Abstract
Saturation mutagenesis of the 2,4-dinitrotoluene dioxygenase (DDO) of Burkholderia cepacia R34 at position valine 350 of the DntAc α-subunit generated mutant V350F with significantly increased activity towards o-nitrophenol (47 times), m-nitrophenol (34 times), and o-methoxyphenol (174 times) as well as an expanded substrate range that now includes m-methoxyphenol, o-cresol, and m-cresol (wild-type DDO had no detectable activity for these substrates). Another mutant, V350M, also displays increased activity towards o-nitrophenol (20 times) and o-methoxyphenol (162 times) as well as novel activity towards o-cresol. Products were synthesized using whole Escherichia coli TG1 cells expressing the recombinant R34 dntA loci from pBS(Kan)R34, and the initial rates of product formation were determined at 1 mM substrate by reverse-phase high-pressure liquid chromatography. V350F produced both nitrohydroquinone at a rate of 0.75 ± 0.15 nmol/min/mg of protein and 3-nitrocatechol at a rate of 0.069 ± 0.001 nmol/min/mg of protein from o-nitrophenol, 4-nitrocatechol from m-nitrophenol at 0.29 ± 0.02 nmol/min/mg of protein, methoxyhydroquinone from o-methoxyphenol at 2.5 ± 0.6 nmol/min/mg of protein, methoxyhydroquinone from m-methoxyphenol at 0.55 ± 0.02 nmol/min/mg of protein, both methylhydroquinone at 1.52 ± 0.02 nmol/min/mg of protein and 2-hydroxybenzyl alcohol at 0.74 ± 0.05 nmol/min/mg of protein from o-cresol, and methylhydroquinone at 0.43 ± 0.1 nmol/min/mg of protein from m-cresol. V350M produced both nitrohydroquinone at a rate of 0.33 nmol/min/mg of protein and 3-nitrocatechol at 0.089 nmol/min/mg of protein from o-nitrophenol, methoxyhydroquinone from o-methoxyphenol at 2.4 nmol/min/mg of protein, methylhydroquinone at 1.97 nmol/min/mg of protein and 2-hydroxybenzyl alcohol at 0.11 nmol/min/mg of protein from o-cresol. The DDO variants V350F and V350M also exhibited 10-fold-enhanced activity towards naphthalene (8 ± 2.6 nmol/min/mg of protein), forming (1R,2S)-cis-1,2-dihydro-1,2-dihydroxynaphthalene. Hence, mutagenesis of wild-type DDO through active-site engineering generated variants with relatively high rates toward a previously uncharacterized class of substituted phenols for the nitroarene dioxygenases; seven previously uncharacterized substrates were evaluated for wild-type DDO, and four novel monooxygenase-like products were found for the DDO variants V350F and V350M (methoxyhydroquinone, methylhydroquinone, 2-hydroxybenzyl alcohol, and 3-nitrocatechol).
Ring-hydroxylating dioxygenases for biocatalyst applications are well studied (5, 6, 13), but these previous reports have been limited to the characterization of the naphthalene, toluene, and biphenyl dioxygenases, not dinitrotoluene dioxygenases (DDO). Dioxygenases are presently used in industrial and pharmaceutical applications, such as the synthesis of cis-(1S,2R)-indandiol from indene with toluene dioxygenase for the human immunodeficiency virus protease inhibitor CRIXIVAN (7), synthesis of indigo from indole with naphthalene dioxygenase (NDO) (4), and synthesis of the heat-stable polymer polyphenylene from benzene by a dioxygenase-catalyzed reaction with Pseudomonas putida (2). These biocatalysts are examples of green chemistry, the synthesis of chemical compounds which reduce the hazardous reagents or by-products that contribute to environmental pollution (1).
Due to exposure to some environmental pollutants, bacteria have responded by evolving degradation pathways with variant dioxygenases catalyzing the initial oxidation steps to gain a selective advantage by utilizing a wider range of synthetic compounds (12, 23). These evolutionarily recent dioxygenases have not been characterized extensively, and therefore, laboratory mutagenesis of these enzymes may lead to broader biocatalyst opportunities for green chemistry applications. Due to the potential for oxygenases to contribute in useful and difficult chemical synthesis processes (45), van Beilen et al. have speculated that 30% of the chemical business may be supplied by biocatalysts by 2050 (45).
Burkholderia cepacia R34 was isolated for its ability to utilize 2,4-dinitrotoluene (2,4-DNT) as the sole source of carbon and nitrogen (29). The enzymes of the B. cepacia R34 2,4-DNT degradation pathway have been identified, and the initial enzyme in the pathway, DDO, belongs to the three-component, Rieske, non-heme-iron class of ring-hydroxylating dioxygenases (21). Recombinant Escherichia coli strains expressing DDO oxidize 2-amino-4,6-DNT at the 3,4 position, releasing nitrite to form 3-amino-4-methyl-5-nitrocatechol, and also have activity towards the methyl group producing 2-amino-4,6-dinitrobenzyl alcohol (22).
The X-ray crystal structure was solved for the alpha subunit (NahAc) of Pseudomonas sp. strain NCIB 9816-4 NDO by Kauppi et al. (25); hence, the amino acids with essential catalytic properties have been identified, such as those with strict interactions with the mononuclear iron active site, aspartate 362, and those which are involved in electron transport, such as aspartate 205 (25, 30, 31). Through site-directed mutagenesis of several NDO alpha subunit active-site amino acids (30, 46), it was determined that the most important amino acid for influencing dioxygenase regiospecificity was phenylalanine 352, which is located at the narrowest region of the active-site channel (diameter of approximately 5 Å) (25). Site-directed mutagenesis of this amino acid was shown to influence the regioselectivity of NDO towards biphenyl and phenanthrene, generating the novel product phenanthrene 9,10-dihydrodiol from phenanthrene (30).
The B. cepacia R34 DDO alpha subunit, DntAc, has 83% amino acid sequence identity to the alpha subunit NahAc of NDO (21). For this reason, the extensive site-directed mutagenesis work involving the NahAc alpha subunit (30-32, 46) has served as a valuable starting point for site-directed mutagenesis of B. cepacia R34 DDO. Valine 350 of the wild-type DDO alpha subunit (DntAc, 448 amino acids) is homologous to NahAc phenylalanine 352 of NDO (450 amino acids).
The use of dioxygenases as biocatalysts relies mainly on the production of cis-dihydrodiols generating chiral centers; however, the Rieske non-heme-iron oxygenases can also hydroxylate substituted aromatic compounds similar to monooxygenase-catalyzed reactions by adding a single hydroxyl group to the substituted aromatic and forming water (36). The formation of phenol from methoxybenzene (anisole) by NDO has been previously reported by Resnick et al. (36); NDO is also capable of other monooxygenase-like reactions such as the oxidation of indene to (+)-(1S)-indenol (36). NDO has been reported to be unable to catalyze the oxidation of the aromatic nucleus (36). The toluene dioxygenase (TDO) from P. putida F1 has been reported to produce 4-methylcatechol from p-cresol and 3-nitrocatechol from m-nitrophenol (41); however, o- and m-cresol or o- and p-nitrophenol have not been reported to be substrates for the TDO enzyme (41). Because of these limitations in TDO and NDO, DDO was studied for this work.
Our goal was to engineer DDO to oxidize substituted phenols into one of the three important classes of dihydroxylated aromatics for applications in the synthesis of various pharmaceuticals and industrial chemicals. Catechol, resorcinol, and hydroquinone are produced worldwide at 25,000, 35,000, and 50,000 tons per year, respectively, and are valuable as precursors for food, pharmaceutical, and industrial compounds (19). Methoxyhydroquinone is used in the synthesis of triptycene quinones for treatment of leukemia (20), and methylhydroquinone is used in the synthesis of (±)-helibisabonol A and puraquinonic acid, which are precursors for herbicides and antileukemia drugs, respectively (17, 27). Nitrohydroquinone is used in the synthesis of dephostatin, an inhibitor of the protein tyrosine phosphatase (16), which holds promise for treating diabetes mellitus, Alzheimer's disease, and Parkinson's disease (44). Applications for 4-nitrocatechol have been discovered in the synthesis of cardiovascular agents (26) and dopaminergic prodrugs used in the treatment of Parkinson's disease (18). Therefore, substituted, dihydroxylated products are valuable precursors for bioactive compounds, and the synthesis of these components by dioxygenases may be advantageous. Herein we report that saturation mutagenesis of the alpha subunit valine 350 of a Rieske non-heme-iron dioxygenase generates DDO mutants V350F and V350M, which form industrially and pharmaceutically relevant substituted dihydroxy aromatics as a result of monooxygenase-like reactions toward substrates o-cresol, m-cresol, o-nitrophenol, m-nitrophenol, o-methoxyphenol, and m-methoxyphenol (previously uncharacterized substrates for wild-type DDO).
MATERIALS AND METHODS
Bacterial strains, growth conditions, and SDS-PAGE.
E. coli strain TG1 (supE hsdΔ5 thi Δ(lac-proAB) F′[traD36 proAB+ lacIq lacZΔM15]) (39) was the host for the cloning vector pBS(Kan) (8) and the recombinant DDO constructs. Strains expressing the vector pBS(Kan) and the recombinant DDO constructs were grown on Luria-Bertani (LB) (39) agar plates containing kanamycin (100 μg/ml) at 37°C. For whole-cell DDO transformations, strains were initially inoculated into 25 ml of LB medium containing 1.0% glucose and kanamycin (100 μg/ml) and grown overnight at 37°C at 250 rpm. Glucose in the culture medium maintained plasmid segregational stability by suppressing dioxygenase expression until enzyme activity was necessary. After 14 to 16 h of growth, 10 ml of overnight-growth culture was inoculated into 250 ml of LB medium containing 1.0 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and kanamycin (100 μg/ml) and grown at 37°C at 250 rpm. The initial optical density at 600 nm (OD) for each regrowth culture was 0.25 to 0.35, as measured by a Beckman (Fullerton, Calif.); DU 640 spectrophotometer the strains were cultured to an OD of 1.8 to 2.0 after approximately 3 h of incubation and then used for substrate transformations. The relative expression of the dntA loci from E. coli TG1/pBS(Kan)R34 (induced with 1.0 mM IPTG) was evaluated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (39) with a 12% acrylamide gel.
Chemicals.
Nitrohydroquinone was purchased from Frinton Laboratories (Vineland, N.J.), and dimethylformamide (DMF) and naphthalene were purchased from Fisher Scientific Co. (Fairlawn, N.J.). Ninety-eight percent 2-methoxyphenol, 99% o-cresol, 97% m-cresol, 98% methoxyhydroquinone, 98% 2,4-DNT, 98% 2,6-DNT, 98% nitrobenzene, 98% 2-nitrotoluene, 98% 4-nitrotoluene, 98% p-cresol, (1R,2S)-cis-1,2-dihydro-1,2-dihydroxynaphthalene, 98% sulfanilamide, and N-(1-naphthyl)ethyldiamine were purchased from Sigma Chemical Co. (St. Louis, Mo.). Ninety-eight percent 2-nitroresorcinol, 97% 4-nitrocatechol, 97% 3-methoxyphenol, 97% 3-hydroxybenzyl alcohol, 97% 2-hydroxybenzyl alcohol, 99% 3-nitrophenol, 99% 2-nitrophenol, 99% p-methoxyphenol, 99% 2-aminophenol, 99% 3-aminophenol, and 99% p-nitrophenol were purchased from Acros Organics (Morris Plains, N.J.). Ninety-seven percent 4-amino-2,6-dinitrotoluene and 97% 2-amino-4,6-dinitrotoluene compounds were purchased from AccuStandard Inc. (New Haven, Conn.). 3-Nitrocatechol was purchased from Vitas-M (Moscow, Russia). 4-Methyl-5-nitrocatechol was supplied by Barth F. Smets, University of Connecticut, Storrs.
Construction of pBS(Kan)R34.
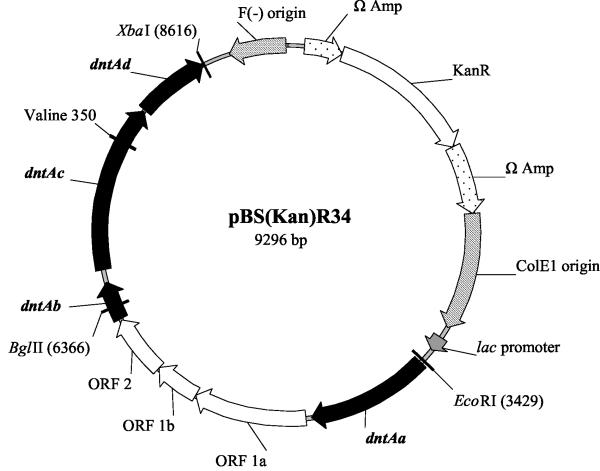
The expression vector pBS(Kan)R34 (Fig. 1) was constructed by PCR amplification the B. cepacia R34 dntA loci from plasmid pJS332 (21) under standard PCR conditions and with 5.0 U of the high-fidelity DNA polymerase Pfu (Stratagene, La Jolla, Calif.). The reaction cycle used consisted of an initial hot start at 96°C for 2 min, after which the Pfu polymerase was added, followed by 30 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 7 min (the final elongation was 72°C for 10 min). The front primer, BSIIR34EcoRI forward (Table 1), contains the restriction site EcoRI and rear primer, BSIIR34Xba reverse, (Table 1), contains the restriction site XbaI; these primers allowed the directional cloning of the 5.3-kb dntA loci into the multiple cloning site, downstream of the lac promoter of the expression vector pBS(Kan), and generated the 9,296-bp plasmid pBS(Kan)R34 (Fig. 1). E. coli TG1 cells were transformed by using a gene pulser (BioRad, Hercules, Calif.) at 15 kV/cm, 25 μF, and 200 Ω and spread on LB agar medium containing 100 μg of kanamycin/ml; Correctly constructed plasmids were selected based on the production of indigoid compounds from recombinant TG1 colonies. The cloned dntA loci were sequenced to confirm correct cloning.
FIG. 1.
Vector pBS(Kan)R34 for constitutive expression of wild-type DDO and mutants. KanR indicates the kanamycin resistance gene, and the relevant restriction enzymes used for cloning (BglII, XbaI, and EcoRI) are shown.
TABLE 1.
Primers used for cloning of R34 dntA loci of B. cepacia strain R34 into pBS(Kan), for saturation mutagenesis of valine 350 of R34 DDO DntAc, and for sequencing of wild-type and mutant insert regions dntAb, dntAc, and dntAd
| Procedure and primer | Sequence |
|---|---|
| dntA loci cloninga | |
| BSIIR34EcoRI forward | 5′-GCATGGGAATTCCAACTGAAAAAAGAGCTTGCATGG-3′ |
| BSIIR34Xba reverse | 5′-GCCAGGTCTAGAAGGGCTCACAGGAAGATTATCAGG-3′ |
| Saturation muta- genesisb | |
| dntAcBgl2 forward | 5′-ATGAGCGAGAACTGGATCG-3′ |
| dntAcXba reverse | 5′-AGGGTTTTCCCAGTCACG-3′ |
| VAL 350 inner forward | 5′-CGCGGTTCAGCGCAGTNNNGGACCAGCAGG-3′ |
| VAL 350 inner reverse | 5′-CCAGTATCCTGCTGGTCCNNNACTGCGCTGAACC-3′ |
| Sequencing | |
| R34ctgB | 5′-CCCCATCACTACCCGTTTTC-3′ |
| R34ctgC | 5′-TTGTACGGAGATGCGATCAA-3′ |
| R34ctgD | 5′-CCTACTCCGGTATCCAGAGC-3′ |
| R34ctgE | 5′-TTTCGGCAAGGACGTCTACG-3′ |
| R34ctgF | 5′-CCCGAGATCAATACCAAGTG-3′ |
Restriction sites used in the cloning of the R34 dntA loci are represented in bold and underlined.
Degenerate positions used in the randomization of position valine 350 are denoted as N (bold and underlined).
Saturation mutagenesis of dntAc.
Saturation mutagenesis at position V350 of the alpha subunit (dntAc) of DDO was performed as described by Sakamoto et al. (38) with the use of the degenerate primers (Table 1) VAL 350 inner forward and VAL 350 inner reverse as well as dntAcBgl2 forward and dntAcXba reverse, which are upstream and downstream of the unique BglII and XbaI restriction sites, respectively (Fig. 1). BglII is a naturally occurring, unique restriction site in dntAb, whereas XbaI is in the multiple cloning site that was conserved during cloning of the dntA loci. Pfu polymerase was used in the PCR to minimize random point mutations outside the desired codon region. A degenerate 1,479-bp PCR fragment was amplified with primers dntAcBgl2 forward and VAL 350 inner reverse, and a 1,023-bp degenerate PCR fragment was amplified with primers VAL 350 inner forward and dntAcXba reverse. After purification of each PCR product from a 0.6% agarose gel, the two fragments were combined at a 1:1 ratio as the templates for the final reassembly PCR with the dntAcBgl2 forward and dntAcXba reverse outer primers. After the initial hot start, a PCR program of 30 cycles of 94°C for 45 s, 50°C for 45 s, and 72°C for 3 min (the final extension was 72°C for 7 min) was executed. The resulting randomized 2,465-bp PCR product (encoding 66% of dntAb and 100% of dntAcAd) was cloned into pBS(Kan)R34 after double digestion of both vector and insert with BglII and XbaI, replacing the wild-type region. The resulting plasmid library was electroporated into exponentially grown E. coli TG1 cells.
Colony screening.
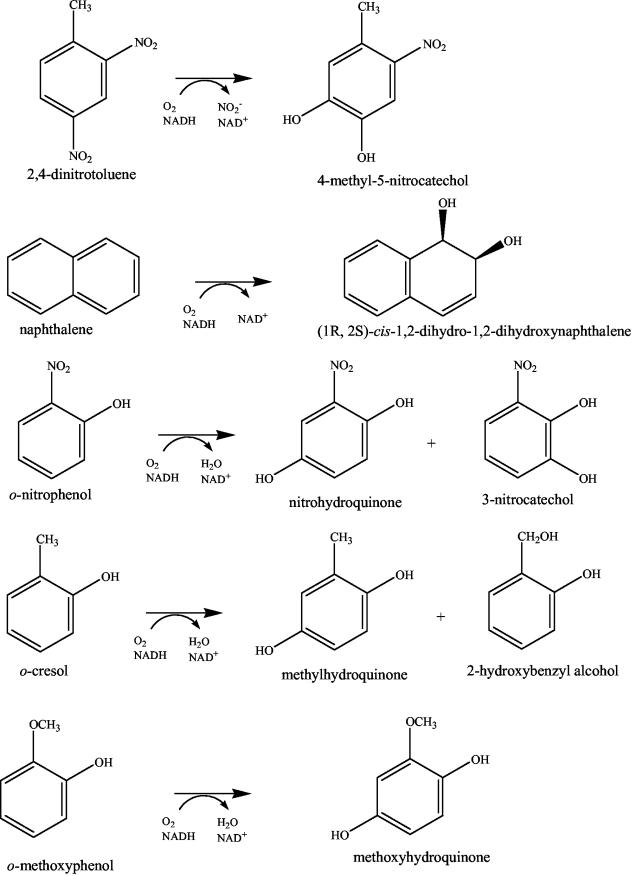
The randomized library was screened for the production of enzyme-mediated products (Fig. 2) that after extracellular oxidation were detected by a solid-phase assay modified from that of Meyer et al. (28) and Barriault et al. (3). The saturation mutant library was first streaked from the original transformation plates onto LB agar plates containing 1.0% glucose and kanamycin (100 μg/ml). Each screening plate contained 50 colonies, which were streaked in replicates to facilitate screening of a large number of substrates. During the screening process, each plate contained both wild-type E. coli TG1/pBS(Kan)R34 and the negative control E. coli TG1/pBS(Kan) since the qualitative nature of the screen required a comparison with both the wild-type reaction and any background reaction caused by the E. coli TG1 host. After 24 to 48 h of growth at 37°C, the colonies were lifted from the original growth plates by using 0.45-μm-pore-size nylon membranes (Osmonics Inc., Minntetonka, Minn.) and placed colony side up onto LB agar plates, for a maximum of 24 h at 37°C, containing 100 μg of kanamycin/ml and 500 to 1000 μM 2,4-DNT, 2,6-DNT, 2-amino-4,6-DNT, 4-amino-2,6-DNT, nitrobenzene, 2-nitrotoluene, 4-nitrotoluene, o-methoxyphenol, o-nitrophenol, m-nitrophenol, or o-cresol from 500 mM DMF stock solutions. To confirm the production of unique oxidation products or enhanced activity, colonies were rescreened with larger amounts of cell mass and fewer clones per plate.
FIG. 2.
Wild-type DDO reactions with 2,4-DNT and naphthalene and the monooxygenase-like reactions of the DDO variants with o-nitrophenol, o-cresol, and o-methoxyphenol.
Product identification and rates of formation via HPLC.
After the second round of solid-phase screening, the positive clones were grown in liquid culture for analysis by reverse-phase high-pressure liquid chromatography (HPLC). The exponentially grown cells were concentrated by centrifugation at 5,400 × g for 7 min at 20°C in a Beckman-Coulter (Palo Alto, Calif.) J2-HS centrifuge and resuspended in either 100 mM sodium phosphate buffer (pH 6.5) or 50 mM Tris-Cl (pH 7.5) to an OD at 600 nm (contact OD) of 20 to 25. Two milliliters of concentrated cells were incubated with 1.0 mM o-cresol, m-cresol, o-nitrophenol, m-nitrophenol, o-methoxyphenol, or m-methoxyphenol (DMF was the diluent; 4 μl of substrate stock solution was added to 2 ml of cell suspension), except 2,4-DNT and naphthalene, which were incubated with 0.1 mM and 5 mM, respectively, in 15.0-ml glass vials sealed with Teflon-lined septa at 37°C and 250 rpm in an IKA Laboratories (Cincinnati, Ohio) KS250 benchtop shaker. After 0 to 60 min, 1.5 ml of contacting suspension was removed and centrifuged at 16,000 × g for 1 to 2 min in a Spectrafuge 16M microcentrifuge (Labnet, Inc., Woodbridge, N.J.). Prior to HPLC analysis, the supernatant was filtered with a Millex-HN (0.45-μm-pore-size and 4-mm-diameter) nylon membrane filter (Millipore Corp., Billerica, Mass.) attached to a Becton Dickinson (Franklin Lakes, N.J.) 1.0-ml syringe. Samples were kept at 4°C immediately prior to HPLC analysis. Substrate and products were separated using a 5-μm, 4.6 by 250 mm Zorbax SB-C8 column (Agilent Technologies, Palo Alto, Calif.). The HPLC pumps (model no. 515), photodiode array detector (model no. 996), and autosampler (model no. 717 plus) were purchased from Waters Corporation (Milford, Mass.). To separate the nitro-substituted substrates and products, a gradient elution was performed with water (0.1% formic acid) and acetonitrile (70:30, 0 to 8 min; 40:60, 8 to 15 min; 70:30, 15 to 20 min) as the mobile phases at a flow rate of 1 ml/min. The resolution of the products 3-nitrocatechol and nitrohydroquinone required the use of a Supelcosil ABZ + Plus HPLC column (Supleco, Bellefonte, Pa.) and an isocratic mobile phase of 80% water (0.1% formic acid) and 20% acetonitrile for 20 min. Separation of the methyl-substituted products and substrates was performed by using a gradient elution with water (0.1% formic acid) and acetonitrile (90:10, 0 to 17 min; 50:50, 17 to 27 min; 90:10, 27 to 30 min) as the mobile phases at a flow rate of 1 ml/min. For the methoxy substrates and products, a gradient elution was performed with water (0.1% formic acid) and acetonitrile (85:15, 0 to 8 min; 65:35, 8 to 13 min; 85:15, 13 to 20 min) as the mobile phases at a flow rate of 1 ml/min. The separation of naphthalene and (1R,2S)-cis-1,2-dihydro-1,2-dihydroxynaphthalene was performed by using a gradient elution with water (0.1% formic acid) and acetonitrile (65:35, 0 to 5 min; 35:65, 5 to 12 min; 65:35, 12 to 20 min) as the mobile phases at a flow rate of 1 ml/min. The products were identified by comparing their retention times and UV-visible spectra to standard chemicals and were confirmed through coelution with authentic standards. To ensure the accuracy of the retention times for the substrates and products, the HPLC columns were equilibrated with the proper elution buffer ratio, for the specific method, prior to evaluation of various transformations. The authentic product standards were also evaluated for their stability to determine when transformation time points should be taken to avoid excessive oxidative breakdown and to identify any potential effects of the breakdown on the UV spectrum of the enzyme-catalyzed oxidation product.
Initial product formation rates were determined by sampling at 5-min intervals for the first 15 min and at 10-min intervals thereafter and were quantified in nanomoles per minute per milligram of protein by converting product peak areas to concentrations with the use of standard curves prepared at the specific absorbance wavelength (Table 2) for each product formed. The initial rates of product formation for wild-type DDO and V350F were determined from at least two independent whole-cell transformation experiments. Protein content was 0.22 mg of protein/ml/1 OD for recombinant E. coli TG1 as determined using the Protein Assay kit (Sigma Diagnostics, Inc., St. Louis, Mo.).
TABLE 2.
Hydroxylation of 2,4-DNT, naphthalene, o-methoxyphenol, m-methoxyphenol, o-cresol, m-cresol, o-nitrophenol, and m-nitrophenol by E. coli TG1 expressing wild-type R34 DDO and dntAc saturation mutants V350F and V350M
| Wild-type DDO | V350F
|
V350M
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Substratea | Productb | Rateb (nmol/ min/mg of protein) | Depletion rateb (nmol of substrate/ min/mg of protein) | Productf | Rate (nmol/ min/mg of protein) | Depletion rate (nmol of substrate/ min/mg of protein) | Product rate relative to that of wild type | Productf | Rate (nmol/ min/mg of protein) | Depletion rate (nmol of substrate/ min/mg of protein) | Product rate relative to that of wild type |
| 2,4-DNT | 4-Methyl-5-nitrocatechol | 1.9 ± 0.2 | —d | ND | 0.00 | 0.00 | 0 | ND | 0.00 | 0.00 | 0 |
| o-Methoxyphenol | Methoxyhydroquinone | 0.0150 ± 0.0002 | 0.19 ± 0.08 | Methoxyhydroquinone | 2.5 ± 0.6 | 2.60 ± 0.0003 | 174 | Methoxyhydroquinone | 2.4 | 2.17 | 162 |
| m-Methoxyphenol | NDc | 0.00 | Methoxyhydroquinone | 0.55 ± 0.02 | 0.65 ± 0.34 | ∞e | Not determined | ||||
| o-Cresol | ND | 0.00 | Methylhydroquinone (67%) | 1.52 ± 0.02 | 3.41 ± 0.55 | ∞ | Methylhydroquinone (90%) | 1.97 | 2.99 | ∞ | |
| 2-Hydroxybenzyl alcohol (33%) | 0.74 ± 0.053 | ∞ | 2-Hydroxybenzyl alcohol (10%) | 0.11 | ∞ | ||||||
| m-Cresol | ND | 0.00 | Methylhydroquinone | 0.43 ± 0.12 | 0.95 ± 0.17 | ∞ | Not determined | ||||
| o-Nitrophenol | Nitrohydroquinone | 0.016 ± 0.002 | — | Nitrohydroquinone (92%) | 0.75 ± 0.15 | 2.55 ± 0.41 | 47 | Nitrohydroquinone (82%) | 0.33 | 2.6 | 20 |
| 3-Nitrocatechol (8%) | 0.069 ± 0.001 | ∞ | 3-Nitrocatechol (18%) | 0.089 | ∞ | ||||||
| m-Nitrophenol | 4-Nitrocatechol | 0.0086 ± 0.002 | — | 4-Nitrocatechol | 0.29 ± 0.02 | — | 34 | Not determined | |||
| Naphthalene | (1R,2S)-cis-1,2-Dihydro-1,2- dihydroxynaphthalene | 0.71 ± 0.3 | — | (1R,2S)-cis-1,2-Dihydro-1,2- dihydroxynaphthalene | 6.6 ± 0.6 | — | 9.0 | (1R,2S)-cis-1,2-Dihydro-1,2- dihydroxynaphthalene | 8.2 ± 2.6 | — | 12 |
Initial concentration, 1 mM, except for 2,4-DNT (0.1 mM) and naphthalene (5 mM).
Determined by HPLC.
ND, not detected.
—, substrate disappearance rates could not be determined due to lack of peak, detectable peak depletion, or substrate peak interference.
∞, infinite improvement relative to wild-type DDO, since the wild-type enzyme had no detectable activity.
Percentages are based on the concentration of each product.
Nitrite assay.
The nitrite generated from the oxidation of 2,4-DNT by DDO was detected following modified standard methods using the reagents sulfanilamide (SUL) and N-(1-naphthyl)ethyldiamine (NAD) (10). Resuspended cells (25 ml) were incubated with 100 μM 2,4-DNT in a 250-ml flask, with shaking at 250 rpm at 37°C. The SUL and NAD reagents were added separately in equal volumes of 100 μl to 1 ml of centrifuged, incubated sample supernatant. The formation of the azo dye was detected at 543 nm after 20 min of incubation at room temperature. During the transformation of 2,4-DNT, the contact OD of recombinant E. coli TG1 strains was kept 3 to 5, as higher OD values led to signal interference.
LC-MS analysis of nitrophenols.
To confirm the results of several of the HPLC analyses, the product of m-nitrophenol and o-nitrophenol oxidation by DntAc mutant V350F was analyzed by reverse-phase liquid chromatography-mass spectrometry (LC-MS) with a Hewlett-Packard (Palo Alto, Calif.) 1090 series II liquid chromatograph with a diode array detector coupled to a Waters Corp. Micromass Q-TOF2 mass spectrometer. Separation was achieved using a Zorbax SB-C18 column (3 μm, 2.1 by 150 mm) with a mobile phase consisting of H2O (0.1% formic acid) and acetonitrile and a gradient elution at 0.3 ml/min starting from 100% H2O (0.1% formic acid) to 0% in 12 min, with a 3-min hold at 100% acetonitrile. The Q-TOF2 mass spectrometer was operated in negative-ion electrospray mode with 3.0 kV applied to the inlet capillary and 75 V applied to the extraction cone. DDO variant V350F was incubated with 1 mM m-nitrophenol or o-nitrophenol for 45 min at a contact OD of 20, after which a supernatant sample was taken for LC-MS analysis. The products generated during the incubation of DDO variant V350F with m-nitrophenol or o-nitrophenol were compared against the retention times, UV spectrum, and molecular mass of the authentic standard compounds under the conditions used for the LC-MS analysis.
DNA sequencing.
The sequencing reactions were performed using the fluorescent labeled BigDye terminators supplied with the ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction kit (Perkin-Elmer, Wellesley, Mass.), and the Perkin-Elmer Biosystems ABI 373 DNA sequencer was used to determine both the wild-type DDO and saturation mutant dntAc nucleotide sequences. The sequencing primer R34ctgD (Table 1) was used to determine codon changes generated by the degenerate primers at the valine 350 codon. The sequencing data were edited, aligned, and translated using the Vector NTI Software (InforMax, Inc., Bethesda, Md.). The sequencing of the cloned dntA locus PCR product revealed an insert sequence of 5′-CCCAACCCAA-3′, which differed from the original template dntA from pJS332 as listed under accession no. AF169302 (21); this region is 60 bp downstream of the stop codon for the dntAb gene and 29 bp upstream of the start of the dntAc gene and is not believed to interfere with enzyme expression or function.
Modeling of DntAc.
The wild-type DDO DntAc alpha subunit was modeled by using SWISS-MODEL Server (15, 33, 40) and was based on the NahAc template 1O7G (24) (polymer chain A) for NDO from Pseudomonas sp. strain NCIB 9816-4. The V350F and V350M mutations were modeled from the generated wild-type DDO model with the DeepView program (Swiss-Pdb Viewer) (15, 33, 40). The DeepView program performed the amino acid substitutions (within the DDO alpha subunit) isosterically for the DDO variants V350F and V350M based on residue-residue interactions, steric hindrance, and energy minimization.
RESULTS
Screening.
A saturation mutagenesis library of 200 clones for R34 dntAc valine 350 was screened for enhanced activity toward 2,4-DNT, 2,6-DNT, 2-amino-4,6-DNT, 4-amino-2,6-DNT, nitrobenzene, 2-nitrotoluene, 4-nitrotoluene, o-methoxyphenol, o-nitrophenol, m-nitrophenol, and o-cresol by the solid-phase assay. Two hundred clones were chosen at random (some produced indigoid compounds) from an initial pool of 2,000 transformation colonies based on our calculation that there is a 96% probability that each of the 64 codons is sampled for a single site and population of this size (37). Mutant VG1 was selected based on its production of uniquely colored oxidation products that diffused from the streaked colony after contact with o-nitrophenol, o-methoxyphenol, and o-cresol (data not shown). Mutant V26-24 was selected based on the production of compounds similar to VG1 on the substrates o-nitrophenol, o-methoxyphenol, and o-cresol. Relative to VG1 during the solid-phase screening, V26-24 appeared to be 25 to 50% less active toward the substrates o-nitrophenol, o-methoxyphenol, and o-cresol. Neither wild-type DDO, VG1, nor V26-24 generated colored oxidation products from nitrobenzene, 2-nitrotoluene, or 4-nitrotoluene. The characterization of the activity of the DDO variants towards these three substrates was not further analyzed; however, the wild-type DDO and variants may have activity not detected by the solid-phase assay. It was also noted that VG1 produced more-intense indigoid-like compounds (the appearance of characteristic blue insoluble precipitate during liquid culture [11]) than wild-type DDO, indicating a potential change in the regiospecific attack of E. coli-derived indole.
Sequencing the complete inserted region of VG1 revealed the codon change from the wild-type GTC (valine 350) to TTT (phenylalanine 350); all other nucleotides encoded by the dntAb, dntAc, and dntAd genes were identical to the wild-type genes; therefore, all other amino acids were conserved. Clone V350F was selected and sequenced on two separate occasions due to its increased indigoid production and the formation of a differently colored product during solid-phase contact with 2,6-DNT compared to wild-type DDO (the TTT codon was found both times). Sequencing V26-24 revealed the codon change at position 350 of GTC (valine 350) to ATG (methionine 350).
Whole-cell transformation of nitrophenols, SDS-PAGE, and LC-MS.
Through comparisons of retention time and the UV-visible spectrum as well as through coelution, the product generated from wild-type DDO oxidation of o-nitrophenol was determined to be nitrohydroquinone (Fig. 2; Table 2). DntAc mutant V350F oxidized o-nitrophenol to nitrohydroquinone with a 47-fold enhancement in the rate. Similarly, variant V350M oxidized o-nitrophenol with a 20-fold rate enhancement relative to that of wild-type DDO. Both variants also produced 3-nitrocatechol, and this product was not formed by the wild-type DDO (infinite rate enhancement). The products 3-nitrocatechol and nitrohydroquinone were the only product peaks present during HPLC analysis of o-nitrophenol oxidation. The negative control, E. coli TG1/pBS(Kan), did not generate nitrohydroquinone or 3-nitrocatechol from o-nitrophenol, so the enhanced oxidations were from the altered DDO.
The discrepancy between the formation rates of 3-nitrocatechol and nitrohydroquinone and the depletion rate of o-nitrophenol (Table 2) may have been due to the reduction of the nitro group of o-nitrophenol to aminophenol; however, a 2-aminophenol peak was not identified by using the Zorbax-C8 or Supelcosil-ABZ column during HPLC analysis. Another source of potential discrepancy in rates may have been due to difficulty in measuring small decreases in substrate concentration (10% of the starting substrate was converted).
For m-nitrophenol, the wild-type DDO formed only 4-nitrocatechol, and V350F formed this product 34-times faster than wild-type DDO (Table 2). The only product peak generated by the wild-type and DDO variants during incubation with m-nitrophenol was the 4-nitrocatechol peak. The negative control, E. coli TG1/pBS(Kan), did not generate 4-nitrocatechol from m-nitrophenol.
The comparison between 4-nitrocatechol production and m-nitrophenol depletion was not possible, due to the inability to accurately measure the baseline of the m-nitrophenol peak. 3-Aminophenol could not be identified by using the Zorbax-C8 or Supelcosil-ABZ column during the current HPLC analysis conditions.
Similar expression levels of the dntA loci from pBS(Kan)R34 for wild-type DDO, V350F, and V350M were seen by SDS-PAGE. The DntAa (37-kDa), DntAc (49-kDa), and DntAd (23-kDa) subunits were visualized on 12% gel since the E. coli TG1 host did not generate proteins having these molecular masses. The DntAb (11-kDa) subunit was not visualized during SDS-PAGE due to the low resolution of low-molecular-mass proteins during this analysis. Hence, the changes in catalytic activity (changed regiospecificity and rates) were not due changes in enzyme expression.
The synthesis of 4-nitrocatechol from m-nitrophenol and nitrohydroquinone from o-nitrophenol were confirmed by LC-MS for DntAc mutant V350F. The products generated matched the expected retention time, UV spectrum, and molecular mass of the authentic standards. The LC-MS identification of 3-nitrocatechol from o-nitrophenol could not be confirmed since nitrohydroquinone and 3-nitrocatechol have identical retention times under the LC-MS conditions used; however, since only nitrohydroquinone was clearly seen with LC-MS, nitrohydroquinone was corroborated to be the major product as determined by the original HPLC analysis. No additional products were identified during the LC-MS analysis to refute any of the conclusions drawn from the HPLC analysis of the whole-cell transformations of the nitrophenols.
Whole-cell transformation of methoxyphenols.
The product generated from wild-type and variant DDO oxidation of o-methoxyphenol was identified as methoxyhydroquinone (Fig. 2; Table 2), but the V350F and V350M mutations increased the rate by 174-fold and 162-fold, respectively. The only product peak generated by DDO variants during incubation with o-methoxyphenol was the methoxyhydroquinone peak. The negative control, E. coli TG1/pBS(Kan), did not generate methoxyhydroquinone from o-methoxyphenol.
For m-methoxyphenol, V350F formed methoxyhydroquinone whereas the wild-type DDO did not form methoxyhydroquinone (Table 2). The only product peak generated by the V350F variant during incubation with m-methoxyphenol was the methoxyhydroquinone peak. The negative control, E. coli TG1/pBS(Kan), did not generate methoxyhydroquinone from m-methoxyphenol, or any other detectable products. For both methoxyphenols, the substrate depletion rates agreed well with the product formation rates.
Whole-cell transformation of cresols.
V350F and V350M oxidized o-cresol to methylhydroquinone and 2-hydroxybenzyl alcohol (Fig. 2; Table 2); in contrast, wild-type DDO did not generate an oxidation product from o-cresol (Table 2). During the o-cresol transformations by V350F, methylhydroquinone was formed at a product molar ratio of 2:1 over 2-hydroxybenzyl alcohol, while for V350M, methylhydroquinone was formed at a product molar ratio of 9:1 over 2-hydroxybenzyl alcohol. The negative control, E. coli TG1/pBS(Kan), did not generate methylhydroquinone or 2-hydroxybenzyl alcohol from o-cresol. The only product peaks present after the incubation of the DDO variants with o-cresol were the methylhydroquinone and 2-hydroxybenzyl alcohol peaks.
The discrepancy between the formation rates of methylhydroquinone and 2-hydroxybenzyl alcohol and the depletion rate of o-cresol (Table 2) may have been due to the instability of the two dihydroxylated methyl compounds. Another source of potential discrepancy in rates may have been due to difficulty in measuring small decreases in substrate concentration (10% of the starting substrate was converted).
For m-cresol, V350F formed methylhydroquinone, whereas wild-type DDO did not generate an oxidation product (Table 2). During the incubation with m-cresol, only the methylhydroquinone product peak was observed. The negative control, E. coli TG1/pBS(Kan), did not generate methylhydroquinone from m-cresol or any other detectable products.
The discrepancy between the formation rates of methylhydroquinone and the depletion rate of m-cresol (Table 2) may have been due to the instability of the methylhydroquinone product. The difficulty in measuring small decreases in substrate concentration (less than 10% of the starting substrate was converted) may have contributed to experimental error in product formation and substrate depletion rates.
Whole-cell transformation of naphthalene.
From naphthalene, wild-type DDO, V350F, and V350M formed (1R,2S)-cis-1,2-dihydro-1,2-dihydroxynaphthalene (Fig. 2; Table 2), but the mutants formed the product at a rate that was enhanced by an order of magnitude. During the incubation with naphthalene, the only product peak present was that of (1R,2S)-cis-1,2-dihydro-1,2-dihydroxynaphthalene; this finding also indicates that the mutant enzymes retain the ability to oxidize aromatics as a true dioxygenase. The depletion rate of naphthalene was not determined during incubation with the DDO variants since the substrate concentration was added to the cell suspensions in excess of its solubility of 0.23 mM (34).
Whole-cell transformation of 2,4-DNT.
Whole cells with wild-type DDO, V350F, and V350M were incubated with 2,4-DNT to determine if the mutant enzymes maintain activity towards the wild-type substrate. Similar rates were found for 4-methyl-5-nitrocatechol formation (Fig. 2) determined by HPLC (Table 2) versus the rate of nitrite production determined by the colorimetric nitrite assay. The initial rate of 2,4-DNT oxidation by wild-type DDO was 2.9 ± 0.8 nmol/min/mg of protein by the nitrite assay and 1.9 ± 0.17 nmol/min/mg of protein by HPLC analysis (Table 2). Neither V350F nor V350M generated a nitrite signal at 543 nm above background by the colorimetric assay, and 4-methyl-5-nitrocatechol was not detected by HPLC (at a contact OD value of 5 or 25). The yellow product generated during the solid-phase screening of V350F and V350M could not be identified during HPLC analysis, and no unique product peaks were identified relative to the wild-type DDO and the negative control, E. coli TG1/pBS(Kan). The negative control, E. coli TG1/pBS(Kan), produced negligible background during the colorimetric detection of nitrite, and no 4-methyl-5-nitrocatechol peak was seen during HPLC analysis. Since the E. coli TG1 host rapidly reduces 2,4-DNT, forming various unidentified reduced products, a correlation between the rate of 4-methyl-5-nitrocatechol formation and the rate of 2,4-DNT removal could not be accurately determined.
Sequencing of V350 saturation mutagenesis library.
The sequencing of clones (at random) from the valine 350 saturation mutagenesis library was conducted to determine if saturation mutagenesis at codon 350 had been accomplished. Other codons identified at V350 (confirming that the saturation mutagenesis technique was accomplished) from this library were TAG (stop), CTC (leucine), and AGT (serine); however, these clones were not further characterized as the activities of these enzymes in E. coli TG1 were not visibly enhanced relative to that of wild-type DDO.
DntAc modeling.
The accuracy of the wild-type DDO DntAc model was determined by evaluating the spatial configuration of the amino acids that coordinate the iron atoms of the Rieske 2Fe-2S center, cysteine 79, cysteine 99, histidine 81, and histidine 102 of DDO (corresponding to cysteine 81, cysteine 101, histidine 83, and histidine 104 in NDO [25]), and the active-site mononuclear iron, aspartate 360, histidine 206, and histidine 211 of the DDO (aspartate 362, histidine 208, and histidine 213 in NDO [25]). By overlapping the three-dimensional structures for these separate regions, it was apparent that the spatial configurations of these key residues between template and model were essentially unchanged. Using the DeepView program, the root-mean-square deviations of the modeled DDO was determined to be 0.07 Å, using 436 Cα atoms of the alpha subunits NahAc and DntAc.
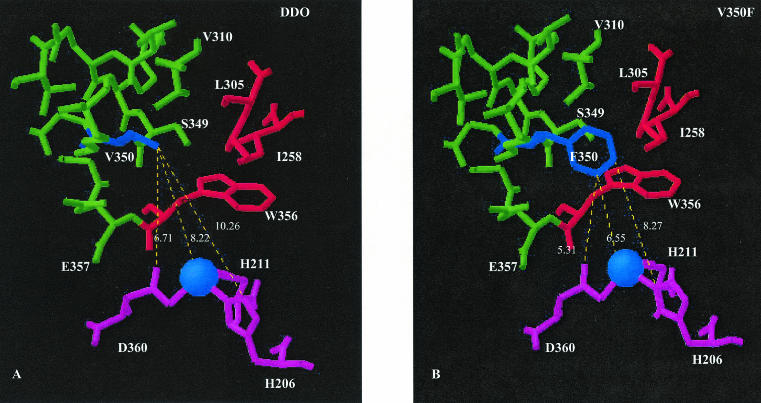
To determine the impact of the amino acid substitutions on the structure of the active sites of the DDO variants, the distance from the position 350 R group to the R groups of each of the amino acids that coordinate the mononuclear iron active site was approximated by using the distance measurement tool of the program DeepView (Fig. 3). The atoms in the R groups, used to determine the distance between amino acids, were chosen to depict the shortest possible distance between position 350 and active-site amino acids. As determined with DeepView, the valine 350 R group of wild-type DDO has an average distance of 8.43 ± 1.83 Å from the R groups of the mononuclear iron-binding active-site amino acids. The R groups of phenylalanine 350 from V350F and methionine 350 from V350M have average distances of 6.59 ± 1.51 and 6.87 ± 1.76 Å, respectively, from these same active-site amino acid R groups. The distance from the valine 350 (DDO), phenylalanine (V350F), and methionine (V350M) R groups to the mononuclear iron atom are 7.81, 5.61, and 6.28 Å, respectively. The substitution of valine 350 with either phenylalanine 350 or methionine 350 does not allow the formation of new hydrogen bonds between position 350 and the neighboring amino acids.
FIG. 3.
(A) Orientation of DntAc valine 350 (V350, dark blue) of wild-type DDO and the approximate distances (expressed in angstroms) of the valine side group to the mononuclear iron-binding active-site amino acids (pink) aspartate 360 (D360), histidine 206 (H206), and histidine 211 (H211). (B) Orientation of phenylalanine 350 of DntAc mutant V350F (F350, dark blue) and the approximate distances (expressed in angstroms) of the phenylalanine side group to the mononuclear iron-binding active-site amino acids. The 10 active-site residues within 4 Å of valine 350, including valine 310 (V310), serine 349 (S349), and glutamate 357 (E357), are shown in green. Active-site residues within 4 Å of only phenylalanine 350, including leucine 305 (L305), isoleucine 258 (I258), and tryptophan 356 (W356), are shown in red. The mononuclear iron is shaded in light blue.
DISCUSSION
It is shown clearly in this study that substitution of the valine 350 residue of the alpha subunit (DntAc) of DDO with phenylalanine or methionine creates enzymes with enhanced reaction rates as well as an expanded substrate range towards naphthalene, o-nitrophenol, m-nitrophenol, o-methoxyphenol, m-methoxyphenol, o-cresol, and m-cresol. Though these variants have lost wild-type DDO activity towards 2,4-DNT, the enhancement in synthesizing the green chemistry precursors will potentially increase the role of Rieske non-heme-iron dioxygenases for industrial and pharmaceutical applications. From the results of this study, it appears that valine at position 350 is critical for removing nitrite from nitroaromatics since mutants with enhanced activity were not detected for 2,4-DNT as well as for 2,6-DNT, 2-amino-4,6-DNT, and 4-amino-2,6-DNT.
This study adds to the list of research (30, 35, 43) in which a single amino acid change can alter dioxygenase activity and regiospecificity to a infinite and novel extent. In previous studies, substitutions of valine and leucine for phenylalanine 352 in NDO, substitution of leucine for phenylalanine 366 in the tetrachlorobenzene dioxygenase (TecA), and the substitutions of phenylalanine 377 for leucine 377, isoleucine 335 for alanine 335, and threonine 376 for asparagine 376 in biphenyl dioxygenase (BphA1) led to the formation of the novel products phenanthrene-9,10-dihydrodiol from phenanthrene for NDO (30), to the formation of 1,2-dihydro-1,2-dihydroxy-1-methyl-3,4,6-trichlorocyclohexa-3,5-diene, 3,4-dichloro-6-methylcatechol, 3,6-dichloro-4-methylcatechol, and 4,6-dichloro-3-methylcatechol from 2,4,5-trichlorotoluene for TecA (35), and to the formation of 2,5,4′-trichloro-3,4-dihydro-3,4-diol from 2,5,2′,5′-tetrachlorobiphenyl for BphA1 (43).
Here, the largest enhancements in DDO V350F activity were for the production of methoxyhydroquinone (2.5 ± 0.6 nmol/min/mg of protein) from o-methoxyphenol and methylhydroquinone (1.52 ± 0.02 nmol/min/mg of protein) from o-cresol (Table 2). These rates for the DDO variants V350F and V350M are similar to the initial transformation rate of 2,4-DNT by the wild-type DDO (1.9 ± 0.2 nmol/min/mg of protein), indicating that the engineered enzymes can have wild-type-like efficiency for previously uncharacterized reactions. Optimizations such as increasing the kcat value of the DDO variants, increasing the expression levels of the enzymes, and designing reactors to remove potentially toxic catechol and hydroquinone products would be required for these DDO mutants to be useful in actual biocatalytic processes (9).
The ratio represented by the regiospecific attack at the methyl group and at the aromatic ring of o-cresol by the DDO variants V350F and V350M is similar to the product distribution obtained for wild-type DDO oxidizing 2-amino 4,6-DNT, where wild-type DDO generates both 3-amino-4-methyl-5-nitrocatechol and 2-amino-4,6-dinitrobenzyl alcohol at approximately a 2:1 ratio (22). While wild-type DDO maintains the ability to activate the aromatic ring with dioxygen, the DDO variants appear to activate only the aromatic ring in a monooxygenase-like reaction (at least for the 1-ring substrates tested). The regiospecific attack at the methyl group of substituted toluenes is a common feature of the Rieske non-heme-iron dioxygenases, and additional amino acid changes at the active site of V350F may be required to promote or prevent the regiospecific attack at the methyl group of substituted toluenes.
By focusing on the synthesized product, instead of trying to predict the effect of a specific amino acid functional group on dioxygenase activity, saturation mutagenesis was used to improve the DDO active site. Screening the saturation mutagenesis library of DntAc valine 350 mutants was facilitated by the use of the nylon membrane in conjunction with agar plates containing substrates and allowed the easy detection of the auto-oxidation products from the substituted catechols and hydroquinones (3, 28). The secreted, auto-oxidized products were necessary for the selection of mutant clones during the solid-phase screening; however, during the determination of initial rates of formation by HPLC, early time points were taken to avoid product breakdown. Though the solid-plate assay is primarily qualitative, the detection of mutant clones expressing enhanced enzymes for substituted phenols was straightforward.
Although there are limitations to homology modeling that arise due to nonconserved loops (14), the use of the NahAc template (83% identity to R34 DntAc) yielded a model which deviates by less than 0.1 Å from the control template. From this model it appears that for the mutants V350F (Fig. 3) and V350M (data not shown), the average distance between the R group of amino acid position 350 and the iron-binding active-site residues aspartate 360, histidine 206, and histidine 211 changes relative to that of the wild-type DDO. The most apparent conformation change in V350F, relative to wild-type DDO, is the shift of the phenylalanine functional group towards histidine 206 and histidine 211 (distance decreased by 2 Å). The electronic space occupied by these mutant R groups may dictate overall active-site pocket size more than the approximated distances, and this effect may be a more difficult impact to quantify. The shift towards histidine 206 and 211 is present in both DDO variants V350F and V350M and potentially blocks the route of 2,4-DNT to the active site, preventing the wild-type reaction, allowing new substrate orientations and new routes to the active site, and perhaps enhancing the stability of the substrates o-cresol, m-cresol, o-methoxyphenol, and m-methoxyphenol within the active site.
In an attempt to explain the changes in active-site topology caused by the substitution of valine 350 with phenylalanine 350 (V350F), the amino acids near position 350 were compared for both R groups of valine and phenylalanine (Fig. 3). At a neighbor distance of 4 Å, the R group of valine 350 is most likely to interact with V310, K312, T326, V346, Q347, R348, S349, G351, G354, and E357 near the outer edge of an alpha helix in the DntAc alpha subunit (Fig. 3). At the same neighbor distance of 4 Å, the R group of phenylalanine 350 appears more likely to interact with all these amino acids as well as I258, L305, and W356, which are located closer to the iron-binding residues and more buried within the active-site pocket (Fig. 3). The nearest-neighbor comparisons imply that the −CH2 group of the phenylalanine 350 R group interacts similarly to the entire valine 350 R group, but it is the large phenyl group that invades the active-site pocket and causes changes in the accessibility of the substrates to the active-site iron and positioning of the substrates relative to the activated iron-dioxygen complex. The structural investigation of the NDO conducted by Karlsson et al. (24) revealed that dioxygen binds side-on to iron, allowing the formation of naphthalene cis-dihydrodiol from naphthalene (24). The phenylalanine R group may allow a potential substrate to orient for side-on or end-on activation, dictating a dioxygenase or monooxygenase reaction mechanism. The relative increase in hydrophobicity in comparing the phenylalaine R group to the valine R group should not be ignored; however, the difference in the activities of the V350F and V350M variants may be due to the sulfur atom of methionine. The role of the R group for active-site amino acids during the interactions between substrates and the mononuclear iron remains to be characterized fully and may enable a better understanding of oxygenase-catalyzed reaction mechanisms.
The determination that variant V350F does not have strong activity towards the p-substituted hydroxylated aromatics implies that steric hindrance at the active site may be dictated by the placement of the hydroxyl group of the substrate rather than the class of compound (i.e., methyl, nitro, or methoxy aromatics). The same relative steric hindrance may also explain why the wild-type enzyme does not have detectable activity towards 4-amino-2,6-DNT (22) (it can be hypothesized that the p-substituted amino group blocks any affinity of the enzyme toward the nitro groups). To overcome the steric hindrance represented by the p-substituted compounds (including 2,4-DNT for V350F), it appears that more than a single amino acid substitution is needed; hence, V350F is being used as a template for future rounds of DNA shuffling (42). Reversing the activity of V350F towards 2,4-DNT will allow the discovery of other amino acids that influence the active-site substrate pocket and substrate selectivity and allow additional insights into engineering DDO for specific applications.
Acknowledgments
This study was supported by the National Science Foundation (BES-0114126) and by the U.S. Department of Education through the Graduate Assistance in Areas of National Need Award in Environmental Biotechnology (P200A000821).
We thank Glenn R. Johnson for plasmid pJS332, Ayelet Fishman for her help with HPLC analyses, and Albert Kind for his assistance with the LC-MS analyses.
REFERENCES
- 1.Anastas, P. T., and M. M. Kirchhoff. 2002. Origins, current status, and future challenges of green chemistry. Accounts Chem. Res. 35:686-694. [DOI] [PubMed] [Google Scholar]
- 2.Ballard, D. G. H., A. Courtis, I. M. Shirley, and S. C. Taylor. 1998. Synthesis of polyphenylene from a cis-dihydrocatechol, a biologically produced monomer. Macromolecules 21:294-304. [Google Scholar]
- 3.Barriault, D., M. M. Plante, and M. Sylvestre. 2002. Family shuffling of a targeted bphA region to engineer biphenyl dioxygenase. J. Bacteriol. 184:3794-3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berry, A., T. C. Dodge, M. Pepsin, and W. Weyler. 2002. Application of metabolic engineering to improve both the production and use of biotech indigo. J. Ind. Microbiol. Biotechnol. 28:127-1333. [DOI] [PubMed] [Google Scholar]
- 5.Boyd, D. R., N. D. Sharma, and C. C. R. Allen. 2001. Aromatic dioxygenases: molecular biocatalysis and applications. Curr. Opin. Biotechnol. 12:564-573. [DOI] [PubMed] [Google Scholar]
- 6.Boyd, D. R., and G. N. Sheldrake. 1998. The dioxygenase-catalysed formation of vicinal cis-diols. Nat. Products Rep. 15:309-324.
- 7.Buckland, B. C., S. W. Drew, N. C. Connors, M. M. Chartrain, C. Lee, P. M. Salmon, K. Gbewonyo, W. Zhou, P. Gailliot, R. Singhvi, R. C. Olewinski, Jr., W.-J. Sun, J. Reddy, J. Zhang, B. A. Jackey, C. Taylor, K. E. Goklen, B. Junker, and R. L. Greasham. 1999. Microbial conversion of Indene to Indandiol: A Key Intermediate in the Synthesis of CRIXIVAN. Metab. Eng. 1:63-74. [DOI] [PubMed] [Google Scholar]
- 8.Canada, K. A., S. Iwashita, H. Shim, and T. K. Wood. 2002. Directed evolution of toluene ortho-monooxygenase for enhanced 1-naphthol synthesis and chlorinated ethene degradation. J. Bacteriol. 184:344-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duetz, W. A., J. B. van Beilen, and B. Witholt. 2001. Using proteins in their natural environment: potential and limitations of microbial whole-cell hydroxylations in applied biocatalysis. Curr. Opin. Biotechnol. 12:419-425. [DOI] [PubMed] [Google Scholar]
- 10.Eaton, A. D., L. S. Clesceri, and A. E. Greenberg (ed.). 1995. Standard methods, 19th ed. American Public Health Association, Washington, D.C.
- 11.Ensley, B. D., B. J. Ratzkin, T. D. Osslund, and M. J. Simon. 1983. Expression of naphthalene oxidation genes in Escherichia coli results in the biosynthesis of indigo. Science 222:167-169. [DOI] [PubMed] [Google Scholar]
- 12.Fuenmayor, S. L., M. Wild, A. L. Boyes, and P. A. Williams. 1998. A gene cluster encoding steps in conversion of naphthalene to gentisate in Pseudomonas sp. strain U2. J. Bacteriol. 180:2522-2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gibson, D. T., and R. E. Parales. 2000. Aromatic hydrocarbon dioxygenases in environmental biotechnology. Curr. Opin. Biotechnol. 11:236-243. [DOI] [PubMed] [Google Scholar]
- 14.Guex, N., A. Diemand, and M. C. Peitsch. 1999. Protein modeling for all. Trends Biochem. Sci. 24:364-367. [DOI] [PubMed] [Google Scholar]
- 15.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-Pdb Viewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 16.Hamel, P., and Y. Girard. 1994. Synthesis of dephostatin, a novel protein tyrosine phosphatase inhibitor. Tetrahedron Lett. 35:8101-8102. [Google Scholar]
- 17.Hisaindee, S., and D. L. J. Clive. 2001. A synthesis of puraquinonic acid. Tetrahedron Lett. 42:2253-2255. [Google Scholar]
- 18.Houwing, H. A., J. C. van Oene, and A. S. Horn. 1983. 3,4-Disubstituted phenyliminoimidazolidines as potential prodrugs of the purported dopamine agonist 3,4-dihydroxyphenylimino-2-imidazolidine (DPI). Pharm. wkbld. 5:177-181. [DOI] [PubMed] [Google Scholar]
- 19.Howe-Grant, M. 1991. Kirk-Othmer encyclopedia of chemical technology, 4th ed., vol. 13. Wiley-Interscience Publishers, New York, N.Y.
- 20.Hua, D. H., M. Tamura, X. Huang, H. A. Stephany, B. A. Helfrich, E. M. Perchellet, B. J. Sperfslage, J.-P. Perchellet, S. Jiang, D. E. Kyle, and P. K. Chiang. 2002. Syntheses and bioactivities of substituted 9,10-dihydro-9,10-[1,2]benzenoanthracene-1,4,5,8-tetrones. unusual reactivities with amines. J. Org. Chem. 67:2907-2912. [DOI] [PubMed] [Google Scholar]
- 21.Johnson, G. R., R. K. Jain, and J. C. Spain. 2002. Origins of the 2,4-dinitrotoluene pathway. J. Bacteriol. 184:4219-4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson, G. R., B. F. Smets, and J. C. Spain. 2001. Oxidative transformation of aminodinitrotoluene isomers by multicomponent dioxygenases. Appl. Environ. Microbiol. 67:5460-5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson, G. R., and J. C. Spain. 2003. Evolution of catabolic pathways for synthetic compounds: bacterial pathways for degradation of 2,4-dinitrotoluene and nitrobenzene. Appl. Microbiol. Biotechnol. 62:110-123. [DOI] [PubMed] [Google Scholar]
- 24.Karlsson, A., J. V. Parales, R. E. Parales, D. T. Gibson, H. Eklund, and S. Ramaswamy. 2003. Crystal structure of naphthalene dioxygenase: side-on binding of dioxygen to iron. Science 299:1039-1042. [DOI] [PubMed] [Google Scholar]
- 25.Kauppi, B., K. Lee, E. Carredano, R. E. Parales, D. T. Gibson, H. Eklund, and S. Ramaswamy. 1998. Structure of an aromatic-ring-hydroxylating dioxygenase-naphthalene 1,2 dioxygenase. Structure 6:571-586. [DOI] [PubMed] [Google Scholar]
- 26.Lumma, W. C., Jr., and G. B. Phillips. 30 October 1990. Preparation of 3,4,5,6-tetrahydro-2H-1,7,4-benzodioxazonines as cardiovascular agents. U.S. patent. 4,966,967.
- 27.Macias, F. A., D. Marin, D. Chinchilla, and J. M. G. Molinillo. 2002. First total synthesis of (+/−)-helibisabonol A. Tetrahedron Lett. 43:6417-6420. [Google Scholar]
- 28.Meyer, A., A. Schmid, M. Held, A. H. Westphal, M. Rothlisberger, H.-P. E. Kohler, W. J. H. van Berkel, and B. Witholt. 2002. Changing the substrate reactivity of 2-hydroxybiphenyl 3-monooxygenase from Pseudomonas azelaica HBP1 by directed evolution. J. Biol. Chem. 277:5575-5582. [DOI] [PubMed] [Google Scholar]
- 29.Nishino, S. F., G. C. Paoli, and J. C. Spain. 2000. Aerobic degradation of dinitrotoluenes and pathway for bacterial degradation of 2,6-dinitrotoluene. Appl. Environ. Microbiol. 66:2139-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parales, R. E., K. Lee, S. M. Resnick, H. Jiang, D. J. Lessner, and D. T. Gibson. 2000. Substrate specificity of naphthalene dioxygenase: effect of specific amino acids at the active site of the enzyme. J. Bacteriol. 182:1641-1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parales, R. E., J. V. Parales, and D. T. Gibson. 1999. Aspartate 205 in the catalytic domain of naphthalene dioxygenase is essential for activity. J. Bacteriol. 181:1831-1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parales, R. E., S. M. Resnick, C.-L. Yu, D. R. Boyd, N. D. Sharma, and D. T. Gibson. 2000. Regioselectivity and enantioselectivity of naphthalene dioxygenase during arene cis-dihydroxylation: control by phenylalanine 352 in the alpha-subunit. J. Bacteriol. 182:5495-5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peitsch, M. C. 1993. Protein modeling by E-mail. Bio/Technology 13:658-660. [Google Scholar]
- 34.Perry, R. H., and C. H. Chilton (ed.). 1973. Chemical engineer's handbook, 5th ed. McGraw-Hill Book Company, New York, N.Y.
- 35.Pollmann, K., V. Wray, H.-J. Hecht, and D. H. Pieper. 2003. Rational engineering of the regioselectivity of TecA tetrachlorobenzene dioxygenase for the transformation of chlorinated toluenes. Microbiology 149:903-913. [DOI] [PubMed] [Google Scholar]
- 36.Resnick, S. M., K. Lee, and D. T. Gibson. 1996. Diverse reactions catalyzed by naphthalene dioxygenase from Pseudomonas sp. strain NCIB 9816. J. Ind. Microbiol. 17:438-457. [Google Scholar]
- 37.Rui, L., Y. M. Kwon, A. Fishman, K. F. Reardon, and T. K. Wood. 2004. Saturation mutagenesis of toluene ortho-monooxygenase for enhanced 1-naphthol synthesis and chloroform degradation. Appl. Environ. Microbiol. 70:3246-3252. [DOI] [PMC free article] [PubMed]
- 38.Sakamoto, T., J. M. Joern, A. Arisawa, and F. H. Arnold. 2001. Laboratory evolution of toluene dioxygenase to accept 4-picoline as a substrate. Appl. Environ. Microbiol. 67:3882-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 40.Schwede, T., J. Kopp, N. Guex, and M. C. Peitsch. 2003. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 31:3381-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spain, J. C., and D. T. Gibson. 1988. Oxidation of substituted phenols by Pseudomonas putida F1 and Pseudomonas sp. strain JS6. Appl. Environ. Microbiol. 54:1399-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stemmer, W. P. C. 1994. DNA shuffling by random fragmentation and reassembly: in vitro recombination for molecular evolution. Proc. Natl. Acad. Sci. USA 91:10747-10751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suenaga, H., T. Watanabe, M. Sato, Ngadiman, and K. Furukawa. 2002. Alteration of regiospecificity in biphenyl dioxygenase by active-site engineering. J. Bacteriol. 184:3682-3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Umezawa, K., M. Kawakami, and T. Watanabe. 2003. Molecular design and biological activities of protein-tyrosine phosphatase inhibitors. Pharmacol. Therapeu. 99:15-24. [DOI] [PubMed] [Google Scholar]
- 45.van Beilen, J. B., W. A. Duetz, A. Schmid, and B. Witholt. 2003. Practical issues in the application of oxygenases. Trends Biotechnol. 21:170-177. [DOI] [PubMed] [Google Scholar]
- 46.Yu, C.-L., R. E. Parales, and D. T. Gibson. 2001. Multiple mutations at the active site of the naphthalene dioxygenase affect regioselectivity and enantioselectivity. J. Ind. Microbiol. Biotechnol. 27:94-103. [DOI] [PubMed] [Google Scholar]