Abstract
Voltage-gated sodium channels (NaVs) provide the initial electrical signal that drives action potential generation in many excitable cells of the brain, heart, and nervous system. For more than 60 years, functional studies of NaVs have occupied a central place in physiological and biophysical investigation of the molecular basis of excitability. Recently, structural studies of members of a large family of bacterial voltage-gated sodium channels (BacNaVs) prevalent in soil, marine, and salt lake environments that bear many of the core features of eukaryotic NaVs have reframed ideas for voltage-gated channel function, ion selectivity, and pharmacology. Here, we analyze the recent advances, unanswered questions, and potential of BacNaVs as templates for drug development efforts.
Introduction
Heartbeats, thoughts, and sensations of pleasure and pain begin with the seemingly simple act of the opening of an ion-selective hole in a cell membrane that allows an inward rush of sodium ions. This influx causes a change in the membrane potential within the timescale of milliseconds and initiates the electrical signaling cascade know as the ‘action potential’ that is the signature electrical behavior of excitable cells such as a neurons and muscle 1. A specialized class of transmembrane proteins, known as voltage-gated sodium channels (NaVs), forms the conduits for this rapid ion influx. Biophysical characterization of NaVs and elucidation of their functional roles in excitable cells have been a pillar of physiological studies for over 60 years 2; 3; 4. The importance of NaVs in human biology is profound. This ion channel class is linked to a multitude of ailments including cardiac arrhythmias, movement disorders, pain, migraine, and epilepsy 5 and is the target for a host of pharmaceuticals and ongoing drug development efforts 6. Moreover, it is becoming increasingly clear that NaVs play a role in many cells that are not traditionally thought of as excitable, such as astrocytes, T cells, macrophages, and cancer cells 7. Hence, the need to understand the mechanics of how such channels function, the molecular basis for their activity, and the development of new tools that can probe and control their function remains exceptionally high.
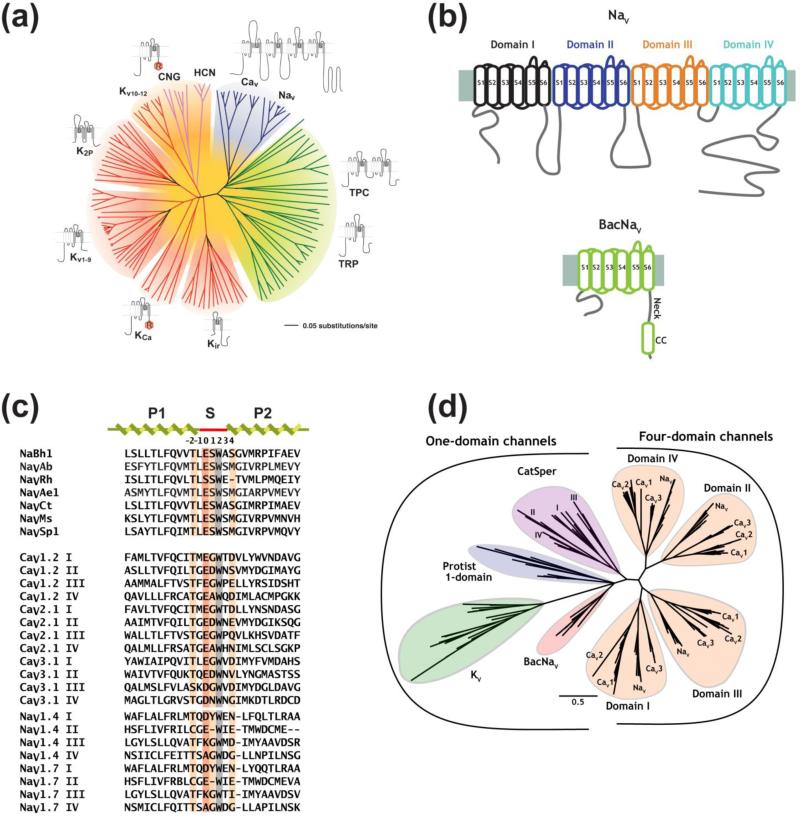
NaVs are found in metazoans from jellyfish to humans and are formed by large polytopic transmembrane proteins that are members voltage-gated ion channel (VGIC) signaling protein superfamily 1; 2. This class encompasses voltage-gated channels for sodium, calcium, and potassium, the large family of transient receptor potential (TRP) channels, and a variety of other ion channels (Fig. 1a). The eukaryotic pore-forming NaV subunit is composed of a single polypeptide chain of ~2,000 amino acids (~260 kDa) comprising four homologous transmembrane domains (Fig. 1b) and, along with that from voltage-gated calcium channels, CaVs, represents the largest pore-forming polypeptide within the superfamily. Nine NaV isoforms are found in humans (NaV1.1-NaV1.9) and have differing pharmacologies, expression patterns, and functional signatures 8. In addition to the pore-forming subunit, native channels associate with a class of single-pass transmembrane NaVβ subunits. These auxiliary subunits affect function, pharmacology, and can carry mutations can cause disease 9.
Figure 1. BacNaV topology and relationships to voltage-gated ion channel (VGIC) superfamily members.
(a) Unrooted tree showing the amino acid sequence relations of the minimal pore regions of VGIC superfamily members (modified from 10). Indicated subfamilies are (clockwise): voltage-gated calcium and sodium channels (CaV and NaV), two-pore (TPC) and transient receptor potential (TRP) channels, inwardly rectifying potassium channels (Kir), calcium-activated potassium channels (KCa), voltage-gated potassium channels (KV1–KV9), K2P channels, voltage-gated potassium channels from the EAG family (KV10–KV12), cyclic nucleotide gated channels (CNG), and hyperpolarization activated channels (HCN). “R” indicates identifiable regulatory domains. (b) Topology diagram comparing eukaryotic NaV (top) and BacNaV pore forming subunits. S1-S6 segments are labeled. Individual NaV six transmembrane repeats of are colored black, blue, orange, and teal. BacNaV neck and coiled coil (CC) domains are indicated. (c) Sequence alignment for selected BacNaVs selectivity filter and pore helices, mammalian CaV subtype exemplars, and mammalian NaV1.4 and NaV1.7. Selectivity filter numbering is indicated. Position (0) is highlighted in dark orange. Residues involved in selectivity are highlighted light orange. Grey highlights the the conserved Trp (+2) anchor position. (d) Unrooted tree showing a comparison based on the selectivity filter sequences for BacNaVs compared with KV channels, CatSper, Protist one-domain channels, and the individual domains of NaVs and CaVs (modified from 43).
Each of the four NaV transmembrane domains (Domains I-IV) has an architecture shared by many VGIC superfamily members 10 (Fig. 1a, b). Transmembrane segments S1-S4 form the voltage sensor domain (VSD), whereas transmembrane segments S5-S6 form the pore-forming domain (PD) that houses the element that defines the ion selectivity properties of the channel, the selectivity filter (SF). The intracellular loops that connect the NaV transmembrane domains have important roles in channel regulation. The best studied are the III-IV loop, which bears a segment known as the inactivation peptide that is essential for the fast inactivation properties of metazoan NaVs 11; 12; 13, and the cytoplasmic C-terminal tail, which forms a hub for binding of a number of regulatory factors including calmodulin 14. Together, these elements endow eukaryotic NaVs with complex functional properties and connect them to various regulatory pathways within the cell.
From the standpoint of ion channel biophysics, studies of NaVs have set a number of paradigms for understanding channel function including: the importance of the S4 segment of the VSD in voltage sensing, the concept of an intracellular ‘inactivation particle’, ideas that some hydrophobic drugs could access the channel pore by lateral access through the membrane hydrophobic bilayer, and the likely physical dimensions of the selectivity filter 1. Yet, without structural data, it has been difficult to place such foundational ideas onto a molecular scaffold. NaVs, similar to many other eukaryotic membrane proteins, have been difficult to obtain in the quantities and qualities required for high-resolution structure determination. Because of their size and complexity, structural understanding of eukaryotic NaVs remains limited to low-resolution electron microscopy images of the complete protein isolated from natural sources, the electric organ of the eel Electrophorus electricus 15. However, there has been steady progress in obtaining structural information for specific domains including: the inactivation peptide 16, portions of the C-terminal cytoplasmic tail alone 17; 18, C-terminal tail complexes with regulatory factors such as calmodulin 14; 19; 20; 21; 22, and extracellular domains for two NaVβ auxiliary subunit isoforms 23; 24. Elucidation of the architecture of these eukaryotic NaV elements begins to flesh out key pieces of the NaV molecular framework but have left the larger question of understanding the molecular structure of the central components of the ion-selective hole unaddressed.
For potassium channels, the biochemical tractability and relative simplicity of bacterial potassium channels was essential for opening the first paths to high-resolution structural studies 25; 26; 27; 28. The discovery of a large family of bacterial NaVs (BacNaVs) 29; 30; 31; 32; 33 gave the NaV field a simplified scaffold to begin to outline key structural principles of NaV function and the substrate for the first structural insights into this channel class. BacNaVs have ~275 residues, making them ~1/8th the size of a eukaryotic NaV pore-forming subunit. Rather than having the twenty-four transmembrane segment architecture of eukaryotic NaVs, BacNaVs are built from a six transmembrane segment architecture comprising a VSD and PD (Fig. 1b) that assembles into homotetramers 34; 35; 36; 37; 38; 39 in a manner similar to many voltage-gated potassium channels 1. Initial studies demonstrated that BacNaVs had an ion selectivity profile that was similar to NaVs 29; 40, even though the actual selectivity BacNaV selectivity filter sequence has more in common with those from CaVs than NaVs 29; 33; 41 (Fig. 1c). It is interesting that although BacNaVs have been posited as ancestors of eukaryotic NaVs 42, clade analysis places them on a different evolutionary branch that is closer to a calcium channel family found in sperm known as CatSper 43 (Fig. 1d) and in a position consistent with the original identification strategy, which was a CatSper-based database search 29. Regardless of the precise evolutionary connections, the initial report of a functional bacterial homolog of NaV/CaV branch of the VGIC superfamily (Fig. 1a), named NaChBac 29, was a critical turning point for the field and held the promise that it would ultimately yield a high-resolution crystal structure that would enlighten understanding of its eukaryotic relatives 29; 44.
Breakthrough BacNav structures
Realization that the BacNaV family is very large, having >500 identifiable members, together with functional characterization a variety of BacNaVs has helped established that these channels share many important features traditionally associated with canonical vertebrate NaVs and CaVs including: voltage-dependent activation, slow inactivation, ion selectivity, and drug block 29; 30; 32; 40; 45; 46; 47; 48. These shared functional characteristics imply a significant structural conservation across 3-4 billion years of evolution and suggest that understanding BacNaV architecture should provide good models for defining core features of the eukaryotic members of the NaV and CaV branch of the VGIC superfamily. Unlike the large, ~2000-3000 residue pore-forming subunits of vertebrate NaV and CaV channels, it has been possible to overexpress and purify large quantities of a variety of bacterial ion channels as stable biochemical samples suitable for crystallization studies 25; 26; 49; 50; 51; 52; 53; 54; 55. The demonstration that some BacNaVs shared these biochemical properties 35; 36; 56, together with the possibility to leverage diversity-based strategies 57 facilitated by the multitude of BacNaV sequences elevated the hopes that studies of this family would yield to structural characterization.
In 2011, a landmark study unveiled the first BacNaV structure, a mutant, I217C, of NaVAb from Arcobacter butzleri at 2.7Å resolution crystallized from a lipid-based bicelle system 37. A virtual explosion of BacNaV structures has since followed, fulfilling the promise that these bacterial proteins would shed light on fundamental relationships within the VGIC superfamily. Three additional full-length BacNaV structures have been reported subsequently: NaVRh from Rickettsiales sp. HIMB114 crystallized from a detergent-lipid mixture in an asymmetric conformation and determined at 3.05Å resolution 39; wild-type NaVAb crystallized in distinct and asymmetric conformations and determined at 3.2Å resolution 38; and NaVCt from Caldalkalibacillus thermarum reconstituted in lipid bilayers and determined at 9Å resolution by electron crystallography 58. A novel protein-engineering strategy akin to surgical removal of the VSDs 35; 36 has also lead to “pore-only” structures crystallized from detergent solutions for NaVMs from Magnetococcus marinus MC-1 and solved at 3.49Å 59 and 2.9Å resolution 60, and NaVAe1p from Alkalilimnicola ehrlichii determined at 3.46Å resolution 41. Crystallographic and physiological studies have been further combined to study a highly Ca2+-selective form of the parental NaVAb channel (nicknamed ‘CaVAb’), which has provided insight into the structural basis for ion selectivity in calcium channels 61. Most recently, crystallographic and computationally-derived models of small molecule drugs bound to the NaVMs channel pore have provided a first glimpse into how some drugs may bind NaV and CaV channels 62. Together these studies highlight the versatility and advantage of employing the relatively “simple” BacNaV channels as a model VGIC system. Considering how far the structural characterization of BacNaVs has advanced in recent years, we anticipate many exciting advances in years to come. Here, we review the available BacNav structures in the context of historical physiological data and ask how these structures might help direct future experiments and on-going drug discovery efforts.
Defining the BacNav architecture
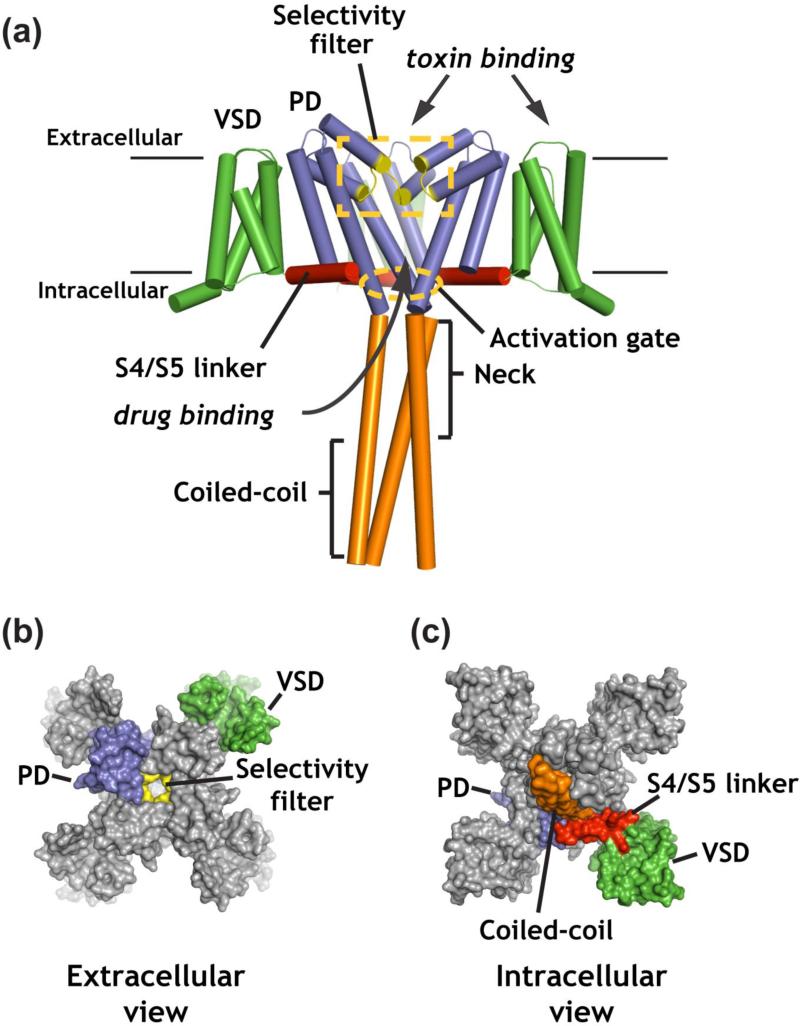
The BacNav structures cement the concept that all VGICs share a conserved architecture (Fig. 2a-c) in which four subunits or homologous domains create a central ion-conducting pore domain (PD) surrounded by four voltage sensor domains (VSDs) 37; 38; 39; 41; 58; 59; 62. The VSDs are composed of the S1-S4 segments, where the S4 places highly conserved arginine residues within the membrane electric field that undergo outward movement upon depolarization, giving rise to the phenomena of the “gating currents” 63; 64; 65; 66; 67; 68; 69. The BacNaV structures also confirm the commonality of the domain-swapped quaternary arrangement, first seen in the KV1.2/2.1 chimera structures 70; 71, whereby the VSD of one subunit packs alongside the PD of the neighboring subunit (Fig. 2b, c). This domain-swapped organization poses a fantastic topological conundrum that must be solved every time a VGIC folds into the membrane. Mechanistically, it also raises the possibility that movement the S4-S5 linker caused by outward translocation of S4 impact more than one pore domain subunit and enhances cooperativity among the channel subunits during gating. In the BacNaV PD (Fig. 2a-b), the S5 helices surround the pore-lining S6 helices and are connected through a critical helix-loop-helix motif. Together, these P1 and P2 helices form the selectivity filter and extracellular vestibule that appears to represent a conserved and defining characteristic shared with eukaryotic NaVs and CaVs (Fig. 1c).
Figure 2. Overall structure of BacNaV channels.
(a) Composite full-length BacNaV structure generated by aligning the NaVAe1p structure containing the neck and coiled-coil region (PDB: 4LTO) 41 onto NaVAb (PDB 3RVY) 37. Key structural and functional features of the BacNaV channels are labeled including the voltage-sensor domain (green) (VSD), pore domain (slate) (PD), S4/S5 linker (red), C-terminal domain (slate), neck and coiled coil (orange), selectivity filter (yellow), and S6 activation gate. General locations of pharmacologically relevant sites in eukaryotic NaV channels are also indicated in italics. Black lines indicate approximate boundaries of the membrane bilayer. For clarity, one pore subunit and voltage sensor domain are not shown. (b) Extracellular and (c) intracellular views of the BacNaV channel highlighting basic functional elements and the domain swapped arrangement of the VSD around the PD of a neighboring subunit.
The cytoplasmic domain (CTD) that follows the pore-lining S6 transmembrane helix has two domains (Figs. 1b and 2a): a membrane proximal region termed the ‘neck’ region 41 and a C-terminal coiled-coil domain (CCD) 33; 41; 72. Although the entire CTD has been present in the protein constructs used for NaVAb 37; 38, CaVAb 61, NaVCt 58, one NaVMs ‘pore-only’ construct 60, and NaVAe1p 41, electron density revealing its structure and relationship to the BacNaV PD has only been reported for NaVAe1p 41. The CTD is unique to the BacNaVs compared to their vertebrate NaV and CaV channel cousins; however analogous structures are seen in other tetrameric channels in the VGIC family, such as Kv7 (KCNQ) 73; 74; 75 and TRP channels 76.
The BacNaV channel pore
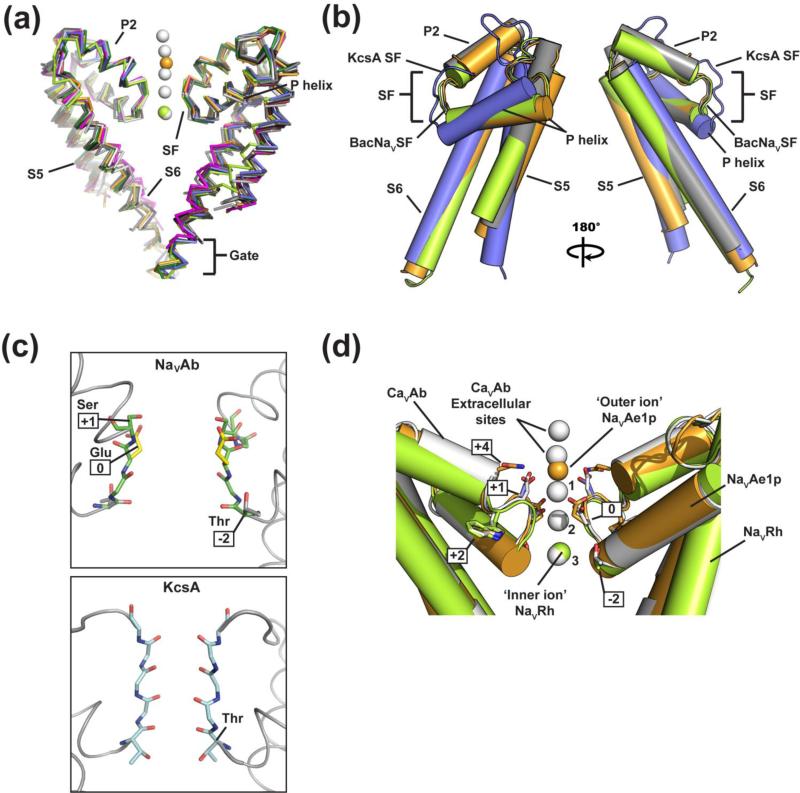
The pore domain (PD) forms the heart of a VGIC that controls ion selectivity and ion passage across the membrane (Fig. 2a). The large collection of BacNaV structures all reveal the same basic PD fold (Fig. 3a). Two transmembrane helices, S5 and S6, are bridged by the pore helices P1 and P2 linked by the selectivity filter (SF). The P1-SF-P2 structure forms the channel ‘active site’ required for engaging and selecting the permeant ions, whereas S6 lines the pore and provides the structure that closes the intracellular activation gate of the channel.
Figure 3. Comparisons of BacNaV PD structures and ion binding sites.
(a) Ribbon diagram of a PD backbone superposition of NaVAb (3RVY) 37 (black), NaVAbA/B (4EKW) 38 (light gray), NaVAbC/D (4EKW) 38 (medium gray), NaVRh (4DXW) 39 (light green), NaVMs (4FLF)59(dark red), NaVMs (3ZJZ) 60 (magenta), NaVAe1p (4LTO) 41 (orange), NaVCt (4BGN) 58(copy A, marine and copy B, slate), and CaVAb (4MS2) 61 (white), Outer ion from NaVAe1p, inner ion from NaVRh, and selectivity filter ions from CaVAb are shown as orange, light green, and white spheres, respectively. Two subunits are shown. Selectivity filter (SF), P1 (P helix), P2, S5, and S6 elements are labeled. Location of the intracellular gate is indicated. (b) Cylinder diagram of the superposition of a single PD subunit from NaVAb (3RVY) 37 (grey), NaVRh (4DXW) 39 (light green), NaVAe1p (4LTO)41 (orange), and KcsA (4EFF) 77 (blue). Selectivity filter (SF), P helix, P2, S5, and S6 elements are indicated. KcsA M1 and M2 correspond to BacNaV S5 and S6, respectively. (c) Equivalent views of the (top) NaVAb (PDB: 3RVY) 37 and (bottom) KcsA (PDB 1K4C) 27 selectivity filters. In contrast to the carbonyl-lined filter in KcsA, the NaVAb selectivity filter is wider and lined by two sidechains: E177 (or site 0, colored yellow) and S178 (site +1) (green). The highly conserved Thr residue at the end of the P1-helix forms part of the Site ‘4’ K+ binding site in potassium channels, but in BacNaVs the equivalent Thr sidechain is oriented to interact with a Trp sidechain in the selectivity filter (not shown). For simplicity, only two subunits are shown and all other sidechain residues are omitted. (d) BacNaV SF crystallographically defined ions. PDs of NaVRh (4DXW) 39 (light green), NaVAe1p (4LTO) 41, and CaVAb (4MS2) 61 (white), are shown. Boxed numbers indicate selectivity filter residue positions. ‘1’, ‘2’, and ‘3’ label the CaVAb SF ions. Outer and Inner ion binding sites are labeled.
Comparison among the BacNaV monomer structures highlights the extremely high similarity in the basic tertiary structure of the individual BacNaV PD subunits (Table 1). For most superpositions, the differences in the Cα positions are well below 1.0 Å RMSD (Fig. 3a). The biggest deviations are with NaVRh 39 and are largely due to a slightly different position of the S5 helix in this structure (Fig. 3a). Notably, the deviation of the NaVRh PD structure from the PD consensus is greater than that observed for PD structures of NaVMs, which have been suggested as models of an open conformation 59; 60.
Table I.
Comparisons of BacNaV Pore domain structures
| PDB | NaVAb I217C | NaVAb WT A/B | NaVAb WT C/D | NaVRh G208S | NavMsp1 | NavMsp2 | NaVAe1p | CaVAb | NaVCt1 | NaVCt2 | KcsA | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NaVAb I217C | 3RVY | Monomer-Monomer | * | 1.14 | 0.98 | 1.57 | 0.85 | 1.27 | 1.04 | 0.32 | 0.79 | 1.12 | 3.22 | Tetramer-Tetramer |
| NaVAb WT A/B | 4EKW | 0.51 | * | 1.84 | 2.11 | 1.53 | 1.98 | 1.49 | 1.24 | 1.34 | 1.74 | 3.61 | ||
| NaVAb WT C/D | 4EKW | 0.56 | 0.69 | * | 1.72 | 0.87 | 1.16 | 1.37 | 0.91 | 1.20 | 1.27 | 3.09 | ||
| NaVRh G208S | 4DXW | 1.09 | 1.03 | 1.19 | * | 1.56 | 1.96 | 2.05 | 1.47 | 1.70 | 1.38 | 2.98 | ||
| NaVMs1 | 4F4L | 0.72 | 0.85 | 0.67 | 1.20 | * | 0.83 | 1.14 | 0.81 | 1.14 | 1.20 | 3.11 | ||
| NaVMs2 | 3ZJZ | 1.01 | 1.19 | 0.91 | 1.70 | 0.59 | * | 1.43 | 1.24 | 1.42 | 1.29 | 3.17 | ||
| NaVAe1p | 4LTO | 0.92 | 1.04 | 1.03 | 1.36 | 0.83 | 0.89 | * | 1.05 | 1.19 | 1.56 | 3.14 | ||
| CaVAb | 4MS2 | 0.30 | 0.48 | 0.59 | 1.01 | 0.70 | 0.99 | 0.87 | * | 0.77 | 1.05 | 3.02 | ||
| NaVCt1 | 4BGN | 0.29 | 0.56 | 0.56 | 1.10 | 0.70 | 0.96 | 0.93 | 0.24 | * | 0.83 | 3.18 | ||
| NaVCt2 | 4BGN | 0.28 | 0.50 | 0.67 | 1.03 | 0.72 | 0.94 | 0.88 | 0.70 | 0.26 | * | 3.23 | ||
| KcsA | 4EFF | 1.86 | 1.73 | 1.73 | 2.08 | 1.86 | 1.92 | 1.86 | 1.66 | 1.65 | 1.62 | * |
Superpositions of the PDs of the indicated BacNaVs based on NaVAb 130-219, NaVAe1p 150-239, and the corresponding residues in each of the entries.
Monomer vs. monomer – orange
Tetramer vs. tetramer – blue
NaVCt1 and NaVCt2 are molecules ‘A’ and ‘B’58, respectively.
Given the presence of the PD in all members of the VGIC superfamily (Fig. 1a), we thought it would be interesting to examine the BacNaV PD tertiary fold in light of the PD of the full-length structure of the prototypical potassium channel, KcsA 77 (Fig. 3b, Table 1). This comparison reveals the striking conservation of the core tertiary fold of an individual PD subunit (Fig. 3b). Although there are some notable differences between BacNaVs and potassium channels, such as the backbone conformation of the selectivity filter and presence of a P2 helix, it is clear from the comparison the essential elements and organization of the PD fold are the same. The S5/S6 transmembrane helix pair forms a platform for the P-helix that leads into the loop forming the selectivity filter. This core structure has also recently been described in the first structure of a TRP channel 78, further establishing that this basic PD fold should be present in all members of the VGIC superfamily. The crossing angle of the P-helix relative to the two transmembrane segments is different between the BacNaV and KcsA exemplars and may be related to the requirement for the filter diameters to be different in order to select different cations. Indeed, the BacNaV extracellular opening is wider than that in potassium channels and its SF is sufficient to hold the potassium channel SF 37 (Fig. 3c). The BacNaV PD P2 helix follows the selectivity filter and is an element absent from known potassium channel structures. Regardless of this difference, it is striking that the core fold of the monomer is so similar, even though the details of the selectivity filters and how they recognize ions (below) are dramatically different.
The shared features of the PD fold point to a common origin and raise the question: ‘Given the apparent constraints of the basic PD fold, what is the range of structural diversity that can be accommodated in the SF region?’ Exploration of proteins having unconventional selectivity filters, such as the related bacterial potassium transporter family TrkH which has the same basic PD fold 79; 80 and channel properties 81, engineered channels designed to test ideas about selectivity 82; 83; 84, or wholesale replacement of the SF in the context of a genetic selection or computational study may help to answer this question and will be important for establishing the ground rules for eventual de novo design of channels having novel properties.
The PD tertiary architecture appears very robust, as experimental and computational studies of KV1.3 potassium channel biogenesis indicate that the basic PD fold can adopt a near native-like tertiary fold in the absence of assembling into a quaternary structure 85. This tantalizing result opens up questions about what happens to the PDs during biogenesis while an individual PD waits to encounter three other PDs to form a complete pore from either disparate chains, as in BacNaV, TRP, and KV channels, or from PDs embedded in very long gene transcripts as in eukaryotic NaVs and CaVs. Are there ways the cell can shield this partially formed hole from misfolding, degradation, or aggregation? Does the apparent stability of the PD tertiary fold accelerate assembly? Further, it raises the question about whether there is a ‘non-channel’ ancestor of the PD fold that has some function outside of the now familiar four-fold arrangement. Can this fold act in a monomeric capacity for some yet uncharacterized function?
When assembled around the central axis that forms the ion conduction pathway, the four subunits of the VGIC PD form a central cavity that is bounded by the selectivity filter on the extracellular side and a constriction made by the S6 pore lining helices on the intracellular side. This second region is thought to form the principal barrier that is controlled by the VSDs and that must be opened in order for ions to pass through the channel. The observed PD conformations in the various BacNaV structures have been suggested to represent closed 37; 41; 58, inactivated 38; 39, and open 58; 59; 60 conformations; however, comparison of the PD quaternary structure 41 (Table 1) reveals that despite some small differences, all of these backbone conformations are very similar, generally having RMSDs for the Cα positions that are ≤1.5Å. By contrast, the strong BacNaV PD monomer similarities with the KcsA fold are washed out if one considers how the BacNaV tetramers compare with an intact potassium channel (Table 1). Hence, even though the core PD subunits are very similar, there are clearly distinctly different ways to arrange the S5 and S6 segments around a closed pore.
When one considers that the quaternary structures of all of the BacNaV PDs are much more similar than they are different, in spite of suggestions about the different possible states that these structures may represent, what is very striking is that none of the channel activation gates are as open as in the KV1.2/2.1 chimera structure 71; 86. It may be that, unlike KV channels 70, BacNaV gating involves relatively subtle changes in the PD conformation. However, it should be noted that, when the BacNaV VSDs are present in the structures, they are in the activated conformation 37; 38; 39; 58. How can activated VSDs yield an open PD in one VGIC case 71; 86, but closed PDs in all the others 37; 38; 39; 58; 87? These observations may indicate something about the stability of the BacNaV PD conformation, which may have a strong bias to close the intracellular gate, or be related to how the BacNaV PDs couple to the VSDs. Clearly, there is a need for further studies of the energetic parameters that govern BacNaV gating to answer these questions. This puzzle also underscores the fact that it maybe hard to identify a truly open BacNaV PD structure unless it is bound to a known opener or contains well-characterized mutations that stabilize the open state.
Ion binding sites and their influence on ion selectivity
One of the main questions that determination of BacNaV structures hoped to address was: ‘What is the origin of ion selectivity?’ Further, as the BacNaV selectivity filter bears features common to both NaVs and CaVs 29; 33; 40 (Fig. 1c), could the homomeric BacNaV structures, having four identical selectivity filter segments, serve as prototypes to inform our understanding of eukaryotic NaVs or CaVs in which the selectivity filters are necessarily heteromeric? Long standing ideas originating in careful biophysical studies of eukaryotic NaVs and CaVs had set the expectation that members of this channel clade should use a selectivity mechanism that was based on sidechain chemistry 1; 88; 89; 90; 91 in contrast to the backbone mediated ion recognition mode used by potassium channels 1; 25; 27. To facilitate comparison among NaV, CaV, and BacNaV SFs, the residue corresponding to the mammalian NaV SF ‘DEKA’ motif 1, the conserved SF ‘EEEE’ motif in CaV 1; 88, and equivalent glutamate in BacNaV SFs, which was also described as forming the high-field strength site in NaVAb 37 , are denoted as position “0” (Fig. 1c). The idea that the sidechains were crucial for selectivity was further supported by the evidence that one could change BacNaV ion selectivity from sodium to calcium by making a triple-aspartate mutant at selectivity filter positions (0), (+1), and (+4) 35; 40; 61 (Fig. 1c). These expectations were all confirmed as the NaVAb SF 37 showed a structure much wider than that of a potassium channel and lined, in part, by sidechains rather than almost entirely backbone carbonyls 25; 27 (Fig. 3C).
Studies of eukaryotic NaVs and CaVs provided strong evidence for a multi-ion mechanism 1; 92; 93; 94; 95; 96, raising the prospect that if ions could be identified in the BacNaV SF structures, there might even be multiple binding sites. However, in contrast to these expectations and the crystallographic results from potassium channel structures 25; 27; 71, the first NaVAb structure lacked identifiable ions in the pore 37, suggesting the possibility of promiscuous ion coordination in the NaV SF. The NaVRh structure, which has a slightly unconventional selectivity filter sequence (Fig. 1c) provided crystallographic evidence for an inner ion binding site (Fig. 3d) formed from the backbone carbonyls of Leu (-1) and Thr (-2) at the C-terminal end of the P-helix that could be occupied by a partly hydrated calcium ion 39, a barium ion 97, or a rubidium ion 97. Electron density for a partially hydrated calcium ion coordinated by the SF (+1) serine in the NaVAe1p structure provided the first direct crystallographic evidence for an outer ion binding site at the mouth of the selectivity filter 41 (Fig. 3d).
The observation of two ion binding sites supports the idea that BacNaVs have multi-ion pores, an idea further validated by the recent structures of a NaVAb mutant bearing the triple aspartate mutation that changes selectivity from sodium to calcium, termed ‘CaVAb’ 61. The CaVAb structures identified a series of three ion binding sites within the SF, denoted Site 1, Site 2, and Site 3, as well as two extracellular sites positioned above the NaVAe1p ‘outer ion’ site (Fig. 3d). The SF in CaVAb revealed two high-affinity hydrated Ca2+-binding sites followed by a third lower-affinity hydrated site. Four carboxyl side chains from SF residue (+1) form Site 1 and have a critical role in determining Ca2+ selectivity 40; 41; 61. Four carboxyls of the ‘DDDD’ motif at SF residue (0) plus four backbone carbonyls from SF residue (-1) form Site 2, a site also targeted by blocking divalent cations (e.g. Mn2+ and Cd2+). The lower-affinity Site 3 is formed by four backbone carbonyls from SF residue (-2) alone and mediates ion exit into the central cavity. In CaVAb, the multi-ion pore architecture is consistent with a conduction occurring by a multi-ion ‘knock-off’ mechanism of ion permeation through a stepwise-binding process that has been suggested to be conserved in CaV channels 61. In addition to this set of crystallographically identified ions in independently determined BacNaV structures, other diffuse electron density has been reported in the SF of some structures that most likely arises from ions but that could not be assigned due to available resolution limits 41; 59. Besides demonstrating that there are multiple ion binding sites in the BacNaV filter, these studies suggest that that many, if not all, of the observed ions are at least partially hydrated. This assertion is in line with prior ideas about how NaV and CaV SFs interact with ions 1, but will require structures to be determined at a much higher resolution than has yet been possible in order to directly visualize these potential permeant ion properties.
The observation of a calcium ion bound to the outer ion site of NaVAe1p also focused attention on a conserved aspartic acid in the selectivity filter of Domain II in all classes of eukaryotic CaVs. Measurement of the effects of mutation of this position in the human cardiac CaV1.2 channel demonstrated that this residue is as important as the (0) position glutamate, which resides deeper in the selectivity filter and is a key determinant of ion selectivity 88, strongly suggesting that it may interact directly with the permeant ion 41. This clear connection between BacNaVs and mammalian CaVs highlights the point that BacNaV filters are actually closer in sequence to CaVs than they are to NaVs (Fig. 1c), and importantly, supports the idea that BacNaVs should be good model systems for understanding how eukaryotic SFs are built 33.
The BacNaV structures have provided an important template for a variety of computational studies directed at trying to understand basic aspects about ion selectivity and permeation behavior. In depth analysis of the many computational studies already reported is beyond the scope of this review, but some key general points are worth noting. Many of the analyses support the multi-ion nature of permeation through the basic BacNaV SF scaffold 98; 99; 100; 101; 102; 103; 104. In general, these studies have revealed a knock-on mechanism of ion permeation characterized by alternating occupancy of the channel by two or three hydrated sodium ions. In extended MD simulations (>μsec) 101; 104, Na+ binding is coupled to an unexpected conformational isomerization of the four key glutamate side chains at position (0) of the selectivity filter to conformations not seen in any of the crystallographic structural studies. The coordination of variable numbers of Na+ ions and carboxylate groups leads to their condensation into ionic clusters of variable charge and spatial arrangement. These structural fluctuations result in a myriad of ion binding modes that foster a highly degenerate, energy landscape propitious to the diffusion of Na+ over K+ and Ca2+ ions. Thus, the resulting proposals for ion selectivity and conduction through the BacNaVs SF's are markedly distinct from the established mechanisms in highly selective K+ channel pores.
BacNaV PD have lateral openings to the bilayer that facilitate hydrophobic modulator access
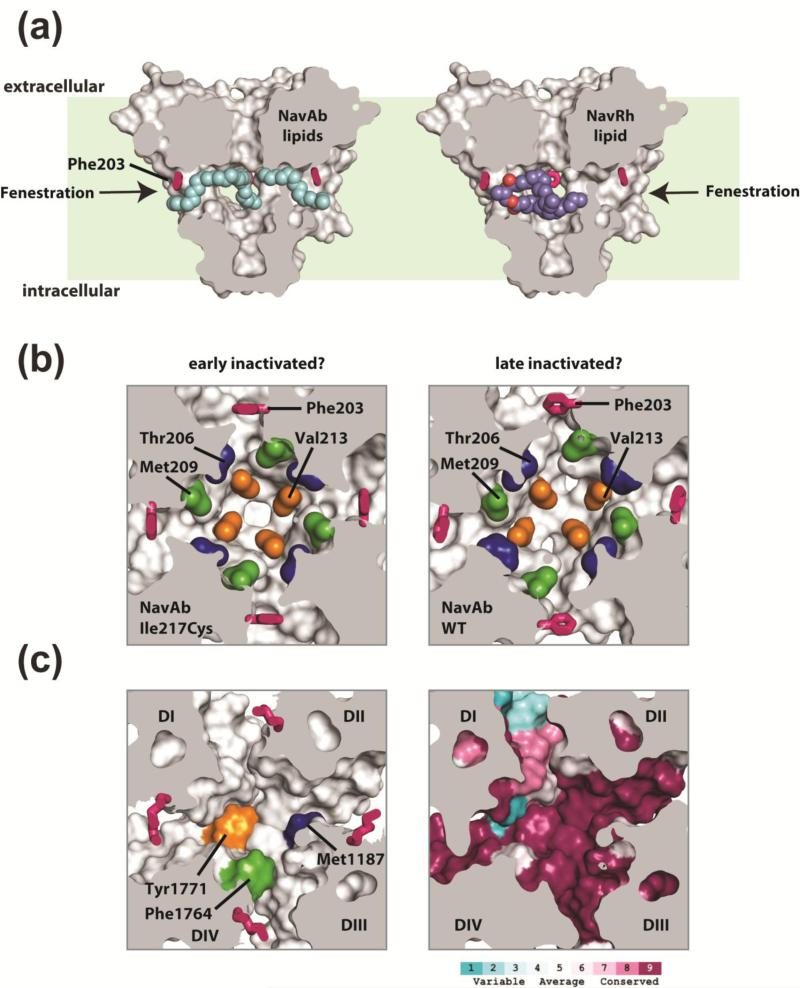
The most striking unanticipated feature of BacNaV PDs was the observation of lateral openings between the PD transmembrane helices that appear to allow access to the hydrophobic core of the lipid bilayer 37 (Fig. 4a). Such openings, which are not present in structures of bacterial potassium channels, inward rectifier potassium channels, or Kv channels, are enticingly consistent with earlier proposals that small molecules such as anesthetics access the channel pore through the lipid bilayer 1; 105; 106; 107; 108; 109. Notably, similar structures have been found in the K2P potassium channel class, a type of potassium channel that is known to be responsive to various types of anesthetics 110; 111; 112.
Figure 4. BacNaV PD fenestrations and pharmacology.
(a) NaVAb PD 37 is shown in surface representation sectioned through the middle. Exemplar side fenestrations are indicated by the arrows. The fenestration “gating” residue (Phe203) is shown as pink sticks. Lipids bound within the central cavity of NaVAb are shown as cyan spheres and are seen penetrating through the pore fenestrations (arrows). For easy comparison, the NaVRh PD (PDB 4DXW 39, not shown) was superimposed onto NaVAb and the bound lipid within the NaVRh pore is shown as purple and red spheres. Light green background indicates approximate bilayer boundaries. (b) A sectioned view of NaVAb I217C 37 (left) and NaVAb WT 38 (right) looking into the central cavity, viewed from below the selectivity filter. Phe203 is shown as pink sticks and select sidechains implicated in drug binding and block in eukaryotic NaV and CaV channels are in space filling color (blue, green and orange). The asymmetric central cavity seen in NaVAb WT (right) has been suggested to represent a slow inactivated conformation of the pore, where a reshaping of the pore fenestrations and putative drug binding sites are seen. (c) Homology model of human NaV1.7 based on the NaVAb (PDB 3RVY). Left: select residues implicated in local anesthetic block in DIV S6 of NaV channels (Phe1764 green and Tyr1771 orange) and a residue implicated in DHP block in DIII S6 of rat CaV1.2 (accession: P22002; Met1187 blue) illustrate a potential composite drug receptor site within the central cavity of eukaryotic NaV and CaV channels. Right: sequence conservation analysis for all human NaV channels (NaV1.1- NaV1.9) is mapped onto the NaV1.7 homology model and demonstrate regions of high and low conservation in and around the central cavity.
The NaVAb and NaVRh structures show that these side portals are filled with lipid or detergent molecules (Fig. 4a) 37; 38; 39 and give support to the idea that such cavities have the capacity to be filled by something hydrophobic. Classic studies demonstrate that small hydrophobic pore blockers can gain access to or leave the central cavity site in the PD of a channel that has not opened 105; 106; 108; 109. Similar experimental results have been obtained with BacNaV block by lidocaine 46 and further suggested by MD simulation 104; 113; 114. Notably, NaVMs has recently been shown to share overlapping pharmacology with human NaV1.1 and crystallographic analysis of the ‘pore-only’ channel has produced plausible models for drug binding near the pore fenestrations within the central cavity 62. Unfortunately, the inherent symmetry of the NaVMs pore and issues with experimentally resolving the drug conformations leaves considerable limitations in these drug-bound structural models. Nevertheless, an entire membrane phospholipid was seen bound within the NaVRh channel central cavity 39 and exemplifies how drug molecules might bind asymmetrically in eukaryotic NaVs (Fig. 4b-d).
Even though the side portals offer an exciting hypothesis for access to the channel cavity, their presence raises some puzzling questions. Given that they can be filled by lipids or lipid like molecules, is there some means to keep this interaction from happening in a physiological membrane bilayer? If they are filled with lipid, how is it that small molecules can pass through? And importantly, do changes in the conformation of the pore domain remodel these elements such that they could be the targets for natural or designed channel modulators (Fig. 4b-c)? A considerable plasticity is observed within the pore fenestrations of the available BacNaV structures, particularly if the sidechain rotamers are considered, and suggests that these portals may be dynamic during gating (Fig. 4b). Intriguingly, mutation of specific pore residues in vertebrate NaVs allows a charged lidocaine derivate (QX-314) to enter the central cavity when applied extracellularly 107. The equivalent BacNaV residues do not obviously connect the central cavity and extracellular milieu, suggesting that pore remodeling must occur during gating. In the absence of an unequivocally open BacNaV PD structure, these questions are sure to remain at the forefront of research. Nevertheless, the available BacNaV PD structures clearly provide meaningful starting points for structure-based drug discovery efforts and unprecedented opportunities to design novel, isoform selective small molecule inhibitors for the treatment of NaV channel related pathologies.
Structure-based sequence comparisons highlight key BacNaV PD positions
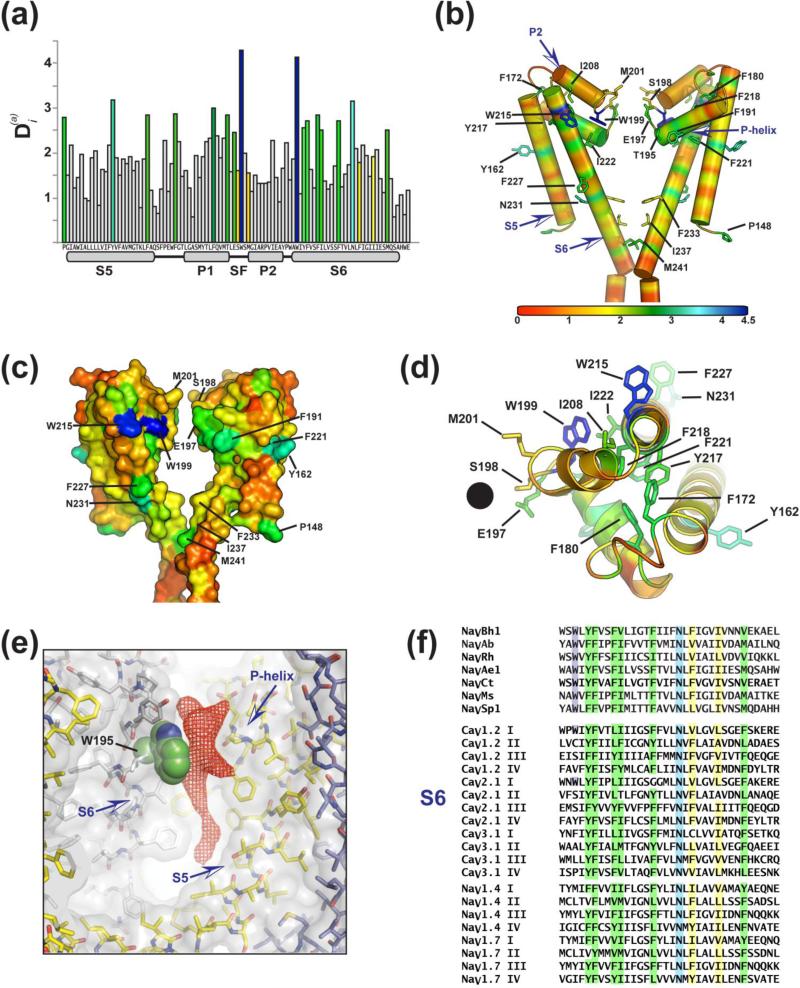
Sequence searches identify the presence of >500 BacNaVs in the current sequence databases from many bacteria, including many soil, marine, and salt lake bacteria and some opportunistic pathogens. This sequence diversity together with the common BacNaV structural framework provides the opportunity to investigate the positional conservation to identify key elements of the structure that have strong conservation and that hence, are important for some aspect of function. We used the positional informational analysis approach 115 to investigate which BacNaV PD positions had the highest degree of statistical information. Because the NaVAe1p structure 41 shows the complete pore, and all BacNaV PDs have similar structures (Fig. 3, Table 1), we used the NaVAe1p structure as the reference.
The two most highly conserved positions revealed by the analysis are two tryptophans (Fig. 5). One of these, Trp (+2) of the selectivity filter, NaVAe1p Trp199, forms the key anchor position for the selectivity filter (Figs. 1c, 5b, 5c). This position makes interactions with the P-helix of the neighboring subunit 37, including a hydrogen bond to the sidechain of Thr195 (-2), which is also highly-conserved (Fig. 5a, b). The Thr195 position is equivalent to the threonine found at the end of the potassium channel P-helix whose sidechain makes important contributions directly to ion binding 27; 83; 116; 117; however, in BacNavs rather than interact with the permeant ions, this sidechain is repositioned to make a direct interaction with the conserved Trp (+2) sidechain. This interaction appears to be essential for buttressing the conformation of the SF in a way that permits the sidechains to be displayed in the ion conduction pathway. Both the SF anchor Trp and the hydrogen bonding capability of the (-2) position are strongly conserved in the eukaryotic NaV and CaV SF sequences (Fig. 1c). The interfacial nature of the interaction between SF positions (+2) and (-2) may allow for it to be more than just an essential buttress for the SF conformation, but may allow SF conformational changes to be influenced by neighboring subunits.
Figure 5. BacNaV PD sequence conservation.
(a) Conservation analysis of the BacNaV PD, measured by the relative statistical entropy at each position, 115, mapped on the NaVAe1p sequence. Highly conserved positions in rank order starting with the most conserved are colored dark blue, light blue, and green. Other positions of interest are colored yellow. (b) Conservation analysis depicted on two subunits of NaVAe1p. Select positions of interest are shown as sticks and are labeled. (c) Spacefilling model of panel (b). (d) Extracellular view of a single PD subunit. Black circle marks the location of the ion conductive pore of the SF. (e) The highly conserved S6 Trp sidechain (Trp195 in NaVAb, shown in green spheres; Trp215 in NaVAe1p) is adjacent to a membrane phospholipid that appears to be well ordered as a lipid or detergent molecule in most available BacNaV structures. Different channel subunits are colored white, yellow, and slate. The fourth subunit is not visible. Red mesh is from an FO-FC omit map calculated at 2.7 Å from PDB 3RVY with the “pore lipid” omitted from the calculation, contoured at 2.5σ. This density likely represents a phosphatidylcholine molecule that is present at high concentrations in the NaVAb crystallization condition. (f) Sequence alignment of S6 segments from BacNaVs, CaV, and NaVs. Colors match those in panels ‘(a)-(d)’. Sequences identities are; NaVBh1 (NaChBac)(NP_343367.1), 207-240; NaVAb (YP_001490668.1), 211-244: NaVRh (PDB:4DXW), 195-228; NaVAe1 (YP_741167.1), 213-246; NaVCt (WP_007502948.1), 210-243; NaVMs (YP_864725.1), 115-129; NaVSp1 (YP_165303.1), 192-225; CaV1.2 (CAA84346), IS6 380-413, IIS6 728-761, IIIS6 1141-1174, IVS6 1451-1484; CaV2.1 (NG_011569.1), IS6 335-368, IIS6 689-722, IIIS6 1488-1521, IVS6 1786-1819; CaV3.1 (O43497), IS6 370-403, IIS6 939-972, IIIS6 1512-1545, IVS6 1826-1860; NaV1.4 (NP000325.4), IS6 422-455, IIS6 777-810, IIIS6 1270-1303, IVS6 1573-1606; NaV1.7 (NM_002977), IS6 377-410, IIS6 932-975, IIIS6 1421-1454, IVS6 1724-1757.
The second most highly conserved PD position is also a Trp, Trp215 in NaVAe1p (Trp195 in NaVAb). This residue is at the extracellular top of the S6 segment (Fig. 5b-d). Strikingly, this residue is not involved in any protein-protein contacts. Why then should it be so strongly conserved? Further inspection of the BacNaV structures shows that the Trp215 equivalent almost invariably has detergent or lipid molecules bound next to it (Fig. 5e). Although this Trp residue not conserved in NaVs (Fig. 5e), it is present in IS6 of high-voltage activated CaVs (Fig. 5f), raising the possibility that this site may be involved in the lipid modulation of channel function which as been functionally described for both BacNaVs 118 and mammalian CaVs 119. Other points of strong conservation are the residues that contribute to the central part of the PD tertiary core and that form intrasubunit contacts between the S5/S6 scaffold (S5, Phe180; S6, Tyr217, Phe218, Phe221, Ile222), the P-helix (Phe191), and loop (Phe180). These same scaffolding residues have also been implicated in transmitting the structural changes observed between the SF and S6 activation gate of NaVAb I217C and NaVAb WT channels, and were suggested to be important in the slow inactivation mechanism of BacNaV and vertebrate NaV channels 38.
The initial set of BacNaV structures did not indicate a common position for the intracellular constriction that should form the channel gate. Resolution of a the full-length NaVAe1p PD structure revealed an extension of the S6 helix that had been absent from prior structures implicated position Met241 as the site of intracellular pore closure, a position corresponding to the suggested NaVAb activation gate 37. When tested in full-length NaVSp1, which is similar to NaVAe1p but more readily investigated using electrophysiology, functional studies showed that alanine mutation of the pore-lining S6 residues in each of the two helical turns above the Met241 equivalent had no effect on channel activation, whereas the M241A equivalent caused a large negative (~-50 mV) shift in the voltage dependence of activation 41. It is notable that the Met241 position shows the same degree of conservation as the Glu (0) position in the SF that is the key determinant of ion selectivity. It has been proposed based on state-dependent accessibility studies of residues in eukaryotic NaV1.4 DIV S6 that the gate is one turn higher (equivalent to Phe233) 120. Whether this apparent difference between NaV1.4 and the BacNaVs reflects the dissimilar approaches used to define the gate, structure based versus chemical reactivity of a mutant channel, or indicates genuine differences between homotetrameric and heterotetrameric pores remains an open question.
The PD conservation analysis highlights two other positions of note. One is Asn231, this S6 position is conserved throughout the BacNaV, NaV, and CaV families (Fig. 5e) but absent from potassium channels 121 and points towards the S4/S5 linker in some of the structures 38. Mutation of this residue to alanine in IS6 of the eukaryotic NaV1.4 shifts the voltage dependence of activation to more depolarized potentials and enhances entry in to the slow inactivated state 122. Effects of mutations in various S6 segments of NaV1.2 further support the importance of this position in slow inactivation and impact channel modulation by kinases 123. The other conserved position is S5 NaVAe1p Tyr162, which makes contacts to the S1 segment of the VSD in NaVAb and NaVRh (as noted below). Given the high information content of both of these positions, investigation of their functional roles merits further experimental attention.
Voltage sensor revisited
VSDs are central to the functioning of our nervous system, form the cornerstone of Hodgkin and Huxley's pioneering physiological studies on VGICs 3, and are one of the most thoroughly studied protein domains in biophysical research 1; 2; 87; 124; 125; 126; 127. Since entering the age of ion channel structural biology, several studies have highlighted the VSDs as modular folding units 28; 128; 129; 130, demonstrated the constraints required for electromechanical coupling to the pore by transferring a VSD onto a non-voltage gated channel 131; 132; 133, and shown the functional transferability of the S3-S4 regions between evolutionary distant VSDs 134; 135; 136.
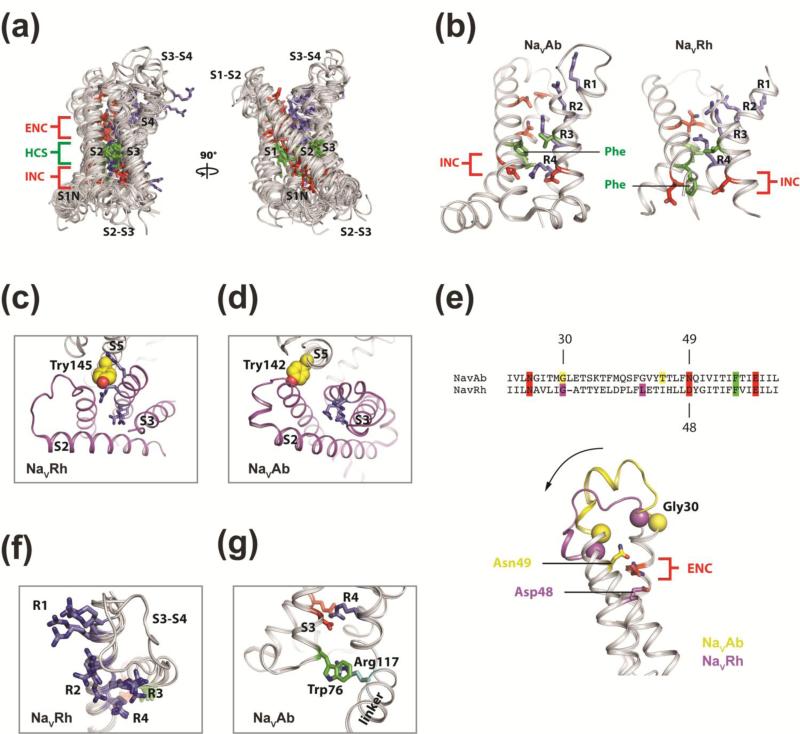
Despite all the fame and attention, the VSD is a four-helix bundle of relatively simple construction (Fig. 6a, b). The S4 segment carries highly conserved positively charged “gating charges” interspaced between two hydrophobic amino acids in a repeating RxxR motif. In response to changes in membrane potential, the gating charges interact with negative charge clusters on the S1-S3 segments to catalyze S4 movement through the electric field 125. These extracellular and intracellular negative charge clusters (ENC and INC) are solvent accessible 127; 137; 138, but separated by a hydrophobic constriction site (HCS) that prevents ion leakage through a hydrophobic bottleneck or “gating pore”. A conserved phenylalanine residue in the hydrophobic constriction site and the intracellular negative charge cluster has been implicated as a “gating charge transfer center (CTC)” in KV139 channels . In NaV1.4, these same residues only appear to be essential for proper VSD folding and membrane trafficking of the channel 140. Nevertheless, all VSDs form an hourglass-shaped structure in the membrane that essentially functions as a voltage-dependent arginine side-chain transporter.
Figure 6. BacNaV VSD structural comparison.
(a) VSDs from representative crystal structures in the PDB were freely superimposed onto the NaVAb VSD (PDB 3RVY) 37. These include VSDs from NaVRh (4DXW) 39, NaVCt (4BGN) 58, KV1.2/2.1 chimera (2R9R) 71, KVAP (1ORS) 28, VSP (4G7V and 4G80) 142, and HV1 (3WKV) 141. Positions of the S1N, S1, S2, S3, and S4 helices and intervening S1-S2, S2-S3, and S3-S4 loops are indicated. Gating charge residues on the S4 segments are shown as purple sticks. Extracellular and intracellular negative charge cluster (ENC and INC) residues are shown as red sticks. Hydrophobic constriction site (HCS) residues are shown as green sticks. In general, structural correspondence between all VSDs is seen, with the largest deviations apparent in the loop (e.g. S1-S2 and S3-S4) and S1N regions. A large sequence insertion in the Kv1.2/2.1 chimera S1-S2 loop has been omitted for clarity. View is from the plane of the membrane bilayer. (b) Taken from the alignment in (a), details of the similarities and differences between the NaVAb and NaVRh VSDs can be appreciated. Gating charges, INC and HCS, a conserved Phe sidechain, resides are highlighted. (c) Extracellular view looking onto the NaVRh VSD and neighboring pore domain. The S5 residue Tyr145 is highly conserved in BacNaV channels and points directly into the VSD extracellular crevice. S4 gating charge residues are shown as purple sticks. (d) Same view as in (c), looking onto the NaVAb VSDs and neighboring pore domain. In contrast to NaVRh, the highly conserved S5 Tyr142 residue points directly at the S1 helix, and not into the VSD extracellular crevice, highlighting the rotation of the NaVAb VSD around the pore domain is with respect to the VSD position in NaVRh. (e) Sequence alignment indicates a simple single residue insertion in the S1-S2 linker of NaVAb (L31) relative to NaVRh. Structure-based alignment however clearly demonstrates “equivalent” residues to be spatially displaces from one another, by up to ~5 Å. This structural plasticity of the S1-S2 loop in NaVRh is, in part, responsible for the intracellular movement of its S2 ENC residue Asp48. (f) Superposition of the four VSDs from the NaVRh structure (4DXW) demonstrates the structural variations seen within the S3-S4 loop and the gating charges of a single VGIC captured in the same crystal. Displacements of equivalent residues up to ~8 Å are seen. (g) A highly conserved non-canonical VSD interaction is observed in NaVAb. Trp76 found at the foot of the S3 is one of the most highly conserved sidechains in all NaV and CaV VSDs. This residue may help anchor the VSD into the membrane. It also makes a π-cation interaction with a highly conserved Arg residue from in the S4/S5 linker, R117.
The BacNaV structures provide an important architectural framework and rich diversity of conformational snap-shots to help understand VGICs. When all of the VSD coordinates are extracted from the BacNav structures and compared to the VSDs from Kv channels 28; 71; 86, the proton gated channel (HV 1) 141, and the voltage-sensitive phosphatase (VSP) 142, a number of general conclusions can be made. First, at a gross level, all VSD structures are essentially identical and share a conserved structural core independent of their amino acid sequence or functional state (Fig. 6a). Second, the S4-S5 linker has a fixed length amongst diverse, but not all, VGICs, which imposes key constraints on the electromechanical coupling mechanism between the PD and VSD. Third, only four gating charges superimpose within the conserved structural core (Fig. 6a), apparently contradicting the notion that vertebrate VSDs may contain 5-7 functional gating charges per VSD 124; 143. Perhaps the “extra” gating charge residues found on the S4-S5 linker (Fig. 7d) may modulate channel gating through interactions with surrounding lipid headgroups or are important for protein folding (also see conservation analysis below). Fourth, the S4 segment is in an entirely α-helical conformation in VSDs that have been cut away from the rest of their native sequence (i.e. KvAP 28, Hv1 141, and Ci-VSP 142). By contrast, VSDs that are still attached to their natural payloads all display some mixture of α-helix and 310-helix along their S4 segments. Finally, the structural correspondence between NaVAb and murine KV1.2 VSDs is absolutely striking and argues that the folding-pathway and voltage-sensing mechanism have been highly conserved over the course of molecular evolution 37.
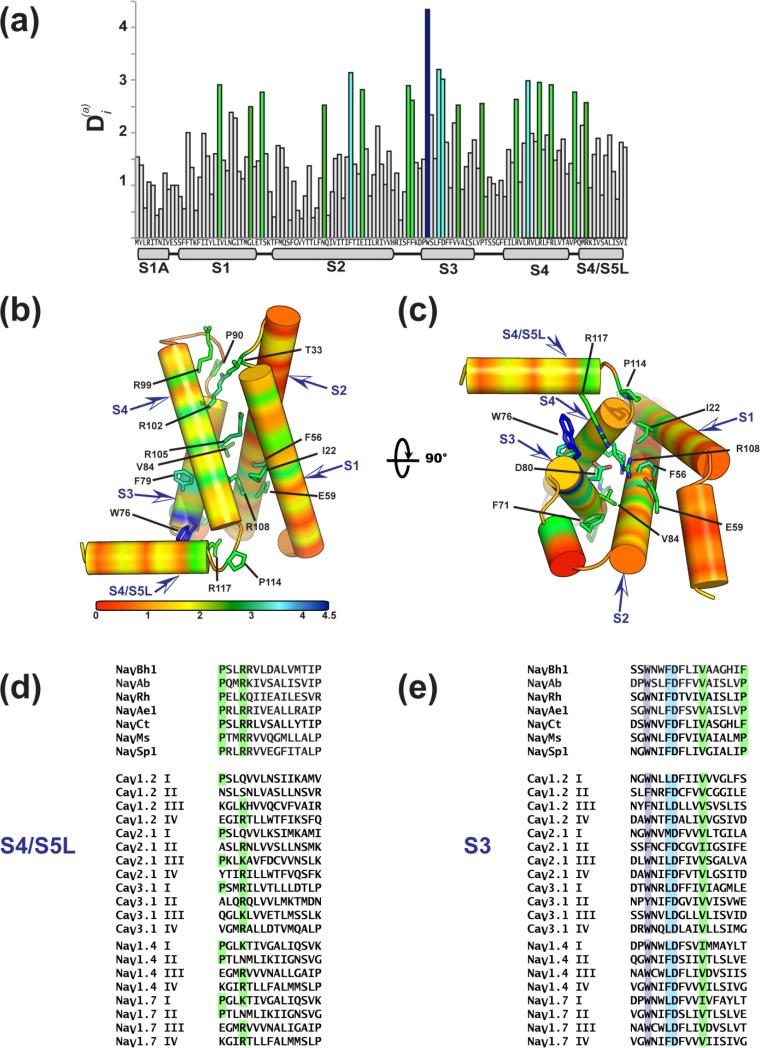
Figure 7. BacNaV VSD sequence conservation.
(a) Conservation analysis of the BacNaV VSD measured by the relative statistical entropy at each position, 115, mapped on the NaVAb sequence. Highly conserved positions are colored dark blue, light blue, and green. (b) and (c) Conservation analysis depicted on the NaVAb VSD. Select positions of interest are shown as sticks and are labeled. (d) Comparison of S4/S5 linker and (e) S3 segments for the indicated sequences: NaVBh1(NaChBac)(NP_343367.1) S3 87-113, S4/S5 linker 128-142; NaVAb (YP_001490668.1) S3 74-90, S4/S5 114-128; NaVRh (PDB:4DXW), S3 75-91, S4/S5117-133; NaVAe1 (YP_741167.1), S3 94-110, S4/S5 134-158; NaVCt (WP_007502948.1) S3 90-105, S4/S5 131-145; NaVMs (YP_864725.1), S3 75-91; S4/S5 linker 115-129; NaVSp1 (YP_165303.1), S3 73-89, S4/S5 112-126; CaV1.2 (CAA84346), IS3 193-209, IS3 586-602, IIIS3 967-983, IVS3 1281-1297, IS4/S5 252-66, IIS4/S5 265-649, IIIS4/S5 1013-1027, IVS4/S5 1344-1358; CaV2.1 (NG_011569.1), IS3 167-183, IIS3 549-565, IIIS3 1311-1327, IVS3 1628-1644, IS4/S5 210-224, IIS4/S5 598-612, IIIS4/S5 1361-1375, IVS4/S5 1676-1690; CaV3.1 (O43497), IS3, 150-166, IIS3 805-821, IIIS3 1344-1360, IVS3 1673-1689, IS4/S5 195-209, IIS4/S5 850-864, IIIS4/S5 1396-1410, IVS4/S5 1727-1741; NaV1.4 (NP_000325.4), IS3 191-207, IIS3 640-656, IIIS3 1095-1111, IVS3 1414-1430, IS4/S5 234-248, IIS4/S5 684-698, IIIS4/S5 1144-1158, IVS4/S5 1466-1480; NaV1.7 (NM_002977), IS3 186-202, IIS3 795-811, IIIS3 1245-1261, IVS3 1565-1581, IS4/S5 229-243, IIS4/S5 839-853,IIIS4/S5 1294-1308, IVS4/S5 1617-1631.
BacNaV VSDs
BacNaVs are small proteins and their “minimal” VSDs can serve to highlight essential features and pinpoint vertebrate-specific functional elaborations. Structure-based alignments indicate that large sequence insertions occur within the extracellular loop regions of vertebrate VSDs 37 but also demonstrate that the intracellular S2-S3 loop can adopt a common conformation. The BacNaV structures call attention to a conserved amphipathic helix that precedes the S1 segments (the S1N helix) and that may be dynamic during the gating process 128; 130. Overall, the BacNaV channels provide important new constraints on this four-helical bundle domain and focus attention on some of the novel details revealed by their structures.
In addition to interactions with negative charge clusters, the NaVAb 37 and NaVRh 39 S4 gating charges show coordination to backbone carbonyl groups and other VSD sidechains. These non-traditional gating charge interactions have since been observed in modeling studies, prompting speculation that they contribute to the S4 activation pathway 67. Amino acid substitutions at these previously considered benign positions might therefore affect VSD function and could predispose certain individuals to pathophysiological states (e.g. migraine or arrhythmias) 124; 143. The BacNaV structures also bring the PDs that buttress the VSDs directly into view (Fig. 2b). PD sidechains such as the conserved Tyr145 in NaVRh, corresponding to NaVAe1p Tyr162 noted above (Fig. 5), help to directly shape the S4 activation pathway by inserting its sidechain into the extracellular crevice of the VSD (Fig. 6c). In NaVAb, however, the equivalent S5 residue, Tyr142, lies outside of this extracellular VSD crevice and instead directly engages the S1 helix (Fig. 6d). These structural observations together indicate how otherwise “distant” PD residues might modulate the function or physiological tuning of VSDs in a VGIC.
On closer inspection, the voltage-sensing S4 segments reveal subtle structural differences between NaVAb and NaVRh 37; 39. In NaVAb, the S4 is found almost exclusively in a 310-helical conformation, placing gating charges from the RxxR motif on the same side of the helix for passage through the gating pore, perhaps through simple translation or tilting. By contrast, the middle portion of the NaVRh S4 is a 310-helix and its N-terminal portion is modeled as an α-helix. This raises the possibility that the S4 might undergo a secondary structure transition during charge transfer through the gating pore. Potentially supporting this view, MD simulations suggest that the NaVAb VSDs have not fully activated 144. However, this later notion is inconsistent with the physiological characterization of NaVAb 61; 145 and ignores the possibility that all available VGIC crystal structures reasonably represent a fully activated or inactivated VSD conformation, considering that a 0 mV condition is maintained over the course of all crystallization experiments. If we assume that the basic details of the voltage sensing mechanism are universally conserved across all VSDs, one parsimonious conclusion is that the S4s from different VSDs are strained (310-helix) or relaxed (α-helix) to varying degrees and that this extent depends primarily upon the amino acid sequence of the crystallized protein construct in question. Future experiments are needed to rigorously interrogate these subtle details of VSD structure and function.
A few VSD surprises
Uniquely, the NaVRh structure presents four simultaneous views of its VSD because one channel tetramer crystallized within the asymmetric unit 39; 146, providing a fortuitous example of the potential structural dynamics that are possible within a VSD. Structural plasticity is seen within the S3-S4 linkers of two subunits (Fig. 6f), and to a lesser extent, in the S1-S2 linkers of all four subunits. These differences highlight the same two focal points of sequence and structural divergence across all VSDs, the S1-S2 and S3-S4 loops (Fig. 6a). The observed structural changes also impact the chemistries available for S4 gating charge interactions (Fig. 6b-f), and the malleability of the NaVRh VSDs suggests the intriguing possibility that inherent structural heterogeneity exists along the S4 activation pathway.
All four gating charges of NaVRh are exposed to the extracellular side of the hydrophobic constriction site, with R4 above the so-called CTC, defined here to include the INC and HCS elements. The R4 of NaVAb remains engaged with the CTC, suggesting that NaVAb may transfer one less gating charge during activation, although equivalent gating charges are nonetheless found at similar depths within the membrane (Fig. 6b). NaVRh achieves R4 transfer above the CTC through the concerted intracellular movements of its S1-S3 segments 146, where its CTC appears “downshifted” within the membrane compared to NaVAb (Fig. 6b). If relevant, these unprecedented observations hint at a new concept in voltage sensor activation and suggest that there may be more then one moving part in VSDs.
Further comparison between NaVRh and NaVAb highlights an unrecognized observation in the S1-S2 linker upon simple sequence alignment and consideration of structurally “equivalent” residues (Fig. 6e). From a structure-based perspective, a portion of the linker sequence appears to toggle between the S1 and S2 segments akin to a slinky toy. The effect in NaVRh is that the S2 translates towards the intracellular side and results in the displacement of the S2 ENC residue by one helical turn (i.e. Asp48 vs. Asn49; Fig. 6e). This implies that a remarkable degree of freedom exists at the S1-S2 junction, which is perhaps consistent the large sequence insertions that are found in Kv channel linkers 71; 86; 147 and the plasticity required for CTC “downshifting” in NaVRh. Subtle structural transitions in the S1-S2 linker may therefore be more commonplace in VSD function then is currently appreciated.
Key BacNaV VSD positions revealed by structure-based sequence comparisons
Similar to the PD analysis described above, we used the large collection of available BacNaV sequences for positional conservation analysis 115 of the VSD residues and mapped these results on the NaVAb VSD (Fig. 7). This analysis highlights well-known conservation patterns, such as the RxxR motif of the S4 segment (Arg99, Arg102, Arg105, and Arg108), two negatively charged residues (S2 Glu59 and S3 Asp80) that make the intracellular negative charge cluster (INC) 37, and the three components of the hydrophobic constriction site (HCS) (S1 Ile22, S2 Phe56, S3 Val84) 37 (Fig. 7a), and shows features common to an analysis of VSDs that included multiple types of bacterial and eukaryotic cation channels148. Notably, two previously unanalyzed residues in the S4/S5 linker stood out. One is the proline (Pro114) that makes the bend between S4 and the S4/S5 linker (Fig. 7b-d). The second is the conserved Arg or Lys (Arg117) that appears one turn into the S4/S5 linker helix. Arg117 is within distance to form a cation-π interaction with the conserved Trp at the base of S3 (Trp76). Strikingly, this Trp is the most conserved feature of the BacNaV VSD (Fig. 7a) and is strongly conserved as an aromatic residue throughout the eukaryotic NaVs and CaVs (Fig. 7e). Mutation to cysteine at the conserved Arg117 position in NaChBac shifts the voltage-dependence of activation in the depolarizing direction (~+30 mV) 65, consistent with the idea that the Arg117-Trp76 interaction may stabilize the active state of the VSD and the interpretation that the VSDs of the BacNaV structures represent an activated state. The positive nature of the equivalent S4/S5 linker position is well conserved in many of the NaV and CaV domains (Fig. 7d). Given the striking conservation of the Trp76 position, and ideas about the down-state of the S4 segment under hyperpolarizing conditions, one wonders if there are interactions with the conserved gating charges of S4 and Trp76 in other VSD states that help to shepherd gating charges into or away from the intracellular negative charge cluster.
On the evolution of VSDs
The discovery of isolated (or pore-less) VSD containing proteins such as CI-VSP 149 and Hv1150; 151 raises questions about the evolution of this domain. Did an ion channel sprout a rudimentary S4 segment that subsequently elaborated into today's VSD, and did this VSD later become detached? Or did VSDs evolve independently and then fuse to the PD of an ion channel? The concept that VSDs are dynamic four-helical bundles, taken with the structural diversity seen in their loops (Fig. 6a), begins to suggest that a covalent linkage may not be an absolute requirement for function. In fact, deleting large portions of the S3-S4 linker still produces a functional Kv channel 152 and genetically cutting the channel into two separate polypeptides through the S3-S4 linker also results in functional expression 153. In preliminary experiments, the co-expression of an S1-only construct along with the S2-S6 region of NaChBac was found to produce a functional VGIC, as did S1-S2 co-expression of with a S3-S6 construct (J.Payandeh, unpublished). Therefore, VSDs do not absolutely require a covalent linkage between the transmembrane helices to function. These observations could provide insight into the enigmatic evolution of this essential domain. Although we can clearly cut and paste a VSD onto a non-voltage gated pore to render it voltage-sensitive 131; 132; 133, perhaps recapitulating an important step in of early evolution within the VGIC superfamily, it remains to be determined how extensively one might be able to cut up a VGIC into pieces and still retain function, where the functional cut sites might be, and how many different cuts can be tolerated at once. Such, ‘deconstruction’ experiments should offer interesting new insights, particularly as the available structural data now offers a clear blueprint for candidate sites.
Insights into gating and inactivation
BacNaVs, just like other voltage-gated channels, respond to membrane potential changes that cause the channel to inhabit non-conductive (closed and inactivated) and conductive (open) states. Thus far, electrophysiological studies have characterized BacNaV homologs from twelve different organisms from the soil, sea, and salt lakes, revealing a broad range of activation and inactivation gating properties 48; 154: NaChBac (Bacillus halodurans) 29; 45; 46; 63; 155; 156; 157, NaVAb (Arcobacter butzleri) 37; 38; 145, NaVAe1 (Alkalilimnicola erlichii) 41, NaVBacL (Bacillus licheniformis) 32, NaVBP (Bacillus pseudofirmus)31, NaVCt (Caldalkalibacillus thermarum) 58, NaVMs (Magentococcus marinus) 62; 98, NaVPz (Paracoccus zeaxanthinifaciens) 30, NaVRosD (Roseobacter denitrificans) 32, NaVSheP (Shewanella putrefaciens) 32, NaVSp1 (Silicibacter pomeroyi) 30; 35; 41, NaVSulP (Sulfitobacter pontiacus) 158. This rich array of functional diversity and systems for investigation suggests that BacNaV structural studies have the potential to uncover the main conformations that underlie important conductive and non-conductive states. However, to do so, as with all structural studies, it is essential to connect the structures with the appropriate functional state 57. Voltage control over the samples is not possible in the context of a crystallization experiment, unlike conformational transitions in membrane proteins that are driven by chemical transformations such as ATP consumption where specific substrates can trap states 159. Hence, it is not as straightforward as it might first seem to know what exact functional states the various BacNaV structures represent absent some explicit tests based on interactions seen in the structures.
For the full-length structures 37; 38; 39; 58, there is agreement that the VSDs are in the activated position due to the long time spent at zero potential once the proteins were purified out of a charged cell membrane; however, despite this general concordance about the VSD state, the structures show conformational variations in both the VSDs and PDs. One common feature of the NaVAb and NaVRh structures is that intracellular access out of the aqueous central cavity below the selectivity filter is blocked by a constriction formed by the pore-lining S6 helices. The first NaVAb structure has been suggested to be ‘pre-open’ 37, whereas the NaVRh was deemed inactivated 39, particularly as the NaVRh selectivity filter is in a conformation that blocks ion transit. The asymmetric pore arrangements observed in the second set of NaVAb structures 38 have been interpreted as inactivated conformations as they match expectation that the so-called ‘slow inactivated’ state should have some conformational change in the pore vestibule 45. It is worth noting that the asymmetric filters of these NaVAb structures are still wide enough for a partial hydrated ion to pass. The two low-resolution conformations described for NaVCt using electron crystallography show pores that are similar and closed, although the small differences between the conformations have been suggested to represent an open and closed inner gate 58. An alternative to these interpretations is that all of the full-length structures are some sort of post-open state, which may not resemble a truly inactivated conformation but may be much closer to how the PD looks when the channels are closed or deactivated.
One other striking difference regarding the VSDs is that, in addition to the differences in S4 interactions described above, the NaVRh VSDs have essentially a rigid body displacement around the PD of ~30° relative to NaVAb (Fig. 6 c-d) 39; 97. This difference is reminiscent of the observed VSD disengagement from the PD upon channel closure in simulations of Kv1.2/2.1 chimera in lipid membranes 160 and would seem to be in line with the independent character of the VSDs with respect to the overall protein structure 28; 129; 131; 142; 161. Such displacements raise interesting questions about the exact functional states of the full length BacNaV structures, how there can be strong coupling between the internal motions of S4 within the VSD in response to voltage, and whether such global VSD motions have a role in voltage sensing 146. Furthermore, upon comparing the available NaVAb I217C and WT structures, significant movements of the VSDs around the PD are also seen that may highlight a novel isoform selective receptor site that is potentially druggable in the eukaryotic NaV channels.
The conformation of NaVAe1p, which has been freed from its VSDs, has all the hallmarks of a closed conformation 41, and moreover, is concordant with the effects of S6 alanine mutations on the energetics of channel opening that were designed to test interactions seen by the structure 41. The PD subunits in first structure of the NaVMs ‘pore-only’ channel had four non-identical conformations, the most variant of which was used to generate a possible open conformation model 59. This model suggests a twisting displacement around the central axis, a motion akin to the iris-like opening of a camera aperture. A similar conformation has since been reported in other NaVMs structures 60; 62. It is notable that in simulations, in the absence of applied restraints, the putative open conformation does not remain open 98, casting doubt upon the functional state represented by the structures.
If the observations about the similarities of the PD structures are put aside and the conformational assignments of the possible states are taken at face value, one might get the impression that the energetic landscape that separates one PD conformation from another is quite flat. Although such a landscape would explain the small conformational differences among the PDs, this interpretation is inconsistent with the large amount of energy that goes into activating a voltage-gated sodium or potassium channel (~14 kcal mol−1) 162; 163. The functional properties of ‘pore-only’ BacNaV versions 35; 36 demonstrate that the PDs can open stochastically without any energetic input from the VSDs. Further, even though the VSDs in all of the full-length BacNaV structures are activated, none have a PD as open as that as found in the KV1.2/2.1 chimera. So, why is it that when the VSDs in the active position one class of channel PDs always looks non-conductive while the other is wide open? Will we ever see a channel structure in which the VSDs are in the ‘down’ state? Additional structures determined using pharmacology that could definitively trap a well-characterized state or of mutants that are known to bias the channel into the open, closed, or inactivated conformations would go a long way to address this puzzle. In this regard it is interesting that PD mutants have been reported that dramatically shift the voltage dependence of opening to more hyperpolarized potentials 164, reverse the voltage-dependence of gating 165, and that selectivity affect channel inactivation, but not activation 41. Investigation of such mutants may help to define conformations of open and inactivated states or of other intermediates in the functional cycle.
The NaVAe1p structure revealed that BacNaV CTD has a continuous helix comprising a ‘neck’ and coiled-coil domains 41. Poly-glycine mutations in the neck shift voltage-dependent opening of NaVAe1 and NaVSp1 to more hyperpolarized potentials consistent with the proposal that this domain undergoes an order to disorder transition upon channel opening 41. In contrast to the order seen in the NaVAe1p neck, studies of NaChBac 72 and NaVMs have suggested that this region can be largely disordered 60. Deletions in the CTDs of different BacNaV isoforms yield loss of function 34; 58; 60, negative shifts 158 or small changes 60 in the voltage-dependence of activation, and varied effects on channel inactivation rates 60; 158. These results may not be as discordant as they first seem. Given that the BacNaV neck has varied composition (Fig. 8), the degree of structure and its impact on channel gating may differ among the BacNaV isoforms. Although exactly how the BacNav neck affects the channel function remains to be understood, one intriguing possibility is that the ‘down’ state of the S4/S5 linker may involve interactions with the neck region. The neck structure has no counterpart in eukaryotic NaVs or CaVs; however, many VGIC superfamily members that are assembled from multiple pore-forming subunits have a C-terminal coiled-coil in the cytoplasmic domain that follows S6 similar to that seen in the BacNaVs 73; 74; 76; 141; 166; 167; 168. Moreover, there is evidence that conformational changes in the regions between S6 and the coiled coil have profound effects on gating, such as in Kv7 channels 75; 169. Hence, elucidation of the mechanisms by which the BacNaV CTD influences voltage-dependent channel function might provide generalizable lessons that will impact our understanding of other VGIC superfamily members.
Figure 8. BacNaV PD and CTD sequence comparison.
S5, selectivity filter (SF), S6, Neck, and coiled-coil regions are indicated. 3-4 hydrophobic repeats of the coiled coils are indicated in grey. Conserved positions in S5, SF, and S6 are highlighted orange. Residues lining the pore are highlighted in blue. SF filter positions are indicated. Blue sequences have structures determined. Purple sequences have reported functional studies.
Pharmacology and fenestrations
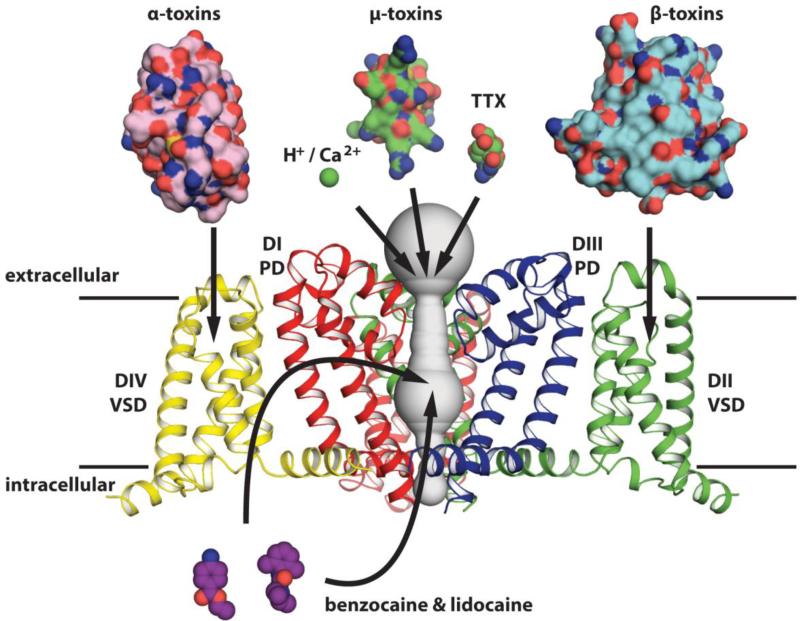
Ion channels represent major drug targets in the human body 170. The rich array of natural and synthetic molecules that comprise the pharmacology of NaVs 124; 171; 172; 173; 174; 175; 176 is one their most interesting and distinguishing characteristics. In line with their complex assortment of functional states, molecules that affect NaV function have complex modes of action including pore block, open state stabilization of the pore, and voltage sensor movement alteration (Fig. 9). Despite this impressive pharmacological artillery, traditional drug discovery efforts have historically failed to find highly selective NaV inhibitors, owing in part to the high sequence identity found amongst the human channel isoforms (Fig. 4c). Despite their inherent limitations, the BacNaV structures now provide our most accurate templates to understand the physical interaction of NaV channel modulators and explore the potential for the rational design of new therapeutics.
Figure 9. Eukaryotic NaV Pharmacology painted onto BacNaVs.
NaVAb subunits are colored to represent the four non-homologous domains of a eukaryotic NaV channels. One pore domain and one voltage-sensor are removed for clarity. Representative structures of channel modulating toxins, small molecules, and cations are depicted to interact with their best-characterized receptor sites on the channel. All structures are rendered to approximately the same scale.
Targeting the extracellular vestibule
The extracellular vestibule is the site of NaV modulation by protons, divalent cations, and small toxins (Fig. 9). Proton block of vertebrae NaVs has been well characterized and occurs predominantly through the protonation of residues lining the extracellular vestibule above the SF 177; 178; 179. The BacNaV structures suggest that protons might neutralize key sidechains required to coordinate and conduct Na+-water complexes through the SF or destabilize interactions required to maintain the SF in a conductive conformation. Tetrodotoxin (TTX) is one of the most famous ion channel blockers and binds directly to the SF region of sensitive NaV channels with low nanomolar affinity to cause its neurotoxic effects (Fig. 9). Models of TTX binding have existed for over four decades and generally envision the TTX guanidinium group interacting with the glutamate side-chain from the DEKA motif 180; 181; 182; 183; 184; 185. Because vertebrate NaVs contain sequence register shifts around the SF region (Fig. 1c), there is uncertainty about how well the bacterial structures recapitulate the eukaryotic NaVs at this site 183; 184. Not surprisingly, BacNaVs have proven to be TTX insensitive 29 (F. Abderemane-Ali., M. Krier, and D. L. Minor, Jr., unpublished). Perhaps a protein engineering approach to create a concatenated BacNaV channel with a heterotetrameric vertebrate-like extracellular vestibule will offer the first tractable structural biology approach to visualize this important pharmacological site.
Access and block in the central cavity
The BacNaV channel structures provide excellent templates to understand one of the most studied receptor sites in ion channel research, the central cavity of NaVs and CaVs. Traditional NaV channel blockers are used clinically as antiepileptic, antiarrhythmic and analgesic drugs that block in the central cavity through at least two different binding modes. Neutral local anesthetic drugs can access the central cavity in resting NaV channels and produce “tonic” block in the micromolar to millimolar range 1; 186. Antiepileptic and antiarrhythmic drugs block opened or inactivated NaV channels in a “use-dependent” way in which affinity is positively correlated with channel activation 1; 187. Although the binding sites for these drugs overlap, different residues contribute to tonic and use-dependent block, possibly reflecting changes in the central cavity associated with channel opening and inactivation 188; 189. A significant structural reshaping of the central NaVAb cavity has been suggested to reflect the characteristics of a slow inactivated state in eukaryotic NaVs (Fig. 4B) and may be relevant to the understanding of these channels 37.
Two highly conserved aromatic residues on the S6 helix of DIV in vertebrate NaVs are major determinants of use-dependent block 186; 189 and equivalent residues in BacNavs line the central cavity (Fig. 4B, C). Mutagenesis studies in CaVs have further implicated an S6 side-chain on DIII 190, suggesting a composite drug-binding site is formed across the central cavity 191; 192. We used the BacNav structures as templates to model the central cavity of vertebrate NaV channels, as illustrated here for human Nav1.7 (Fig. 4C). Homology modeling and conservation analysis immediately rationalizes why this traditional receptor site lacks significant isoform selectivity (Fig. 4C). Remarkably, an adjacent interface between DI and DII represents an intriguing and potentially highly isoform selective site (Fig. 4C). Because the NaVMs channel has already undergone crystallographic analysis to produce models of drug binding within the central cavity 62, the available BacNaV structures can clearly provide meaningful starting points for structure-based drug discovery efforts and opportunities to design novel, isoform selective small molecule inhibitors for the treatment of NaV channel related pathologies. One intriguing result from recent pharmacological and MD studies of NaChBac is the conclusion that drug binding may occur at multiple sites, in multiple binding modes113. There is little doubt that further higher-resolution studies of such compounds bound to BacNaVs in different conformations will be essential for defining how the PD can interact with small molecules that perturb function and for generating new ideas about modulator design.
Interfering with the VSD
Diverse organisms in nature target the VSDs of vertebrate NaVs by deploying small protein toxins 193. These “gating modifying” neurotoxins are classically described to bind to the VSDs of DII or DIV (Fig. 9) and confound the activation or inactivation properties of the channel through different voltage sensor trapping mechanisms, either by enhancing or suppressing activation of the S4 segment 135; 171; 176. Importantly, some gating modifying toxins are known to display significant NaV channel isoform selectivity 194 and have major binding determinants found in the extracellular S1-S2 and S3-S4 loops. Feasible structural models of the CssIV toxin-VSD2 interaction and the Lhq2 toxin-VSD4 complex have already appeared using a composite VSD homology model drawn from the S1-S2 and S3-S4 regions of the BacNaV and KV channels, respectively 195; 196. It is hoped that these protein toxins may represent suitable scaffolds for pharmaceutical development and that their receptor sites can be exploited for isoform-selective drug discovery.
A recent landmark study has described an exciting new series of small molecules that appear to bind selectively to VSD4 of NaV1.7 or NaV1.3 and antagonize these channels 197. These drug-like molecules seem to stabilize an inactivated state of the VSD. In another breakthrough report, a monoclonal antibody directed against the S3-S4 loop of VSD2 has been shown to selectively stabilize the closed form of Nav1.7 and have profound efficacy in pain studies in vivo 198. Together, these incredible findings not only validate the pharmacological inhibition of NaV1.7, but they promise that we are on the verge of truly isoform selective NaV channel inhibitors showing utility in the clinic.
Prospects
The tractability of BacNaVs for structural and functional studies sets the stage for rapid advances in outlining the structural basis for the conformational transitions that underlie VGIC function. The diversity of forms available for investigation will undoubtedly be a major advantage and are likely to reveal the source of functional differences within the family. If we were making a wish list for the next five years, one would hope to see structures at high resolution of ions in the selectivity filter and small molecules bound to the PD or VSD, structures of mutants that trap well-defined functional states, and possibly structures of engineered BacNaVs that bear key features of their eukaryotic counterparts, such as an asymmetric selectivity filter. Because there are lipids intimately bound with parts of the channel and some specificity in how lipids modulate BacNaV function 118, it seems important to further characterize these sites and understand their relationships with the way hydrophobic drugs interact with and affect channel function. Given that native eukaryotic NaVs have been the subject of electron microscopy studies for many years15 and that efforts on expression of complex eukaryotic membrane proteins bear ever more successes, it seems likely that the recent breakthroughs enabling single particle analysis of sub-megadalton complexes at a resolution rivaling X-ray crystallographic structures determined at ~3.0 Å resolution 199 are poised to give the first view of a eukaryotic NaV. If one compares the state of the field with that of kinases, it is obvious that even a few structures are only the beginning. As more and more members of the VGIC superfamily are revealed in three-dimensions, the future for connecting the details of channel biophysics with meaningful biological outcomes seems very, very bright indeed.
Highlights.
Bacterial sodium channels (BacNaVs) offer unprecedented structural insights into voltage gated ion channel function
BacNaV structures reveal the details of sodium and calcium-selective pores
BacNaV structures offer plausible models to understand the action of drugs
Acknowledgements
We thank M. Grabe for help with the conservation analysis, C. Arrigoni and F. Findeisen for help with the alignments and figures, B. Liebeskind and H. Zakon for figure assistance, and C. Arrigoni, F. Findeisen, and M. Grabe for comments on the manuscript. This work was supported by NIH R01-HL080050, R01-DC007664, and U54-GM094625 grants to D.L.M.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing financial interests statement
The authors declare no competing financial interests.
References
- 1.Hille B. Ion Channels of Excitable Membranes. 3rd edit Sinauer Associates, Inc.; Sunderland, MA.: 2001. [Google Scholar]
- 2.Catterall WA. From ionic currents to molecular mechanisms: the structure and function of voltage-gated sodium channels. Neuron. 2000;26:13–25. doi: 10.1016/s0896-6273(00)81133-2. [DOI] [PubMed] [Google Scholar]
- 3.Hodgkin AL, Huxley AF. A quantitative description of membrane current and its application to conduction and excitation in nerve. J Physiol. 1952;117:500–44. doi: 10.1113/jphysiol.1952.sp004764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Catterall WA, Raman IM, Robinson HP, Sejnowski TJ, Paulsen O. The Hodgkin-Huxley heritage: from channels to circuits. J Neurosci. 2012;32:14064–73. doi: 10.1523/JNEUROSCI.3403-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mantegazza M, Curia G, Biagini G, Ragsdale DS, Avoli M. Voltage-gated sodium channels as therapeutic targets in epilepsy and other neurological disorders. Lancet Neurol. 2010;9:413–24. doi: 10.1016/S1474-4422(10)70059-4. [DOI] [PubMed] [Google Scholar]
- 6.Cregg R, Momin A, Rugiero F, Wood JN, Zhao J. Pain Channelopathies. J Physiol. 2010;588:1897–1904. doi: 10.1113/jphysiol.2010.187807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Black JA, Waxman SG. Noncanonical roles of voltage-gated sodium channels. Neuron. 2013;80:280–91. doi: 10.1016/j.neuron.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 8.Catterall WA, Goldin AL, Waxman SG. International Union of Pharmacology. XLVII. Nomenclature and structure-function relationships of voltage-gated sodium channels. Pharmacol Rev. 2005;57:397–409. doi: 10.1124/pr.57.4.4. [DOI] [PubMed] [Google Scholar]
- 9.Brackenbury WJ, Isom LL. Na Channel beta Subunits: Overachievers of the Ion Channel Family. Front Pharmacol. 2011;2:53. doi: 10.3389/fphar.2011.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu FH, Catterall WA. The VGL-chanome: a protein superfamily specialized for electrical signaling and ionic homeostasis. Sci STKE. 2004;2004:re15. doi: 10.1126/stke.2532004re15. [DOI] [PubMed] [Google Scholar]
- 11.Vassilev PM, Scheuer T, Catterall WA. Identification of an intracellular peptide segment involved in sodium channel inactivation. Science. 1988;241:1658–61. doi: 10.1126/science.241.4873.1658. [DOI] [PubMed] [Google Scholar]
- 12.Stuhmer W, Conti F, Suzuki H, Wang XD, Noda M, Yahagi N, Kubo H, Numa S. Structural parts involved in activation and inactivation of the sodium channel. Nature. 1989;339:597–603. doi: 10.1038/339597a0. [DOI] [PubMed] [Google Scholar]
- 13.Ulbricht W. Sodium channel inactivation: molecular determinants and modulation. Physiol Rev. 2005;85:1271–301. doi: 10.1152/physrev.00024.2004. [DOI] [PubMed] [Google Scholar]
- 14.Van Petegem F, Lobo PA, Ahern CA. Seeing the forest through the trees: towards a unified view on physiological calcium regulation of voltage-gated sodium channels. Biophys J. 2012;103:2243–51. doi: 10.1016/j.bpj.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sato C, Ueno Y, Asai K, Takahashi K, Sato M, Engel A, Fujiyoshi Y. The voltage-sensitive sodium channel is a bell-shaped molecule with several cavities. Nature. 2001;409:1047–51. doi: 10.1038/35059098. [DOI] [PubMed] [Google Scholar]
- 16.Rohl CA, Boeckman FA, Baker C, Scheuer T, Catterall WA, Klevit RE. Solution structure of the sodium channel inactivation gate. Biochemistry. 1999;38:855–61. doi: 10.1021/bi9823380. [DOI] [PubMed] [Google Scholar]
- 17.Chagot B, Potet F, Balser JR, Chazin WJ. Solution NMR structure of the C-terminal EF-hand domain of human cardiac sodium channel NaV1.5. J Biol Chem. 2009;284:6436–45. doi: 10.1074/jbc.M807747200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miloushev VZ, Levine JA, Arbing MA, Hunt JF, Pitt GS, Palmer AG., 3rd Solution structure of the NaV1.2 C-terminal EF-hand domain. J Biol Chem. 2009;284:6446–54. doi: 10.1074/jbc.M807401200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chagot B, Chazin WJ. Solution NMR structure of Apo-calmodulin in complex with the IQ motif of human cardiac sodium channel NaV1.5. J Mol Biol. 2011;406:106–19. doi: 10.1016/j.jmb.2010.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feldkamp MD, Yu L, Shea MA. Structural and energetic determinants of apo calmodulin binding to the IQ motif of the Na(V)1.2 voltage-dependent sodium channel. Structure. 2011;19:733–47. doi: 10.1016/j.str.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sarhan MF, Tung CC, Van Petegem F, Ahern CA. Crystallographic basis for calcium regulation of sodium channels. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:3558–3563. doi: 10.1073/pnas.1114748109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang C, Chung BC, Yan H, Lee SY, Pitt GS. Crystal structure of the ternary complex of a NaV C-terminal domain, a fibroblast growth factor homologous factor, and calmodulin. Structure. 2012;20:1167–76. doi: 10.1016/j.str.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilchrist J, Das S, Van Petegem F, Bosmans F. Crystallographic insights into sodium-channel modulation by the beta4 subunit. Proc Natl Acad Sci U S A. 2013;110:E5016–24. doi: 10.1073/pnas.1314557110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Namadurai S, Balasuriya D, Rajappa R, Wiemhofer M, Stott K, Klingauf J, Edwardson JM, Chirgadze DY, Jackson AP. Crystal Structure and Molecular Imaging of the Nav Channel beta3 Subunit Indicates a Trimeric Assembly. J Biol Chem. 2014 doi: 10.1074/jbc.M113.527994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doyle DA, Morais Cabral J, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 26.Jiang Y, Lee A, Chen J, Cadene M, Chait BT, MacKinnon R. Crystal structure and mechanism of a calcium-gated potassium channel. Nature. 2002;417:515–22. doi: 10.1038/417515a. [DOI] [PubMed] [Google Scholar]
- 27.Zhou Y, Morais-Cabral JH, Kaufman A, MacKinnon R. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 A resolution. Nature. 2001;414:43–8. doi: 10.1038/35102009. [DOI] [PubMed] [Google Scholar]
- 28.Jiang Y, Lee A, Chen J, Ruta V, Cadene M, Chait BT, MacKinnon R. X-ray structure of a voltage-dependent K+ channel. Nature. 2003;423:33–41. doi: 10.1038/nature01580. [DOI] [PubMed] [Google Scholar]
- 29.Ren D, Navarro B, Xu H, Yue L, Shi Q, Clapham DE. A prokaryotic voltage-gated sodium channel. Science. 2001;294:2372–5. doi: 10.1126/science.1065635. [DOI] [PubMed] [Google Scholar]
- 30.Koishi R, Xu H, Ren D, Navarro B, Spiller BW, Shi Q, Clapham DE. A superfamily of voltage-gated sodium channels in bacteria. J Biol Chem. 2004;279:9532–8. doi: 10.1074/jbc.M313100200. [DOI] [PubMed] [Google Scholar]
- 31.Ito M, Xu H, Guffanti AA, Wei Y, Zvi L, Clapham DE, Krulwich TA. The voltage-gated Na+ channel NaVBP has a role in motility, chemotaxis, and pH homeostasis of an alkaliphilic Bacillus. Proc Natl Acad Sci U S A. 2004;101:10566–71. doi: 10.1073/pnas.0402692101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Irie K, Kitagawa K, Nagura H, Imai T, Shimomura T, Fujiyoshi Y. Comparative study of the gating motif and C-type inactivation in prokaryotic voltage-gated sodium channels. J Biol Chem. 2010;285:3685–94. doi: 10.1074/jbc.M109.057455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Durell SR, Guy HR. A putative prokaryote voltage-gated Ca(2+) channel with only one 6TM motif per subunit. Biochem Biophys Res Commun. 2001;281:741–6. doi: 10.1006/bbrc.2001.4408. [DOI] [PubMed] [Google Scholar]
- 34.Mio K, Mio M, Arisaka F, Sato M, Sato C. The C-terminal coiled-coil of the bacterial voltage-gated sodium channel NaChBac is not essential for tetramer formation, but stabilizes subunit-to-subunit interactions. Prog Biophys Mol Biol. 2010;103:111–21. doi: 10.1016/j.pbiomolbio.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 35.Shaya D, Kreir M, Robbins RA, Wong S, Hammon J, Bruggemann A, Minor DL., Jr. Voltage-gated sodium channel (NaV) protein dissection creates a set of functional pore-only proteins. Proc Natl Acad Sci U S A. 2011;108:12313–8. doi: 10.1073/pnas.1106811108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCusker EC, D'Avanzo N, Nichols CG, Wallace BA. A simplified bacterial “Pore” provides insight into the assembly, stability and structure of sodium channels. J Biol Chem. 2011;286:16386–91. doi: 10.1074/jbc.C111.228122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Payandeh J, Scheuer T, Zheng N, Catterall WA. The crystal structure of a voltage-gated sodium channel. Nature. 2011;475:353–8. doi: 10.1038/nature10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Payandeh J, Gamal El-Din TM, Scheuer T, Zheng N, Catterall WA. Crystal structure of a voltage-gated sodium channel in two potentially inactivated states. Nature. 2012;486:135–9. doi: 10.1038/nature11077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang X, Ren W, DeCaen P, Yan C, Tao X, Tang L, Wang J, Hasegawa K, Kumasaka T, He J, Clapham DE, Yan N. Crystal structure of an orthologue of the NaChBac voltage-gated sodium channel. Nature. 2012;486:130–4. doi: 10.1038/nature11054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yue L, Navarro B, Ren D, Ramos A, Clapham DE. The cation selectivity filter of the bacterial sodium channel, NaChBac. J Gen Physiol. 2002;120:845–53. doi: 10.1085/jgp.20028699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shaya D, Findeisen F, Abderemane-Ali F, Arrigoni C, Wong S, Nurva SR, Loussouarn G, Minor DL., Jr. Structure of a prokaryotic sodium channel pore reveals essential gating elements and an outer ion binding site common to eukaryotic channels. J Mol Biol. 2014;426:467–83. doi: 10.1016/j.jmb.2013.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Charalambous K, Wallace BA. NaChBac: the long lost sodium channel ancestor. Biochemistry. 2011;50:6742–52. doi: 10.1021/bi200942y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liebeskind BJ, Hillis DM, Zakon HH. Independent acquisition of sodium selectivity in bacterial and animal sodium channels. Curr Biol. 2013;23:R948–9. doi: 10.1016/j.cub.2013.09.025. [DOI] [PubMed] [Google Scholar]
- 44.Catterall WA. Physiology. A one-domain voltage-gated sodium channel in bacteria. Science. 2001;294:2306–8. doi: 10.1126/science.1067417. [DOI] [PubMed] [Google Scholar]
- 45.Pavlov E, Bladen C, Winkfein R, Diao C, Dhaliwal P, French RJ. The pore, not cytoplasmic domains, underlies inactivation in a prokaryotic sodium channel. Biophys J. 2005;89:232–42. doi: 10.1529/biophysj.104.056994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee S, Goodchild SJ, Ahern CA. Local anesthetic inhibition of a bacterial sodium channel. J Gen Physiol. 2012;139:507–16. doi: 10.1085/jgp.201210779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ouyang W, Jih TY, Zhang TT, Correa AM, Hemmings HC., Jr. Isoflurane inhibits NaChBac, a prokaryotic voltage-gated sodium channel. J Pharmacol Exp Ther. 2007;322:1076–83. doi: 10.1124/jpet.107.122929. [DOI] [PubMed] [Google Scholar]
- 48.Corry B, Lee S, Ahern CA. Pharmacological insights and quirks of bacterial sodium channels. Handb Exp Pharmacol. 2014;221:251–67. doi: 10.1007/978-3-642-41588-3_12. [DOI] [PubMed] [Google Scholar]
- 49.Chang G, Spencer RH, Lee AT, Barclay MT, Rees DC. Structure of the MscL homolog from Mycobacterium tuberculosis: a gated mechanosensitive ion channel. Science. 1998;282:2220–6. doi: 10.1126/science.282.5397.2220. [DOI] [PubMed] [Google Scholar]
- 50.Bocquet N, Nury H, Baaden M, Le Poupon C, Changeux JP, Delarue M, Corringer PJ. X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation. Nature. 2009;457:111–4. doi: 10.1038/nature07462. [DOI] [PubMed] [Google Scholar]
- 51.Corringer PJ, Poitevin F, Prevost MS, Sauguet L, Delarue M, Changeux JP. Structure and pharmacology of pentameric receptor channels: from bacteria to brain. Structure. 2012;20:941–56. doi: 10.1016/j.str.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 52.Hilf RJ, Bertozzi C, Zimmermann I, Reiter A, Trauner D, Dutzler R. Structural basis of open channel block in a prokaryotic pentameric ligand-gated ion channel. Nat Struct Mol Biol. 2010;17:1330–6. doi: 10.1038/nsmb.1933. [DOI] [PubMed] [Google Scholar]
- 53.Hilf RJ, Dutzler R. X-ray structure of a prokaryotic pentameric ligand-gated ion channel. Nature. 2008;452:375–9. doi: 10.1038/nature06717. [DOI] [PubMed] [Google Scholar]
- 54.Hilf RJ, Dutzler R. Structure of a potentially open state of a proton-activated pentameric ligand-gated ion channel. Nature. 2009;457:115–8. doi: 10.1038/nature07461. [DOI] [PubMed] [Google Scholar]
- 55.Dutzler R, Campbell EB, Cadene M, Chait BT, MacKinnon R. X-ray structure of a ClC chloride channel at 3.0 A reveals the molecular basis of anion selectivity. Nature. 2002;415:287–94. doi: 10.1038/415287a. [DOI] [PubMed] [Google Scholar]
- 56.Nurani G, Radford M, Charalambous K, O'Reilly AO, Cronin NB, Haque S, Wallace BA. Tetrameric bacterial sodium channels: characterization of structure, stability, and drug binding. Biochemistry. 2008;47:8114–21. doi: 10.1021/bi800645w. [DOI] [PubMed] [Google Scholar]
- 57.Minor DL., Jr. The neurobiologist's guide to structural biology: a primer on why macromolecular structure matters and how to evaluate structural data. Neuron. 2007;54:511–33. doi: 10.1016/j.neuron.2007.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tsai CJ, Tani K, Irie K, Hiroaki Y, Shimomura T, McMillan DG, Cook GM, Schertler GF, Fujiyoshi Y, Li XD. Two alternative conformations of a voltage-gated sodium channel. J Mol Biol. 2013;425:4074–88. doi: 10.1016/j.jmb.2013.06.036. [DOI] [PubMed] [Google Scholar]
- 59.McCusker EC, Bagneris C, Naylor CE, Cole AR, D'Avanzo N, Nichols CG, Wallace BA. Structure of a bacterial voltage-gated sodium channel pore reveals mechanisms of opening and closing. Nat Commun. 2012;3:1102. doi: 10.1038/ncomms2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bagneris C, Decaen PG, Hall BA, Naylor CE, Clapham DE, Kay CW, Wallace BA. Role of the C-terminal domain in the structure and function of tetrameric sodium channels. Nat Commun. 2013;4:2465. doi: 10.1038/ncomms3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tang L, Gamal El-Din TM, Payandeh J, Martinez GQ, Heard TM, Scheuer T, Zheng N, Catterall WA. Structural basis for Ca2+ selectivity of a voltage-gated calcium channel. Nature. 2014;505:56–61. doi: 10.1038/nature12775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bagneris C, DeCaen PG, Naylor CE, Pryde DC, Nobeli I, Clapham DE, Wallace BA. Prokaryotic NavMs channel as a structural and functional model for eukaryotic sodium channel antagonism. Proc Natl Acad Sci U S A. 2014;111:8428–33. doi: 10.1073/pnas.1406855111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kuzmenkin A, Bezanilla F, Correa AM. Gating of the bacterial sodium channel, NaChBac: voltage-dependent charge movement and gating currents. J Gen Physiol. 2004;124:349–56. doi: 10.1085/jgp.200409139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.DeCaen PG, Yarov-Yarovoy V, Sharp EM, Scheuer T, Catterall WA. Sequential formation of ion pairs during activation of a sodium channel voltage sensor. Proc Natl Acad Sci U S A. 2009;106:22498–503. doi: 10.1073/pnas.0912307106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chahine M, Pilote S, Pouliot V, Takami H, Sato C. Role of arginine residues on the S4 segment of the Bacillus halodurans Na+ channel in voltage-sensing. J Membr Biol. 2004;201:9–24. doi: 10.1007/s00232-004-0701-z. [DOI] [PubMed] [Google Scholar]
- 66.Blanchet J, Chahine M. Accessibility of four arginine residues on the S4 segment of the Bacillus halodurans sodium channel. J Membr Biol. 2007;215:169–80. doi: 10.1007/s00232-007-9016-1. [DOI] [PubMed] [Google Scholar]
- 67.Yarov-Yarovoy V, DeCaen PG, Westenbroek RE, Pan CY, Scheuer T, Baker D, Catterall WA. Structural basis for gating charge movement in the voltage sensor of a sodium channel. Proc Natl Acad Sci U S A. 2012;109:E93–102. doi: 10.1073/pnas.1118434109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Armstrong CM, Bezanilla F. Currents related to movement of the gating particles of the sodium channels. Nature. 1973;242:459–61. doi: 10.1038/242459a0. [DOI] [PubMed] [Google Scholar]
- 69.Bezanilla F, Armstrong CM. Gating currents of the sodium channels: three ways to block them. Science. 1974;183:753–4. doi: 10.1126/science.183.4126.753. [DOI] [PubMed] [Google Scholar]
- 70.Long SB, Campbell EB, Mackinnon R. Voltage sensor of Kv1.2: structural basis of electromechanical coupling. Science. 2005;309:903–8. doi: 10.1126/science.1116270. [DOI] [PubMed] [Google Scholar]
- 71.Long SB, Tao X, Campbell EB, MacKinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007;450:376–82. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- 72.Powl AM, O'Reilly AO, Miles AJ, Wallace BA. Synchrotron radiation circular dichroism spectroscopy-defined structure of the C-terminal domain of NaChBac and its role in channel assembly. Proc Natl Acad Sci U S A. 2010;107:14064–9. doi: 10.1073/pnas.1001793107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Howard RJ, Clark KA, Holton JM, Minor DL., Jr. Structural insight into KCNQ (Kv7) channel assembly and channelopathy. Neuron. 2007;53:663–75. doi: 10.1016/j.neuron.2007.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wiener R, Haitin Y, Shamgar L, Fernandez-Alonso MC, Martos A, Chomsky-Hecht O, Rivas G, Attali B, Hirsch JA. The KCNQ1 (Kv7.1) COOH terminus, a multitiered scaffold for subunit assembly and protein interaction. J Biol Chem. 2008;283:5815–30. doi: 10.1074/jbc.M707541200. [DOI] [PubMed] [Google Scholar]
- 75.Haitin Y, Attali B. The C-terminus of Kv7 channels: a multifunctional module. J Physiol. 2008;586:1803–10. doi: 10.1113/jphysiol.2007.149187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tsuruda PR, Julius D, Minor DL., Jr. Coiled coils direct assembly of a cold-activated TRP channel. Neuron. 2006;51:201–12. doi: 10.1016/j.neuron.2006.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Uysal S, Vasquez V, Tereshko V, Esaki K, Fellouse FA, Sidhu SS, Koide S, Perozo E, Kossiakoff A. Crystal structure of full-length KcsA in its closed conformation. Proc Natl Acad Sci U S A. 2009;106:6644–9. doi: 10.1073/pnas.0810663106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liao M, Cao E, Julius D, Cheng Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature. 2013;504:107–12. doi: 10.1038/nature12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cao Y, Jin X, Huang H, Derebe MG, Levin EJ, Kabaleeswaran V, Pan Y, Punta M, Love J, Weng J, Quick M, Ye S, Kloss B, Bruni R, Martinez-Hackert E, Hendrickson WA, Rost B, Javitch JA, Rajashankar KR, Jiang Y, Zhou M. Crystal structure of a potassium ion transporter, TrkH. Nature. 2011;471:336–40. doi: 10.1038/nature09731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vieira-Pires RS, Szollosi A, Morais-Cabral JH. The structure of the KtrAB potassium transporter. Nature. 2013;496:323–8. doi: 10.1038/nature12055. [DOI] [PubMed] [Google Scholar]
- 81.Cao Y, Pan Y, Huang H, Jin X, Levin EJ, Kloss B, Zhou M. Gating of the TrkH ion channel by its associated RCK protein TrkA. Nature. 2013;496:317–22. doi: 10.1038/nature12056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Derebe MG, Zeng W, Li Y, Alam A, Jiang Y. Structural studies of ion permeation and Ca2+ blockage of a bacterial channel mimicking the cyclic nucleotide-gated channel pore. Proc Natl Acad Sci U S A. 2011;108:592–7. doi: 10.1073/pnas.1013643108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Derebe MG, Sauer DB, Zeng W, Alam A, Shi N, Jiang Y. Tuning the ion selectivity of tetrameric cation channels by changing the number of ion binding sites. Proc Natl Acad Sci U S A. 2011;108:598–602. doi: 10.1073/pnas.1013636108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sauer DB, Zeng W, Raghunathan S, Jiang Y. Protein interactions central to stabilizing the K+ channel selectivity filter in a four-sited configuration for selective K+ permeation. Proc Natl Acad Sci U S A. 2011;108:16634–9. doi: 10.1073/pnas.1111688108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gajewski C, Dagcan A, Roux B, Deutsch C. Biogenesis of the pore architecture of a voltage-gated potassium channel. Proc Natl Acad Sci U S A. 2011;108:3240–5. doi: 10.1073/pnas.1017097108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Long SB, Campbell EB, Mackinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 87.Isacoff EY, Jan LY, Minor DL., Jr. Conduits of life's spark: a perspective on ion channel research since the birth of neuron. Neuron. 2013;80:658–74. doi: 10.1016/j.neuron.2013.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yang J, Ellinor PT, Sather WA, Zhang JF, Tsien RW. Molecular determinants of Ca2+ selectivity and ion permeation in L-type Ca2+ channels. Nature. 1993;366:158–61. doi: 10.1038/366158a0. [DOI] [PubMed] [Google Scholar]
- 89.Ellinor PT, Yang J, Sather WA, Zhang JF, Tsien RW. Ca2+ channel selectivity at a single locus for high-affinity Ca2+ interactions. Neuron. 1995;15:1121–32. doi: 10.1016/0896-6273(95)90100-0. [DOI] [PubMed] [Google Scholar]
- 90.Heinemann SH, Terlau H, Stuhmer W, Imoto K, Numa S. Calcium channel characteristics conferred on the sodium channel by single mutations. Nature. 1992;356:441–3. doi: 10.1038/356441a0. [DOI] [PubMed] [Google Scholar]
- 91.Favre I, Moczydlowski E, Schild L. On the structural basis for ionic selectivity among Na+, K+, and Ca2+ in the voltage-gated sodium channel. Biophys J. 1996;71:3110–25. doi: 10.1016/S0006-3495(96)79505-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Begenisich T. Molecular properties of ion permeation through sodium channels. Annu Rev Biophys Biophys Chem. 1987;16:247–63. doi: 10.1146/annurev.bb.16.060187.001335. [DOI] [PubMed] [Google Scholar]
- 93.Hess P, Tsien RW. Mechanism of ion permeation through calcium channels. Nature. 1984;309:453–6. doi: 10.1038/309453a0. [DOI] [PubMed] [Google Scholar]
- 94.Tsien RW, Hess P, McCleskey EW, Rosenberg RL. Calcium channels: mechanisms of selectivity, permeation, and block. Annu Rev Biophys Biophys Chem. 1987;16:265–90. doi: 10.1146/annurev.bb.16.060187.001405. [DOI] [PubMed] [Google Scholar]
- 95.Begenisich TB, Cahalan MD. Sodium channel permeation in squid axons. II: Non-independence and current-voltage relations. J Physiol. 1980;307:243–57. doi: 10.1113/jphysiol.1980.sp013433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Almers W, McCleskey EW, Palade PT. A non-selective cation conductance in frog muscle membrane blocked by micromolar external calcium ions. J Physiol. 1984;353:565–83. doi: 10.1113/jphysiol.1984.sp015351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhang X, Xia M, Li Y, Liu H, Jiang X, Ren W, Wu J, DeCaen P, Yu F, Huang S, He J, Clapham DE, Yan N, Gong H. Analysis of the selectivity filter of the voltage-gated sodium channel Na(v)Rh. Cell Res. 2012;23:409–22. doi: 10.1038/cr.2012.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ulmschneider MB, Bagneris C, McCusker EC, Decaen PG, Delling M, Clapham DE, Ulmschneider JP, Wallace BA. Molecular dynamics of ion transport through the open conformation of a bacterial voltage-gated sodium channel. Proc Natl Acad Sci U S A. 2013;110:6364–9. doi: 10.1073/pnas.1214667110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Furini S, Domene C. On conduction in a bacterial sodium channel. PLoS Comput Biol. 2012;8:e1002476. doi: 10.1371/journal.pcbi.1002476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Corry B, Thomas M. Mechanism of ion permeation and selectivity in a voltage gated sodium channel. J Am Chem Soc. 2012;134:1840–6. doi: 10.1021/ja210020h. [DOI] [PubMed] [Google Scholar]
- 101.Chakrabarti N, Ing C, Payandeh J, Zheng N, Catterall WA, Pomes R. Catalysis of Na+ permeation in the bacterial sodium channel Na(V)Ab. Proc Natl Acad Sci U S A. 2013;110:11331–6. doi: 10.1073/pnas.1309452110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Stock L, Delemotte L, Carnevale V, Treptow W, Klein ML. Conduction in a biological sodium selective channel. J Phys Chem B. 2013;117:3782–9. doi: 10.1021/jp401403b. [DOI] [PubMed] [Google Scholar]
- 103.Ke S, Zangerl EM, Stary-Weinzinger A. Distinct interactions of Na+ and Ca2+ ions with the selectivity filter of the bacterial sodium channel Na(V)Ab. Biochem Biophys Res Commun. 2013;430:1272–6. doi: 10.1016/j.bbrc.2012.12.055. [DOI] [PubMed] [Google Scholar]
- 104.Boiteux C, Vorobyov I, Allen TW. Ion conduction and conformational flexibility of a bacterial voltage-gated sodium channel. Proc Natl Acad Sci U S A. 2014;111:3454–9. doi: 10.1073/pnas.1320907111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hille B. The pH-dependent rate of action of local anesthetics on the node of Ranvier. J Gen Physiol. 1977;69:475–96. doi: 10.1085/jgp.69.4.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hondeghem LM, Katzung BG. Time- and voltage-dependent interactions of antiarrhythmic drugs with cardiac sodium channels. Biochim Biophys Acta. 1977;472:373–98. doi: 10.1016/0304-4157(77)90003-x. [DOI] [PubMed] [Google Scholar]
- 107.Ragsdale DS, McPhee JC, Scheuer T, Catterall WA. Molecular determinants of state-dependent block of Na+ channels by local anesthetics. Science. 1994;265:1724–8. doi: 10.1126/science.8085162. [DOI] [PubMed] [Google Scholar]
- 108.Hille B. Local anesthetics: hydrophilic and hydrophobic pathways for the drug-receptor reaction. J Gen Physiol. 1977;69:497–515. doi: 10.1085/jgp.69.4.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Schwarz W, Palade PT, Hille B. Local anesthetics. Effect of pH on use-dependent block of sodium channels in frog muscle. Biophys J. 1977;20:343–68. doi: 10.1016/S0006-3495(77)85554-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Miller AN, Long SB. Crystal structure of the human two-pore domain potassium channel K2P1. Science. 2012;335:432–6. doi: 10.1126/science.1213274. [DOI] [PubMed] [Google Scholar]
- 111.Brohawn SG, Campbell EB, MacKinnon R. Domain-swapped chain connectivity and gated membrane access in a Fab-mediated crystal of the human TRAAK K+ channel. Proc Natl Acad Sci U S A. 2013;110:2129–34. doi: 10.1073/pnas.1218950110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Brohawn SG, del Marmol J, MacKinnon R. Crystal structure of the human K2P TRAAK, a lipid- and mechano-sensitive K+ ion channel. Science. 2012;335:436–41. doi: 10.1126/science.1213808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Raju SG, Barber AF, LeBard DN, Klein ML, Carnevale V. Exploring volatile general anesthetic binding to a closed membrane-bound bacterial voltage-gated sodium channel via computation. PLoS Comput Biol. 2013;9:e1003090. doi: 10.1371/journal.pcbi.1003090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kaczmarski JA, Corry B. Investigating the size and dynamics of voltage-gated sodium channel fenestrations: A molecular dynamics study. Channels (Austin) 2014;8 doi: 10.4161/chan.28136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Halabi N, Rivoire O, Leibler S, Ranganathan R. Protein sectors: evolutionary units of three-dimensional structure. Cell. 2009;138:774–86. doi: 10.1016/j.cell.2009.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhou M, MacKinnon R. A mutant KcsA K(+) channel with altered conduction properties and selectivity filter ion distribution. J Mol Biol. 2004;338:839–46. doi: 10.1016/j.jmb.2004.03.020. [DOI] [PubMed] [Google Scholar]
- 117.Chatelain FC, Gazzarrini S, Fujiwara Y, Arrigoni C, Domigan C, Ferrara G, Pantoja C, Thiel G, Moroni A, Minor DL., Jr. Selection of inhibitor-resistant viral potassium channels identifies a selectivity filter site that affects barium and amantadine block. PLoS One. 2009;4:e7496. doi: 10.1371/journal.pone.0007496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.D'Avanzo N, McCusker EC, Powl AM, Miles AJ, Nichols CG, Wallace BA. Differential lipid dependence of the function of bacterial sodium channels. PLoS One. 2013;8:e61216. doi: 10.1371/journal.pone.0061216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Roberts-Crowley ML, Mitra-Ganguli T, Liu L, Rittenhouse AR. Regulation of voltage-gated Ca2+ channels by lipids. Cell Calcium. 2009;45:589–601. doi: 10.1016/j.ceca.2009.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Oelstrom K, Goldschen-Ohm MP, Holmgren M, Chanda B. Evolutionarily conserved intracellular gate of voltage-dependent sodium channels. Nat Commun. 2014;5:3420. doi: 10.1038/ncomms4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Guda P, Bourne PE, Guda C. Conserved motifs in voltage-sensing and pore-forming modules of voltage-gated ion channel proteins. Biochem Biophys Res Commun. 2007;352:292–8. doi: 10.1016/j.bbrc.2006.10.190. [DOI] [PubMed] [Google Scholar]
- 122.Wang SY, Wang GK. A mutation in segment I-S6 alters slow inactivation of sodium channels. Biophys J. 1997;72:1633–40. doi: 10.1016/S0006-3495(97)78809-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chen Y, Yu FH, Surmeier DJ, Scheuer T, Catterall WA. Neuromodulation of Na+ channel slow inactivation via cAMP-dependent protein kinase and protein kinase C. Neuron. 2006;49:409–20. doi: 10.1016/j.neuron.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 124.Moreau A, Gosselin-Badaroudine P, Chahine M. Biophysics, pathophysiology, and pharmacology of ion channel gating pores. Front Pharmacol. 2014;5:53. doi: 10.3389/fphar.2014.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Vargas E, Yarov-Yarovoy V, Khalili-Araghi F, Catterall WA, Klein ML, Tarek M, Lindahl E, Schulten K, Perozo E, Bezanilla F, Roux B. An emerging consensus on voltage-dependent gating from computational modeling and molecular dynamics simulations. J Gen Physiol. 2012;140:587–94. doi: 10.1085/jgp.201210873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Okamura Y. Biodiversity of voltage sensor domain proteins. Pflugers Arch. 2007;454:361–71. doi: 10.1007/s00424-007-0222-6. [DOI] [PubMed] [Google Scholar]
- 127.Tombola F, Pathak MM, Isacoff EY. How does voltage open an ion channel? Annu Rev Cell Dev Biol. 2006;22:23–52. doi: 10.1146/annurev.cellbio.21.020404.145837. [DOI] [PubMed] [Google Scholar]
- 128.Butterwick JA, MacKinnon R. Solution structure and phospholipid interactions of the isolated voltage-sensor domain from KvAP. J Mol Biol. 2010;403:591–606. doi: 10.1016/j.jmb.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Minor DL., Jr. A sensitive channel family replete with sense and motion. Nat Struct Mol Biol. 2006;13:388–90. doi: 10.1038/nsmb0506-388. [DOI] [PubMed] [Google Scholar]
- 130.Shenkarev ZO, Paramonov AS, Lyukmanova EN, Shingarova LN, Yakimov SA, Dubinnyi MA, Chupin VV, Kirpichnikov MP, Blommers MJ, Arseniev AS. NMR structural and dynamical investigation of the isolated voltage-sensing domain of the potassium channel KvAP: implications for voltage gating. J Am Chem Soc. 2010;132:5630–7. doi: 10.1021/ja909752r. [DOI] [PubMed] [Google Scholar]
- 131.Arrigoni C, Schroeder I, Romani G, Van Etten JL, Thiel G, Moroni A. The voltage-sensing domain of a phosphatase gates the pore of a potassium channel. J Gen Physiol. 2013;141:389–95. doi: 10.1085/jgp.201210940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lu Z, Klem AM, Ramu Y. Ion conduction pore is conserved among potassium channels. Nature. 2001;413:809–13. doi: 10.1038/35101535. [DOI] [PubMed] [Google Scholar]
- 133.Lu Z, Klem AM, Ramu Y. Coupling between voltage sensors and activation gate in voltage-gated K+ channels. J Gen Physiol. 2002;120:663–76. doi: 10.1085/jgp.20028696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Alabi AA, Bahamonde MI, Jung HJ, Kim JI, Swartz KJ. Portability of paddle motif function and pharmacology in voltage sensors. Nature. 2007;450:370–5. doi: 10.1038/nature06266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bosmans F, Martin-Eauclaire MF, Swartz KJ. Deconstructing voltage sensor function and pharmacology in sodium channels. Nature. 2008;456:202–8. doi: 10.1038/nature07473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bosmans F, Puopolo M, Martin-Eauclaire MF, Bean BP, Swartz KJ. Functional properties and toxin pharmacology of a dorsal root ganglion sodium channel viewed through its voltage sensors. J Gen Physiol. 2011;138:59–72. doi: 10.1085/jgp.201110614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Catterall WA. Ion channel voltage sensors: structure, function, and pathophysiology. Neuron. 2010;67:915–28. doi: 10.1016/j.neuron.2010.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bezanilla F. The voltage sensor in voltage-dependent ion channels. Physiol Rev. 2000;80:555–92. doi: 10.1152/physrev.2000.80.2.555. [DOI] [PubMed] [Google Scholar]
- 139.Tao X, Lee A, Limapichat W, Dougherty DA, MacKinnon R. A gating charge transfer center in voltage sensors. Science. 2010;328:67–73. doi: 10.1126/science.1185954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Pless SA, Elstone FD, Niciforovic AP, Galpin JD, Yang R, Kurata HT, Ahern CA. Asymmetric functional contributions of acidic and aromatic side chains in sodium channel voltage-sensor domains. J Gen Physiol. 2014;143:645–56. doi: 10.1085/jgp.201311036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Takeshita K, Sakata S, Yamashita E, Fujiwara Y, Kawanabe A, Kurokawa T, Okochi Y, Matsuda M, Narita H, Okamura Y, Nakagawa A. X-ray crystal structure of voltage-gated proton channel. Nat Struct Mol Biol. 2014;21:352–7. doi: 10.1038/nsmb.2783. [DOI] [PubMed] [Google Scholar]
- 142.Li Q, Wanderling S, Paduch M, Medovoy D, Singharoy A, McGreevy R, Villalba-Galea CA, Hulse RE, Roux B, Schulten K, Kossiakoff A, Perozo E. Structural mechanism of voltage-dependent gating in an isolated voltage-sensing domain. Nat Struct Mol Biol. 2014;21:244–52. doi: 10.1038/nsmb.2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Cannon SC. Voltage-sensor mutations in channelopathies of skeletal muscle. J Physiol. 2010;588:1887–95. doi: 10.1113/jphysiol.2010.186874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Amaral C, Carnevale V, Klein ML, Treptow W. Exploring conformational states of the bacterial voltage-gated sodium channel NavAb via molecular dynamics simulations. Proc Natl Acad Sci U S A. 2012;109:21336–41. doi: 10.1073/pnas.1218087109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Gamal El-Din TM, Martinez GQ, Payandeh J, Scheuer T, Catterall WA. A gating charge interaction required for late slow inactivation of the bacterial sodium channel NavAb. J Gen Physiol. 2013;142:181–90. doi: 10.1085/jgp.201311012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zhang X, Yan N. The conformational shifts of the voltage sensing domains between Na(v)Rh and Na(v)Ab. Cell Res. 2013;23:444–7. doi: 10.1038/cr.2012.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Chen X, Wang Q, Ni F, Ma J. Structure of the full-length Shaker potassium channel Kv1.2 by normal-mode-based X-ray crystallographic refinement. Proc Natl Acad Sci U S A. 2010;107:11352–7. doi: 10.1073/pnas.1000142107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Palovcak E, Delemotte L, Klein ML, Carnevale V. Evolutionary imprint of activation: the design principles of VSDs. J Gen Physiol. 2014;143:145–56. doi: 10.1085/jgp.201311103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Murata Y, Iwasaki H, Sasaki M, Inaba K, Okamura Y. Phosphoinositide phosphatase activity coupled to an intrinsic voltage sensor. Nature. 2005;435:1239–43. doi: 10.1038/nature03650. [DOI] [PubMed] [Google Scholar]
- 150.Sasaki M, Takagi M, Okamura Y. A voltage sensor-domain protein is a voltage-gated proton channel. Science. 2006;312:589–92. doi: 10.1126/science.1122352. [DOI] [PubMed] [Google Scholar]
- 151.Ramsey IS, Moran MM, Chong JA, Clapham DE. A voltage-gated proton-selective channel lacking the pore domain. Nature. 2006;440:1213–6. doi: 10.1038/nature04700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Xu Y, Ramu Y, Lu Z. A shaker K+ channel with a miniature engineered voltage sensor. Cell. 2010;142:580–9. doi: 10.1016/j.cell.2010.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Priest MF, Lacroix JJ, Villalba-Galea CA, Bezanilla F. S3-S4 linker length modulates the relaxed state of a voltage-gated potassium channel. Biophys J. 2013;105:2312–22. doi: 10.1016/j.bpj.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Scheuer T. Bacterial sodium channels: models for eukaryotic sodium and calcium channels. Handb Exp Pharmacol. 2014;221:269–91. doi: 10.1007/978-3-642-41588-3_13. [DOI] [PubMed] [Google Scholar]
- 155.Blunck R, Starace DM, Correa AM, Bezanilla F. Detecting rearrangements of shaker and NaChBac in real-time with fluorescence spectroscopy in patch-clamped mammalian cells. Biophys J. 2004;86:3966–80. doi: 10.1529/biophysj.103.034512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Finol-Urdaneta RK, Wang Y, Al-Sabi A, Zhao C, Noskov SY, French RJ. Sodium channel selectivity and conduction: prokaryotes have devised their own molecular strategy. J Gen Physiol. 2014;143:157–71. doi: 10.1085/jgp.201311037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Lee S, Goodchild SJ, Ahern CA. Molecular and functional determinants of local anesthetic inhibition of NaChBac. Channels (Austin) 2012;6:403–6. doi: 10.4161/chan.21807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Irie K, Shimomura T, Fujiyoshi Y. The C-terminal helical bundle of the tetrameric prokaryotic sodium channel accelerates the inactivation rate. Nat Commun. 2012;3:793. doi: 10.1038/ncomms1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Moller JV, Olesen C, Winther AM, Nissen P. The sarcoplasmic Ca2+-ATPase: design of a perfect chemi-osmotic pump. Q Rev Biophys. 2010;43:501–66. doi: 10.1017/S003358351000017X. [DOI] [PubMed] [Google Scholar]
- 160.Jensen MO, Jogini V, Borhani DW, Leffler AE, Dror RO, Shaw DE. Mechanism of voltage gating in potassium channels. Science. 2012;336:229–33. doi: 10.1126/science.1216533. [DOI] [PubMed] [Google Scholar]
- 161.Chakrapani S, Sompornpisut P, Intharathep P, Roux B, Perozo E. The activated state of a sodium channel voltage sensor in a membrane environment. Proc Natl Acad Sci U S A. 2010;107:5435–40. doi: 10.1073/pnas.0914109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Zagotta WN, Hoshi T, Aldrich RW. Shaker potassium channel gating. III: Evaluation of kinetic models for activation. J Gen Physiol. 1994;103:321–62. doi: 10.1085/jgp.103.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Chowdhury S, Chanda B. Estimating the voltage-dependent free energy change of ion channels using the median voltage for activation. J Gen Physiol. 2012;139:3–17. doi: 10.1085/jgp.201110722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zhao Y, Yarov-Yarovoy V, Scheuer T, Catterall WA. A gating hinge in Na+ channels; a molecular switch for electrical signaling. Neuron. 2004;41:859–65. doi: 10.1016/s0896-6273(04)00116-3. [DOI] [PubMed] [Google Scholar]
- 165.Zhao Y, Scheuer T, Catterall WA. Reversed voltage-dependent gating of a bacterial sodium channel with proline substitutions in the S6 transmembrane segment. Proc Natl Acad Sci U S A. 2004;101:17873–8. doi: 10.1073/pnas.0408270101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Yu Y, Ulbrich MH, Li MH, Dobbins S, Zhang WK, Tong L, Isacoff EY, Yang J. Molecular mechanism of the assembly of an acid-sensing receptor ion channel complex. Nat Commun. 2012;3:1252. doi: 10.1038/ncomms2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Fujiwara Y, Kurokawa T, Okamura Y. Long alpha helices projecting from the membrane as the dimer interface in the voltage-gated H(+) channel. J Gen Physiol. 2014;143:377–86. doi: 10.1085/jgp.201311082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Fujiwara Y, Kurokawa T, Takeshita K, Kobayashi M, Okochi Y, Nakagawa A, Okamura Y. The cytoplasmic coiled-coil mediates cooperative gating temperature sensitivity in the voltage-gated H(+) channel Hv1. Nat Commun. 2012;3:816. doi: 10.1038/ncomms1823. [DOI] [PubMed] [Google Scholar]
- 169.Xu Q, Chang A, Tolia A, Minor DL., Jr. Structure of a Ca(2+)/CaM:Kv7.4 (KCNQ4) B-helix complex provides insight into M current modulation. J Mol Biol. 2013;425:378–94. doi: 10.1016/j.jmb.2012.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Bagal SK, Brown AD, Cox PJ, Omoto K, Owen RM, Pryde DC, Sidders B, Skerratt SE, Stevens EB, Storer RI, Swain NA. Ion channels as therapeutic targets: a drug discovery perspective. J Med Chem. 2013;56:593–624. doi: 10.1021/jm3011433. [DOI] [PubMed] [Google Scholar]
- 171.Catterall WA, Cestele S, Yarov-Yarovoy V, Yu FH, Konoki K, Scheuer T. Voltage-gated ion channels and gating modifier toxins. Toxicon. 2007;49:124–41. doi: 10.1016/j.toxicon.2006.09.022. [DOI] [PubMed] [Google Scholar]
- 172.Knapp O, McArthur JR, Adams DJ. Conotoxins targeting neuronal voltage-gated sodium channel subtypes: potential analgesics? Toxins (Basel) 2012;4:1236–60. doi: 10.3390/toxins4111236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Nardi A, Damann N, Hertrampf T, Kless A. Advances in targeting voltage-gated sodium channels with small molecules. ChemMedChem. 2012;7:1712–40. doi: 10.1002/cmdc.201200298. [DOI] [PubMed] [Google Scholar]
- 174.Klint JK, Senff S, Rupasinghe DB, Er SY, Herzig V, Nicholson GM, King GF. Spider-venom peptides that target voltage-gated sodium channels: pharmacological tools and potential therapeutic leads. Toxicon. 2012;60:478–91. doi: 10.1016/j.toxicon.2012.04.337. [DOI] [PubMed] [Google Scholar]
- 175.Thottumkara AP, Parsons WH, Du Bois J. Saxitoxin. Angew Chem Int Ed Engl. 2014;53:5760–84. doi: 10.1002/anie.201308235. [DOI] [PubMed] [Google Scholar]
- 176.Quintero-Hernandez V, Jimenez-Vargas JM, Gurrola GB, Valdivia HH, Possani LD. Scorpion venom components that affect ion-channels function. Toxicon. 2013;76:328–42. doi: 10.1016/j.toxicon.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Sun YM, Favre I, Schild L, Moczydlowski E. On the structural basis for size-selective permeation of organic cations through the voltage-gated sodium channel. Effect of alanine mutations at the DEKA locus on selectivity, inhibition by Ca2+ and H+, and molecular sieving. J Gen Physiol. 1997;110:693–715. doi: 10.1085/jgp.110.6.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Khan A, Romantseva L, Lam A, Lipkind G, Fozzard HA. Role of outer ring carboxylates of the rat skeletal muscle sodium channel pore in proton block. J Physiol. 2002;543:71–84. doi: 10.1113/jphysiol.2002.021014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Jones DK, Ruben PC. Proton modulation of cardiac I Na: a potential arrhythmogenic trigger. Handb Exp Pharmacol. 2014;221:169–81. doi: 10.1007/978-3-642-41588-3_8. [DOI] [PubMed] [Google Scholar]
- 180.Hille B. The receptor for tetrodotoxin and saxitoxin. A structural hypothesis. Biophys J. 1975;15:615–9. doi: 10.1016/S0006-3495(75)85842-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Lipkind GM, Fozzard HA. A structural model of the tetrodotoxin and saxitoxin binding site of the Na+ channel. Biophys J. 1994;66:1–13. doi: 10.1016/S0006-3495(94)80746-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Penzotti JL, Fozzard HA, Lipkind GM, Dudley SC., Jr. Differences in saxitoxin and tetrodotoxin binding revealed by mutagenesis of the Na+ channel outer vestibule. Biophys J. 1998;75:2647–57. doi: 10.1016/S0006-3495(98)77710-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Walker JR, Novick PA, Parsons WH, McGregor M, Zablocki J, Pande VS, Du Bois J. Marked difference in saxitoxin and tetrodotoxin affinity for the human nociceptive voltage-gated sodium channel (Nav1.7) [corrected]. Proc Natl Acad Sci U S A. 2012;109:18102–7. doi: 10.1073/pnas.1206952109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Tikhonov DB, Zhorov BS. Architecture and pore block of eukaryotic voltage-gated sodium channels in view of NavAb bacterial sodium channel structure. Mol Pharmacol. 2012;82:97–104. doi: 10.1124/mol.112.078212. [DOI] [PubMed] [Google Scholar]
- 185.Chen R, Chung SH. Mechanism of tetrodotoxin block and resistance in sodium channels. Biochem Biophys Res Commun. 2014;446:370–4. doi: 10.1016/j.bbrc.2014.02.115. [DOI] [PubMed] [Google Scholar]
- 186.Ragsdale DS, Scheuer T, Catterall WA. Frequency and voltage-dependent inhibition of type IIA Na+ channels, expressed in a mammalian cell line, by local anesthetic, antiarrhythmic, and anticonvulsant drugs. Mol Pharmacol. 1991;40:756–65. [PubMed] [Google Scholar]
- 187.Ragsdale DS, Avoli M. Sodium channels as molecular targets for antiepileptic drugs. Brain Res Brain Res Rev. 1998;26:16–28. doi: 10.1016/s0165-0173(97)00054-4. [DOI] [PubMed] [Google Scholar]
- 188.Li HL, Galue A, Meadows L, Ragsdale DS. A molecular basis for the different local anesthetic affinities of resting versus open and inactivated states of the sodium channel. Mol Pharmacol. 1999;55:134–41. doi: 10.1124/mol.55.1.134. [DOI] [PubMed] [Google Scholar]
- 189.Ragsdale DS, McPhee JC, Scheuer T, Catterall WA. Common molecular determinants of local anesthetic, antiarrhythmic, and anticonvulsant block of voltage-gated Na+ channels. Proc Natl Acad Sci U S A. 1996;93:9270–5. doi: 10.1073/pnas.93.17.9270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Hockerman GH, Peterson BZ, Johnson BD, Catterall WA. Molecular determinants of drug binding and action on L-type calcium channels. Annu Rev Pharmacol Toxicol. 1997;37:361–96. doi: 10.1146/annurev.pharmtox.37.1.361. [DOI] [PubMed] [Google Scholar]
- 191.Yarov-Yarovoy V, Brown J, Sharp EM, Clare JJ, Scheuer T, Catterall WA. Molecular determinants of voltage-dependent gating and binding of pore-blocking drugs in transmembrane segment IIIS6 of the Na(+) channel alpha subunit. J Biol Chem. 2001;276:20–7. doi: 10.1074/jbc.M006992200. [DOI] [PubMed] [Google Scholar]
- 192.Yarov-Yarovoy V, McPhee JC, Idsvoog D, Pate C, Scheuer T, Catterall WA. Role of amino acid residues in transmembrane segments IS6 and IIS6 of the Na+ channel alpha subunit in voltage-dependent gating and drug block. J Biol Chem. 2002;277:35393–401. doi: 10.1074/jbc.M206126200. [DOI] [PubMed] [Google Scholar]
- 193.Kalia J, Milescu M, Salvatierra J, Wagner J, Klint JK, King GF, Olivera BM, Bosmans F. From foe to friend: Using animal toxins to investigate ion channel function. J Mol Biol. 2014 doi: 10.1016/j.jmb.2014.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Liu Z, Cai T, Zhu Q, Deng M, Li J, Zhou X, Zhang F, Li D, Li J, Liu Y, Hu W, Liang S. Structure and function of hainantoxin-III, a selective antagonist of neuronal tetrodotoxin-sensitive voltage-gated sodium channels isolated from the Chinese bird spider Ornithoctonus hainana. J Biol Chem. 2013;288:20392–403. doi: 10.1074/jbc.M112.426627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Wang J, Yarov-Yarovoy V, Kahn R, Gordon D, Gurevitz M, Scheuer T, Catterall WA. Mapping the receptor site for alpha-scorpion toxins on a Na+ channel voltage sensor. Proc Natl Acad Sci U S A. 2011;108:15426–31. doi: 10.1073/pnas.1112320108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Zhang JZ, Yarov-Yarovoy V, Scheuer T, Karbat I, Cohen L, Gordon D, Gurevitz M, Catterall WA. Mapping the interaction site for a beta-scorpion toxin in the pore module of domain III of voltage-gated Na(+) channels. J Biol Chem. 2012;287:30719–28. doi: 10.1074/jbc.M112.370742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.McCormack K, Santos S, Chapman ML, Krafte DS, Marron BE, West CW, Krambis MJ, Antonio BM, Zellmer SG, Printzenhoff D, Padilla KM, Lin Z, Wagoner PK, Swain NA, Stupple PA, de Groot M, Butt RP, Castle NA. Voltage sensor interaction site for selective small molecule inhibitors of voltage-gated sodium channels. Proc Natl Acad Sci U S A. 2013;110:E2724–32. doi: 10.1073/pnas.1220844110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Lee JH, Park CK, Chen G, Han Q, Xie RG, Liu T, Ji RR, Lee SY. A Monoclonal Antibody that Targets a NaV1.7 Channel Voltage Sensor for Pain and Itch Relief. Cell. 2014;157:1393–404. doi: 10.1016/j.cell.2014.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 199.Henderson R. Structural biology: Ion channel seen by electron microscopy. Nature. 2013;504:93–4. doi: 10.1038/504093a. [DOI] [PubMed] [Google Scholar]