatory protein NifA. It has been shown that NifA is produced constitutively even under ex planta conditions, but since it is intrinsically oxygen-sensitive, it is active only under microaerobic or symbiotic conditions (25).
In order to acquire higher expression levels of nitrogenase while preserving its NifA-dependent regulation, we modified such expression by placing one of the reiterated nifHDK operons under the control of the stronger nifHc promoter region. It is important that all sequences used in this work are derived from R. etli's own symbiotic plasmid and that no exogenous DNA other than that of the vector was added.
As reported above, the chimeric construct pr c nifHcDK was functional under the tested conditions of a low-oxygen atmosphere and in symbiosis (Table 2). It is important that the nifH sequence was not altered by the substitution of the promoter region (Fig. 1).
The expression of the chimeric pr c nifHcDK operon, either on a Rhizobium stably replicating plasmid or incorporated into pSym, produced a better symbiotic performance with P. vulgaris plants. The parameters used to assess the symbiosis were nitrogenase activity, dry plant weight, seed yield, and nitrogen content in plants and seeds as described above (Fig. 3 and 5). Furthermore, plant appearance confirmed the enhancement of the symbiotic ability of modified strains HP55 (Fig. 3D) and HP310 (data not shown).
The role of PHB in rhizobial symbiosis is still controversial. The symbiotic relationship between S. meliloti and alfalfa (Medicago sativa) is very successful, given that the plant derives 80% of its nitrogen requirement from symbiotic nitrogen fixation (15). Since S. meliloti does not accumulate PHB in symbiosis (16), reductive power not used for PHB synthesis could be used for nitrogen fixation. However, R. etli produces PHB in free life and also in symbiosis (8, 10). An R. etli PHB− mutant produced increased nitrogenase activity in symbiosis and a moderate augmentation in seed yield in comparison with wild-type strain CFN42 (8). Apparently, in this case, part of the reducing power present in the strain was channeled to nitrogenase. By this token, in order to further increase the symbiotic performance of an R. etli strain expressing the pr c nifHcDK construct, we intended to derive the reducing power excess, produced by the phaC mutation, to energize nitrogenase catalysis. As observed above, by combining the latter two characteristics, we obtained a strongly enhanced symbiotic relationship, which gave the highest values of nitrogen fixation reported to date in R. etli (Fig. 3). Apparently, this nitrogen fixation effectiveness is the sum obtained by nitrogenase overexpression plus the phaC mutation.
According to the results presented, carbon supply to the bacteroid is always in excess under normal nitrogenase activity. The rest of the processes involved in the synthesis of the nitrogenase structural proteins and their assembly are not limited. In addition, it is possible to derive carbon and reductive power to obtain energy for nitrogenase catalysis by abolishing the synthesis of the polymer PHB.
Field testing of the modified strains presented in this work may determine their potential use as a biofertilizer, which could reduce the cost incurred with the application of chemical fertilizers.
Acknowledgments
This work was supported by grants 3309PB, 29025B and 33575N from CONACyT-México. H.P. was the recipient of a Cátedra Patrimonial II award from CONACyT.
B. Valderrama and A. Mendoza participated in initial experimental planning. We thank M. Dunn for critical reviewing of the manuscript; A. Dávalos for help in constructing the pHP789 plasmid; V. Bustos, I. Alvear, and J. L. Zitlalpopoca for support in laboratory and greenhouse work; and A. Leija for light microscopy observations. We acknowledge S. Contreras, R. Santamaría, and P. Bustos for support with sequencing and P. Gaytán and E. López (IBt-UNAM) for oligonucleotide synthesis.
REFERENCES
- 1.Bascones, E., J. Imperial, T. Ruiz-Argueso, and J. M. Palacios. 2000. Generation of new hydrogen-recycling Rhizobiaceae strains by introduction of a novel hup minitransposon. Appl. Environ. Microbiol. 66:4292-4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bergersen, F. J., M. B. Peoples, and G. L. Turner. 1991. A role for poly-β-hydroxybutyrate in bacteroids of soybean root nodules. Proc. R. Soc. Lond. Ser. B Biol. Sci. 245:59-64. [Google Scholar]
- 3.Bosworth, A. H., M. K. Williams, K. A. Albrecht, R. Kwiatkowski, J. Beynon, T. R. Hankinson, C. W. Ronson, F. Cannon, T. J. Wacek, and E. W. Triplett. 1994. Alfalfa yield response to inoculation with recombinant strains of Rhizobium meliloti with an extra copy of dctABD and/or modified nifA expression. Appl. Environ. Microbiol. 60:3815-3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyer, H. W., and D. Roulland-Dussoix. 1969. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 41:459-472. [DOI] [PubMed] [Google Scholar]
- 5.Bravo, A., and J. Mora. 1988. Ammonium assimilation in Rhizobium phaseoli by the glutamine synthetase-glutamate synthase pathway. J. Bacteriol. 170:980-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brom, S., A. Garcia-de los Santos, L. Cervantes, R. Palacios, and D. Romero. 2000. In Rhizobium etli symbiotic plasmid transfer, nodulation competitivity and cellular growth require interaction among different replicons. Plasmid 44:34-43. [DOI] [PubMed] [Google Scholar]
- 7.Castellanos, J. 1997. Hábitos preferenciales de los consumidores de frijol Phaseolus vulgaris en México. Arch. Latinoam. Nutr. 47:163-167. [PubMed] [Google Scholar]
- 8.Cevallos, M. A., S. Encarnación, A. Leija, Y. Mora, and J. Mora. 1996. Genetic and physiological characterization of a Rhizobium etli mutant strain unable to synthesize poly-β-hydroxybutyrate. J. Bacteriol. 178:1646-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, H., A. E. Richardson, E. Gartner, M. A. Djordjevic, R. J. Roughley, and B. G. Rolfe. 1991. Construction of an acid-tolerant Rhizobium leguminosarum biovar trifolii strain with enhanced capacity for nitrogen fixation. Appl. Environ. Microbiol. 57:2005-2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Encarnación, S., M. Dunn, K. Willms, and J. Mora. 1995. Fermentative and aerobic metabolism in Rhizobium etli. J. Bacteriol. 177:3058-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freiberg, C., R. Fellay, A. Bairoch, W. J. Broughton, A. Rosenthal, and X. Perret. 1997. Molecular basis of symbiosis between Rhizobium and legumes. Nature 387:394-401. [DOI] [PubMed] [Google Scholar]
- 13.Girard, M. L., M. Flores, S. Brom, D. Romero, R. Palacios, and G. Dávila. 1991. Structural complexity of the symbiotic plasmid of Rhizobium leguminosarum bv. phaseoli. J. Bacteriol. 173:2411-2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.González, V., P. Bustos, M. A. Ramírez-Romero, A. Medrano-Soto, H. Salgado, I. Hernández-González, J. C. Hernández-Célis, V. Quintero, G. Moreno-Hagelsieb, L. Girard, O. Rodríguez, M. Flores, M. A. Cevallos, J. Collado-Vides, D. Romero, and G. Dávila. 2003. The mosaic structure of the symbiotic plasmid of Rhizobium etli CFN42 and its relation to other symbiotic genome compartments. Genome Biol. 4:R36. [online posting date, 13 May 2003.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hardarson, G. 1993. Methods for enhancing symbiotic nitrogen fixation. Plant Soil 152:1-17. [Google Scholar]
- 16.Hirsch, A. M., M. Bang, and F. M. Ausubel. 1983. Ultrastructural analysis of ineffective alfalfa nodules formed by nif::Tn5 mutants of Rhizobium meliloti. J. Bacteriol. 155:367-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hynes, M. F., and N. F. McGregor. 1990. Two plasmids other than the nodulation plasmid are necessary for formation of nitrogen-fixing nodules by Rhizobium leguminosarum. Mol. Microbiol. 4:567-574. [DOI] [PubMed] [Google Scholar]
- 18.Kadouri, D., S. Burdman, E. Jurkevitch, and Y. Okon. 2002. Identification and isolation of genes involved in poly(β-hydroxybutyrate) biosynthesis in Azospirillum brasilense and characterization of a phbC mutant. Appl. Environ. Microbiol. 68:2943-2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karr, D. B., J. K. Waters, F. Suzuki, and D. W. Emerich. 1984. Enzymes of the poly-β-hydroxybutyrate and citric acid cycles of Rhizobium japonicum bacteroids. Plant Physiol. 75:1158-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kokotek, W., and W. Lotz. 1989. Construction of a lacZ-kanamycin-resistance cassette, useful for site-directed mutagenesis and as a promoter probe. Gene 84:467-471. [DOI] [PubMed] [Google Scholar]
- 21.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 22.Marsh, J. L., M. Erfle, and E. J. Wykes. 1984. The plC plasmid and phage vectors with versatile cloning sites for recombinant selection by insertional inactivation. Gene 32:481-486. [DOI] [PubMed] [Google Scholar]
- 23.Martínez, E., M. A. Pardo, R. Palacios, and M. A. Cevallos. 1985. Reiteration of nitrogen fixation gene sequences and specificity of Rhizobium in nodulation and nitrogen fixation in Phaseolus vulgaris. J. Gen. Microbiol. 131:1779-1786. [Google Scholar]
- 24.Mendoza, A., B. Valderrama, A. Leija, and J. Mora. 1998. NifA-dependent expression of glutamate dehydrogenase in Rhizobium etli modifies nitrogen partitioning during symbiosis. Mol. Plant-Microbe Interact. 11:83-90. [DOI] [PubMed] [Google Scholar]
- 25.Morett, E., H. M. Fischer, and H. Hennecke. 1991. Influence of oxygen on DNA binding, positive control, and stability of the Bradyrhizobium japonicum NifA regulatory protein. J. Bacteriol. 173:3478-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quinto, C., H. de la Vega, M. Flores, J. Leemans, M. A. Cevallos, M. A. Pardo, R. Azpiroz, M. de L. Girard, E. Calva, and R. Palacios. 1985. Nitrogenase reductase: a functional multigene family in Rhizobium phaseoli. Proc. Natl. Acad. Sci. USA 82:1170-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Romero, D., P. W. Singleton, L. Segovia, E. Morett, B. B. Bohlool, R. Palacios, and G. Dávila. 1988. Effect of naturally occurring nif reiterations on symbiotic effectiveness in Rhizobium phaseoli. Appl. Environ. Microbiol. 54:848-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Segovia, L., J. P. W. Young, and E. Martínez-Romero. 1993. Reclassification of American Rhizobium leguminosarum biovar phaseoli type I strains as Rhizobium etli sp. nov. Int. J. Syst. Bacteriol. 43:374-377. [DOI] [PubMed] [Google Scholar]
- 30.Selbitschka, W., S. Niemann, and A. Pühler. 1993. Construction of the gene replacement vectors for gram negative bacteria using a genetically modified sacRB gene as a positive selection marker. Appl. Microbiol. Biotechnol. 38:615-618. [Google Scholar]
- 31.Spaink, H. P., R. J. H. Okker, C. A. Wijffelman, T. Tak, L. Goosen-de Roo, E. Pees, A. A. N. van Brussel, and B. J. J. Lugtenberg. 1989. Symbiotic properties of rhizobia containing a flavonoid-independent hybrid nodD product. J. Bacteriol. 171:4045-4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stam, H., W. de Vries, A. H. Stouthamer, and H. W. van Verseveld. 1986. Utilization of poly-β-hydroxybutyrate in free-living cultures of Rhizobium ORS571. FEMS Mirobiol. Lett. 35:215-220. [Google Scholar]
- 33.Steel, R. G., and J. H. Torrie. 1980. Principles and procedures of statistics: a biometrical approach, 2nd ed. McGraw-Hill Book Co., New York, N.Y.
- 34.Tombolini, R., and M. P. Nuti. 1989. Poly (β-hydroxyalkanoate) biosynthesis and accumulation by different Rhizobium species. FEMS Microbiol. Lett. 60:299-304. [Google Scholar]
- 35.Valderrama, B., A. Dávalos, L. Girard, E. Morett, and J. Mora. 1996. Regulatory proteins and cis-acting elements involved in the transcriptional control of Rhizobium etli reiterated nifH genes. J. Bacteriol. 178:3119-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weinstein, M., R. Roberts, and D. Helinski. 1992. A region of the broad-host-range plasmid RK2 causes stable in planta inheritance of plasmids in Rhizobium meliloti cells isolated from alfalfa root nodules. J. Bacteriol. 174:7486-7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weisburg, W., S. Burns, D. Pelletier, and D. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wong, P. P., and H. J. Evans. 1971. Poly-β-hydroxybutyrate utilization by soybean (Glycine max Merr.) nodules and assessment of its role in maintenance of nitrogenase activity. Plant Physiol. 47:750-755. [DOI] [PMC free article] [PubMed] [Google Scholar]