Abstract
Purpose
Determine the frequency of discordant gene expression profile (GEP) classification of posterior uveal melanomas sampled at two tumor sites by fine needle aspiration biopsy (FNAB).
Design
Prospective single institution longitudinal study performed in conjunction with a multi-center validation study of the prognostic value of GEP class of posterior uveal melanoma cells for metastasis and metastatic death.
Methods
FNAB aspirates of 80 clinically diagnosed primary choroidal and ciliochoroidal melanomas were obtained from two tumor sites prior to or at the time of initial ocular tumor treatment and submitted for independent GEP testing and classification. Frequency of discordant GEP classification of these specimens was determined.
Results
Using the support vector machine learning algorithm favored by the developer of the GEP test employed in this study, 9 of the 80 cases (11.3% [95% confidence interval: 9.0% to 13.6%]) were clearly discordant. If cases with a failed classification at one site or a low confidence class assignment by the support vector machine algorithm at one or both sites are also regarded as discordant, then this frequency rises to 13 of the 80 cases (16.3% [95% confidence interval: 13.0% to 19.6%]).
Conclusion
Sampling of a clinically diagnosed posterior uveal melanoma at a single site for prognostic GEP testing is associated with a substantial probability of misclassification. Two-site sampling of such tumors with independent GEP testing of each specimen may be advisable to lessen the probability of underestimating an individual patient’s prognostic risk of metastasis and metastatic death.
Introduction
Gene expression profiling (GEP) is a transcriptional method of cellular analysis1–3 that has been applied to human uveal melanomas for the purpose of prognostic classification.4,5 The prognostic value of a standardized 15-gene assay developed by Harbour and coworkers has been validated in a multi-center clinical trial.6 Patients having a primary uveal melanoma categorized by this standardized assay as GEP (gene expression profile) class 2 experienced a substantially higher rate of post-biopsy metastasis than did patients whose tumor was categorized as GEP class 1. Most of the tumor specimens evaluated in the multi-center validation trial were cellular aspirates obtained by fine needle aspiration biopsy (FNAB)7 at the time of or shortly prior to initial tumor treatment. In almost all of these cases, a single tumor site was sampled by FNAB for gene expression profile testing and classification and was assumed to be representative of the tumor as a whole. But as recent publications have shown in other tumors, intratumor genetic heterogeneity has been documented in a range of tumor types, affecting clinically relevant parameters such as gene-expression signatures that reflect prognosis, and response to therapeutic agents.8 Recently, researchers showed that many patients with glioblastomas displayed multiple transcriptomic subtypes within different biopsies from the same tumor.8 Although the heterogeneous distribution of chromosome 3 in uveal melanomas has been reported,9 limited information is available on the frequency of discordant GEP class assignments to paired tumor cells specimens obtained by FNAB from slightly different sites within clinically diagnosed primary uveal melanomas.
The principal purpose of this study was to determine the frequency of discordant GEP class assignment of tumor cells obtained from two adjacent tumor sites within clinically diagnosed primary posterior uveal melanomas by FNAB performed prior to or at the time of initial tumor treatment. Secondary purposes were to determine the relationships between discordant GEP class assignment and other evaluated clinical and pathological variables and to determine the actuarial rates of uveal melanoma metastasis and metastatic death in patients with discordant GEP results at the two sampled tumor sites.
Patients and Methods
Our study was conducted as an IRB-approved inter-institutional prospective longitudinal clinical and laboratory study of gene expression profile (GEP) testing of fine needle aspiration biopsy (FNAB) obtained specimens of primary posterior uveal (choroidal or ciliochoroidal) melanomas (ClinicalTrials.gov Identifier: NCT00406120). This study was approved and monitored by the University of Cincinnati Institutional Review Board. Informed consent was obtained from each patient and the study adhered to the tenets of the Declaration of Helsinki. All work using patient information was performed in compliance with the Health Insurance Portability and Accountability Act.
Patients with a clinically diagnosed primary choroidal or ciliochoroidal melanoma evaluated in the Oncology Service of the University of Cincinnati Medical Center who consented to participate in this study underwent investigational FNAB of their intraocular tumor prior to or at the time of planned initial treatment of that tumor. Patient and tumor variables determined at each patient’s pre-biopsy clinical examination included patient age, largest basal diameter of the intraocular tumor (estimated by indirect ophthalmoscopy with fundus mapping and ocular transillumination [when appropriate]) in millimeters to the nearest 0.5 mm, maximal thickness of the tumor (by A-scan ultrasonographic biometry) in millimeters to the nearest 0.1 mm, and intraocular tumor location (exclusively choroidal versus involving ciliary body).
Procurement of tumor specimens by fine needle aspiration biopsy (FNAB)
All tumor biopsies were performed by one of the authors (JJA, ZMC). The instrumentation and techniques employed by the authors have been reported previously.10,11 At least three and usually four tumor sites within each tumor were sampled. These sites were selected on the basis of tumor thickness, intraocular tumor location and clinical features of the tumor. Special caution was taken to sample comparable areas of the tumor for cytology and GEP. A separate biopsy needle (25 gauge) was used to sample each tumor site. Two of the aspirates (usually the second and fourth aspirates) were suspended immediately in a vial containing tissue culture medium, snap frozen in a styrofoam box containing dry ice, and delivered via overnight courier service to the Harbour laboratory in the Department of Ophthalmology and Vision Science of Washington University, St. Louis, Missouri, for subsequent GEP testing. The specimens were kept frozen until the day of analysis.
Gene Expression Profile (GEP) testing and classification
The methods of preparation of RNA samples, testing of samples by real-time polymerase chain reaction (PCR), and analysis of gene expression data for prognostic classification have been reported previously.5,7 In short, the RNA from each specimen was analyzed by microarray technology, and a typical heatmap of up-regulation and down-regulation of key discriminating genes was generated. Two different machine learning algorithms (the weighted voting [WV] algorithm12 and the support vector machine [SVM] algorithm13) that had been calibrated against 30 uveal melanomas of known prognostic category were used to translate the heatmap results for each individual specimen into a prognostic GEP class (GEP class 1 [low risk of future emergence of distant metastasis] or GEP class 2 [relatively high risk of short term emergence of distant metastasis]) in the Harbour laboratory. Each GEP class assignment by the weighted voting algorithm was associated with a probability value between −1.0 (strong probability of GEP Class 1) and +1.0 (strong probability of Glass 2), and each GEP class assignment by the support vector machine algorithm was associated with a discriminant function score that ranged from −1.53 (strong probability of GEP Class 1) to +1.57 (strong probability of GEP Class 2) in this series of cases. The gene chips employed in this test contained 12 classifying genes (CDH1, ECM1, EIF1B, FXR1, HTR2B, ID2, LMCD1, LTA4H, MTUS1, RAB31, ROBO1, and SATB1) and 3 control genes (MRPS21, RBM23, and SAP130). Control genes were consistently expressed in all cases but not differentially up regulated or down regulated in class 1 versus class 2 cases.5 GEP classification is determined by the combined expression of these genes. An individual gene was considered undetectable if its amplification product did not reach the expression threshold after 40 cycles of quantitative PCR, and a sample was considered a technical failure if one or more control genes or at least 3 of the 12 discriminating genes were undetectable.6 Failed aspirates were categorized as GEP Class 0 with an associated weighted voting probability value of 0.0 and a support vector machine discriminant score of 0.0 for the purposes of this study.
Cytopathological analysis and classification
The remaining aspirates were flushed into a transport syringe containing a 50:50 mixture of balanced salt solution and absolute alcohol and submitted via courier to the pathology laboratory of the Cincinnati Children’s Hospital Medical Center for cytopathological processing, analysis and classification as previously published by our group.11 The pathologist assigned to the case issued an official report that included the cytopathological category of the cells in the aspirate. Immunocytochemistry was performed on specific cases that yielded non-pigmented cells and those with cytologic features considered borderline. HMB 45 was successfully used to confirm the melanocytic nature of the biopsied tumor in these situations.14 No attempt was made to retrieve the cytopathology slides and reclassify the specimens prospectively in this study.
Assessment of concordance versus discordance of Gene Expression Profile (GEP) classification of paired aspirates
Agreement between the GEP class assignment of the tumor cells obtained from the first site sampled for GEP testing (designated the “a specimen”) and the second site sampled for GEP testing (designated the “b specimen”) was evaluated both graphically (in the form of scatter plots of weight voting probability values and support vector machine discriminant function scores on the “a” specimens [x-axis] versus the “b”specimens [y-axis]) and in side-to-side tabular comparisons for both the weight voting and support vector machine assignments. A concordant result was defined as a GEP class assignment of the same sign for the tumor cells obtained from each of the two evaluated tumor sites. A discordant result was defined as a GEP class assignment of different sign for the tumor cells obtained from the two sites or a failed GEP test (assigned the value of 0.0) for the tumor cells obtained from one but not both tumor sites.
Evaluation of associations between baseline study variables and discordant Gene Expression Profile (GEP) class
Associations between categorical baseline patient and tumor variables and concordance versus discordance of GEP class assignment to the tumor aspirates obtained from two tumor sites were evaluated by cross-table analysis. Strength of associations determined by this analysis was assessed using the chi-squared test.
Evaluation of actuarial rates of metastasis and metastatic death in study patients by Gene Expression Profile (GEP) category
The Kaplan-Meier product limit method of actuarial survival analysis was used to compute cumulative probabilities of uveal melanoma metastasis and metastatic death in patients with concordant versus discordant GEP results. The principal reason for this analysis was to determine whether patients with discordant GEP class at the two sites had a survival experience more like that of concordant Class 2 patients, concordant Class 1 patients, or intermediate between the two concordant Class patients.
Results
Study patients and tumors
We identified 80 patients who had a clinically diagnosed primary posterior uveal melanoma that was sample by fine needle aspiration biopsy (FNAB) at three or more tumor sites and whose aspirates from two of these sites were submitted for gene expression profile (GEP) testing and classification during the years 2007 through 2009. The baseline characteristics of the patients and their tumors are summarized in Table 1. The patients ranged in age at the time of FNAB from 17.6 years to 86.9 years (mean age = 62.1 years, standard deviation = 14.7 years). Their tumors ranged in largest basal diameter (LBD) from 6.0 mm to 22.0 mm (mean LBD = 12.3 mm, standard deviation = 3.3 mm) and in thickness from 1.8 mm to 14.5 mm (mean thickness = 5.8 mm, standard deviation = 2.7 mm). Cytopathologically, over half of the tumors (44 of 80 cases, 55.0%) were classified as containing epithelioid melanoma cells. However, an insufficiently cellular aspirate for cytopathological classification was observed in 15 of the 80 cases (18.8%). In spite of this latter result, none of the tumors in this series yielded an insufficient aspirate for GEP classification in both of the two sampled sites. GEP testing did fail, however, in one of the two aspirates in two of the 80 cases (2 of 160 aspirates, 1.25%).
Table 1.
Baseline characteristics of 80 study patients and their intraocular posterior uveal melanoma.
| Variable | ||
|---|---|---|
| categories of variable | number | (%) |
| Age (years) at time of FNAB | ||
| younger (<= 50) | 12 | (15.0) |
| intermediate (> 50 but <= 70) | 43 | (53.8) |
| older (> 70) | 25 | (31.3) |
| Largest basal diameter (mm) of intraocular tumor | ||
| small (<= 10) | 28 | (35.0) |
| medium (> 10 but <= 15) | 38 | (47.5) |
| large (> 15) | 14 | (17.5) |
| Maximal thickness (mm) of intraocular tumor | ||
| thinner (<= 3.5) | 21 | (26.3) |
| intermediate (> 3.5 but <= 7) | 36 | (45.0) |
| thicker (> 7) | 23 | (28.8) |
| Intraocular location category of tumor | ||
| exclusively choroidal | 59 | (73.8) |
| involving ciliary body | 21 | (26.3) |
FNAB for procurement of tumor cells in this series was performed at the time of radioactive I-125 plaque implantation in 53 cases, immediately post-enucleation in 15 patients, as a separate procedure for pathologic diagnosis prior to any treatment in 10 cases, and immediately following transcleral en bloc tumor resection in 2 patients. Initial patient management consisted of I-125 plaque radiotherapy in 55 patients, primary enucleation in 20 patients, proton beam irradiation in 2 patients, stereotactic radiation therapy in 1 patient, and observation without treatment in 2 patients. One of the two patients managed by observation had an insufficiently cellular aspirate for cytopathological classification but was categorized as Class 1 by GEP testing. The other patient managed by observation had a tumor classified as benign uveal nevus by cytopathological analysis and was also categorized as Class 1 by GEP testing.
Cytopathological classification of aspirates
Table 2 summarizes the cytopathological classification of the tumor aspirates by the pathologists who analyzed the specimens. Note that 15 of the 80 aspirates (18.8%) submitted for cytopathological analyses were judged to be insufficiency cellular to be classified pathologically. Note also that the pathologists classified 6 tumors (7.5%) as “malignant uveal melanoma” but did not specify a cell type and 4 tumors (5.0%) as “borderline” between benign uveal nevus and malignant melanoma.
Table 2.
Cytopathological classification of aspirated tumor cells from 80 clinically diagnosed choroidal and ciliochoroidal melanomas in this series.
| Cytopathological classification of aspirated tumor cells | number | (%) |
|---|---|---|
| insufficiently cellular specimen for classification | 15 | (18.8) |
| melanocytic uveal nevus | 1 | (1.3) |
| borderline melanocytic uveal tumor | 4 | (5.0) |
| uveal melanoma - spindle cell type | 16 | (20.0) |
| uveal melanoma - unspecified cell type | 6 | (7.5) |
| uveal melanoma - mixed cell type | 20 | (25.0) |
| uveal melanoma - epithelioid & necrotic | 18 | (22.5) |
GEP class assignment and failed tests
Using the weighted voting algorithm on the “a” (first site sampled) aspirates, 53 of the 80 evaluated tumors (66.3%) were categorized as GEP class 1, twenty-five (31.3%) were categorized as GEP class 2, and two (2.5%) were categorized as failed GEP tests. Using the weighted voting algorithm on the “b” (second site sampled) aspirates, 53 of the tumors were again categorized as GEP class 1 while the remaining 27 tumors (33.8%) were categorized as GEP class 2. Using the support vector machine algorithm (the machine learning algorithm favored by the developer of the GEP test we used and employed by the company that now offers this test commercially) on the “a” specimen, 52 of the 80 tumors (65.0%) were categorized as GEP class 1, twenty-six (32.5%) were categorized as GEP class 2, and two were categorized as failed GEP tests. Using the support vector machine algorithm on the “b” aspirates, 55 of the tumors (68.8%) were categorized as GEP class 1 while the remaining 25 (31.3%) were classified as GEP class 2.
Frequency of concordant and discordant Gene Expression Profile (GEP) class assignments of paired tumor aspirates
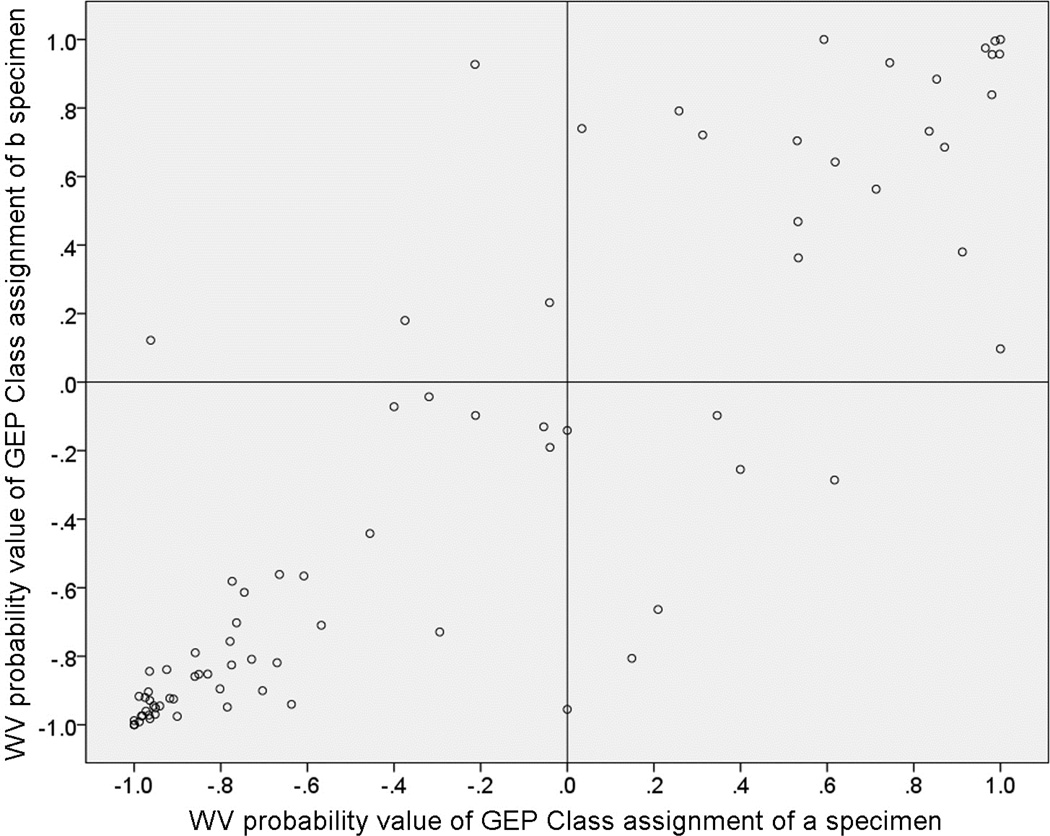
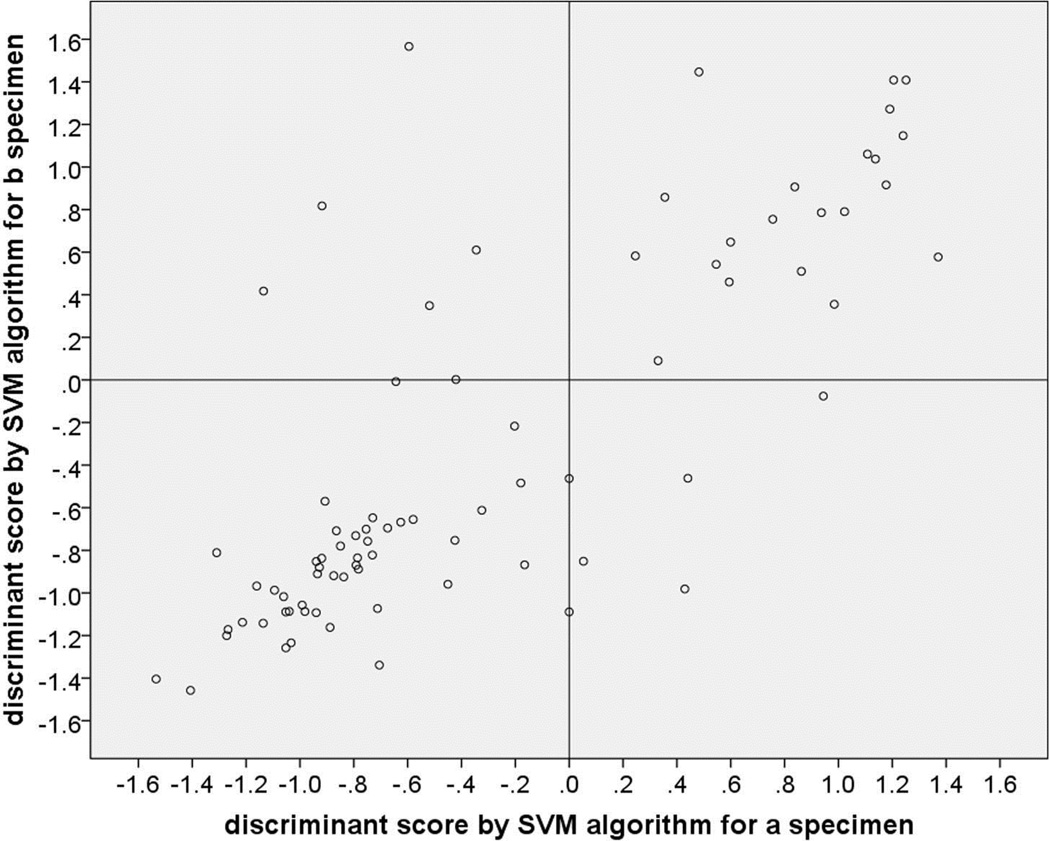
Figure 1 is a scatter plot of the weighted voting algorithm probability values associated with the GEP class assignments on that “a” versus “b” aspirates, and Figure 2 is a corresponding scatter plot of the support vector machine algorithm discriminant function scores associated with the GEP class assignments on the two aspirates. In each of these figures, cases with a concordant GEP class assignment appear in the lower left and upper right quadrants while discordant cases appear in the upper left and lower right quadrants. Each scatter plot shows 48 cases (60.0%) categorized as concordant GEP class 1 and 21 cases (26.3%) categorized as concordant GEP class 2. Each scatter plot shows 9 of the 80 cases (11.3% [95% confidence interval of proportion: 9.0% to 13.6%]) as discordant. In each of the scatter plots, 4 of the discordant cases were categorized as Class 1 on the “a” aspirate and Class 2 on the “b” aspirate (upper left quadrant of the figure) and 5 were categorized as Class 2 on the “a” aspirate and Class 1 on the “b” aspirate. In addition, the plotted location of the paired GEP class assignments fell on the vertical line through the 0.0 value of the x-axis (i.e., was associated with a failed GEP test on the “a” specimen or an extremely low confidence value of the classification for one of the specimens) for 2 cases on the weighted voting scatter plot and 4 cases on the support vector machine scatter plot. Interestingly, two cases were classified as discordant by one of the algorithms and concordant by the other algorithm (one discordant by each of the two algorithms).
Figure 1.
Scatterplot of probability scores associated with gene expression profile (GEP) class assignment of “a” and “b” aspirates of 80 posterior uveal melanoma sampled in two sites by fine needle aspiration biopsy (FNAB) and classified using the weighted voting (WV) algorithm. Dots in the lower left box are concordant GEP class 1 cases, those in the upper right box are concordant GEP class 2 cases, and those in the upper left and lower right boxes are discordant cases.
Figure 2.
Scatterplot of discriminant function scores associated with gene expression profile (GEP) class assignment of “a” and “b” aspirates of 80 posterior uveal melanoma sampled in two sites by FNAB and classified using the support vector machine (SVM) algorithm. Dots in the lower left box are concordant GEP class 1 cases, those in the upper right box are concordant GEP class 2 cases, and those in the upper left and lower right boxes are discordant cases.
In each case, the correct GEP class assignment was regarded as the class assigned to both specimens in the concordant cases, the identified GEP class in the two cases in which GEP testing failed on one specimen, and class 2 in all discordant cases.
Associations between Gene Expression Profile (GEP) class discordance and other study variables
Cross-table analysis of categorical values of baseline patient and tumor variables and GEP discordance in this series showed a trend between thickness of the tumor and GEP discordance (Table 2). In this analysis, discordant GEP class assignment occurred in 5 of 21 (23.8%) tumors <= 3.5 mm in thickness, 6 of 36 (16.7%) tumors >3.5 mm thick but <= 7 mm thick, and 1 of 23 (4.3%) tumors > 7.0 mm thick. Given the limited number of cases in our series, this association was not statistically significant (P = 0.18) by chi-squared testing. There was no similar association with patient age, largest basal tumor diameter or intraocular tumor location (cross-tables not shown).
Actuarial rates of uveal melanoma metastasis and metastatic death in Gene Expression Profile (GEP) class subgroups
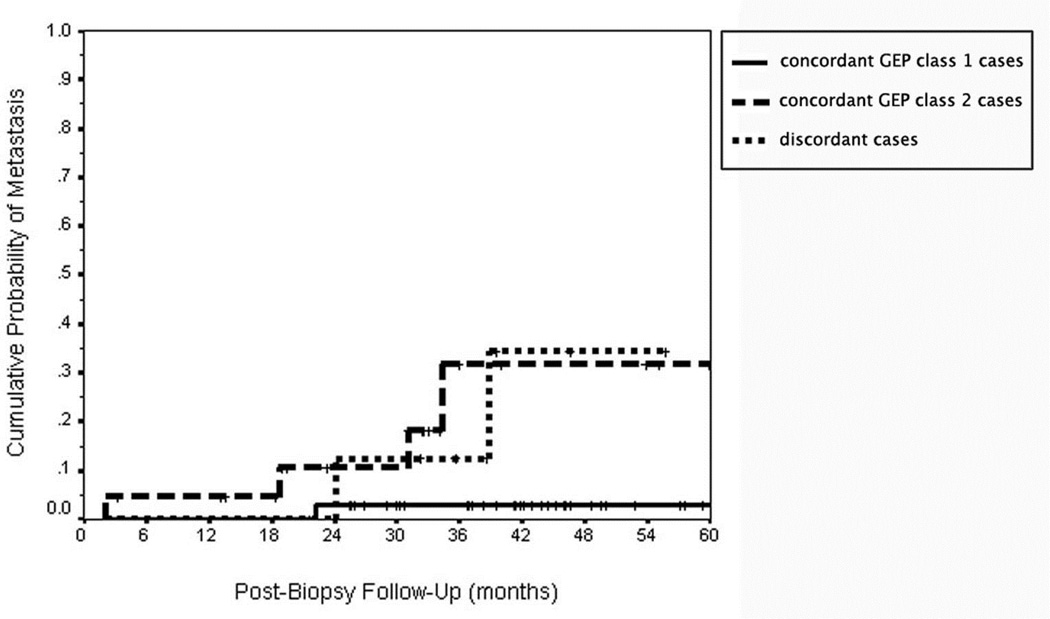
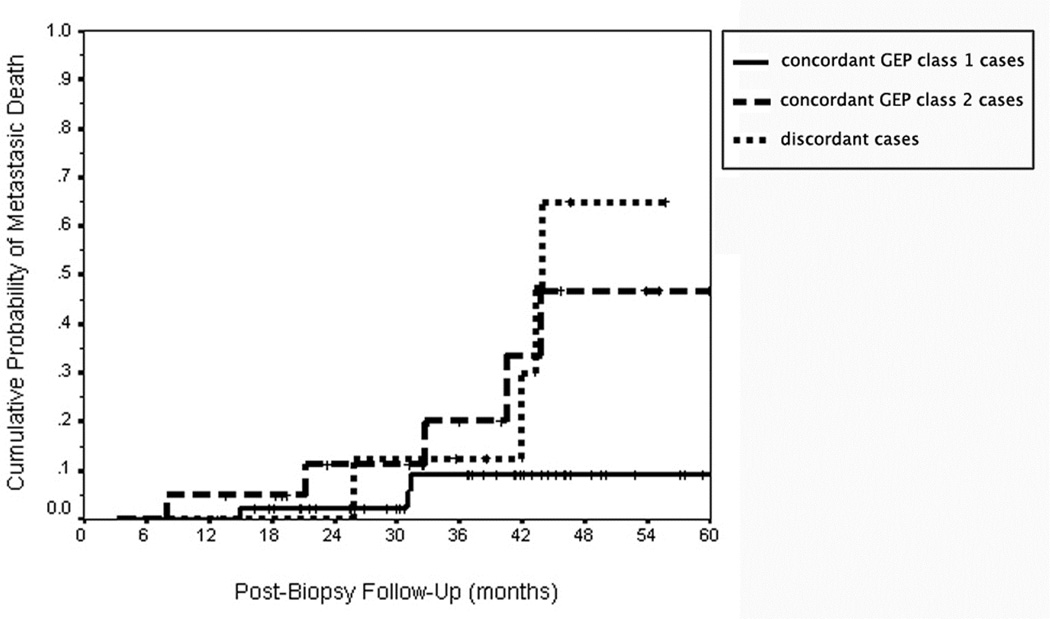
Figures 3 and 4 shows the Kaplan-Meier curve of cumulative probability of uveal melanoma metastasis and metastatic death respectively for 46 cases classified as concordant GEP class 1 by the support vector machine algorithm, 21 cases classified as GEP class 2 by the support vector machine algorithm, and nine clearly discordant cases identified by this algorithm. The outcomes of the two patients with failed GEP testing on the “a” specimen (both of which were GEP class 2 on the “b” specimen) and those of two other patients whose “b” specimen was associated with an extremely low confidence value (i.e., those having an absolute value of their discriminant score < 0.05) are not shown on this graph. As anticipated, the patients categorized as concordant GEP class 2 had a much higher metastatic rate than did the patients categorized as concordant GEP class 1. The curve for the discordant cases appears more similar to that of the concordant Class 2 cases than to that of the concordant Class 1 cases.
Figure 3.
Kaplan-Meier cumulative actuarial event rate curves for probability of uveal melanoma metastasis in 80 patients with posterior uveal melanoma as a function of concordant versus discordant subgroup. Legend: continuous line = concordant gene expression profile (GEP) class 1 cases; dash line = concordant gene expression profile (GEP) class 2 cases; small-dash line = discordant gene expression profile (GEP) cases.
Figure 4.
Kaplan-Meier cumulative actuarial event rate curves for uveal melanoma death from metastasis in 80 patients with posterior uveal melanoma as a function of concordant versus discordant subgroup. Legend: continuous line = concordant gene expression profile (GEP) class 1 cases; dash line = concordant gene expression profile (GEP) class 2 cases; small-dash line = discordant gene expression profile (GEP) cases.
Complications from Fine Needle Aspiration Biopsy (FNAB) performed in this series of patients
Out of the 53 patients treated by plaque, a total of 32 patients (60.4%) had some vitreous hemorrhage. Only 8 (15.1%) of those required pars plana vitrectomy because of a nonclearing vitreous hemorrhage (n=5 (9.4%)) or non-clearing vitreous hemorrhage and rhegmatogenous retinal detachment (n=3 (5.7%)). Two of these 53 patients later required enucleation but neither of them was because of a complication related to the FNAB procedure but secondary to radiation vasculopathy complications.
Discussion
Our results indicate (and provide corresponding probability evidence) that single-site FNAB of posterior uveal melanomas for GEP testing and prognostic classification entails a not insignificant risk of misclassification of a GEP class 2 tumor as a GEP class 1 tumor or an inconclusive GEP class assignment because of a low confidence result plus a small risk of a failed GEP test (i.e., a biopsy for which GEP testing of the tumor cells does not result in prognostic classification). The more pertinent result may be our finding that if we had aspirated and evaluated tumor cells from only one (the first sampled) site, the GEP testing of those aspirates would have identified the tumor cells as discordant with the final classification in 7.5% of the cases. If we included those cases with a “low confidence” GEP class assignment for one or both aspirates (those plotted in the concordant quadrants lie very close to either the horizontal or vertical line through the 0.0 value on one of the axe) and the two cases with a failed GEP test on one aspirate are also classified as “discordant”, as many as 13 cases (16.3%) by the weighted voting algorithm and 15 cases (18.8%) by the support vector machine algorithm could have been classified as discordant. Realizing that the advantage of GEP is to capture a functional “snapshot” of the tumor's microenvironment and that is not expected to vary as much across most tumors,6 it has been reported within other systemic tumors that subclonal diversity may be observed and these tumor subclones may show differential gene expression due to both genetic and epigenetic heterogeneity.8
Based on our results, we anticipate the following potential misinterpretation of sequential FNABs for GEP testing and classification in the future. Consider the following scenario. A melanocytic choroidal tumor in the “nevus versus melanoma” category11 is biopsied by single-site FNAB shortly after its detection. GEP testing of the aspirated cells yields a class 1 assignment. The tumor is left untreated initially but is noted to have enlarged unequivocally by the patient’s 6-month post-biopsy follow-up evaluation. The tumor is rebiopsied by single-site FNAB at that time. GEP testing of this specimen yields a class 2 assignment. The ophthalmologists involved in the patient’s care are likely to report this case as convincing evidence of “transformation” of a GEP class 1 tumor to a GEP class 2 tumor. Based on this hypothetical case, clinicians are likely to advocate for aggressive treatment of GEP class 1 melanocytic uveal tumors to prevent such transformation. In this scenario, one will never know whether the tumor would have yielded GEP class 2 cells at the initial biopsy if a second tumor site had been sampled.
To date, we have rebiopsied 3 melanocytic posterior uveal tumors in the “nevus versus melanoma” category (none of which occurred in the group of patients reported in the present study) that proved to be concordant GEP class 1 tumors on their baseline 2-site tumor FNAB, were left untreated initially, and enlarged during follow-up. To date, all of these tumors have proved to be concordant GEP class 1 tumors on repeat two-site tumor FNAB at that time. While we do not deny that transformation from GEP class 1 to GEP class 2 is possible, we anticipate on the basis of our experience that such transformation may be rather uncommon. A case such as that described in the preceding paragraph should not be accepted as convincing or compelling evidence of such transformation and should certainly not be regarded as evidence that class 1 to class 2 transformation occurs frequently or routinely.
In this study, we considered the “correct” GEP class of all discordant tumors that were associated with satisfactory probability scores for the GEP class 2 aspirate to be class 2 tumors. One could argue that such discordant tumors should have been regarded as “borderline” and not GEP class 2 and that patients having such tumors should have been regarded as a separate subset with intermediate prognosis. As shown by the actuarial curves for metastasis and metastatic death (Figures 3 and 4) in this subgroup, however, the curves for the discordant cases resembled those of the patients with class 2 tumors much more closely than those of the patients with class 1 tumors. Because of this, we believe our interpretation to be appropriate.
Additionally, because we sampled two tumor sites for GEP testing (and one or more additional sites for cytopathological testing) in each case, it is possible to speculate that if more than two tumor sites were sampled for GEP testing in each case, we might have identified a few additional tumors that were concordant GEP class 1 at two sites but GEP class 2 at the third sampled site. Although sampling multiple sites does not increase the surgery time by more that 5 minutes and the cost of cytology of 1 or 3 specimens of the same patient is similar, the cost of GEP testing of 3 tumor specimens is likely to be prohibitive in most clinical situations. Moreover, the greater the number of tumor sites sampled in an individual patient, the greater the cumulative risk of complications (principally vitreous hemorrhage, retinal tears, and rhegmatogenous retinal detachment in eyes biopsied by a transvitreal FNAB technique and tract seeding leading to subsequent development of implantation tumors in eyes sampled by either method) that could affect the patient’s ocular-visual outcome in the affected eye adversely. While limited vitreous hemorrhage occurs during most transvitreal FNABs, major vitreous hemorrhage or retinal detachment that prompts subsequent posterior vitrectomy and/or a scleral buckling procedure is a relatively uncommon event in our experience.11 In this series, our rate of retinal detachment and need for surgical re-intervention is comparable to other reported series.14 To date, we have not had a single patient with posterior uveal melanoma who developed a clinically apparent implantation tumor at any eye wall puncture site for FNAB or had to remove an eye due to FNAB complication. At present, there is no reported scientifically valid evidence that FNAB of a posterior uveal melanoma worsens a patient’s risk of metastasis. Nevertheless, the potential for ocular complications associated with multiple FNABs should always discussed with the patient prior to the procedure.
Although the mathematics behind machine learning algorithms used for classification of GEP data describe the classification process as occurring in “multidimensional space”, many authors have attempted to show the separation between cases in 3-dimensional space.7,15–18 In our opinion, such 3-dimensional plotting of results is no more valid than two-dimensional plotting19 of weight voting probability values and support vector machine discriminant function scores, which we employed for data summarization and analysis in this study.
Genetic testing of uveal melanoma cells by a variety of methods (including karyotyping, CGH, FISH, SNP, MLPA, etc.) has identified a number of non-random abnormalities (principally loss of one chromosome 3 [monosomy 3], especially if coupled with a loss or duplication of the short arm of chromosome 8, and gain of chromosome 6p) that are associated with increased probabilities of subsequent emergence of distant metastasis or better prognosis. Because genetic analysis is performed routinely for a variety of conditions in many laboratories and because such testing is generally much less expensive than the commercially available, validated prognostic GEP testing performed by a CLIA certified laboratory (DecisionDx-UM™)20, many uveal melanoma subspecialists around the world current arrange for genetic testing and classification of uveal melanoma cells obtained at the time of initial ocular tumor treatment as an alternative to GEP testing and classification. Studies reporting chromosomal heterogeneity are mostly performed on enucleation specimens rather than in FNAB harvested specimens.9,21 Although the reported intratumoral heterogeneity using fluorescence in situ hybridization (FISH) in a smaller number of enucleated eyes is 14%,9 the relative merit of the different approaches is beyond the scope of this work. Interested readers are referred to a published editorial-opinion of this topic for further information.6,22
Comparing GEP testing and the various currently available methods of genetic analysis in the context of our study, GEP is the only genetic test that is associated with a probability score corresponding to its prognostic subgroup assignment. This probability score is a function of many features, including the homogeneity versus heterogeneity of the tumor cells in the evaluated specimen, the relative proportion of different cellular clones within the evaluated specimen, the extent of cellular necrosis that exists in the sampled region of the tumor, the density of any lymphocytic infiltration of the tumor in the specimen, and the amount of blood in the specimen. The greater the number of tumor cells an evaluated specimen contains, the greater the likelihood that those cells will be representative of the tumor as a whole. An FNAB-derived tumor aspirate may contain only a few cells (a feature usually indicative of cohesiveness and relative benignity of those cells in the case of uveal melanomas),19 and this increases the chance of obtaining a sample that is not representative of the entire tumor. With chromosomal testing, an aberration must be present in a substantial proportion of the evaluated tumor cells for it to be detected with reasonable assurance; however, the precise rules for categorizing a specimen of tumor cells as having or not having a particular mutation can vary greatly from lab to lab and even over time within the same laboratory.9,21 In general, chromosomal testing usually requires a substantially greater number of cells to obtain a successful result. With tumor specimens that are hypocellular, we anticipate a substantially greater frequency of failed tests (i.e., ones that do not provide a definitive classification associated with a reasonable degree of certainly) when they are evaluated by chromosomal tests.
There are clearly some relevant limitations to routine sampling of 2 (or more) tumor sites for GEP testing and prognostic classification in clinical practice. When the testing reported for this study was being performed, all GEP testing and classification was done in the developer’s research laboratory, and the costs of the testing were paid with grant funds. Now the test is available commercially as the DecisionDx-UM™ (Castle Biosciences, Inc., Phoenix, Arizona)20 and unfortunately each sample test incurs in a cost sometimes not covered by insurance carriers in the United States even for a single sample, let alone two samples. Conversely, our experience has shown minimal if any measurable increase in surgical time when we sample a tumor 4 opposed to 2 times.
In summary, our study suggests that two-site sampling may be advantageous to reduce the risk of prognostic underclassification of clinically diagnosed posterior uveal melanomas biopsied prior to or at the time of initial ocular tumor treatment for GEP testing and prognostic classification of the tumor cells. The statement is not a criticism of GEP testing in general, the DecisionDx-UM test in specific, or of the accuracy or validity of the test result on the evaluated cells. It is really a criticism of the limitations of FNAB for obtaining a truly representative sample of the intraocular tumor. In spite of these results, clinicians who perform single-site FNAB of their patients’ posterior uveal melanomas for prognostic GEP testing should be reassured that this testing is likely to result in the correct prognostic classification of the tumor about 85% of the time in smaller tumors. Consequently, prognostic misclassification may explain a substantial proportion of the instances of metastasis that are currently observed in GEP class 1 tumor patients based on single-site FNAB specimen testing.
Table 3.
Cross tabulation of association between thickness of tumor at the time of treatment and discordant Gene Expression Profile (GEP) class assignment in paired tumor aspirates from 80 patients with posterior uveal melanoma.
| Concordance versus Discordance of
GEP Class in Paired Tumor Aspirates by Support Vector Machine Algorithm |
||||
|---|---|---|---|---|
| Concordant | Discordant | |||
| Number | (%) | Number | (%) | |
| Thickness category of tumor | ||||
| thinner (<= 3.5 mm thick) | 16 | (76.2) | 5 | (23.8) |
| intermediate (>3.5 mm but <= 7.0 mm thick) | 30 | (83.3) | 6 | (16.7) |
| thicker (>7.0 mm thick) | 22 | (95.7) | 1 | (4.3) |
chi-squared = 3.4 df = 2 P = 0.18
Acknowledgements
FUNDING/SUPPORT: This work was supported in part by an unrestricted grant from Research to Prevent Blindness, Inc., New York, New York, to the Department of Ophthalmology, University of Cincinnati College of Medicine (James J. Augsburger, M.D., Chairman) and the Quest for Vision Fund of the Department of Ophthalmology, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA. Dr. Harbour and his laboratory were supported in part by grants from the National Eye Institute (P30 EY02687c), the National Cancer Institute (RO1 CA125970), the Barnes-Jewish Hospital Foundation, the Kling Family Foundation, the Tumori Foundation, the Horncrest Foundation, and Research to Prevent Blindness (David F. Weeks Professorship) during the time the laboratory testing described in this paper was performed.
OTHER ACKNOWLEDGMENTS: The authors acknowledge J. William Harbour MD, Michael D. Onken PhD, and Lori A. Worley BS (all affiliated with the Department of Ophthalmology and Visual Science, Washington University School of Medicine, St. Louis, Missouri, USA when the laboratory testing described in this study was performed) for their preparation of submitted RNA samples, testing of those samples, and prognostic classification of the aspirates described in this report. Dr. Harbour is currently affiliated with the Bascom Palmer Eye Institute, University of Miami, Miami, FL.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
FINANCIAL DISCLOSURES: the authors have no financial disclosures or conflicts to disclose.
CONTRIBUTION OF AUTHORS: concept and design of study (JJA, ZMC), data collection (ZMC, BAA), data analysis (JJA, ZMC, BAA) and interpretation of findings (JJA, ZMC), statistical expertise (JJA, BAA), provision of material, patients, etc (JJA, ZMC), literature search (JJA, ZMC), writing the article and critical revision (ZMC, JJA), final approval of article (ZMC), funding (JJA), Administrative support (ZMC).
References
- 1.Liotta L, Petricoin E. Molecular profiling of human cancer. Nat Rev Genet. 2000;1(1):48–56. doi: 10.1038/35049567. [DOI] [PubMed] [Google Scholar]
- 2.Alizadeh AA, Ross DT, Perou CM, van de Rijn M. Toward a novel classification of human malignancies based on gene expression patterns. J Pathol. 2001;195(1):41–52. doi: 10.1002/path.889. [DOI] [PubMed] [Google Scholar]
- 3.Kallioniemi OP, Wagner U, Kononen J, Sauter G. Tissue microarray technology for high-throughput molecular profiling of cancer. Hum Mol Genet. 2001;10(7):657–662. doi: 10.1093/hmg/10.7.657. [DOI] [PubMed] [Google Scholar]
- 4.Onken MD, Worley LA, Ehlers JP, Harbour JW. Gene expression profiling in uveal melanoma reveals two molecular classes and predicts metastatic death. Cancer Res. 2004;64(20):7205–7209. doi: 10.1158/0008-5472.CAN-04-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Onken MD, Worley LA, Tuscan MD, Harbour JW. An accurate, clinically feasible multi-gene expression assay for predicting metastasis in uveal melanoma. J Mol Diagn. 2010;12(4):461–468. doi: 10.2353/jmoldx.2010.090220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Onken MD, Worley LA, Char DH, et al. Collaborative Ocular Oncology Group report number 1: prospective validation of a multi-gene prognostic assay in uveal melanoma. Ophthalmology. 2012;119(8):1596–1603. doi: 10.1016/j.ophtha.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Onken MD, Worley LA, Davila RM, et al. Prognostic testing in uveal melanoma by transcriptomic profiling of fine needle biopsy specimens. J Mol Diagn. 2006;8(5):567–573. doi: 10.2353/jmoldx.2006.060077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burrell RA, McGranahan N, Bartek J, Swanton C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature. 2013;501(7467):338–345. doi: 10.1038/nature12625. [DOI] [PubMed] [Google Scholar]
- 9.Maat W, Jordanova ES, van Zelderen-Bhola SL, et al. The heterogeneous distribution of monosomy 3 in uveal melanomas: implications for prognostication based on fine-needle aspiration biopsies. Arch Pathol Lab Med. 2007;131(1):91–96. doi: 10.5858/2007-131-91-THDOMI. [DOI] [PubMed] [Google Scholar]
- 10.Augsburger JJ, Shields JA. Fine needle aspiration biopsy of solid intraocular tumors: indications, instrumentation and techniques. Ophthalmic Surg Lasers. 1984;15(1):34–40. [PubMed] [Google Scholar]
- 11.Augsburger JJ, Corrêa ZM, Schneider S, et al. Diagnostic transvitreal fine-needle aspiration biopsy of small melanocytic choroidal tumor in nevus versus melanoma category. Trans Am Ophthalmol Soc. 2002;100:225–232. [PMC free article] [PubMed] [Google Scholar]
- 12.Golub TR, Slonim DK, Tamayo M, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286(5439):531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 13.Gist. Support vector machine. Overview of the SVM algorithm. svm.edsc.edu/svm-overview.html (9/19/1999) [Google Scholar]
- 14.Eide N, Walaas L. Fine-needle aspiration biopsy and other biopsies in suspected intraocular malignant disease: a review. Acta Ophthalmol. 2009;87(6):588–601. doi: 10.1111/j.1755-3768.2009.01637.x. [DOI] [PubMed] [Google Scholar]
- 15.Yeoh EJ, Ross ME, Shurtleff SA, et al. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell. 2002;1(2):133–143. doi: 10.1016/s1535-6108(02)00032-6. [DOI] [PubMed] [Google Scholar]
- 16.Bullinger L, Döhner K, Bair E, et al. Use of gene-expression profiling to identify prognostic subclasses in adult acute myeloid leukemia. N Engl J Med. 2004;350(16):1605–1616. doi: 10.1056/NEJMoa031046. [DOI] [PubMed] [Google Scholar]
- 17.Ross ME, Mahfouz R, Onciu M, et al. Gene expression profiling of pediatric acute myelogenous leukemia. Blood. 2004;104(12):3679–3687. doi: 10.1182/blood-2004-03-1154. [DOI] [PubMed] [Google Scholar]
- 18.Freije WA, Castro-Vargas FE, Fang Z, et al. Gene expression profiling of gliomas strongly predicts survival. Cancer Research. 2004;64(18):6503–6510. doi: 10.1158/0008-5472.CAN-04-0452. [DOI] [PubMed] [Google Scholar]
- 19.Harbour JW, Chen R. The DecisionDx-UM gene expression profile test provides risk stratification and individualized patient care in uveal melanoma. PLoS Curr. 2013 Apr 9; doi: 10.1371/currents.eogt.af8ba80fc776c8f1ce8f5dc485d4a618. Published online 2013 April 9. doi:10.1371/currents.eogt.af8ba80fc776c8f1ce8f5dc485d4a618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harbour JW. Molecular prognostic testing and individualized patient care in uveal melanoma. Am J Ophthalmol. 2009;148(6):823–829. doi: 10.1016/j.ajo.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mensink HW, Vaarwater I, Kilic E, et al. Chromosome 3 Intratumor Heterogeneity in Uveal Melanoma. Invest Ophthalmol Vis Sci. 2009;50(2):500–504. doi: 10.1167/iovs.08-2279. [DOI] [PubMed] [Google Scholar]
- 22.Augsburger JJ, Corrêa ZM, Trichopoulos N. Prognostic implications of cytopathologic classification of melanocytic uveal tumors evaluated by fine-needle aspiration biopsy. Arq Bras Oftalmol. 2013;76(2):72–79. doi: 10.1590/s0004-27492013000200004. [DOI] [PubMed] [Google Scholar]