Abstract
As part of a large international project for standardization of PCR (Food-PCR; www.pcr.dk), a multiplex, multiplatform, ready-to-go enrichment followed by a real-time PCR method, including an internal amplification control, was developed for detection of food-borne thermotolerant campylobacters in chickens. Chicken rinse samples were enriched in Bolton broth for 20 h, a simple and rapid (1-h) resin-based DNA extraction was performed, and DNA samples were then tested with two instrument platforms: ABI-PRISM 7700 and RotorGene 3000. The method was validated against an International Standard Organization (ISO)-based culture method by testing low, medium, and high levels of 12 spiked and 66 unspiked, presumably naturally contaminated, chicken rinse samples. In the RotorGene, a positive PCR response was detected in 40 samples of the 66. This was in complete agreement with the enriched ISO culture. The ABI-PRISM 7700 missed one culture-positive sample. Positive samples contained 102 to 107 CFU/ml after enrichment in Bolton broth. In the enriched samples a detection probability of 95% was obtained at levels of 1 × 103 and 2 × 103 CFU/ml in the RotorGene and ABI-PRISM, respectively. The amplification efficiency in both platforms was 90%, although the linear range of amplification of purified genomic DNA was 1.5 × 101 to 1 × 107 (R2 = 1.00) for the RotorGene and 103 to 107 (R2 = 0.99) for the ABI-PRISM. In RotorGene and ABI-PRISM the levels of precision of detection as determined by standard deviation (coefficients of variation) of 6-carboxyfluorescein (FAM) threshold cycle (Ct) values were 0.184 to 0.417 (0.65 to 2.57%) and 0.119 to 0.421 (0.59 to 1.82%), respectively. The results showed a correlation (R2) of 0.94 between the target FAM Ct values and CFU per milliliter of enriched naturally contaminated chicken samples, which indicates PCR's additional potential as a tool for quantitative risk assessment. Signal from the internal amplification control was detected in all culture-negative samples (VIC Ct: 23.1 to 28.1). The method will be taken further and validated in an international collaborative trial with regard to standardization.
Thermotolerant campylobacters especially Campylobacter jejuni and C. coli are recognized worldwide as a leading cause of human food-borne infections (19). They are zoonotic bacteria, with many wild and domesticated animals serving as potential reservoirs. Sources of sporadic campylobacteriosis are seldom identified, but water, pets, and especially poultry products are known to be associated with the illness (14). There is a need for rapid detection methods during chicken production, in order to prevent distribution of infected chicken products to consumers. Rapid methods, as currently practiced in Denmark, can be used on-site to quickly identify infected flocks prior to and after slaughter and to provide the consumers with Campylobacter-free chickens. The implementation of the so-called “strategic slaughter” seems to have contributed to the recent decline of campylobacteriosis in Denmark (www.dfvf.dk). In addition, quantitative detection tools are needed for estimation of the level of Campylobacter in flocks and meat products, as part of risk assessment studies.
A limited number of real-time PCR methods have been reported for the specific detection of C. jejuni (5, 15, 18, 20). The present study deals with detection of all food-borne thermotolerant campylobacters (C. jejuni, C. coli, and C. lari), which would prepare laboratories for an unforeseen shift of prevalence in poultry from C. jejuni to currently less frequently isolated species. In addition we have included an internal amplification control (IAC), which is mandatory according to the draft international standard (4). Furthermore, the real-time PCR method reported here is based on a specific and robust conventional gel-based PCR method validated in an international collaborative trial (10, 11). Closed-tube real-time PCR assays can be more specific than gel-based PCR due to the inclusion of a target sequence-recognizing probe. They can also be faster and more sensitive and involve less manual handling than conventional PCR. Finally, the risk of carryover contamination is substantially reduced, which makes them more suitable for implementation in end use laboratories.
To our knowledge, this is the first study to report an enrichment multiplatform real-time PCR, including an IAC, which deals with the detection of all three food-borne thermotolerant campylobacters and to provide data to support its potential use as a quantitative tool in risk assessment studies.
MATERIALS AND METHODS
Spiked and naturally contaminated samples. (i) Spiked samples.
The PCR method was validated on 12 frozen slaughtered chickens (Danpo A/S, Aars, Denmark), purchased at local retailers as Campylobacter free. Initial suspensions of chicken carcass rinse were prepared as recommended in the ISO/CD 6887-2 protocol (1). A whole thawed chicken was placed in a large sterile plastic bag, 500 ml of physiological saline (0.9% NaCl) was added, and the bag was closed and shaken gently by hand for a minimum of 60 s (8). Then 25 ml of chicken carcass rinse was transferred to 225 ml of Bolton broth (BB) prepared according to the recommendations of the Bacteriological Analytical Manual Online (6). The broth samples were inoculated with C. jejuni strain 1677, which represents a genotype frequently isolated from chickens in Denmark. Three broth samples were not inoculated; three were inoculated with approximately 1 to 10 CFU/250 ml, three were inoculated with approximately 10 to 100 CFU/250 ml, and three were inoculated with approximately 100 to 1,000 CFU/250 ml. They were incubated at 42.0 ± 1.0°C under microaerobic conditions (6% O2, 7% CO2, 7% H2, and 80% N2) for 20 h. Following the enrichment, the concentrations of C. jejuni 1677 in the spiked samples were approximately 0, 104, 105, and 106 CFU/ml. After 20 h of enrichment, 1-ml samples were drawn for PCR and stored at −80°C. To verify that the chicken carcass rinse was initially Campylobacter free, prespiking samples were drawn for traditional microbiological analysis before inoculation. The microbiological analysis was conducted in accordance with recommendations of the International Standard Organization (3) and the Nordic Committee on Food Analysis (2). The CFU levels postenrichment were determined by plating 100 μl of a 10-fold dilution series (10−1, 10−2, 10−3, 10−4, 10−5, 10−6, and 10−7) onto modified charcoal cefoperazone deoxycholate agar (Oxoid, Basingstoke, United Kingdom) and Preston agar (Oxoid). The agar plates were incubated for 48 h at 42.0 ± 1.0°C under microaerobic conditions. Five typical colonies from each plate were verified by applying the following tests: Gram reaction by the KOH 3% method, oxidase test, catalase test, and microscopic inspection of motility and morphology (8).
(ii) Naturally contaminated samples.
Sixty-six chickens, including 26 Danish and 40 French free-range or intensively reared chickens, were purchased on six different occasions at local retailers. The chicken carcass rinses were prepared as described above. Twenty-five milliliters of the chicken carcass rinse was transferred to 225 ml of BB and enriched for 20 h, after which 1-ml samples were drawn for PCR and stored at −80°C. Microbiological analysis was conducted as described in the section for spiked samples.
Preparation of template DNA.
A simple and rapid resin-based sample treatment was performed essentially as described by Malorny et al. (13). Briefly, the 1-ml frozen enriched BB samples were thawed and centrifuged for 5 min at 10,000 × g and 4°C. The pellets were resuspended in 300 μl of 6% Chelex 100 resin suspension (Bio-Rad Laboratories, Hercules, Calif.) and incubated for 20 min in a 56°C water bath. The samples were vortexed for 10 s and incubated in a 95°C water bath for 8 min, followed by immediate chilling on ice. The samples were centrifuged for 5 min at 14,000 × g and 4°C, and 5 μl of the supernatants was used as the template in the PCR. The spiked and naturally contaminated samples were run in duplicate in an ABI-PRISM 7700 sequence detector (Applied Biosystems, Foster City, Calif.) and a RotorGene 3000 (Corbett Research, Mortlake, Australia). Preparation of the IAC template (hemorrhagic septicemia virus mRNA for envelope protein; GenBank accession no. X66134) was performed as previously described (10).
Campylobacter real-time PCR.
The real-time PCR was performed in the ABI-PRISM using 0.2-ml thermostrips (ABgene House, Surrey, United Kingdom) or MicroAmp Optical 96-well reaction plates (Applied Biosystems) closed with MicroAmp Optical caps (Applied Biosystems) and in the RotorGene using 0.2-ml capped tubes (Corbett Research). The 25-μl real-time PCR mixture contained 1× PCR buffer for Tth DNA polymerase (Roche A/S, Hvidovre, Denmark), 1 U of Tth DNA polymerase (Roche A/S), 0.4 mM deoxynucleoside triphosphate mixture (Amersham Pharmacia Biotech, Buckinghamshire, United Kingdom), 0.44 μM forward primer 5′ CTG CTT AAC ACA AGT TGA GTA GG 3′, 0.48 μM reverse primer 5′ TTC CTT AGG TAC CGT CAG AA 3′ (DNA Technology, Århus, Denmark; C. jejuni 16S rRNA; GenBank accession no. Y19244), 2.5 mM MgCl2 (Applied Biosystems), 30 μg of bovine serum albumin (BSA) for chicken samples and 5 μg of BSA for pure DNA (Roche A/S), 20 nM target Campylobacter probe labeled with 6-carboxyfluorescein (FAM; reporter dye) and 6-carboxytetramethylrhodamine (TAMRA; quencher dye) (5′ FAM-TGT CAT CCT CCA CGC GGC GTT GCT GC-TAMRA 3′; DNA Technology), 50 nM IAC probe (5′ VIC-TTC ATG AGG ACA CCT GAG TTG A-TAMRA 3′; Applied Biosystems), 5 × 103 copies of IAC (124 bp), and 5 μl of DNA sample. The cycle profile was as follows: initial denaturation at 95°C for 3 min, followed by 40 cycles of 95°C for 15 s and 58°C for 60 s. Fluorescence measurements were obtained online and analyzed on the ABI-PRISM with the SDS software (version 1.7a; Applied Biosystems) and on the RotorGene with the version 4.6 software (Corbett Research). Note that no passive reference fluorescence was assigned on the ABI-PRISM, since the PCR mixture did not contain any 6-carboxy-X-rhodamine. The real-time PCR method is based on a specific and robust conventional gel-based PCR method validated in an international collaborative trial. No attempt was made to analyze the specificity of the test since the primers used previously have been validated against 150 related and nonrelated species (10, 11).
Determination of cutoff.
The FAM threshold was assigned to a fixed value intersecting the amplification curves in the linear region of the logarithmic plot (a normalized reporter signal, ΔRn, of 230 for the ABI-PRISM and a normalized fluorescence of 0.02 for the RotorGene). The fluorescence was normalized to the background signal, as no 6-carboxy-X-rhodamine was present in the master mixture. Any sample showing a fluorescence signal above this value was regarded as positive. The VIC threshold was assigned a normalized reporter signal (ΔRn) of 100 for the ABI-PRISM and a normalized fluorescence of 0.01 for the RotorGene. To exclude possible weak signals from samples or nontemplate controls with a slight end point drift upward, a 10% quantitation setting on the RotorGene was employed, meaning that the software would reject any reaction not increasing more than 10% in fluorescence relative to the samples producing the highest fluorescence increase in the real-time run.
Detection probability.
The probability of detection was examined essentially as described by Knutsson et al. (9). Six chicken carcass rinse samples containing approximately 101 to 106 Campylobacter CFU/ml were tested. Five microliters of extracted DNA from each sample was added to five separate real-time PCR tubes and was run as described above on both the ABI-PRISM and the RotorGene. This experiment was repeated five times in the same PCR run on six separate occasions, resulting in a total of 30 responses for each chicken sample. Master mixture was prepared on each of the six occasions and divided among five PCR tubes. DNA was added to the tubes individually. The real-time PCR response was regarded as binary, giving either a positive or a negative signal, according to the cutoff criteria described earlier. The detection probability in spiked chicken carcass rinse samples was obtained by plotting the relative percentage of positive PCR responses against the cell concentration in the samples tested. A sigmoid line fitting was performed with ORIGIN, version 4.0 (Microcal Software, Northampton, Mass.).
Precision of detection.
Chicken carcass rinse samples spiked with 0, 1 to 10, 10 to 100, and 100 to 1,000 CFU/250 ml, in the presence of 5 × 103 copies of the IAC, were used to establish the precision of detection. Five microliters of DNA extracted from three spiked samples from each level was added to 10 separate real-time PCR tubes, and samples were run as described above on both the ABI-PRISM and the RotorGene. For statistical analysis the results for the precision of detection were plotted into the Online Calculator for Standard Deviation (www.compute.uwlax.edu/stats/), and the standard deviation (SD), sample variance (s2), and the coefficient of variation (CV) were computed.
Amplification efficiency and linear range.
The amplification efficiency and linear range of the real-time PCR method were determined in quadruple by using purified DNA from plate colonies in the concentration range of 5 × 100 to 1 × 107 copies of genomic DNA from type strain C. jejuni CCUG 11284. The analysis was performed on both the ABI-PRISM and the RotorGene. By plotting the number of genomic DNA copies/25 μl of PCR sample against the threshold cycle value (Ct; the fractional PCR cycle at which the fluorescence signal of a sample rises above the determined baseline signal), a linear relationship was formed. The slope of this curve was used to determine the amplification efficiency from the equation amplification efficiency = 10−1/slope − 1. The DNA was extracted with a DNeasy tissue kit (QIAGEN GmbH, Hilden, Germany) according to the manufacturer's instructions. The DNA concentration was determined by PicoGreen quantitation of double-stranded DNA with a TD-360 minifluorometer (Turner Designs, Sunnyvale, Calif.) (7). The number of genomic copies of the purified DNA was calculated from the equation m = n(1.013 × 10−21), where m is the mass, and n is the number of base pairs in the genome (12). The genomic sequence of C. jejuni NCTC11168 has been determined by Parkhill et al. (16) to be 1,641,481 bp. Entering this in the above equation yields the result that one C. jejuni genome weighs approximately 1.7 fg. The number of genomic DNA copies/milliliter was adjusted with 1× Tris-EDTA buffer-0.1 M EDTA.
RESULTS
Amplification efficiency and linear range.
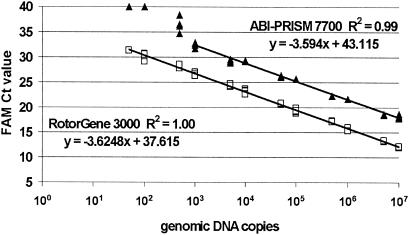
The standard curves produced directly by software for both platforms are shown in Fig. 1. The linear range of amplification for the PCR method was between 5 × 101 and 1 × 107 copies of purified genomic DNA from C. jejuni CCUG 11284 in the RotorGene and 103 to 107 copies in the ABI-PRISM (Fig. 2). The amplification efficiency was computed from the slope of the linear relationship (R2 = 1.00) and was 90.6% for the RotorGene. In the ABI-PRISM the results were 89.9% for amplification efficiency and a linear relationship of R2 = 0.99.
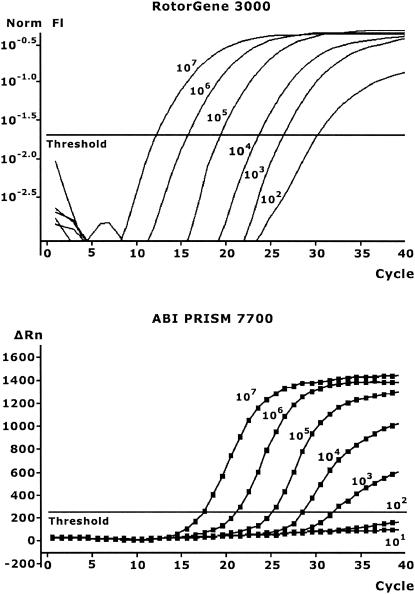
FIG. 1.
Standard curves produced from purified DNA in the range of 101 to 107 copies of genomic DNA from type strain C. jejuni CCUG 11284 on the ABI-PRISM 7700 and the RotorGene 3000. The lowest level, 101 copies, is not shown for the RotorGene since it was eliminated by the quantification settings of 10%. Norm fl, normalized fluorescence.
FIG. 2.
The linear range of the real-time PCR method when detecting purified DNA from C. jejuni CCUG 11284 on the RotorGene 3000 (5 × 101 to 1 × 107 copies; □) and the ABI-PRISM 7700 (103 to 107 copies; ▴). A real-time PCR sample containing below 5 × 101 genomic DNA copies gave either no positive response or was removed by the detection software because of the 10% quantification correction on the RotorGene. Levels above 107 genomic DNA copies were not tested.
Detection probability.
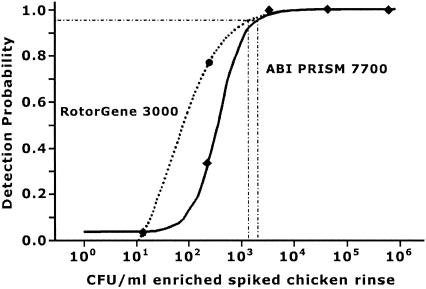
Figure 3 shows the probability of detecting C. jejuni 1677 in chicken carcass rinse samples by the real-time PCR method. On the ABI-PRISM the detection probability for 103 CFU/ml was 85% and for 4 × 103 CFU/ml it was 100%. On the RotorGene the probability of detecting 103 CFU/ml was 90%, while it was 100% for detecting 4 × 103 CFU/ml. The levels of detection at 95% probability were determined to be 2 × 103 CFU/ml for the ABI-PRISM and 103 CFU/ml for the RotorGene.
FIG. 3.
Detection probability on ABI-PRISM 7700 and RotorGene 3000 for thermotolerant campylobacters with spiked chicken rinse samples containing 101 to 106 CFU/ml. Thirty responses for each sample were generated on six separate occasions. Thick dotted line, RotorGene 3000; solid line, ABI-PRISM 7700; intersecting thin dotted lines, 95% detection probability. The detection probability was determined in the presence of 5 × 103 copies of the IAC.
Precision of detection in artificially contaminated chicken rinse samples.
To evaluate the precision of detection with the two instrument platforms, a range of identical chicken carcass rinse samples were run simultaneously in the same real-time PCR. As shown in Table 1, the SD, s2, and CV were low for both the ABI-PRISM and the RotorGene, reflecting a high degree of precision of detection. For the ABI-PRISM the SD and CV ranged from 0.119 to 0.421 and 0.59 to 1.82%, respectively, and for the RotorGene the SD and CV ranged from 0.184 to 0.417 and 0.65 to 2.57%, respectively.
TABLE 1.
Results from the determination of the precision of detection of C. jejuni 1677 in spiked chicken rinse samplesa
| Spiking level (CFU/250 ml) | ABI-PRISM 7700
|
RotorGene 3000
|
||||||
|---|---|---|---|---|---|---|---|---|
| Mean Ct value | SD | s2 | CV (%) | Mean Ct value | SD | s2 | CV (%) | |
| 0 | 40 | 0 | 0 | 0 | 40 | 0 | 0 | 0 |
| 0 | 40 | 0 | 0 | 0 | 40 | 0 | 0 | 0 |
| 0 | 40 | 0 | 0 | 0 | 40 | 0 | 0 | 0 |
| 1-10 | 27.89 | 0.290 | 0.084 | 1.04 | 22.61 | 0.365 | 0.133 | 1.61 |
| 1-10 | 26.42 | 0.388 | 0.151 | 1.60 | 20.64 | 0.293 | 0.086 | 1.42 |
| 1-10 | 27.33 | 0.421 | 0.178 | 1.54 | 21.09 | 0.283 | 0.080 | 1.34 |
| 10-100 | 21.50 | 0.148 | 0.022 | 0.69 | 14.85 | 0.184 | 0.034 | 1.24 |
| 10-100 | 21.94 | 0.223 | 0.050 | 1.02 | 16.29 | 0.417 | 0.175 | 2.57 |
| 10-100 | 22.61 | 0.261 | 0.068 | 1.15 | 16.51 | 0.371 | 0.138 | 2.25 |
| 100-1,000 | 19.45 | 0.219 | 0.048 | 1.12 | 13.34 | 0.328 | 0.108 | 2.46 |
| 100-1,000 | 20.24 | 0.119 | 0.014 | 0.59 | 13.66 | 0.286 | 0.082 | 2.09 |
| 100-1,000 | 19.64 | 0.358 | 0.128 | 1.82 | 13.46 | 0.296 | 0.088 | 0.65 |
Samples contained 0, 104, 105, and 106 CFU/ml after enrichment.
Spiked and naturally contaminated samples.
The samples spiked with 0, 1 to 10, 10 to 100, and 100 to 1,000 CFU/250 ml reached levels of approximately 0, 104, 105, and 106 CFU/ml, respectively, after 20 h of enrichment. PCR detection was possible on both instruments at all spiking levels. However, in general the RotorGene gave lower FAM Ct values than the ABI-PRISM. Based on all spiking levels, the average FAM Ct value was 6 U lower on RotorGene (Table 2).
TABLE 2.
Results of real-time PCR for detection of food-borne thermotolerant campylobacters on spiked chicken rinse samples tested in duplicate on ABI-PRISM 7700 and RotorGene 3000a
| Level of spiking (CFU/250 ml) | Ct value for:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| ABI-PRISM 7700
|
RotorGene 3000
|
|||||||
| Duplicate 1
|
Duplicate 2
|
Duplicate 1
|
Duplicate 2
|
|||||
| FAM | VIC | FAM | VIC | FAM | VIC | FAM | VIC | |
| 0 | 40 | 26.5 | 40 | 27.1 | 40 | 27.2 | 40 | 26.9 |
| 0 | 40 | 27.0 | 40 | 27.0 | 40 | 28.4 | 40 | 26.6 |
| 0 | 40 | 26.9 | 40 | 26.7 | 40 | 28.3 | 40 | 26.1 |
| 1-10 | 27.5 | 27.2 | 27.3 | 26.8 | 21.7 | 26.9 | 22.2 | 26.2 |
| 1-10 | 26.6 | 29.3 | 25.4 | 26.9 | 20.1 | 26.1 | 18.9 | 26.7 |
| 1-10 | 26.7 | 27.8 | 25.3 | 26.6 | 19.8 | 26.6 | 20.7 | 25.4 |
| 10-100 | 21.3 | 29.4 | 21.1 | 26.8 | 15.2 | 26.3 | 15.0 | 25.1 |
| 10-100 | 22.2 | 28.9 | 21.2 | 27.6 | 14.7 | 26.0 | 15.5 | 25.0 |
| 10-100 | 22.5 | 28.1 | 21.2 | 26.8 | 16.0 | 26.1 | 15.1 | 25.6 |
| 100-1,000 | 20.7 | 31.7 | 19.0 | 28.1 | 12.6 | 27.0 | 12.6 | 25.7 |
| 100-1,000 | 19.5 | 29.7 | 18.6 | 28.4 | 13.0 | 26.3 | 13.1 | 25.5 |
| 100-1,000 | 20.0 | 27.7 | 19.0 | 28.5 | 13.3 | 26.8 | 12.7 | 28.3 |
After enrichment the spiked samples contained approximately 0, 104, 105, and 106 CFU/ml.
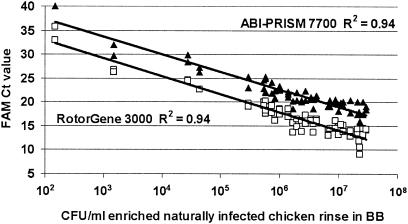
Thermotolerant campylobacters were found in 40 out of the 66 presumably naturally contaminated chicken rinse samples by the culture-based method. The same 40 culture-positive samples were also found positive with the RotorGene, whereas the ABI-PRISM gave 39 positive responses. The only sample not detected by the ABI-PRISM contained 1.5 × 102 CFU of thermotolerant campylobacters/ml. There was 100% agreement between the duplicate responses from the 66 naturally contaminated samples. Figure 4 illustrates the linear correlation between the Ct values for the target probe and the number of CFU in naturally contaminated chicken samples enriched in BB in both platforms (data available at www.pcr.dk/fig.4.doc). Comparison of real-time PCR with the culture-based method, as applied to the 66 naturally contaminated samples, indicated no difference between the two methods, independent of the real-time platform used. The 26 culture Campylobacter-negative samples all showed a FAM Ct value of 40, while the IAC (VIC) signal was detected in all negative samples. VIC Ct values ranged from 26.9 to 28.1 for the ABI-PRISM and 23.1 to 27.9 for the RotorGene.
FIG. 4.
Linear correlation between target Ct values and CFU in naturally contaminated chicken samples enriched in BB. Samples were tested in duplicate on ABI-PRISM 7700 (▴) and RotorGene 3000 (□).
Analytical accuracy.
No attempt was made to analyze the specificity of the tests, since the primers used previously have been validated against 150 related and nonrelated species (11). A complete “ready-to-go mixture” was stored at −20°C up to 1 month and tested on several occasions against freshly prepared master mixture. DNA material from spiked chicken rinse samples was added to the master mixtures, and the resulting FAM Ct values showed no difference throughout the month (data not shown). Furthermore a protection against carryover contamination was achieved by exchanging 2.5 mM dTTP in the nucleotide mixture with 5 mM dUTP (12.5 mM nucleotide mixture including dUTP; Applied Biosystems) and 1/4 U of heat-labile uracil-DNA glycosylase (Invitrogen A/S, Tåstrup, Denmark). For genomic DNA this resulted in FAM Ct values that were between 1 and 3 cycles lower (stronger signal). However, no FAM response was detected for enriched naturally contaminated chicken samples containing less than 4.3 × 104 CFU/ml of BB. Adding 7% (vol/vol) glycerol (Merck, Darmstadt, Germany) to the PCR mixture reestablished the FAM Ct to values identical to levels seen with dTTP (data not shown).
DISCUSSION
In general, the main advantage of PCR over culture methods is its potential for rapid screening of negative samples. Positive samples, however, still need to be verified by culture to obtain isolates for subtyping, antimicrobial resistance testing, and so on. In contrast to the end point detection by conventional PCR, the real-time PCR provides the opportunity to monitor the accumulation of PCR product during amplification. By generating standard curves from Ct values from samples of known DNA concentrations, information on the linear range of amplification and amplification efficiency of the assay can be determined (17). These features are important when optimizing the amplification, studying PCR inhibition, and investigating the effect of pre-PCR processing. If a real-time PCR method is intended for quantitative measurements, it is a prerequisite to obtain information on both linear range and amplification efficiency in order to ensure correct quantification. In the present study the amplification efficiency of the real-time PCR assay on the ABI-PRISM (89.9%) was similar to that on the RotorGene (90.6%). However, the linear range of amplification was wider on the RotorGene, beginning at 5 × 101 copies of genomic DNA compared to the ABI-PRISM linear range, beginning at 103 copies. The PCR results (Fig. 4) indicate that the method described may be used to estimate the level of Campylobacter in flocks or meat products, although exact quantification requires use of PCR directly on samples without any preenrichment. However, the available technology for sample treatment and DNA extraction does not provide us with a detection limit of 1 CFU per 25-g sample, which is required by international standards.
The limitation in current PCR detection is partly due to loss of Campylobacter during the DNA extraction step and partly due to the low sample volume (5 μl) tested. The only publication available on quantitative C. jejuni PCR testing of nonenriched chicken samples reports the use of a commercial DNA extraction kit on highly contaminated chicken samples from the Chinese market, containing >105 CFU of Campylobacter/ml before enrichment (20). The Campylobacter contamination level of chicken flocks and chicken products in countries with more-hygienic production methods is known to be much lower, making enrichment of most samples necessary to obtain PCR-detectable results. Cheng and Griffiths have previously observed that a minimum of 100 Campylobacter CFU per ml is necessary for PCR detection by Triton X-100 DNA extraction. Therefore they used a 12-h enrichment step before PCR detection, although they did not test any naturally contaminated samples (5).
The enrichment followed by real-time PCR data presented here indicates that even chicken flocks with low levels of infection would be detected, in particular by the RotorGene platform. The PCR-based method showed the same responses as the culture-based method, indicating that it is as good as the existing “gold standard.” Testing the spiked and naturally contaminated samples on RotorGene resulted in lower FAM Ct values (stronger signal) and produced one more positive result than the ABI-PRISM, reflecting a greater sensitivity of this platform.
The terms sensitivity and detection limit are often used interchangeably in diagnostic PCR and real-time PCR. However, both sensitivity and detection limit can be considerably affected by several factors, including presence of inhibitory substances and quality of DNA. Although a given concentration of cells can be detected by a real-time PCR, it will not necessarily be detected in every real-time PCR. For this reason, the detection limit of any PCR and real-time PCR should always be assessed in association with the detection probability to illustrate the reliability of the assay (17).
In conclusion, the real-time PCR method described seems to be sensitive and robust both for detection and quantification in enriched samples. The method is intended for further validation in an international collaborative trial with regard to standardization.
Acknowledgments
This work was supported in part by EC grant no. QLK1-CT-1999-00226 and by Danish Directorate for Food, Fisheries and Agri Business grant no. 3401-66-03-5.
We thank Mette Skafte Thomsen, and Stefan Jensen for excellent technical assistance, Burkhard Malorny for assistance with preparation of Fig. 3, and Claire Harrington for critical discussions during preparation of the manuscript.
REFERENCES
- 1.Anonymous. 2000. Microbiology of food and animal feeding stuffs. Preparation of test samples, initial suspension and decimal dilution for microbiological examination. Part 2. Specific rules for the preparation of the initial suspension and decimal dilutions of meat and meat products. International Standard Organisation ISO/CD 6887-2. AFNOR, Paris, France.
- 2.Anonymous. 2002. Microbiology of food and animal feeding stuffs. Horizontal method for the detection and enumeration of Campylobacter growing at 41.5°C. Part 1. Detection method. Document ISO/TC 34/SC 9 N 553, revision 2. Result of voting on ISO/CD 10272-1. International Standard Organisation, Geneva, Switzerland.
- 3.Anonymous. 2002. Campylobacter. Detection and enumeration of thermotolerant Campylobacter in foods. Draft version. Nordic Committee on Food Analysis, Oslo, Norway.
- 4.Anonymous. 2002. Microbiology of food and animal feeding stuffs. Polymerase chain reaction (PCR) for the detection of foodborne pathogens. General method specific requirements. Draft International Standard ISO/DIS 22174. DIN, Berlin, Germany.
- 5.Cheng, Z., and M. W. Griffiths. 2003. Rapid detection of Campylobacter jejuni in chicken rinse water by melting peak analysis of amplicons in real-time polymerase chain reaction. J. Food Prot. 66:1343-1352. [DOI] [PubMed] [Google Scholar]
- 6.Hunt, J. M., C. Abeyta, and T. Tran. January 2001, posting date. Campylobacter. In Bacteriological analytical manual online, 8th ed., revision A. [Online.] http://www.cfsan.fda.gov/∼ebam/bam-toc.html. U.S. Food and Drug Administration Washington, D.C.
- 7.Jensen, A. N., and J. Hoorfar. 2003. Optimal purification and sensitive quantification of DNA from fecal samples. J. Rapid Methods Auto. Microbiol. 10:231-244. [Google Scholar]
- 8.Josefsen, M. H., P. S. Lübeck, B. Aalbæk, and J. Hoorfar. 2002. Preston and Park-Sanders protocols adapted for semi-quantitative isolation of thermotolerant campylobacter from chicken rinse. Int. J. Food Microbiol. 80:177-183. [DOI] [PubMed] [Google Scholar]
- 9.Knutsson, R., Y. Blixt, H. Grage, E. Borch, and P. Rådström. 2002. Evaluation of selective enrichment PCR procedures for Yersinia enterocolitica. Int. J. Food Microbiol. 73:35-46. [DOI] [PubMed] [Google Scholar]
- 10.Lübeck, P. S. L., P. Wolffs, S. L. W. On, P. Ahrens, P. Rådström, and J. Hoorfar. 2003. Toward an international standard for PCR-based detection of food-borne thermotolerant campylobacters. Part 1. Assay development and analytical validation. Appl. Environ. Microbiol. 69:5664-5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lübeck, P. S. L., N. Cook, M. Wagner, P. Fach, and J. Hoorfar. 2003. Toward an international standard for PCR-based detection of food-borne thermotolerant campylobacters. Part 2. Validation of the PCR assay in a multicenter collaborative trial. Appl. Environ. Microbiol. 69:5670-5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malorny, B., J. Hoorfar, C. Bunge, and R. Helmuth. 2003. Multicenter validation of the analytical accuracy of Salmonella PCR: towards an international standard. Appl. Environ. Microbiol. 69:290-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malorny, B., J. Hoorfar, M. Hugas, A. Heuvelink, P. Fach, L. Ellerbroek, C. Bunge, C. Dorn, and R. Helmut. 2003. Inter-laboratory diagnostic accuracy of a Salmonella-specific PCR-based method. Int. J. Food Microbiol. 89:241-249. [DOI] [PubMed] [Google Scholar]
- 14.Nachamkin, I., H. Ung, and M. Li. 2002. Increasing fluoroquinolone resistance in Campylobacter jejuni, Pennsylvania, USA, 1982-2001. Emerg. Infect. Dis. 8:1501-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nogva, H. K., A. Bergh, A. Holck, and K. Rudi. 2000. Application of the 5′-nuclease PCR assay in evaluation and development of methods for quantitative detection of Campylobacter jejuni. Appl. Environ. Microbiol. 66:4029-4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M.-A. Rajandream, K. M. Rutherford, A. H. M. van Vliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 17.Rådström, P., C. Löfstöm, M. Lövenklev, R. Knutsson, and P. Wolffs. 2003. Strategies for overcoming PCR inhibition, p. 149-161. In C. W. Dieffenbach and G. S. Dveksler (ed.), PCR primer: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 18.Sails, A. D., A. J. Fox, F. J. Bolton, D. R. A. Wareing, and D. L. A. Greenway. 2003. A real-time PCR assay for the detection of Campylobacter jejuni in foods after enrichment culture. Appl. Environ. Microbiol. 69:1383-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Solomon, E. B., and D. G. Hoover. 1999. Campylobacter jejuni: a bacterial paradox. J. Food Safety 19:121-136. [Google Scholar]
- 20.Yang, C., Y. Jiang, K. Huang, C. Zhu, and Y. Yin. 2003. Application of real-time PCR for quantitative detection of Campylobacter jejuni in poultry, milk and environmental water. FEMS Immnol. Med. Microbiol. 38:265-271. [DOI] [PubMed] [Google Scholar]