Abstract
Cells growing on surfaces in biofilms exhibit properties distinct from those of planktonic cells, such as increased resistance to biocides and antimicrobial agents. In spite of increased interest in biofilms, very little is known about alterations in cell physiology that occur upon attachment of cells to a surface. In this study we have investigated the changes induced in the protein synthesis by contact of Streptococcus mutans with a surface. Log-phase planktonic cells of S. mutans were allowed to adhere to a glass slide for 2 h in the presence of a 14C-amino acid mixture. Nonadhered cells were washed away, and the adhered cells were removed by sonication. The proteins were extracted from the nonadhered planktonic and the adhered biofilm cells and separated by two-dimensional gel electrophoresis followed by autoradiography and image analysis. Image analysis revealed that the relative rate of synthesis of 25 proteins was enhanced and that of 8 proteins was diminished ≥1.3-fold in the biofilm cells. Proteins of interest were identified by mass spectrometry and computer-assisted protein sequence analysis. Of the 33 proteins associated with the adhesion response, all but 10 were identified by mass spectrometry and peptide mass fingerprinting. The most prominent change in adhered cells was the increase in relative synthesis of enzymes involved in carbohydrate catabolism indicating that a redirection in protein synthesis towards energy generation is an early response to contact with and adhesion to a surface.
Bacterial cells attached to, and growing on, surfaces in mature biofilms are physiologically distinct from their planktonic counterparts (7, 11, 12). One of the earliest observations of such altered behavior was the increased resistance of biofilm cells to antimicrobial agents, biocides, and adverse environmental conditions, including nutrient deprivation (5, 7, 17). On approaching a surface, bacteria encounter environmental conditions different from those in the bulk liquid phase, a situation known to induce significant alterations in gene expression (8, 38). In Pseudomonas aeruginosa, new protein synthesis is required during the initial stage of biofilm formation (6), and the protein expression profile changes 5 min after attachment. Alginate production (algC) was also up-regulated within 15 min of cell attachment to a glass surface (9). It has been shown similarly that in Bacillus cereus 15 uniquely expressed proteins were synthesized within 2 h after surface attachment, compared to only 7 uniquely expressed proteins in 18-h-old biofilm compared to planktonic controls (23).
Considerable biofilm research is now focused on the cellular functions that are modified during cellular transition from the planktonic to the biofilm state. In this process, it is clear that environmental conditions—for example, nutrient, pH, osmolarity, and temperature—signal the altered regulation of multiple genes determined by the organism and the nature of the surface involved (8, 15, 24, 37). In Escherichia coli the Cpx two-component signal transduction pathway plays a key role in regulating adhesion-induced genes. Surface contact of E. coli with hydrophobic surfaces for 1 h induces the transcriptional activity of Cpx-regulated promoters. This on the other hand did not occur when the bacteria adhered to a hydrophilic surface (25). Analysis of E. coli mutants derived by insertional mutagenesis has demonstrated that motility and type 1 (pili) fimbriae are required for the initial attachment to a surface (30). Initial attachment was accompanied by alterations in the composition of outer membrane proteins, which has led to the suggestion that type 1 fimbrial surface contact is a surface-sensing mechanism (26).
Considerable information is known regarding the adhesion and coaggregation of oral bacteria in plaque development (8, 41), but little is known of the proteins involved in initial attachment of oral bacteria to surfaces. Using a microtiter plate assay, biofilms of the early colonizer of the dentition, Streptococcus gordonii, were grown for 16 h. Biofilm-defective mutants generated by transposon mutagenesis were identified, and the genes inactivated included those associated with osmoregulation, adhesion, nutrient sensing, and quorum sensing (comD) (22). A later study of the same organism showed that the adc operon involved in manganese homeostasis is also involved in biofilm formation (21).
Oral streptococci are important components of the complex oral biofilm known as dental plaque, and members of the Streptococcus genus, such as Streptococcus mutans, are associated with dental caries (13). S. mutans has recently been shown to possess a peptide pheromone quorum-sensing signaling system involving a histidine two-component regulatory mechanism that induces genetic competence and which functions more efficiently in biofilms than in planktonic cultures (19). It has also been shown that the brpA gene of S. mutans encoding a novel protein of 406 amino acid residues is important for the maturation of the biofilm (40). We have previously demonstrated that S. mutans H7 grown in a biofilm for 3 days expresses an array of proteins different from those demonstrated by their planktonic counterparts when grown in steady state under the same conditions in a biofilm chemostat on hydroxyapatite rods (34). These cells not only expressed different proteins but also were physiologically different in terms of acid tolerance, with 41.5% of the biofilm cells surviving an acid shock of pH 3.0 compared to only 4 × 10−5% of the planktonic cells (39). S. mutans is know to induce an acid tolerance response (ATR) at sublethal pH values (∼5.5) that enhances survival at lower pH values (36). Exposing the 3-day biofilm cells to pH 5.5 for 2 h only induced a negligible ATR and didn't increase the number of survivors, suggesting that surface growth itself triggered an ATR in biofilm cells sometime during biofilm development (39). In this study we were interested in whether initial surface contact per se induced the same alterations in protein expression as in mature biofilms.
MATERIALS AND METHODS
Bacterial strain and media.
S. mutans H7, the strain used in this study, was isolated from dental plaque above a carious enamel surface following plating directly onto a nonselective, solid agar medium buffered at pH 5.0 (33). For the labeling experiments, single colonies on blood agar were used to inoculate minimal medium containing 40 mM phosphate-citrate buffer (pH 7.5) and 20 mM glucose (MM4) (14). Wash medium used during the experiments was MM4 devoid of glucose and phosphate-citrate buffer.
Biofilm formation and labeling of proteins.
Glass slides (Menzel) were used for biofilm formation, with each slide cut in half resulting in a total area of 7 cm2. Prior to use, each slide was washed in concentrated hydrochloric acid, 8 M sodium hydroxide, and ethanol to remove contamination. The glass slides were maintained in each solution for 24 h and washed between each step with sterile ultra-high-quality-water before being sterilized at 121°C for 2 min.
Cells were grown overnight in MM4 in triplicate with each culture inoculated from a different colony on blood agar. On the day of the experiment, the cells from each culture were inoculated into 100 ml of fresh MM4 and grown to mid-log phase (optical density at 600 nm = 0.7), harvested by centrifugation (5,000 × g for 5 min), and washed twice in wash medium. The cells were then resuspended in 1 ml of equal parts double-strength MM4 and 14C-amino acid mixture (25 μCi; Amersham Pharmacia Biotech). One milliliter of cell suspension was immediately added to each glass slide, and the cells were allowed to adhere for 2 h at 37°C in a humid chamber. At 2 h, protein synthesis was stopped by immersion of the glass slide into wash medium containing chloramphenicol (0.8 mg/ml). The cells that had not adhered to the glass slide during the incubation time (planktonic cells) were removed by immersion of the glass slide in 10 ml of wash medium. The rinsing procedure was repeated five times, each time in fresh medium, to remove all nonadherent cells. The cells were collected by centrifugation and resuspended in TEM buffer (10 mM Tris-HCl [pH 6.8], 1 mM EDTA, 5 mM MgSO4) and stored at −20°C. After washing the glass slide, the cells that had adhered (biofilm cells) were immediately removed by sonication (10 pulses of 1 s at amplitude 40) (Sonics Vibra cell VC130; Sonics & Materials Inc.), collected by centrifugation, resuspended in TEM buffer, and stored at −20°C. To ensure that adhering cells were removed during sonication, a Gram stain of the glass slide was carried out. Viable cell counts were carried out by plating on blood agar.
Fluorescent staining.
To ascertain the viability of the biofilm and planktonic cells, they were stained using a LIVE/DEAD BacLight Bacterial Viability kit (Molecular Probes) following the washing steps. The stained cells were photographed using a fluorescence microscope (magnification, ×1,000). The living and dead cells were counted in 10 areas, and the proportion of viable cells was calculated as a percentage of all the cells.
Protein extraction and 2DE.
Proteins were extracted by ultrasonication in lysis buffer as previously described (34), and the protein concentration was determined by the method of Bradford (4) with the appropriate concentration of lysis buffer in the standards as described by Fey et al. (10). Proteins in the cell extracts were separated by two-dimensional gel electrophoresis (2DE) essentially as described previously (34, 35), with the exception that the procedure was modified for the Mini PROTEAN II electrophoresis system (Bio-Rad). The same amount of radioactivity (20,000 cpm corresponding to approximately 2 μg of protein) was applied to each gel strip. The cell extracts were diluted with lysis buffer containing 10 mM dithiothreitol and applied in a reswelling cassette with the 7-cm Immobiline Dry Strip (IPG) (pH 4 to 7; Amersham Pharmacia Biotech) on top. Rehydration was allowed to proceed at room temperature, and after 4 h another 25 μl of buffer was added and rehydration continued overnight under silicone oil. The isoelectric focusing was carried out using the Multiphor II (Amersham Pharmacia Biotech) with cooling water at 15°C supplied by Pharmacia Multitemp II. The focusing was initiated at 150 V for 1 h and continued at 300 V for 1 h and 600 V for 1 h, with the voltage increased to 3,500 V over 13 h. After focusing, the IPG gel strips were stored at −80°C. Before the second dimension, the IPG gel strips were equilibrated as previously described (34). The equilibrated IPG strips were embedded on top of 14% polyacrylamide gradient gels (60 by 80 by 1.0 mm) using 0.5% (wt/vol) molten agarose. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis was performed at 120 V for 120 min in a PROTEAN II Minicell (Bio-Rad) with 14C-labeled molecular mass standards run on the acidic side of the IPG gel strips. Sodium dodecyl sulfate-polyacrylamide gels were stained with silver nitrate as described by the manufacturer (Pharmacia Biotech). After staining, the gels were vacuum dried at 60°C for 3 h and exposed to X-ray film (Hyperfilm β-max; Amersham) for 12 weeks.
Image analysis.
Autoradiograms were scanned with a UMAX transmission scanner, and images were analyzed by using the Bio Image software on a Sun Sparc workstation as described previously (34, 35). The integrated optical density for each spot, as a percentage of the total blackening of the image attributed to proteins (IOD%), was measured for each protein spot. The IOD% gives a value for the relative rate of synthesis (RRS) for each protein, i.e., how much of that protein has been synthesized compared to the total protein synthesis. For each protein spot, the average IOD% and coefficient of variation (CV) were calculated from three autoradiograms of each set of planktonic and biofilm cells. The CV was obtained by dividing the standard deviation by the mean and multiplying by 100. The CV aids in adjusting for the tendency of the variability to increase as the mean increases. Proteins with enhanced expression in biofilm cells were determined by comparing protein expression in biofilm cells with those in planktonic cells using the following criteria: (i) the protein was present in at least two of the three autoradiograms of the biofilm cells, (ii) the average IOD% for the protein in the biofilm cells was >1.3-fold higher than the average IOD% in the planktonic cells, and (iii) the percentage increase of protein expression in biofilm cells was higher than the CV for the protein in planktonic and biofilm cells, respectively. Proteins with diminished expression in biofilm cells were determined in a similar manner, but based on the average IOD% of the protein in planktonic versus biofilm cells.
Protein identification.
Identification of proteins on the small gels was extrapolated from identifications of proteins from large (18-cm IPG strip) gels from previous experiments with S. mutans H7 (34, 35, 39). Comparison of protein spot positions was made on silver-stained gels. Protein spots had previously been identified by manually cutting spots from large (18-cm IPG strip) Coomassie blue-stained or dried gels and subjected to in-gel digestion with trypsin as described previously (34, 35, 39). The resulting protein peptides were analyzed by mass spectrometry at two locations: Centre for Proteome Analysis, Odense, Denmark, and Department of Microbiology, GKT Dental Institute, London, England.
RESULTS AND DISCUSSION
Biofilm formation.
The aim of this study was to get a better understanding of the changes taking place in cells of S. mutans going from the planktonic phase to the biofilm phase. Cells of S. mutans H7 were allowed to adhere to a glass surface in the presence of 14C-amino acids for 2 h. The cells that didn't adhere during the 2-h incubation time were treated as planktonic cells and were compared to the biofilm cells. The possibility that a proportion of the planktonic cells had experienced surface contact and subsequently detached cannot be excluded. Therefore, these data may underrepresent differences between planktonic and biofilm cells in the initial stage of biofilm formation.
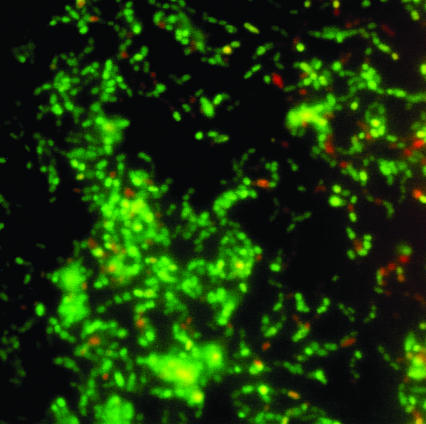
After 2 h the cell density, by viable counting, was 2 × 107 cells/cm2. The 2-h biofilm visualized using the LIVE/DEAD fluorescent stain consisted primarily of a single layer of cells with few clusters, covering approximately 20% of the glass surface (Fig. 1). The proportions of viable cells in the biofilm and planktonic cells populations were 73% ± 4% and 78% ± 4%, respectively (P > 0.1), indicating there was no difference in the affinity of live or dead cells for the glass surface.
FIG. 1.
LIVE/DEAD BacLight fluorescent staining of cells of S. mutans H7 adhering to a glass slide after 2-h incubation and rinsing.
Previous studies (20) of S. mutans in biofilms subjected to glucose limitation described phases of adhesion, cell division, and accumulation, with the adhesion phase involving adhesion (∼1 h) and a period resembling a lag phase (1 to 4 h), while P. aeruginosa biofilms were reported to develop through five stages, with the first signs of transition from initial attachment to irreversible attachment commencing 2 h after surface contact (31). From these two diverse studies it is apparent that the 2-h incubation time used here can be considered representative of initial attachment phase for S. mutans.
Alterations in relative rate of protein synthesis after surface contact.
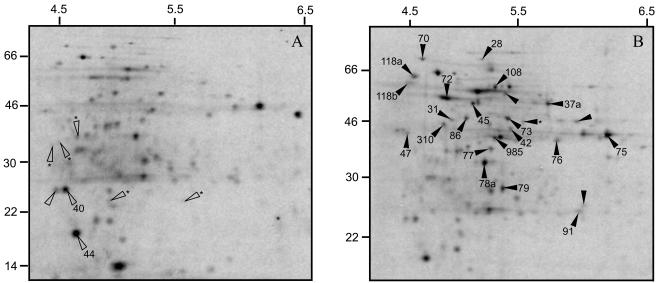
Of the 124 protein spots analyzed, 25 showed enhanced RRS and only one was de novo in the biofilm cells (Table 1). Diminished RRS was apparent in 8 protein spots expressed in the biofilm cells, while 5 protein spots were observed only in the planktonic cells and the expression of 91 protein spots was not significantly altered. These data confirm those obtained with Pseudomonas spp. and B. cereus showing that surface adherence results in altered protein expression (6, 23, 32). An altered RRS of 33 protein spots, 27% of the number of protein spots analyzed, in these 2-h-old biofilms is a proportion similar to that in a 3-day-old S. mutans biofilm, where 19% (135 of 694) of the analyzed protein spots were modulated (34). To evaluate the level of reproducibility of protein expression, the CVs of spots were calculated for the spots on the autoradiograms from the biofilm and planktonic cells separately, and the mean values were 28.2 ± 19.4 and 30.1 ± 20.7, respectively (P > 0.1), which were within an acceptable range (3). The results of this study clearly show that when planktonic cells of S. mutans H7 adhere to a glass surface, changes in RRS occur within 2 h. The approach of comparing the RRS has the advantage that differences in 14C-amino acid incorporation, and thereby total protein synthesis between planktonic and biofilm cells, that could occur if the cells were metabolically different will be compensated for. The use of radioactive labeling will only reveal proteins that were synthesized during the incubation time. Responses scored via the comparison of relative values are potentially more informative, especially when the goal is to explain cellular adjustments under different conditions (2).
TABLE 1.
Protein spots in S. mutans H7 cellsa
| Protein spot category | No. of protein spots |
|---|---|
| Total analyzed | 124 |
| Enhanced | 25 |
| Diminished | 8 |
| Novel in biofilm cells | 1 |
| Not expressed in biofilm cells | 5 |
Number of protein spots in cells of S. mutans H7 exhibiting RRS enhanced or diminished by 1.3-fold or more in 2-h-old biofilm cells compared to nonadherent planktonic cells. Data obtained from the analysis of the autoradiograms in Fig. 4.
Protein identification.
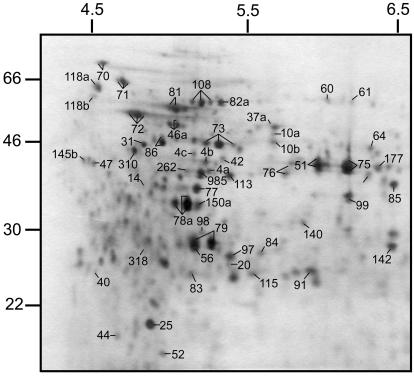
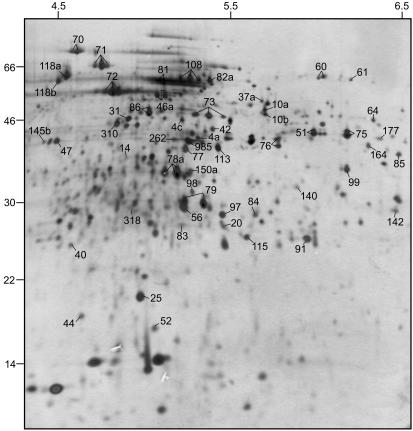
Protein separation was performed in a minigel system (Fig. 2), and silver staining revealed 200 protein spots, a protein spot pattern that was very similar to that observed using 18-cm IPG strips, and greater protein loading under the same conditions (Fig. 3). Due to the lower protein load on the minigels, only protein spots with the highest expression level were revealed and protein spots of very low expression were not detectable (34). However, the expression levels of the 20 most abundant proteins were within the same range on both gel types. Therefore, the similarity of protein spot patterns on the two gel types enable protein identifications from the large gels on the basis of spot matching.
FIG. 2.
Silver-stained 2DE-minigel protein profile of S. mutans H7 extracted from cells after adhesion. Protein spots marked with a number have been identified, and multiple spots marked with the same number represent isoforms of the same protein. Isoelectric focusing was carried out on a 7-cm IPG strip.
FIG. 3.
Silver-stained 2DE protein profile of batch-grown S. mutans H7. Protein spots marked with a number have been identified. Several spots marked with the same number represent isoforms of the same protein. Isoelectric focusing was carried out on an 18-cm IPG strip.
Twenty-three of the proteins modulated by adherence of S. mutans to the glass surface were identified (Table 2). The greatest number of changes occurred in proteins involved with carbohydrate catabolism with 7,6-phosphofructokinase, fructose bisphosphate aldolase, glyceraldehyde-3-phosphate dehydrogenase, phosphoglycerate kinase, phosphoglycerate mutase, enolase, and pyruvate kinase belonging to the glycolytic pathway. All but 6-phosphofructokinase were also among the 20 most abundant proteins in biofilm cells. The glycolytic enzymes constituted 24.6 and 15.7% of the total protein synthesized in the biofilm and planktonic cells, respectively (P = 0.02). In the biofilm cells there is therefore a redirection of the protein synthesis towards more energy-generating reactions, which supports the observation that lactic acid production was increased in S. mutans when adsorbed to hydroxyapatite (1). While enteric bacteria adsorbed to glass surfaces were also more metabolically active than the same organisms in suspension (16). In E. coli the demand for ATP controls the glycolytic flux through the cell (18), and the majority of the control of growth in E. coli resides in the cellular anabolic reactions in the cell and the fact that a central metabolic pathway can be controlled by processes outside that pathway. The stimulation of the glycolysis observed in the biofilm cells (Fig. 4; Table 2) could be due to the enhanced activity in ATP-consuming, biosynthetic reactions (Table 2). The decrease in the intracellular level of ATP would be expected to lead to an increase in glycolytic activity. One could speculate that the enhanced synthesis in glycolytic enzymes may also be a response to a higher growth rate; although if that were the case, increased growth rate would, in S. mutans, be a response to surface attachment since the planktonic and biofilm cells experienced the same conditions.
TABLE 2.
Partial list of protein spots in biofilm cells showing RRS enhanced and diminished more than 1.3-fold compared to planktonic cellsa
| Process | Spot no. | Changeb (fold) | Protein synthesis
|
|
|---|---|---|---|---|
| Enhanced | Diminished | |||
| Carbohydrate metabolism | 28 | 1.7 | Glucan 1,6-alpha-glucosidase | |
| 37a | 2.1 | NADH oxidase | ||
| 72 | 1.9 | Enolase | ||
| 73 | 1.5 | Phosphoglycerate kinase | ||
| 75 | 1.7 | Glyceraldehyde-3-phosphate dehydrogenase | ||
| 76 | 1.4 | 6-Phosphofructokinase | ||
| 78a | 1.7 | Fructose bisphosphate aldolase | ||
| 79 | 1.4 | Phosphoglycerate mutase | ||
| 108 | 1.4 | Pyruvate kinase | ||
| 985 | 1.7 | l-Lactate dehydrogenase | ||
| Protein folding | 70 | 1.6 | DnaK protein | |
| 118a | 1.4 | Trigger factor, ppiase | ||
| 310 | 1.6 | DnaK protein | ||
| Transcription or | 31 | 1.4 | DNA-directed RNA polymerase | |
| translation | 45 | 1.5 | SSU ribosomal protein S1P | |
| 77 | 1.5 | SSU ribosomal protein S2P | ||
| 86 | 1.3 | Elongation factor Ts | ||
| 91 | 1.4 | Ribosome recycling factor | ||
| Cell division | 47 | 1.8 | Minicell-associated protein Div IVA | |
| 118b | 2.0 | Cell division protein FTSZ | ||
| Amino acid biosynthesis | 42 | 1.7 | Phospho-2-dehydro-3-deoxyheptonate aldolase | |
| Antioxidation | 40 | 1.7 | Alkylhydroperoxidase | |
| 44 | 2.0 | Thiol peroxidase | ||
Data obtained from the analysis of the autoradiograms in Fig. 4.
Change calculated as the IOD% of the protein in the planktonic and biofilm cells.
FIG. 4.
Autoradiograms of 2DE protein profiles of S. mutans H7. (A) Protein expression in planktonic cells; (B) protein expression during initial stage of adhesion. The proteins indicated with filled arrowheads are enhanced and proteins indicated with open arrowheads are diminished 1.3-fold or more in biofilm cells compared to the planktonic cells. Proteins indicated with an asterisk at the base of the arrowhead were only expressed in planktonic cells (A) or were only expressed in biofilm cells (B). The gels are representatives of three independent experiments.
Glycolytic enzymes were considered, because of their highly conserved nature, to be related simply to substrate turnover (27) but more recently other functions have been ascribed to certain glycolytic enzymes. Enolase and glyceraldehyde-3-phosphate dehydrogenase in Streptococcus pyogenes are found at the cell surface and bind plasmin, plasminogen, and fibronectin, which assist the bacteria in generating an inflammatory response (28, 29). Streptococcus oralis, a member of the oral biofilm but also associated with extraoral diseases, including endocarditis and bacteremia in neutropenic patients, also has enolase and glyceraldehyde-3-phosphate dehydrogenase at its cell surface as well as two other glycolytic enzymes, phosphoglycerate kinase and fructose bisphosphate aldolase (42). The expression of these enzymes was enhanced in the biofilm cells and if these are also surface associated in S. mutans they could be involved in the adhesion process. Two other proteins, elongation factor Ts and the ribosome recycling factor, surface associated in S. oralis, exhibited enhanced expression in biofilm cells; further studies will be directed to determine whether these proteins are surface associated, and their likely function, in S. mutans. The proteins associated with protein folding, including DnaK, of which five were involved with transcription and translation, two were involved with cell division, and one was involved with amino acid biosynthesis (Table 2), were also enhanced in biofilm cells. The expression of these proteins was also enhanced in 3-day-old biofilm cells of S. mutans (34). One novel protein was discovered in biofilm cells; this protein could not be identified. Of the eight proteins exhibiting diminished protein synthesis in biofilm cells, two were identified as the antioxidants alkylhydroperoxidase and thiol peroxidase, and the other six were not identified.
We have previously demonstrated that the protein profiles of three-day-old biofilm cells of S. mutans H7 growing in a chemostat were significantly different from those of their planktonic counterparts (34). Of interest to the present study was the observation that the RRS of enzymes involved in carbohydrate catabolism was less than in the planktonic cells. Thus, during initial attachment to the surfaces, proteins associated with carbohydrate catabolism were enhanced, but as the biofilm community matured, the level of these proteins diminished. This indicates that there is a specific pattern of protein expression associated with the initial adhesion response in S. mutans and that a different pattern is observed in older biofilm cells as has been observed in both P. putida (32) and P. aeruginosa (31).
Acknowledgments
This study was supported by grants to G.S. from the KK foundation and the Medical Research Council of Sweden (K2001-24X-12266-05C). J.C.W. was supported by The Pathological Society of Great Britain and Ireland.
REFERENCES
- 1.Berry, C. W., and C. A. Henry. 1977. The effect of adsorption on the acid production of caries and non-caries-producing streptococci. J. Dent. Res. 56:1193-1200. [DOI] [PubMed] [Google Scholar]
- 2.Blomberg, A. 1997. Osmoresponsive proteins and functional assessment strategies in Saccharomyces cerevisiae. Electrophoresis 18:1429-1440. [DOI] [PubMed] [Google Scholar]
- 3.Blomberg, A., L. Blomberg, J. Norbeck, S. J. Fey, P. Mose-Larsen, P. Roepstorff, H. Degan, M. Boutry, A. Posch, and A. Görg. 1995. Interlaboratory reproducibility of yeast protein patterns analysed by immobilised pH gradient two-dimensional gel electrophoresis. Electrophoresis 16:1935-1945. [DOI] [PubMed] [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-354. [DOI] [PubMed] [Google Scholar]
- 5.Brown, M. R. W., and P. Gilbert. 1993. Sensitivity of biofilms to antimicrobial agents. J. Appl. Bacteriol. 74(Suppl.):87-97. [DOI] [PubMed] [Google Scholar]
- 6.Brözel, V., G. M. Strydom, and E. T. Cloete. 1995. A method for the study of de novo protein synthesis in Pseudomonas aeruginosa after attachment. Biofouling 8:195-201. [Google Scholar]
- 7.Costerton, J. W., K.-J. Cheng, G. G. Geesey, T. I. Ladd, J. D. Nickel, M. Dagupta, and T. J. Marrie. 1987. Bacterial biofilms in nature and disease. Annu. Rev. Microbiol. 41:435-464. [DOI] [PubMed] [Google Scholar]
- 8.Davey, E. M., and G. A. O'Toole. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64:847-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davies, D. G., and G. G. Geesey. 1995. Regulation of the alginate biosynthesis gene algC in Pseudomonas aeruginosa during biofilm development in continuous culture. Appl. Environ. Microbiol. 61:860-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fey, S. J., A. Nawrocki, M. R. Larsen, A. Gorg, P. Roepstorff, G. N. Skews, R. Williams, and P. M. Larsen. 1997. Proteome analysis of Saccharomyces cerevisiae: a methodological outline. Electrophoresis 18:1361-1372. [DOI] [PubMed] [Google Scholar]
- 11.Fletcher, M. 1991. The physiological activity of bacteria attached to solid surfaces. Adv. Microb. Physiol. 32:53-85. [DOI] [PubMed] [Google Scholar]
- 12.Goodman, A. E., and K. C. Marshall. 1995. Genetic responses of bacteria at surfaces, p. 80-98. In J. W. Costerton and H. M. Lappin-Scott (ed.), Microbial biofilms. Cambridge University Press, Cambridge, United Kingdom.
- 13.Hamilton, I. R., and G. H. Bowden. 2000. Oral microbiology, p. 466-480. In J. Lederberg (ed.), Encyclopedia of microbiology. Academic Press, San Diego, Calif.
- 14.Hamilton, I. R., and G. Svensäter. 1998. Acid-regulated proteins induced by Streptococcus mutans and other oral bacteria during acid shock. Oral Microbiol. Immunol. 13:292-300. [DOI] [PubMed] [Google Scholar]
- 15.Heilmann, C., C. Gerke, F. Perdreau-Remington, and F. Gotz. 1996. Characterization of Tn917 insertion mutants of Staphylococcus epidermidis affected in biofilm formation. Infect. Immun. 64:277-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hendricks, C. W. 1974. Sorption of heterotropic and enteric bacteria to glass surfaces in the continuous culture of river water. Appl. Microbiol. 28:572-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoyle, B. D., and J. W. Costerton. 1991. Bacterial resistance to antibiotics: the role of biofilms. Prog. Drug Res. 37:91-105. [DOI] [PubMed] [Google Scholar]
- 18.Koebmann, B. J., H. V. Westerhoff, J. L. Snoep, D. Nilsson, and P. R. Jensen. 2002. The glycolytic flux in Escherichia coli is controlled by the demand for ATP. J. Bacteriol. 184:3909-3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, Y.-H, P. C. Y. Lau, J. H. Lee, R. P. Ellen, and D. G. Cvitkovitch. 2001. Natural genetic transformation of Streptococcus mutans growing in biofilms. J. Bacteriol. 183:897-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, Y. H., and G. H. Bowden. 1994. Characteristics of accumulation of oral gram-positive bacteria on mucin-conditioned glass surfaces in a model system. Oral Microbiol. Immunol. 9:1-11. [DOI] [PubMed] [Google Scholar]
- 21.Loo, C. Y., K. Mitrakul, I. B. Voss, C. V. Hughes, and N. Ganeshkumar. 2003. Involvement of the adc operon and manganese homeostasis in Streptococcus gordonii biofilm formation. J. Bacteriol. 185:2887-2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loo, C. Y., D. A Corliss, and N. Ganeshkumar. 2000. Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. J. Bacteriol. 182:1374-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oosthuizen, M. C., B. Steyn, J. Theron, P. Cosette, D. Lindsay, A. von Holy, and V. S. Brözel. 2002. Proteomic analysis reveals differential protein expression by Bacillus cereus during biofilm formation. Appl. Environ. Microbiol. 68:2770-2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Toole, G. A., and R. Kolter. 1998. The initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 25.Otto, K., and T. Silhavy. 2002. Surface sensing and adhesion of Escherichia coli controlled by the Cpx-signaling pathway. Proc. Natl. Acad. Sci. USA 99:2287-2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Otto, K., J. Norbeck, T. Larsson, K.-A. Karlsson, and M. Hermansson. 2001. Adhesion of type 1-fimbriated Escherichia coli to abiotic surfaces leads to altered composition of outer membrane proteins. J. Bacteriol. 183:2445-2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pancholi, V. 2001. Multifunctional α-enolase: its role in disease. Cell. Mol. Life Sci. 58:902-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pancholi, V., and V. A. Fischetti. 1998. α-enolase, a novel strong plasmin(ogen) binding protein on the surface of pathogenic streptococci. J. Biol. Chem. 273:14503-14515. [DOI] [PubMed] [Google Scholar]
- 29.Pancholi, V., and V. A. Fischetti. 1992. A major surface protein on group A streptococci is a glyceraldehyde-3-phosphate-dehydrogenase with multiple binding activity. J. Exp. Med. 176:415-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pratt, L. A., and R. Kolter. 1998. Genetic analysis of Escherichia coli biofilm formation: defining the roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30:285-294. [DOI] [PubMed] [Google Scholar]
- 31.Sauer, K., A. K. Camper, G. D. Ehrlich, J. W. Costerton, and D. G. Davies. 2002. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 184:1140-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sauer, K., and A. K. Camper. 2001. Characterization of phenotypic changes in Pseudomonas putida in response to surface-associated growth. J. Bacteriol. 183:6579-6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Svensäter, G., M. Borgström, G. H. W. Bowden, and S. Edwardssson. 2003. The acid-tolerance microbiota associated with plaque from initial caries and healthy tooth surfaces. Caries Res. 37:395-403. [DOI] [PubMed] [Google Scholar]
- 34.Svensäter, G., J. Welin, J. C. Wilkins, D. Beighton, and I. R. Hamilton. 2001. Protein expression by planktonic and biofilm cells of Streptococcus mutans. FEMS Microbiol. Lett. 27:139-146. [DOI] [PubMed] [Google Scholar]
- 35.Svensäter, G., B. Sjögreen, and I. R. Hamilton. 2000. Multiple stress responses in Streptococcus mutans and the induction of general and stress-specific proteins. Microbiology 146:107-117. [DOI] [PubMed] [Google Scholar]
- 36.Svensäter, G., U.-B. Larsson, E. C. G. Greif, D. G. Cvitkovitch, and I. R. Hamilton. 1997. Acid tolerance response and survival by oral bacteria. Oral Microbiol. Immunol. 12:266-273. [DOI] [PubMed] [Google Scholar]
- 37.Wang, J., S. Lory, R. Ramphal, and S. Jin. 1996. Isolation and characterization of Pseudomonas aeruginosa genes inducible by respiratory mucus derived from cystic fibrosis patients. Mol. Microbiol. 22:1005-1012. [DOI] [PubMed] [Google Scholar]
- 38.Watnick, P. I., and R. Kolter. 1999. Biofilm, city of microbes. J. Bacteriol. 182:2675-2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Welin, J., J. C. Wilkins, D. Beighton, K. Wrzesinski, S. J. Fey, P. Mose-Larsen, I. R. Hamilton, and G. Svensäter. 2003. Effect of acid shock on protein expression by biofilm cells of Streptococcus mutans. FEMS Microbiol. Lett. 227:287-293. [DOI] [PubMed] [Google Scholar]
- 40.Wen, Z. T., and R. A. Burne. 2002. Functional genomics approach to identifying gene required for biofilm development by Streptococcus mutans. Appl. Environ. Microbiol. 68:1196-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whitaker, C. J., C. M. Klier, and P. E. Kolenbrander. 1996. Mechanisms of adhesion by oral bacteria. Annu. Rev. Microbiol. 50:513-552. [DOI] [PubMed] [Google Scholar]
- 42.Wilkins, J. C., D. Beighton, and K. A. Homer. 2003. Effect of acid shock on surface-associated proteins of Streptococcus oralis. Appl. Environ. Microbiol. 69:5290-5296. [DOI] [PMC free article] [PubMed] [Google Scholar]