Abstract
Recombinant DNA technologies enable the direct isolation and expression of novel genes from biotopes containing complex consortia of uncultured microorganisms. In this study, genomic libraries were constructed from microbial DNA isolated from insect intestinal tracts from the orders Isoptera (termites) and Lepidoptera (moths). Using a targeted functional assay, these environmental DNA libraries were screened for genes that encode proteins with xylanase activity. Several novel xylanase enzymes with unusual primary sequences and novel domains of unknown function were discovered. Phylogenetic analysis demonstrated remarkable distance between the sequences of these enzymes and other known xylanases. Biochemical analysis confirmed that these enzymes are true xylanases, which catalyze the hydrolysis of a variety of substituted β-1,4-linked xylose oligomeric and polymeric substrates and produce unique hydrolysis products. From detailed polyacrylamide carbohydrate electrophoresis analysis of substrate cleavage patterns, the xylan polymer binding sites of these enzymes are proposed.
The arthropod gut is a differentiated organ harboring a complex biotope comprising both resident and transient members from protozoal, bacterial, and archaeal genera. Many of these organisms are symbionts that contribute in a concerted way to a complex chemical cycle sustaining the metabolic competence of the host. These symbiotic relationships help to define the metabolic traits of the insect, contributing to efficient sharing of the derived nutrients. In addition to carbon metabolism, data have demonstrated that the microbial consortia contribute to nitrogen cycling, methano- and acetogenesis, and prevention of foreign microbial pathogenesis (29).
A complete understanding of the complexities of the gut biotopes is lacking, however, due to the difficulties encountered in culturing the myriad of contributing microbes and in describing the physiologies of the individual species. Studies aimed at a description of microbial complexity have recently been undertaken in termites by using 16S ribosomal DNA analysis and have shown that a number of unique lineages of microorganism inhabit the hindgut (28). These data support the idea that the insect gut represents a contained biome wherein unique species and chemistries evolve.
Polysaccharide hydrolysis is a key element in insect nutrition. Since the diet of most arthropods comprises primarily plant matter, digestion of the structural polysaccharides cellulose and hemicellulose is essential for energy metabolism and the ability to obtain carbon from these sources contributes materially to the success of the order. These polysaccharides are resistant to degradation, and the insects themselves do not secrete all of the digestive enzymes to hydrolyze β-linkages in the polymer. Rather, much of the hydrolysis of these polysaccharides is carried out by enzymes produced by the microbial symbionts (4, 5, 36, 41).
Hemicellulose consists primarily of xylan and it is the second most abundant polymer type in plant material after cellulose. Xylan is made up of a main chain of β-1,4-linked xylopyranose residues that is most often replaced by α-linked units of arabinofuranose and methylglucuronic acid. When ingested by the insect, this complex polysaccharide must be hydrolyzed to sugar monomers to realize metabolic input. Hemicellulolytic biomass degradation and recapture of carbon in nature result from the concerted action of a number of hydrolytic enzymes that sequentially reduce the sugar polymer to smaller metabolizable units. The main enzyme in the progression of xylan breakdown is an endo-1,4-β-xylanase that attacks the nonhydrolyzed polymer. This enzyme class is of particular interest because of its role in the initial steps of polymer breakdown and its potential for use in industrial biomass utilization processes. Endo-acting polymer-hydrolyzing enzymes like xylanases have binding site arrangements wherein the polymer binds to a set of subsites and cleavage occurs in a position-specific manner. This positional specificity, analogous to that of proteases, leads to specific polymer hydrolysis patterns. Enzymes derived from unusual biotopes, because of extant physical conditions or specificities, may have attractive characteristics for targeted industrial use.
Recent advances in direct DNA cloning methodology have enabled the comprehensive expression of genes from the majority of genomes collected from complex consortia (31, 33-35). Direct DNA extraction and generation of environmental DNA (eDNA) libraries provide a format for high-throughput expression screening for targeted activities and capture of unique enzymes. These new methods have resulted in the discovery of thousands of new enzymes with novel sequences and often with novel activities, stabilities, and operational optima. In this study, direct DNA extraction from insect guts was used to target the discovery of new xylanases with a view to understand their evolutionary relationships to other known xylanases and to explore their potential for application in industrial hemicellulose degradation.
MATERIALS AND METHODS
Materials.
Reagents used for insect extraction, p-nitrophenyl-linked substrates, xylose, xylan from beechwood, and urea were purchased from Sigma-Aldrich Corp., St. Louis, Mo. Polysaccharide substrates (arabinoxylan, azo dye-linked xylan, β-1-3, β-1-4 glucan, and carboxymethylcellulose) and oligoxylans were purchased from Megazyme Corp. Wicklow, Ireland. The fluorescent probe, 8-aminonaphthalene-1,3,6-trisulfonic acid (ANTS) was purchased from Molecular Probes (Leiden, The Netherlands). Polyacrylamide containing a 29:1 ratio of acrylamide to N,N′-methylenebisacrylamide was obtained from Bio-Rad (Hertfordshire, United Kingdom). The cloning vector pASK-IBA2 was obtained from IBA (Goettingen, Germany), and the pET101/D-TOPO vector and E. coli strain BL21 Star were purchased from Invitrogen (Carlsbad, Calif.).
Insect collection.
Insects were collected under permit 215.980 FAU (Costa Rica). No voucher specimens were collected. Approximately 100 termites (family Termitidae, subfamily Nasutitermitidae) were collected from an aerial nest, located in El Rodeo Protected Zone, Costa Rica, a transitional wet forest-to-premountain life zone (20). Two different sets of samples of lepidopterans (family Saturniidae) were collected in the Barra Honda National Park (Costa Rica), a transitional dry tropical forest according to Holdridge classification of life zones (20). The first set of samples was collected by using light traps during the night and consisted of 15 large adult moths belonging to different genera. The second set of samples consisted of 6 lepidopteran larvae, collected while feeding on food plants during the day. The three sample sets were placed separately in plastic bags, kept at 4°C during transportation, and brought alive to the laboratory.
Insect evisceration and microbial DNA extraction.
Intestinal tracts were carefully dissected for each set of samples, and a pool for each set was processed as follows. In some cases (for the moth samples in particular), small amounts of surrounding tissue were extracted with the intestinal tract. As such, a trace of exogenous microbial DNA may have been extracted also. The gut samples were homogenized in 6.0 ml of phosphate-buffered saline (PBS) (8-g/liter NaCl, 0.2-g/liter KCl, 1.44-g/liter Na2HPO4, 0.24-g/liter of KH2PO4 adjusted to pH 7.4 with HCl) with a glass mortar. A Percoll gradient was prepared in a 15-ml Corex tube by centrifugation of 4.5 ml of Percoll in PBS at 10,000 × g for 20 min at room temperature. In order to ensure the clean isolation of microbial DNA, the opalescent microbial DNA layer was carefully removed and inspected under a microscope. The layer was then transferred to a microcentrifuge tube, and an equal volume of PBS was added. The tube was centrifuged for 10 min at 10,000 × g, and the pellet was resuspended and washed six times. The resulting pellet was resuspended in 10 mM Tris-HCl (pH 8.0) containing 100 mM NaCl and 100 mM EDTA and mixed with an equal volume of 1% molten agarose. The mixture was drawn into a plastic 1-ml syringe and then placed on ice for 10 min. The solidified agarose containing the DNA was then extruded with the 1-ml syringe. The extrusion was transferred to a 15-ml tube and incubated with agitation for 1 h at 37°C in 12 ml of 10 mM Tris-HCl (pH 8) containing 50 mM NaCl, 100 mM EDTA, 1% Sarkosyl, and 2-mg/ml lysozyme. Following incubation, the extrusion was transferred to a clean tube and incubated overnight in a water bath at 50°C in a mixture containing 1% Sarkosyl, 100 mM EDTA, and 4-mg/ml proteinase K. This incubation was then repeated for 2 to 24 h in 10 ml of 1 mM HEPES (pH 8), 1 mM EDTA, and 2-mg/ml subtilisin. The extrusions were then washed three times for 20 min in a solution of 1 mM HEPES (pH 8) and 1 mM EDTA. The extrusions, containing isolated microbial genomic DNA, were then used for the preparation of environmental genomic DNA libraries as previously described (31, 33-35).
Genomic DNA libraries and xylanase discovery.
Xylanase genes were discovered by screening approximately 106 plaques from the genomic microbial DNA libraries. Plaques were generated from λ DNA on semisolid medium by established procedures (25), except that top agar containing 0.1 to 0.3% azo dye-linked xylan was used. The disappearance of the dye-linked substrate surrounding a plaque was used to identify positive clones. Genomic DNA from positive plaques was recovered and sequenced. Insert sizes ranged from 3,000 to 6,000 bp.
Subcloning and expression of xylanase genes.
Open reading frames that encode xylanases were identified in the genomic clones by sequence alignment with known xylanase genes along with sequence analysis to identify ribosome binding sites and promoter regions. Xylanase genes XYL6807, XYL6806, and XYL6805 were subcloned into the expression vector pET101/D-TOPO. The XYL6806 gene was cloned without the N-terminal domain of unknown function (the gene product did not fold stably in E. coli with the N-terminal extension attached). An ATG codon was inserted in front of codon 655 in the full-length gene. XYL6807 and XYL6805 were cloned without their secretion signals with start codons inserted in front of codon 20 for both genes. XYL6419 was subcloned into the vector pASK-IBA2, also without its secretion signal, and an ATG codon was inserted in front of codon 21.
Overnight cultures of BL21 Star cells carrying the appropriate gene in pET101/D-TOPO or pASK-IBA2 were used to inoculate 1.5 liters of Terrific broth (12-g/liter Bacto Tryptone, 24-g/liter yeast extract, 4-ml/liter glycerol, 12.5-g/liter K2HPO4, 2.3-g/liter KH2PO4) containing 100-μg/ml ampicillin in a 6-liter shake flask. The cultures were incubated at 30°C and 250 rpm until an A600 between 0.4 and 0.9 was reached and were then induced with 1 mM isopropyl-1-thio-β-galactopyranoside (pET101/D-TOPO clones) or 100-μg/liter anhydrous tetracycline (pASK-IBA2 clone). Following 16 to 18 h of induction at 30°C, the cells were harvested by centrifugation at 10,000 × g for 20 min at 4°C. The pellet was resuspended in lysis buffer (50 mM potassium phosphate, pH 7.0, 100 mM KCl, 7.5 mM β-mercaptoethanol), and the cells were lysed by French press (15,000 lb/in2). The cell lysate was treated with 0.2% protamine sulfate and centrifuged at 100,000 × g for 30 min to remove cell debris and precipitate nucleic acids. Protein concentrations were estimated with the assay described by Bradford (3).
Xylanase activity assays.
For pH profile determination, enzymatic activities were measured with 400 μl of 2% azo dye-linked xylan as a substrate in 550 μl of 100 mM Britton-Robinson buffer at pH 3.0, 5.0, 6.0, 7.0, 8.0, 9.0, 10.0, and 11.0. The buffer was prepared by mixing solutions of 0.1 M phosphoric acid, 0.1 M boric acid, and 0.1 M acetic acid followed by pH adjustment with 1 M sodium hydroxide. Reactions were initiated by adding 50 μl of diluted clarified cell lysate (2- to 10-mg/ml total protein). Time point samples (150 μl each) were taken during the initial rate period and quenched into 540 μl of 100% ethanol prealiquoted into a deep 96-well plate. The samples were mixed and allowed to sit for approximately 10 min at room temperature followed by centrifugation at 3,000 × g for 10 min. After centrifugation, 300 μl of supernatant was transferred into a clear 96-well plate and A590 was measured.
For the analysis of substrate specificities, a 2,2′-bicinchoninic acid (BCA) reducing sugar assay was used (39). Assays were performed at 37°C in 250 μl of a 1% polysaccharide substrate solution buffered in 50 mM sodium acetate (pH 5.3). Reactions were initiated by adding 10 μl of diluted clarified cell lysate (1- to 2-mg/ml total protein). Time point samples (10 μl each) were taken during the initial rate period and quenched into 100 μl of BCA working reagent (13) in a 96-well PCR plate on ice. Once the last time point sample was taken, the 96-well plate was incubated for 30 min at 80°C in a thermal cycler equipped with a hot top. Following incubation, 100 μl from each well was transferred to a 96-well plate and A560 was measured.
PACE.
For polysaccharide analysis by carbohydrate gel electrophoresis (PACE), xylan and arabinoxylan (0.5 mg ml−1, 100 μl) or oligoxylans (1 mM, 20 μl) were treated with enzyme (0.075 U for 2 h or 0.15 U overnight where 1 U of xylanase activity is defined as the release of 1 μmol xylose reducing equivalents min−1 from medium-viscosity arabinoxylan at 37°C and pH 5.3). Reactions were carried out in 0.1 M ammonium acetate (adjusted with acetic acid to pH 5.5) in a total volume of 250 μl at 37°C. A short incubation using 0.075 U for 30 min was also performed to aid interpretation of digestion intermediates (data not shown). Controls without substrates or enzymes were performed under the same conditions to identify any nonspecific compounds in the enzymes, polysaccharides, oligosaccharides, or labeling reagents. The reactions were stopped by boiling for 20 min, and the samples were dried with a centrifugal vacuum evaporator. Assays were independently performed at least two times for each condition.
Derivatization using the ANTS fluorophore was carried out in the tubes containing dried polysaccharides, oligosaccharides, or monosaccharides as described previously (15). Once derivatized, samples (1 μl) were separated under conditions previously described (15). Gels were scanned with a MasterImager charge-coupled device camera system (Amersham, Buckinghamshire, United Kingdom) with an excitation filter at 400 nm and a detection filter at 530 nm. An image of the gel (resolution of 100 μm) was obtained and exported in an 8-bit file to PowerPoint.
To prepare ANTS-labeled substrates, oligoxylan standards were derivatized by ANTS as described previously (15) and dialyzed with dialysis tubing (molecular mass cutoff of 500 Da) to eliminate all free ANTS and salt from the derivatization process. Each standard (4 μl, 1 mM) was incubated in ammonium acetate buffer (0.1 M, pH 5.5) at 37°C with the enzyme (0.015 U for 10 min or 2 h or 0.15 U for 1 day). Following incubation, the samples were boiled for 10 min, and after drying, 100 μl of 6 M urea was added and the samples were analyzed with a polyacrylamide gel as described in the previous paragraph.
Xylanase sequence and phylogeny analysis.
All sequences were aligned by using ClustalW (38), and alignments were restricted to include only catalytic domains and were manually corrected with Bioedit software (18). In cases in which sequence pairs from the public domain shared more than 97% identity, only one sequence from the pair was used for the analysis. As such, no two sequences in the alignment shared more than 97% sequence identity. Phylogenetic analysis was performed with the PHYLIP package (10), and trees were drawn by using the program TreeView (30). The family 11 xylanase phylogenetic tree was generated by the neighbor-joining method (32) and for the family 8 sequences, a maximum-likelihood (9) tree was constructed from all known family 8 catalytic domain sequences.
Nucelotide sequence accession numbers.
The GenBank accession numbers for XYL6806, XYL6419, XYL6807, and XYL6805 are AY542134, AY542135, AY542136, and AY542137, respectively.
RESULTS
Insect specimens.
The termites collected from the colony nest in El Rodeo fall into the Nasutitermitidae family following the classification key proposed by Myles (26). Termites are characterized by “reduced mandibles, cephalic gland with separate reservoir bulb and pipette like shaft.” Dissection of collected specimens resulted in the clean separation of guts for the isolation of microbial DNA.
Lepidopterans are attracted to light and therefore can be collected by hand very easily when they rest on an illuminated white sheet during the night. Although taxonomic identification of the pool of moths collected was not conducted, the sample consisted of large lepidopterans (moths) of the Saturniidae family, whereby Rotschildia lebaeu, a common insect in Barra Honda National Park, could be easily identified within the sample pool. Caterpillars were collected when feeding from unidentified plants. Barra Honda National Park possesses approximately 40% of the expected genera of Saturniidae in Costa Rica, and the adult specimens collected belong to that category. The adult Saturniidae do not have a proboscis and cannot feed at this particular life stage. During their larval stages, these species typically feed from the leaves of a wide range of host plants (21). Nutrients needed for the adult and for reproduction are ingested during larval stages and utilized during the remaining lifetime of the moth.
Xylanase discovery and phylogeny.
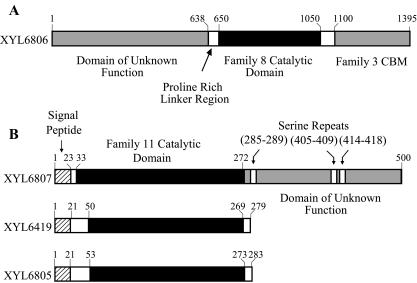
The genomic microbial DNA libraries generated from the insect gut samples were screened for genes that encode xylanases. Microbial DNA was isolated from the gut samples by using a Percoll gradient (see Materials and Methods), and as such, the libraries do not contain any host genes. In addition, eukaryotic genes with introns would not be expressed in the genomic library. Four unique clones that generated clearing zones on azo dye-linked xylan agar plates were isolated and sequenced. Three xylanases were discovered from lepidopteran intestinal tract samples (XYL6806, XYL6807, and XYL6805), and one was discovered from a termite sample (XYL6419). Open reading frames that encode xylanase catalytic domains were identified for each clone by sequence comparison to known xylanases. Two of the protein sequences (XYL6806 and XYL6807) contain domains that show (Fig. 1A and 1B) no detectable homology to described domains by BLAST searches. When compared to the sequences of published genes, the overall sequence identities of the xylanase catalytic domains were relatively low (25 to 39% pairwise amino acid sequence identities). Alignments also revealed significant insertions and gaps; however, signature motifs and catalytic residues for xylanases from known glycosyl hydrolase families (6, 7, 17, 40) were identifiable, suggesting that XYL6806 (from adult lepidopteran moth) belongs to glycosyl hydrolase family 8 and that XYL6419 (from a termite), XYL6807 (from caterpillars), and XYL6805 (from an adult lepidopteran moth) belong to family 11 (Fig. 2).
FIG. 1.
Schematic diagrams of xylanase primary sequences isolated from insect gut microbial libraries. (A) XYL6806 is a very unique xylanase belonging to family 8. It has a large N-terminal domain that returns no BLAST hits against the National Center for Biotechnology Information database. It is linked to the catalytic domain with a proline-rich linker and has a C-terminal carbohydrate-binding module. (B) Schematic diagrams of the domain structures of three family 11 xylanases. All three are very unique enzymes and have very low identity to known xylanases but clearly contain all family 11 signature motifs and catalytic residues. XYL6807 has a C-terminal domain that returns no BLAST hits against the National Center for Biotechnology Information database.
FIG. 2.
Sequence alignments showing the conserved regions in the family 8 (XYL6806) and family 11 (XYL6807, XYL6419, and XYL6805) xylanases discovered from insect gut samples. Regions of significant sequence identity or similarity are shown in boldface. Catalytic residues are shown with white text on black, and residues known to be involved in substrate binding are boxed. All published family 8 sequences (family 8 includes glucanase, chitosanase, and xylanase sequences) were used to construct the family 8 alignment. However, for illustration purposes, only the xylanase sequences are shown (B. hal., Bacillus halodurans; B. sp., Bacillus sp. strain KK-1; P. hal, Pseudoalteromonas haloplanktis). For the family 11 alignment, all published family 11 xylanases were used, and the most similar sequences to XYL6419, XYL6807, and XYL6805 are shown (P. co., Piromyces communis; N. fr., Neocallimastix frontalis; O. sp, Orpinomyces sp. strain PC-2; R. al., Ruminococcus albus) as well as some selected more common xylanases (A. ni, Aspergillus niger; T. re., Trichoderma reesei; B. st., Bacillus stearothermophilus; S. co., Streptomyces coelicolor).
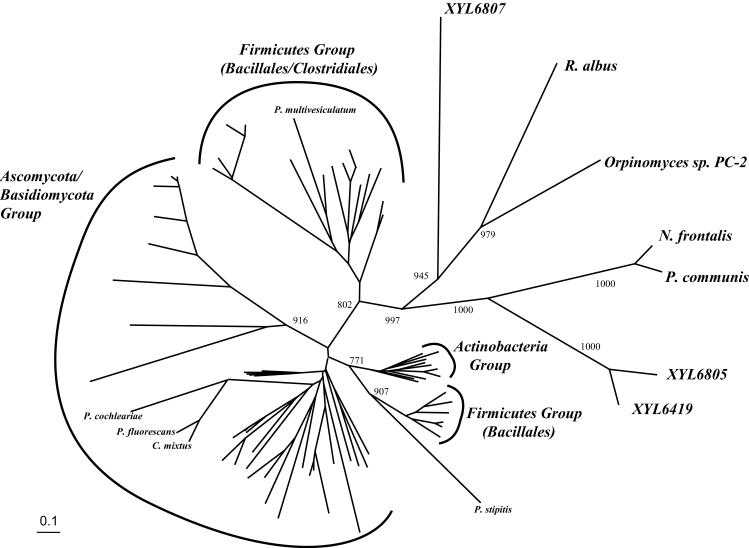
The protein sequences of the four xylanases were compared to published xylanases by phylogenetic analysis. The family 11 phylogenetic tree (Fig. 3) shows that these insect gut-derived xylanases are only distantly related to their family 11 counterparts. Interestingly, although they are significantly different (identities to published sequences range from 33 to 39%), they are most closely related to xylanases that were also isolated from gut samples. They branch with xylanases from the rumen fungi Neocallimastix frontalis, Orpinomyces sp. strain PC-2, and Piromonas communis and the rumen bacterium Ruminococcus albus. However, they differ significantly from the family 11 xylanase from the xylose-fermenting yeast, Pichia stipitis, which has been shown to inhabit the gut of wood-ingesting beetles (36).
FIG. 3.
Phylogenetic tree of family 11 catalytic domain sequences generated by the neighbor-joining method. At major nodes, bootstrap values for 1,000 resamplings are shown. One Firmicutes group contains organisms of the orders Bacillales and Clostridiales (GenBank accession no. U76545, X00660, AF326785, AF220528, X59059, AB010958, Z11127, U43089, AJ272430, AJ132472, Z49970, AF036925, M31726, and D13325) and two sequences from the ciliate P. multivesiculatum (accession no. AJ009828 and AB011274). The other Firmicutes group contains only sequences from the order Bacillales (GenBank accession no. U15985, X59059, AP001510, U51675, AF195421, and D32065) and are branched with the yeast xylanase from Pichia stipitis (AF151379). All of the actinobacterial xylanase sequences form a clade (GenBank accession no. X76729, AF120156, X81045, X98518, AJ292317, AL109949, AL133220, AF198618, AF194025, and U01242). The fungal xylanases form a large group containing the phyla Ascomycota and Basidiomycota (GenBank accession no. AJ238895, X76047, U35436, AF169630, U58915, U58916, L13596, AB003085, U39784, S45138, L26988, O43097, AJ278385, Z50050, AB035540, Z68891, Y16969, X69573, X69574, AF246831, AF246830, P48793, AJ012718, D49850, D49851, L37529, Z81317, A44597, AF301904, and D63382) and two bacterial xylanase sequences (Pseudomonas fluorescens [Z48927] and Cellvibrio mixtus [Z48925]) and the xylanase from the beetle Phaedon cochleariae (Y17908). The three insect gut xylanases discovered in this study (XYL6807, XYL6805, and XYL6419) branch with xylanases from the rumen fungi Neocallimastix frontalis (X82439), Orpinomyces sp. strain PC-2 (U57819), and Piromonas communis (AF297649) and the rumen bacterium Ruminococcus albus (AB057588).
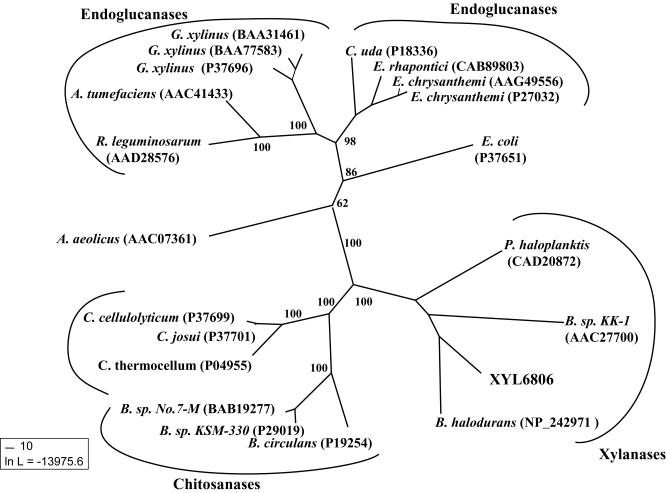
Glycosyl hydrolase family 8 is primarily made up of endoglucanases, and there are only three known family 8 xylanases in the public domain. XYL6806 clearly falls within the xylanase clade of the family 8 tree (Fig. 4), but it is also an unusual sequence sharing only 40% sequence identity with its closest neighbor, Bacillus halodurans xylanase Y.
FIG. 4.
Maximum-likelihood tree of all family 8 catalytic domain sequences. Numbers at nodes are bootstrap values for 100 resamplings. The GenBank accession numbers for the sequences used to calculate the tree are shown in parentheses. The tree is formed of two endoglucanase clades: a xylanase clade containing XYL6806 and a clade consisting of chitosanase sequences.
Biochemical properties.
All four enzymes possess endoxylanase activity and were tested on β-1,4-linked xylose polysaccharide substrates, including birchwood azo-xylan (data not shown), high-viscosity rye arabinoxylan (average molecular mass of >300 kDa), and medium-viscosity (∼278 kDa) and low-viscosity (∼66 kDa) wheat arabinoxylan (Table 1). The enzymes hydrolyze the three xylans tested with a preference for high-viscosity xylan (greatest molecular mass).
TABLE 1.
Biochemical characteristics of XYL6806, XYL6419, XYL6807, and XYL6805
| Xylanase | Source | Glycosyl hydrolase family | pH optimuma | Arabinoxylan activity (%) at viscosityb:
|
||
|---|---|---|---|---|---|---|
| High | Medium | Low | ||||
| XYL6806 | Adult lepidopteran | 8 | 5 | 100 | 68.4 | 79.6 |
| XYL6419 | Adult isopteran | 11 | 6 | 100 | 94.7 | 69.8 |
| XYL6807 | Caterpillar lepidopteran | 11 | 6 | 100 | 96.3 | 92.9 |
| XYL6805 | Adult lepidopteran | 11 | 6 | 100 | 89.6 | 91.1 |
The temperature optimum (50°C for each) and pH optimum were determined from initial rates by using Azo-xylan as a substrate as described in Materials and Methods.
The activities shown are relative initial rates normalized to activity on high- viscosity arabinoxylan and were measured as described in Materials and Methods.
The ability of these enzymes to hydrolyze β-glucose-linked polysaccharides (carboxymethyl cellulose, β-1,3, β-1,4 glucan and xyloglucan) was also tested, and no activity was detectable even at a 10-fold enzyme concentration relative to the concentration used to measure xylanase activity. No hydrolysis of pNP-cellobiose was detected, indicating that the enzymes do not have any β-glucosidase activity. All four enzymes had initial rate temperature optima at 50°C and initial rate pH optima between 5 and 6.
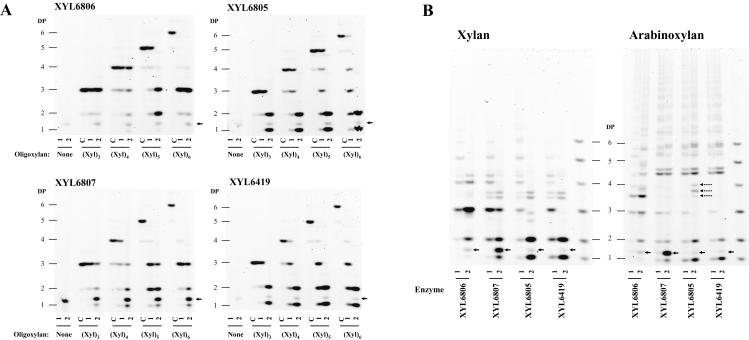
The activities of the enzymes on various xylan substrates were studied in more detail by using PACE (15). This technique involves the derivatization of oligosaccharides and sugars at their reducing ends with a fluorophore followed by polyacrylamide electrophoresis under optimized conditions. Uncharged oligosaccharides (such as xylooligosaccharides) are derivatized with a charged fluorophore, enabling size-specific separation. Oligosaccharides from (Xyl)2 to (Xyl)6 and the polysaccharides beechwood xylan and wheat arabinoxylan were incubated with the xylanases at both low and high activity to reveal digestion intermediates and final products (conditions 1 and 2, respectively). The ANTS fluorophore-derivatized products were analyzed on polyacrylamide gels (Fig. 4). None of the xylanases could hydrolyze (Xyl)2 (data not shown). To further characterize the substrate binding site, we also hydrolyzed xylo-oligosaccharides after derivatization by ANTS. In this PACE experiment, only the “reducing end” oligosaccharide product containing the ANTS was visible on the polyacrylamide gels. The results were consistent with the interpretation that the ANTS-derivatized xylose was not accommodated in a xylose-binding subsite of the enzymes (data not shown).
The family 8 enzyme XYL6806 did not hydrolyze (Xyl)3, and the (Xyl)4 substrate was hydrolyzed only poorly (Fig. 5A). The (Xyl)5 and (Xyl)6 substrates, however, were very good substrates, and the enzyme had a preference for the larger (Xyl)6 oligosaccharide. This indicated that the XYL6806 enzyme active site has at least six positions for xylose residue binding and requires at least five positions to be occupied for substantial activity. Since the products of (Xyl)6 digestion were largely (Xyl)3, the hydrolysis occurred mainly between sites 3 and 4. Digestion of (Xyl)5 produced mainly (Xyl)3 and (Xyl)2, and (Xyl)6-ANTS yielded (Xyl)4-ANTS. The ANTS-derivatized xylose was probably not accommodated in a xylose-binding subsite of the enzyme, and this suggests that the (Xyl)5 was cut at the (Xyl)2-(Xyl)3 linkage and occupies sites 2 to 6.
FIG. 5.
Hydrolysis product fingerprints of xylanases XYL6806, XYL6807, XYL6805, and XYL6419. DP, degree of polymerization. (A) Oligosaccharides obtained after hydrolysis of oligoxylans (Xyl)3, (Xyl)4, (Xyl)5, and (Xyl)6. All reactions were performed at pH 5.5 and 37°C. Two conditions were used: 7.5 mU of enzyme for 2 h (lanes 1) or 150 mU for 1 day (lanes 2). Controls without enzymes (lanes c) (to determine the purity of the oligosaccharides) or without substrate oligoxylans (to define unspecific products from the enzyme samples) were performed. Arrows indicate a nonspecific band from the cell lysates that is present in the absence of substrate. (B) Fingerprints of oligosaccharides obtained after hydrolysis of beechwood xylan and wheat arabinoxylan. All reactions were performed at pH 5.5 and 37°C. The oligoxylan (DP 1 through 6) mixture was used to characterize oligoxylan bands. Two conditions were used: 7.5 mU of enzyme for 2 h (lanes 1) or 150 mU for 1 day (lanes 2). Arrows indicate a nonspecific band from the cell lysates that is present in the absence of substrate. The dashed arrows show specific oligosaccharides produced from arabinoxylan hydrolysis by XYL6805 but not by XYL6419 and XYL6807.
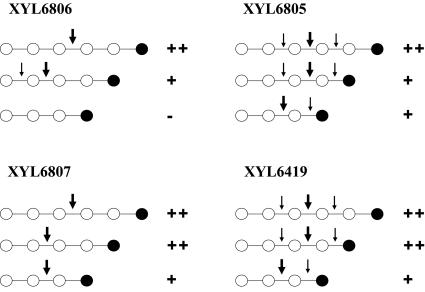
The family 11 xylanases XYL6419, XYL6807, and XYL6805 all hydrolyzed oligoxylans ranging from (Xyl)3 through (Xyl)6, but at very different rates (Fig. 5A). XYL6805 has at least six sites, since (Xyl)6 was a better substrate than (Xyl)5, whereas the closely related XYL6419 already had high activity when five sites were occupied. However, both require only three to be filled for substantial activity. In contrast, the more distantly related XYL6807, which also has at least six sites, needs four sites to be filled for substantial activity. Thus, the main oligosaccharide hydrolysis difference between these enzymes was that XYL6807 hydrolyzed (Xyl)3 only very slowly, whereas XYL6419 and XYL6805 were able to hydrolyze (Xyl)3 completely. The PACE results with oligosaccharide substrates and positional specificities are summarized schematically in Fig. 6.
FIG. 6.
Schematic illustration showing the substrates bound and hydrolyzed by the four xylanases XYL6419, XYL6807, XYL6806, and XYL6805 as determined by PACE. Open circles represent xylosyl residues, and solid circles indicate xylosyl residues at the reducing ends. Small arrows show possible hydrolysis positions deduced from the PACE results, and large arrows show main hydrolysis positions identified by PACE (see Fig. 5). ++, complete hydrolysis at a low enzyme dose; +, partial hydrolysis at a low dose but complete hydrolysis at higher doses; −, no or very little hydrolysis.
All four enzymes hydrolyzed both beechwood xylan and wheat arabinoxylan. They appeared more active on xylan than on arabinoxylan, producing a higher proportion of small polysaccharide fragments (Fig. 5B). Xylan was hydrolyzed into xylose and (Xyl)2, with additional oligosaccharides that migrated between the oligoxylose standards. Oligosaccharides have a specific migration in PACE based on degree of polymerization and composition (15, 19). These oligosaccharides are therefore likely oligoglucuronoxylans, as xylan from beechwood is replaced with glucuronosyl residues that can be methylated. Arabinoxylan digestion also yielded some xylose and (Xyl)2, but many other bands migrated between, or slower than, the oligoxylose standards, corresponding to oligoarabinoxylans.
On both the polysaccharide substrates, the family 8 xylanase, XYL6806, produced unique fingerprints very different from the family 11 enzymes. The oligoglucuronoxylan products had different mobility from the products of all three family 11 enzymes. On the arabinoxylan substrate, the family 8 enzyme also produced additional enzyme-specific products. There was some (Xyl)3 in the final fingerprint from the xylan and arabinoxylan hydrolyzed by XYL6806, consistent with the oligosaccharide hydrolysis results.
The xylan and arabinoxylan hydrolysis fingerprints for XYL6807, XYL6805, and XYL6419 were largely similar but with a few significant differences. First, (Xyl)3 was present in the XYL6807 digests even at high doses of enzyme, consistent with the oligosaccharide hydrolysis results. Second, XYL6805 hydrolysis of arabinoxylan produced some unique bands (dashed arrows in Fig. 5B). The intermediate fingerprints (condition 1) were similar, but the final fingerprints (condition 2) were different, indicating that these unique oligoarabinoxylans were produced by XYL6805 very slowly. Thus, even though all three enzymes are in family 11 and XYL6419 and XYL6805 share substantial sequence similarity and xylooligosaccharide hydrolysis patterns, they hydrolyze the arabinoxylan substrate differently.
DISCUSSION
As with other species that ingest a diet rich in the plant structural polysaccharides cellulose and hemicellulose, insects of the orders Isoptera and Lepidoptera depend on glycosyl hydrolases produced by gastrointestinal symbionts to digest the polysaccharides and liberate sugars that can be utilized by the insect host. This report describes the first use of direct environmental cloning to isolate eDNA and, by expression, the identification of genes encoding novel xylanases from the gut contents of tropical insects. Previous work suggests that microorganisms inhabiting the guts of insects may be unique to the particular ecosystem (28, 36, 44). Our demonstration that xylanases produced by these microbial symbionts are only distantly related to known xylanases further supports the view that the insect gut is a closed ecosystem in which glycosyl hydrolases have evolved independently. Although from the same glycosyl hydrolase family, the enzymes discovered in this work are only distantly similar to the xylanase from the beetle-gut yeast, Pichia stipitis (2, 36). Interestingly, xylanases most closely related to three of the four enzymes described here are found in species of anaerobic bacteria and fungi that normally inhabit the gastrointestinal tract of ruminant and nonruminant herbivores, another closed ecosystem (although very different from the insect gut) inhabited by commensal and symbiotic species of obligately anaerobic microorganisms, typically not found elsewhere in nature.
In common with other xylanases, XYL6806 and XYL6807 are modular in architecture and contain multiple domains separated by linkers rich in proline and serine residues, respectively. In addition to catalytic domains of glycosyl hydrolase families 8, 10, and 11, modular xylanases can contain noncatalytic carbohydrate binding modules (CBM) (14, 27), thermostabilizing domains (12, 42), surface layer homology domains that promote binding to the cell envelope (24), domains homologous with the Rhizobium-derived NodB protein (23), and dockerin domains that mediate assembly of large multienzyme cellulosome complexes via binding to a scaffolding protein (11). Based on sequence alignments, the C-terminal domain of XYL6806 belongs to family CBM3. Similar domains have been described predominantly in bacterial endoglucanases but also occur in several other xylanases (1, 27, 37, 43), including xylanases produced by the rumen protozoan Polyplastron multivesiculatum (8) and the rumen bacterium Fibrobacter succinogenes (22). The lack of similarity between the sequence of the N-terminal domain of XYL6806 and sequences contained in protein sequence databases suggests that this has a unique function, not previously ascribed to domains in any of the known modular xylanases. Similarly, the lack of sequence similarity between the C-terminal half of XYL6807 and known proteins is indicative of either a novel function or a new sequence that is able to fulfill a function already attributed to auxiliary domains found in modular xylanases. Based on the fact that the gene encoding XYL6807 was isolated from an anaerobic environment, and the observation that the C-terminal domain of unknown function in XYL6807 is divided by serine repeats, it is tempting to speculate that the domain serves as a dockerin to integrate XYL6807 into a cellulosome structure. Dockerins have been described in modular xylanases from anaerobic bacteria (11), but similarity between reiterated dockerin domains is typical in such cases, and the two domains in XYL6807 exhibit little similarity.
It is also interesting that XYL6807 contains a large loop between the conserved tyrosine substrate-binding residues that is not present in XYL6805 or XYL6419. XYL6807 does not hydrolyze small substrates such as (Xyl)3 and (Xyl)4 as effectively as XYL6805 and XYL6419, and it may be that this loop prevents the productive binding of the smaller substrates.
The sequences of these four enzymes are interesting for their distant locations in the xylanase phylogenetic tree. Clearly, these enzymes evolved under influences unique to their origin and far from the selective influences felt by other, previously described enzymes of either glycosyl hydrolase family 8 or 11, derived from cultured microorganisms. Additionally, the modular arrangements and the unknown origins or functions of some modules are evidence for an isolated evolutionary origin and possibly for unique activities in the insect gut. The gut environment of animals contains commensal or symbiotic microbes that conduct permanent or transient lifestyles in this biotope. The species-specific origin of the xylanases is unknown at this time; however, they appear to confine their activities to endospecific hydrolysis of xylan polymer, and though they exhibit different cleavage patterns, they utilize the well-described, position-specific polymer cleavage mechanism to effect primary hemicellulose degradation (16).
Acknowledgments
We thank the Diversa Library, Sequencing and Subcloning groups, and Marielos Mora for initial work in Percoll gradient method development. Special thanks go to Mircea Podar for advice on phylogenetic analysis.
PACE studies were supported by grants from the Biotechnology and Biological Sciences Research Council (United Kingdom).
REFERENCES
- 1.Abou, H. M., K. E. Nordberg, E. Bartonek-Roxa, S. Raghothama, P. J. Simpson, H. J. Gilbert, M. P. Williamson, and O. Holst. 2000. Carbohydrate-binding modules from a thermostable Rhodothermus marinus xylanase: cloning, expression and binding studies. Biochem. J. 345:53-60. [PMC free article] [PubMed] [Google Scholar]
- 2.Basaran, P., Y. D. Hang, N. Basaran, and R. W. Worobo. 2001. Cloning and heterologous expression of xylanase from Pichia stipitis in Escherichia coli. J. Appl. Microbiol. 90:248-255. [DOI] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Brune, A., and M. Friedrich. 2000. Microecology of the termite gut: structure and function on a microscale. Curr. Opin. Microbiol. 3:263-269. [DOI] [PubMed] [Google Scholar]
- 5.Cazemier, A. E., J. C. Verdoes, F. A. Reubsaet, J. H. Hackstein, C. van der Drift, and H. J. Op den Camp. 2003. Promicromonospora pachnodae sp. nov., a member of the (hemi)cellulolytic hindgut flora of larvae of the scarab beetle Pachnoda marginata. Antonie Leeuwenhoek 83:135-148. [DOI] [PubMed] [Google Scholar]
- 6.Coutinho, P. M., and B. Henrissat. 1999. Carbohydrate-active enzymes: an integrated database approach, p. 3-12. In H. J. Gilbert, G. Davies, B. Henrissat, and B. Svensson (ed.), Recent advances in carbohydrate bioengineering. The Royal Society of Chemistry, Cambridge, United Kingdom.
- 7.Coutinho, P. M., and B. Henrissat. 1999. The modular structure of cellulases and other carbohydrate-active enzymes: an integrated database approach, p. 15-23. In K. Ohmiya, K. Hayashi, K. Sakka, Y. Kobayashi, S. Karita, and T. Kimura (ed.), Genetics, biochemistry and ecology of cellulose degradation. Uni Publishers Co., Tokyo, Japan.
- 8.Devillard, E., C. Bera-Maillet, H. J. Flint, K. P. Scott, C. J. Newbold, R. J. Wallace, J. P. Jouany, and E. Forano. 2003. Characterization of XYN10B, a modular xylanase from the ruminal protozoan Polyplastron multivesiculatum, with a family 22 carbohydrate-binding module that binds to cellulose. Biochem. J. 373:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Felsenstein, J. 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17:368-376. [DOI] [PubMed] [Google Scholar]
- 10.Felsenstein, J. 1989. PHYLIP—phylogeny inference package version 3.5. Cladistics 5:164-166. [Google Scholar]
- 11.Fernandes, A. C., C. M. Fontes, H. J. Gilbert, G. P. Hazlewood, T. H. Fernandes, and L. M. Ferreira. 1999. Homologous xylanases from Clostridium thermocellum: evidence for bi-functional activity, synergism between xylanase catalytic modules and the presence of xylan-binding domains in enzyme complexes. Biochem. J. 342:105-110. [PMC free article] [PubMed] [Google Scholar]
- 12.Fontes, C. M., G. P. Hazlewood, E. Morag, J. Hall, B. H. Hirst, and H. J. Gilbert. 1995. Evidence for a general role for non-catalytic thermostabilizing domains in xylanases from thermophilic bacteria. Biochem. J. 307:151-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fox, J. D., and J. F. Robyt. 1991. Miniaturization of three carbohydrate analyses using a microsample plate reader. Anal. Biochem. 195:93-96. [DOI] [PubMed] [Google Scholar]
- 14.Gilkes, N. R., B. Henrissat, D. G. Kilburn, R. C. Miller, Jr., and R. A. J. Warren. 1991. Domains in microbial β-1,4-glycanases: sequence conservation, function, and enzyme families. Microbiol. Rev. 55:303-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goubet, F., P. Jackson, M. J. Deery, and P. Dupree. 2002. Polysaccharide analysis using carbohydrate gel electrophoresis: a method to study plant cell wall polysaccharides and polysaccharide hydrolases. Anal. Biochem. 300:53-68. [DOI] [PubMed] [Google Scholar]
- 16.Gruber, K., G. Klintschar, M. Hayn, A. Schlacher, W. Steiner, and C. Kratky. 1998. Thermophilic xylanase from Thermomyces lanuginosus: high-resolution X-ray structure and modeling studies. Biochemistry 37:13475-13485. [DOI] [PubMed] [Google Scholar]
- 17.Guerin, D. M., M. B. Lascombe, M. Costabel, H. Souchon, V. Lamzin, P. Beguin, and P. M. Alzari. 2002. Atomic (0.94 A) resolution structure of an inverting glycosidase in complex with substrate. J. Mol. Biol. 316:1061-1069. [DOI] [PubMed] [Google Scholar]
- 18.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 19.Hogg, D., G. Pell, P. Dupree, F. Goubet, S. M. Martin-Orue, S. Armand, and H. J. Gilbert. 2003. The modular architecture of Cellvibrio japonicus mannanases in glycoside hydrolase families 5 and 26 points to differences in their role in mannan degradation. Biochem. J. 371:1027-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holdridge, L. R. 1967. Life zone ecology. Tropical Science Center, San Jose, Costa Rica.
- 21.Janzen, D. H. 2003. How polyphagous are Costa Rican dry forest saturniid moth caterpillars?, p. 369-380. In Y. Basset, V. Novotny, S. E. Miller, and R. Kitching (ed.), Arthropods of tropical forests: spatio-temporal dynamics and resource use in the canopy. Cambridge University Press, Cambridge, United Kingdom.
- 22.Jun, H. S., J. K. Ha, L. M. Malburg, Jr., G. A. Verrinder, and C. W. Forsberg. 2003. Characteristics of a cluster of xylanase genes in Fibrobacter succinogenes S85. Can. J. Microbiol. 49:171-180. [DOI] [PubMed] [Google Scholar]
- 23.Laurie, J. I., J. H. Clarke, A. Ciruela, C. B. Faulds, G. Williamson, H. J. Gilbert, J. E. Rixon, J. Millward-Sadler, and G. P. Hazlewood. 1997. The NodB domain of a multidomain xylanase from Cellulomonas fimi deacetylates acetylxylan. FEMS Microbiol. Lett. 148:261-264. [Google Scholar]
- 24.Lemaire, M., I. Miras, P. Gounon, and P. Beguin. 1998. Identification of a region responsible for binding to the cell wall within the S-layer protein of Clostridium thermocellum. Microbiology 144:211-217. [DOI] [PubMed] [Google Scholar]
- 25.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 26.Myles, T. G., B. H. Strack, and B. Forschler. 1998. Distribution and abundance of Antennopsis gallica (Hyphomycetes: gleohaustoriales), an ectoparasitic fungus, on the eastern subterranean termite in Canada. J. Invertebr. Pathol. 72:132-137. [DOI] [PubMed] [Google Scholar]
- 27.Notenboom, V., A. B. Boraston, S. J. Williams, D. G. Kilburn, and D. R. Rose. 2002. High-resolution crystal structures of the lectin-like xylan binding domain from Streptomyces lividans xylanase 10A with bound substrates reveal a novel mode of xylan binding. Biochemistry 41:4246-4254. [DOI] [PubMed] [Google Scholar]
- 28.Ohkuma, M., T. Iida, and T. Kudo. 1999. Phylogenetic relationships of symbiotic spirochetes in the gut of diverse termites. FEMS Microbiol. Lett. 181:123-129. [DOI] [PubMed] [Google Scholar]
- 29.Ohkuma, M., and T. Kudo. 1996. Phylogenetic diversity of the intestinal bacterial community in the termite Reticulitermes speratus. Appl. Environ. Microbiol. 62:461-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Page, R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 31.Robertson, D. E., E. J. Mathur, R. V. Swanson, B. L. Marrs, and J. M. Short. 1996. The discovery of new biocatalysts from microbial diversity. SIM News 46:3-8. [Google Scholar]
- 32.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 33.Short, J. M. 1997. Recombinant approaches for accessing biodiversity. Nat. Biotechnol. 15:1322-1323. [DOI] [PubMed] [Google Scholar]
- 34.Short, J. M. September 1999. Protein activity screening of clones having DNA from uncultured microorganisms. U.S. patent 5,958,672.
- 35.Short, J. M. August 2001. Gene expression library produced from DNA from uncultivated microorganisms and methods for making the same. U.S. patent 6,280,926.
- 36.Suh, S. O., C. J. Marshall, J. V. McHugh, and M. Blackwell. 2003. Wood ingestion by passalid beetles in the presence of xylose-fermenting gut yeasts. Mol. Ecol. 12:3137-3145. [DOI] [PubMed] [Google Scholar]
- 37.Sunna, A., M. D. Gibbs, and P. L. Bergquist. 2000. The thermostabilizing domain, XynA, of Caldibacillus cellulovorans xylanase is a xylan binding domain. Biochem. J. 346:583-586. [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Waffenschmidt, S., and L. Jaenicke. 1987. Assay of reducing sugars in the nanomole range with 2,2′-bicinchoninate. Anal. Biochem. 165:337-340. [DOI] [PubMed] [Google Scholar]
- 40.Wakarchuk, W. W., R. L. Campbell, W. L. Sung, J. Davoodi, and M. Yaguchi. 1994. Mutational and crystallographic analyses of the active site residues of the Bacillus circulans xylanase. Protein Sci. 3:467-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wenzel, M., I. Schonig, M. Berchtold, P. Kampfer, and H. Konig. 2002. Aerobic and facultatively anaerobic cellulolytic bacteria from the gut of the termite Zootermopsis angusticollis. J. Appl. Microbiol. 92:32-40. [DOI] [PubMed] [Google Scholar]
- 42.Winterhalter, C., P. Heinrich, A. Candussio, G. Wich, and W. Liebl. 1995. Identification of a novel cellulose-binding domain within the multidomain 120 kDa xylanase XynA of the hyperthermophilic bacterium Thermotoga maritima. Mol. Microbiol. 15:431-444. [DOI] [PubMed] [Google Scholar]
- 43.Xie, H., H. J. Gilbert, S. J. Charnock, G. J. Davies, M. P. Williamson, P. J. Simpson, S. Raghothama, C. M. Fontes, F. M. Dias, L. M. Ferreira, and D. N. Bolam. 2001. Clostridium thermocellum Xyn10B carbohydrate-binding module 22-2: the role of conserved amino acids in ligand binding. Biochemistry 40:9167-9176. [DOI] [PubMed] [Google Scholar]
- 44.Zhang, N., S. O. Suh, and M. Blackwell. 2003. Microorganisms in the gut of beetles: evidence from molecular cloning. J. Invertebr. Pathol. 84:226-233. [DOI] [PubMed] [Google Scholar]