Abstract
Quantitative PCR (Q-PCR) is a fast and efficient tool to quantify target genes. In eukaryotic cells, quantitative reverse transcription-PCR (Q-RT-PCR) is also used to quantify gene expression, with stably expressed housekeeping genes as standards. In bacteria, such stable expression of housekeeping genes does not occur, and the use of DNA standards leads to a broad underestimation. Therefore, an accurate quantification of RNA is feasible only by using appropriate RNA standards. We established and validated a Q-PCR method which enables the quantification of not only the number of copies of target genes (i.e., the number of bacterial cells) but also the number of RNA copies. The genes coding for InvA and the 16S rRNA of Salmonella enterica serovar Typhimurium were selected for the evaluation of the method. As DNA standards, amplified fragments of the target genes were used, whereas the same DNA standards were transcribed in vitro for the development of appropriate RNA standards. Salmonella cultures and environmental water samples inoculated with bacteria were then employed for the final testing. Both experimental approaches led to a sensitive, accurate, and reproducible quantification of the selected target genes and RNA molecules by Q-PCR and Q-RT-PCR. It is the first time that RNA standards have been successfully used for a precise quantification of the number of RNA molecules in prokaryotes. This demonstrates the potential of this approach for determining the presence and metabolic activity of pathogenic bacteria in environmental samples.
The presence of pathogenic bacteria in water represents a major health concern (21, 28). However, the detection of bacterial contamination still relies on cultivation-based methods. The use of these conventional approaches is time-consuming and does not allow the detection of viable but nonculturable bacteria. The use of PCR has emerged as a new approach to overcome these problems. Different protocols have been developed for the detection of pathogenic bacteria in water samples by PCR (7, 9, 13). However, the exploitation of gene targets for the evaluation of presence or absence of bacteria is still a matter of controversy, since the long persistence of DNA, even after bacterial death, may lead to false-positive results (6). Therefore, the detection of RNA species that are much less stable in the environment is considered as an attractive alternative (20). In this context, carefully chosen RNA species not only may allow the presence of bacteria to be proved but also would give information on gene expression (i.e., metabolic activity). This would also allow an evaluation of the risk derived from the presence of waterborne bacterial pathogens in the environment.
Real-time quantitative PCR (Q-PCR) and reverse transcription PCR (Q-RT-PCR) are methods which allow an efficient quantification of genes and gene products, even in environmental samples (16, 24). However, accurate quantification of RNA species is still hampered by the absence of a reliable standardization. In eukaryotic cells, stably expressed housekeeping genes (e.g., the β-actin gene) can be used as standards to perform a relative quantification of gene expression (3). Unfortunately, for bacteria no such stably expressed gene is known. Some groups have proposed that the expression of a functional gene be compared with the total amount of 16S rRNA (12, 14). However, the expression of rRNA is tightly dependent on the physiological status of bacteria (5, 18). When the measurement of gene expression in environmental samples is considered, this physiological status (i.e., the expression levels of 16S rRNA per cell) would be unknown. Furthermore, this approach would not allow quantification of the exact number of copies of the target gene. Other groups have used either genomic DNA or plasmids containing genomic DNA fragments as standards for Q-RT-PCR (17, 22, 23). However, the use of DNA standards does not take into consideration the efficiency of the reverse transcription step, which leads to reaction efficiencies reduced by 84 to 98.6% compared to the real values (15). Therefore, the use of DNA standards results in a broad underestimation of the target molecules. This problem has been overcome in eukaryotes by using in vitro-transcribed RNA standards (25). However, similar approaches still need to be implemented for bacteria.
The aim of the present study was to develop and evaluate an analytical approach based on the use of Q-PCR to achieve an accurate quantification of bacterial genes, as well as RNA species from pathogens relevant to health in environmental water samples. Salmonella enterica serovar Typhimurium was selected as a model organism, and the genes coding for InvA and the 16S rRNA were selected as targets. The method was validated by pure cultures and water samples inoculated with bacteria. The obtained results demonstrated, for the first time, that RNA standards obtained by in vitro transcription could be successfully used for a precise quantification of the number of RNA copies in bacteria. This Q-PCR-based approach would allow a sensitive and accurate determination of the presence and metabolic activity of pathogenic bacteria in environmental water samples.
MATERIALS AND METHODS
Bacterial strain and growth conditions.
The S. enterica serovar Typhimurium strain ATCC 14028 was grown in brain heart infusion (BHI; Difco) broth at 37°C with shaking (100 rpm). Bacterial growth was monitored by determining the optical density at 600 nm (OD600). For establishing the number of viable bacteria, samples were serially diluted in BHI broth and 50 μl was spread plated on BHI agar plates in replicates. Plates were incubated at 37°C for 24 h. For calculation of the CFU per milliliter, only dilutions showing between 20 and 300 colonies were used.
Nucleic acid extraction.
DNA and RNA extractions were performed with the DNeasy and RNeasy kits, as recommended by the producer (QIAGEN, Hilden, Germany). For RNA extractions, the lysis was performed with 100 μl of lysozyme (500 μg ml−1) for 5 min at room temperature. DNase (QIAGEN) treatment was performed for 30 min at room temperature.
Design of primers and standards for Q-PCR.
All primers (Table 1) were purchased from MWG Biotech (Ebersberg, Germany). Targets for Q-PCR and Q-RT-PCR were the genes coding for the Salmonella 16S rRNA and InvA, as well as the corresponding RNAs. To enable an accurate quantification of the targets, DNA and RNA standards were generated. For this purpose a second primer set was designed for each target gene in which (i) the primers were located up- and downstream of the sequences recognized by the first set (i.e., longer amplification product) and (ii) the forward primer contained the sequences of the T7 promoter (Table 1). These primers were used to amplify the genomic DNA of S. enterica by PCR, according to the following protocol: initial denaturation 4 min at 94°C; 30 cycles of 1 min at 94°C, 1 min at 58 (invA) or 60°C (16S rRNA gene), and 1 min 72°C; and then a final elongation at 72°C for 6 min. The PCR mixture (50 μl per sample) contained 5 μl of 10× PCR buffer (QIAGEN), 3 mM MgCl2, 200 μM deoxynucleoside triphosphates (Invitrogen, Karlsruhe, Germany), 400 nM (each) primer, 0.2 μl of Taq polymerase (QIAGEN), and 1 ng of genomic DNA. PCR products were purified with the PCR purification kit (QIAGEN) and subsequently used as DNA standards for Q-PCR. To obtain RNA standards, the purified PCR products were transcribed in vitro with T7 polymerase by using the Riboprobe System-T7 (Promega). This was followed by a digestion with DNase I (15 min, 37°C) and a subsequent purification by the RNA cleanup protocol of the RNeasy minikit (QIAGEN), which included a second DNase I digestion on the column for 15 min at room temperature during purification. The transcripts were analyzed in agarose gels (1%) containing 0.65% formaldehyde. DNA and RNA standards were quantified with PicoGreen (double-stranded DNA quantitation kit) and RiboGreen (RNA quantitation kit) from Molecular Probes (27). The standards were diluted in nuclease free water and stored at −20 (DNA) or −70°C (RNA).
TABLE 1.
Primers used to generate DNA and RNA standards and PCR primers for Q-PCR
| Target gene | Primer name | Sequence (5′→3′)a | Position on geneb | Product size (bp) | Accession no. |
|---|---|---|---|---|---|
| Generation of standards | |||||
| 16S rRNA | Salm16S-T7F | TAATACGACTCACTATAGGGTGATGGAGGGGGATAACTACTGGA | 130-153 | 705 | Z49264 |
| Salm16S-R2 | CATCGTTTACGGCGTGGACTACCA | 791-814 | Z49264 | ||
| invA | invA-T7-F2 | TAATACGACTCACTATAGGGAACAGTGCTCGTTTACG | 103-119 | 885 | M90846 |
| invA-T7-R1 | GCAGAGTTCCCATTGAAATGGTC | 945-967 | M90846 | ||
| PCR primers | |||||
| 16S rRNA | Salm16S-F | CGGGGAGGAAGGTGTTGTG | 436-454 | 178 | Z49264 |
| Salm16S-R1 | GAGCCCGGGGATTTCACATC | 594-613 | Z49264 | ||
| invA | invA2-F | GATTCTGGTACTAATGGTGATGATC | 132-156 | 288 | M90846 |
| invA2-R | GCCAGGCTATCGCCAATAAC | 400-419 | M90846 |
Sequences corresponding to the T7 promoter are underlined.
Positions of genes are given according to the accession numbers.
Calculation of copy numbers.
The numbers of copies of the Q-PCR standards were calculated by assuming average molecular masses of 660 Da for 1 bp of double-stranded DNA and 340 Da for 1 nucleotide of single-stranded RNA (PCR applications manual, 2nd ed., Roche Diagnostics GmbH, Mannheim, Germany, 1999). The calculation was done with the following equation: copies per nanogram = (n × mw)/(NL × 10−9), were n is the length of the standard in base pairs or nucleotides, mw is the molecular weight per bp or nucleotide, and NL is the Avogadro constant (6.02 × 1023 molecules per mol).
Q-PCR and Q-RT-PCR.
For Q-PCR, 5 μl of diluted sample was added to 20 μl of a PCR mixture prepared from 2× Sybr Green PCR Master Mix (Applied Biosystems), which contained each primer at a concentration of 400 nM. The cycle parameters were as follows: 10 min at 95°C and 50 cycles of 20 s at 95°C and 1 min at 60 (invA) or 65°C (16S rRNA gene). This was followed by 20 s at 95°C and 1 min at 60 (invA) or 65°C (16S rRNA gene). For the determination of the melting curve, the temperature was increased 1°C every 20 s from 65 to 94°C.
For Q-RT-PCR, cDNA was generated prior to Q-PCR with the Sybr Green RT-PCR reagents (Applied Biosystems) and random hexamers for priming, according to the instructions of the manufacturer. RNA standards were similarly treated. In each reverse transcription reaction, some RNA samples were not supplemented with reverse transcriptase to rule out DNA contamination. One-step Q-RT-PCR performed using Sybr Green RT-PCR reagents and sequence-specific primers was compared with two-step Q-RT-PCR. No differences in amplification efficiency were detected; thus, the two-step Q-RT-PCR was used in all further studies for practical reasons (e.g., long-term storage of cDNA, use of one set of cDNA for quantification of different targets). Q-PCR and Q-RT-PCR were performed on a RotorGene 2000 (Corbett).
Data analysis.
To generate a calibration curve, the serially diluted DNA or RNA standard (1,000 pg to 0.001 fg) was quantified in each Q-PCR run. The calibration curves were generated by the RotorGene software, version 4.6. For each standard, the concentration was plotted against the cycle number at which the fluorescence signal increased above the background or threshold value (Ct value). The slope of each calibration curve was used in the following equation to determine the reaction efficiency: efficiency = 10−1/slope − 1. According to this, an efficiency of 1 means a doubling of product in each cycle. Using the calibration curve, the RotorGene software calculated the initial number of target copies in the measured samples. From these values, the number of copies per milliliter of culture or inoculated water was calculated.
Experimental design.
For the determination of the S. enterica serovar Typhimurium growth kinetics, 80 ml of BHI broth was inoculated with 2 ml of overnight culture (final OD600 = 0.065) in 250-ml Erlenmeyer flasks. Three replicate flasks were incubated at 37°C with shaking (100 rpm), and samples were obtained at different time intervals to determine the OD600 and the number of viable bacteria (colony counts). For nucleic acid extraction, between 0.2 and 1 ml of liquid culture (maximum of 109 bacterial cells) was transferred to a 1.5-ml reaction tube. Samples for RNA extraction were then homogenized with 2 volumes of RNAprotect solution (QIAGEN) and further incubated for 5 min at room temperature to stabilize bacterial RNA (1). Reaction tubes were centrifuged at 13,000 × g for 10 min, and the supernatant fluids were stored at −20°C until nucleic acid extraction.
For spiking experiments, fresh BHI broth was inoculated with an overnight culture of the Salmonella strain. After 4.5 h of growth (37°C, 100 rpm, final OD600 = 1.7), bacterial cultures were centrifuged for 10 min at 3,000 × g. The pellet was resuspended in 0.25× Ringer solution (Oxoid), and the OD600 was adjusted to 0.2 to obtain a cell density of approximately 108 bacteria per milliliter. One milliliter of bacterial suspension was added to 100 ml of either tap water or water collected from the fishpond on the grounds of our institute in 100-ml glass bottles (approximately 106 cells per milliliter). Samples of inoculated water were then transferred to bottles containing 90 ml of the respective water to obtain bottles containing between 106 cells and 1 cell per ml. For DNA and RNA extraction, two separate 10-ml samples were obtained after homogenization and were transferred into 15-ml tubes. After centrifugation (10 min, 3,000 × g), supernatant fluids were removed and the pellets were stored at −20°C until extraction. Each experiment was carried out in triplicate, and nonspiked control flasks were also processed.
RESULTS
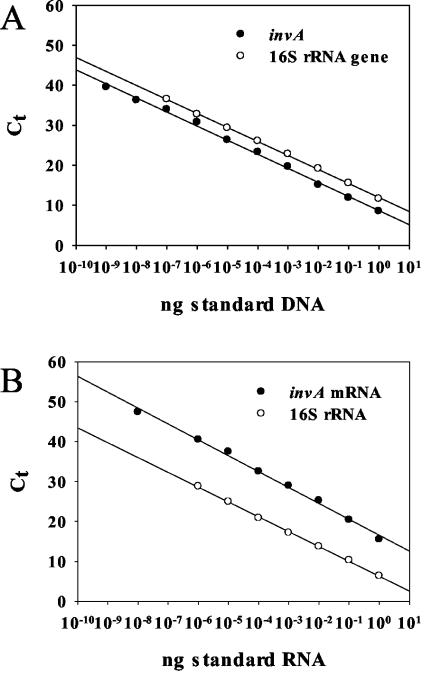
Establishment of Q-PCR and Q-RT-PCR protocols. The newly designed DNA and RNA standards were evaluated by Q-PCR and Q-RT-PCR using the PCR primers specific for the invA and 16S rRNA genes of Salmonella (Table 1). In all cases we obtained linear calibration curves, which had a correlation coefficient (R2) of ≥0.995, with linear ranges of ≥8 orders of magnitude for invA and ≥7 orders of magnitude for the 16S rRNA gene and 16S rRNA (Fig. 1). This corresponds to a detection limit of 2 copies for invA and up to 1,000 copies for 16S rRNA (Table 2). The efficiencies of the reactions ranged between 0.8 and 1.0 (Table 2). The melting curve analysis that was performed for each Q-PCR showed a clear melting peak (Table 2) and no formation of unspecific products.
FIG. 1.
Calibration curves generated using the DNA (A) and RNA (B) standards for invA and 16S rRNA. The target nucleic acid concentrations are plotted against the Ct values. The Ct is the cycle number at which the fluorescence in the sample increases above a defined threshold fluorescence, which is calculated by the software.
TABLE 2.
Main properties of the primer sets used for Q-PCR analysis
| Target | Reaction efficiencya | Melting temperature (°C) | Detection limitb |
|---|---|---|---|
| invA gene | 0.80 ± 0.04 | 81.6 | 2 |
| invA RNA | 0.85 ± 0.06 | 80.4 | 20 |
| 16S rRNA gene | 1.00 ± 0.08 | 87.1 | 100 |
| 16S rRNA | 0.91 ± 0.04 | 86.1 | 1,000 |
Reaction efficiencies are mean values of three or more determinations ± standard deviations.
Number of copies.
Validation of the experimental protocols with pure cultures.
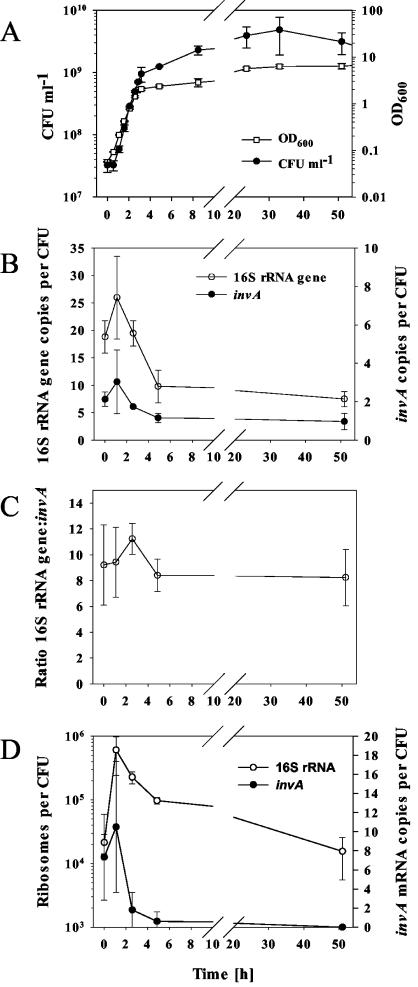
The growth in BHI broth of S. enterica serovar Typhimurium was monitored for 5 days. The logarithmic growth phase started 30 min after inoculation and lasted for 2.5 h. After 3 days, the number of CFU started to decrease (Fig. 2A). Samples for nucleic acid extraction were taken at different time points (i.e., immediately after inoculation, early and late logarithmic phases, and early and late stationary phases). Then, the invA and 16S rRNA genes and the corresponding gene transcripts (RNAs) were quantified by Q-PCR and Q-RT-PCR, respectively. An increment in the number of 16S rRNA and invA gene copies per CFU was observed in the early logarithmic phase (i.e., after 1.1 h of incubation), which might be explained by the presence of multiple replicating chromosomes (Fig. 2B). Then, the copy number decreased to 1 and 5 to 10 copies/CFU for invA and 16S rRNA, respectively. These results are in agreement with the fact that invA is a single-copy gene in Salmonella spp., whereas there are seven rrn operons (11). The ratio of the 16S rRNA gene to the invA gene was stable during the course of the experiment at approximately eight copies of the 16S rRNA gene per copy of invA (Fig. 2C), which was close to the expected value. The number of RNA copies for the 16S rRNA increased from approximately 2 × 104 after inoculation to a maximum of 5 × 105 in the early logarithmic phase (Fig. 2D). Thereafter, the 16S rRNA copy number decreased to values similar to those observed at the beginning of the incubation. The virulence gene invA was also expressed in the early logarithmic phase (maximum of 10 RNA copies per CFU; Fig. 2D).
FIG. 2.
Growth curve of S. enterica serovar Typhimurium (ATCC 14028) in BHI broth at 37°C. (A) OD600s and CFU of samples taken at different time points. (B) Number of 16S rRNA or invA gene copies per CFU, as determined by Q-PCR analysis. (C) Ratio of 16S rRNA gene to invA, as determined by Q-PCR. (D) Number of copies of ribosomes (16S rRNA) or invA mRNA per CFU, as determined by Q-RT-PCR analysis. All data are mean values of triplicates; standard deviations are indicated by vertical lines. Please note the broken x axes.
Evaluation of the experimental protocols in water samples inoculated with Salmonella.
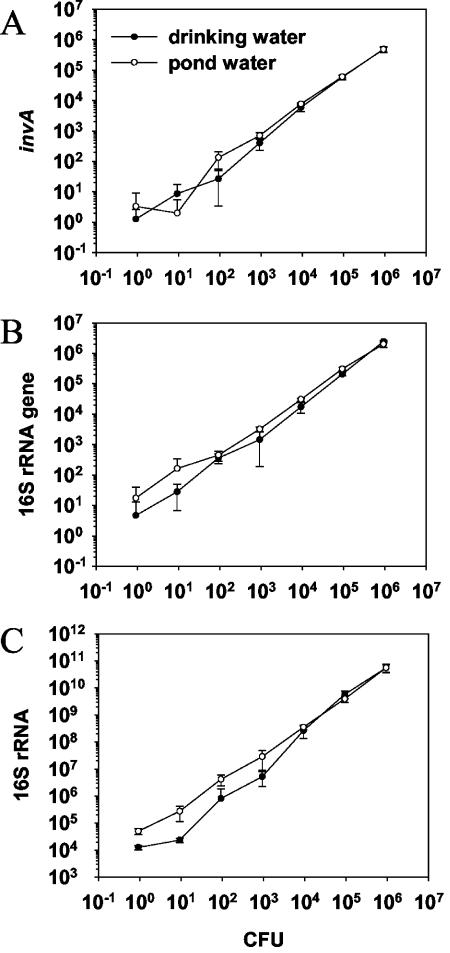
Q-PCR analysis of drinking and fishpond water samples inoculated with 1 to 106 Salmonella cells per ml showed similar DNA gene copy numbers per CFU for invA and 16S rRNA (Fig. 3A and B). Low cell numbers resulted in a higher variation, suggesting that the precision of the method drops as the level of contamination decreases. The mean numbers of invA and 16S rRNA gene copies per CFU were 0.69 and 2.9 for drinking water and 0.74 and 5.8 for pond water, respectively.
FIG. 3.
Q-PCR and Q-RT-PCR analysis of S. enterica serovar Typhimurium inoculated in drinking or fishpond water. (A) The number of CFU per milliliter is plotted against the number of copies of invA, as determined by Q-PCR. (B) The number of CFU per milliliter is plotted against the number of copies of the 16S rRNA gene, as determined by Q-PCR. (C) The number of CFU per milliliter is plotted against the number of copies of ribosomes (16S rRNA), as determined by Q-RT-PCR analysis. All data are mean values of triplicates per milliliter of inoculated water samples; standard deviations are indicated by vertical lines.
16S RNA expression in drinking water was similar to that in pond water when the water was inoculated with 104 to 106 cells (Fig. 3C). However, lower cell numbers resulted in lower expression in drinking water samples. Overall, the mean numbers of RNA copies per CFU for 16S rRNA were 2.6 × 104 and 4.2 × 104 in drinking and pond water, respectively. The mean numbers of copies of ribosomes (16S rRNA) per invA gene copy were 5 × 104 and 5.5 × 104 in drinking and pond water, respectively. There was no PCR signal in noninoculated controls when the invA genes were amplified, whereas the background noise for 16S rRNA (RNA and DNA) was similar to the values obtained by spiking water with 1 CFU per ml (data not shown).
DISCUSSION
Q-PCR has emerged as a promising tool for the detection and quantification of bacteria in environmental samples (16, 19). The quantification is based on the determination of the gene copy number, which is mainly carried out with genomic DNA as the standard. However, the use of this standardization approach is not feasible for a quantitative determination of transcripts corresponding to specific target genes. For eukaryotic cells, this has been solved by using stably expressed housekeeping genes (3). However, no such stably expressed gene is known in bacteria. We developed a Q-PCR-based quantification approach, which allows a robust and precise quantification of not only bacterial DNA, but also RNA targets in environmental samples in the absence of stably expressed housekeeping genes. To this end, DNA and RNA standards were designed for Q-PCR and Q-RT-PCR analysis.
To our knowledge, this is the first time that this type of approach has been successfully used for the quantification of RNA targets in prokaryotic cells. A similar strategy was exploited to generate RNA standards for the quantification of mRNA corresponding to the rbcL and rbcL genes of diatoms and pelagophytes (25). In vitro-transcribed RNA has also been used as a standard for competitive RT-PCR (29). However, the use of random hexamers as primers led to an overestimation of the RNA copy number, as a result of the synthesis of a truncated cDNA standard lacking binding sites for the PCR primers. We have observed a similar effect using the same primer sets to produce the DNA and RNA standards and for Q-PCR. In this case the results were still suitable for Q-PCR analysis, but poor amplification of the standards in the Q-RT-PCR led to a significant overestimation of the RNA copy numbers, making this approach unacceptable for accurate quantification (data not shown). This could be due to the lack of binding sites for sequence-specific primers, as a result of the formation of truncated cDNA fragments (29).
To avoid this problem, a separate set of primers was used to generate RNA standards, which amplified a much larger fragment than the primers that were then used in the Q-PCR step (Table 1; Fig. 1). It was also critical to leave an overlap at both ends, to enable priming of random hexamers during the synthesis of cDNA. An additional factor to be considered was the presence of residual DNA template after DNase treatment (10). To minimize this problem, the DNA concentration in the RNA standards was reduced by 4 orders of magnitude by a second DNase treatment.
As shown in pure culture and spiking experiments (Fig. 2 and 3), the DNA targets were accurately quantified. In pure cultures we found up to four times more DNA copies of 16S rRNA and invA per cell than expected (Fig. 2B). This can be explained by the presence of multiple chromosomes during the bacterial logarithmic-growth phase. In fact, up to 36 rrn genes have been reported for Enterobacteriaceae (2). After this phase of rapid replication, one copy of invA and approximately eight copies of the 16S rRNA gene were detected. This matches the expected values, considering that invA is a single-copy gene and that seven rrn operons have been described in Salmonella (11). Thus, Salmonella DNA was recovered with nearly 100% efficiency throughout the complete procedure (i.e., sampling, centrifugation, cell lysis, DNA extraction, and DNA purification). In inoculated samples, the efficiency was approximately 70% (i.e., 0.7 copies for invA and 2.9 to 5.8 copies for the 16S rRNA gene). It is important to highlight that in a recent study efficiencies not higher than 52% were reported when extracting DNA from indigenous gram-negative bacteria in the same fishpond water (26).
Concerning the determination of the number of ribosomal 16S rRNA copies, a strong increment was observed at the beginning of the logarithmic phase in pure cultures as earlier described (18), whereas the number decreases towards the end of the experiment. The number of 16S rRNA copies per CFU was between 20,000 and 500,000. These values are close to the number of ribosomes per bacterial cell described for Escherichia coli, 6,700 to 71,000 depending on the growth rate and the physiological state (2, 5, 8). An overestimation in the number of 16S rRNA copies seems unlikely, since pure small RNA molecules were used as external standards here. In fact, the efficiency of reverse transcription and the subsequent Q-PCR would be higher for the standards than for the samples (i.e., we would underestimate rather than overestimate). Thus, rapid growth in a complex medium (i.e., BHI) might account for the slightly higher number of copies observed in the present work. A maximum of only 35 16S rRNA copies per 16S rRNA gene were detected when genomic DNA was used as the standard to determine staphylococcal 16S rRNA gene expression in pure cultures (23). However, the use of a DNA standard for the Q-RT-PCR has not taken into consideration the efficiency of the reverse transcription, which might vary between 1.4 and 90% (4, 15). This probably led to a large underestimation of the target RNA molecules.
The expression rates for 16S rRNA in inoculated water samples were similar to the values observed during the stationary phase in pure cultures (Fig. 3C). However, the expression levels in drinking water samples inoculated with a small number of bacterial cells were lower. This might be due to the fact that bacteria are under stress as a result of the suboptimal conditions in drinking water. Alternatively, the efficacy of the filtration and/or nucleic acid extraction procedures may constitute a limiting factor.
The Q-PCR-based methods that were developed and validated here allow achieving an accurate quantification of not only DNA, but also bacterial RNA targets with high sensitivity and accuracy. There is a clear relationship between the physiological status of bacteria and the number of 16S rRNA molecules per cell. Thus, the developed protocols can be easily exploited to quantify bacteria in environmental samples, as well as to monitor their activity and, by choosing appropriate targets, their physiological status and/or pathogenic potential.
Acknowledgments
We thank Stephan Stubner for critical discussions and Claudia Höltje for outstanding technical help.
This work was supported by funds from the European Commission (Aqua-Chip project; QLK4-2000-00764).
The authors are solely responsible for the content of this publication. It does not represent the opinion of the European Commission. The European Commission is not responsible for any use that might be made of data appearing therein.
REFERENCES
- 1.Bhagwat, A. A., R. P. Phadke, D. Wheeler, S. Kalantre, M. Gudipati, and M. Bhagwat. 2003. Computational methods and evaluation of RNA stabilization reagents for genome-wide expression studies. J. Microbiol. Methods 55:399-409. [DOI] [PubMed] [Google Scholar]
- 2.Bremer, H., and P. P. Dennis. 1996. Modulation of chemical composition and other parameters of the cell by growth rate, p. 1553-1569. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaecter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 3.Bustin, S. A. 2000. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 25:169-193. [DOI] [PubMed] [Google Scholar]
- 4.Freeman, W. M., S. J. Walker, and K. E. Vrana. 1999. Quantitative RT-PCR: pitfalls and potential. BioTechniques 26:112-125. [DOI] [PubMed] [Google Scholar]
- 5.Gourse, R. L., T. Gaal, M. S. Bartlett, J. A. Appleman, and W. Ross. 1996. rRNA transcription and growth rate-dependent regulation of ribosome synthesis in Escherichia coli. Annu. Rev. Microbiol. 50:645-677. [DOI] [PubMed] [Google Scholar]
- 6.Keer, J. T., and L. Birch. 2003. Molecular methods for the assessment of bacterial viability. J. Microbiol. Methods 53:175-183. [DOI] [PubMed] [Google Scholar]
- 7.Kingombe, C., G. Huys, M. Tonolla, M. Albert, J. Swings, R. Peduzzi, and T. Jemmi. 1999. PCR detection, characterization, and distribution of virulence genes in Aeromonas spp. Appl. Environ. Microbiol. 65:5293-5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kjeldgaard, N. O., and K. Gausing. 1974. Regulation of biosynthesis of ribosomes, p. 369-392. In M. Nomura, A. Tissières, and P. Lengyel (ed.), Ribosomes. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 9.Malorny, B., J. Hoorfar, C. Bunge, and R. Helmuth. 2003. Multicenter validation of the analytical accuracy of Salmonella PCR: towards an international standard. Appl. Environ. Microbiol. 69:290-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matthews, J., M. Chung, and J. Matyas. 2002. Persistent DNA contamination in competitive RT-PCR using cRNA internal standards: identity, quantity, and control. BioTechniques 32:1412-1417. [DOI] [PubMed] [Google Scholar]
- 11.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 12.Neretin, L. N., A. Schippers, A. Pernthaler, K. Hamann, R. Amann, and B. B. Jorgensen. 2003. Quantification of dissimilatory (bi)sulphite reductase gene expression in Desulfobacterium autotrophicum using real-time RT-PCR. Environ. Microbiol. 5:660-671. [DOI] [PubMed] [Google Scholar]
- 13.Paton, A., and J. Paton. 1998. Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfbO111, and rfbO157. J. Clin. Microbiol. 36:598-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [Online.] [DOI] [PMC free article] [PubMed]
- 15.Pfaffl, M. W., and M. Hageleit. 2001. Validities of mRNA quantification using recombinant RNA and recombinant DNA external calibration curves in real-time RT-PCR. Biotechnol. Lett. 23:275-282. [Google Scholar]
- 16.Rodriguez-Lazaro, D., M. Hernandez, T. Esteve, J. Hoorfar, and M. Pla. 2003. A rapid and direct real time PCR-based method for identification of Salmonella spp. J. Microbiol. Methods 54:381-390. [DOI] [PubMed] [Google Scholar]
- 17.Rokbi, B., D. Seguin, B. Guy, V. Mazarin, E. Vidor, F. Mion, M. Cadoz, and M.-J. Quentin-Millet. 2001. Assessment of Helicobacter pylori gene expression within mouse and human gastric mucosae by real-time reverse transcriptase PCR. Infect. Immun. 69:4759-4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruimy, R., V. Breittmayer, V. Boivin, and R. Christen. 1994. Assessment of the state of activity of individual bacterial cells by hybridization with a ribosomal RNA targeted fluorescently labelled oligonucleotidic probe. FEMS Microbiol. Ecol.. 15:207-213. [Google Scholar]
- 19.Sharma, V. K., E. A. Dean-Nystrom, and T. A. Casey. 1999. Semi-automated fluorogenic PCR assays (TacMan) for rapid detection of Escherichia coli O157:H7 and other Shiga toxigenic E. coli. Mol. Cell. Probes 13:291-302. [DOI] [PubMed] [Google Scholar]
- 20.Sheridan, G. E. C., C. I. Masters, J. A. Shallcross, and B. M. Mackey. 1998. Detection of mRNA by reverse transcription-PCR as an indicator of viability in Escherichia coli cells. Appl. Environ. Microbiol. 64:1313-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Straub, T. M., and D. P. Chandler. 2003. Towards a unified system for detecting waterborne pathogens. J. Microbiol. Methods 53:185-197. [DOI] [PubMed] [Google Scholar]
- 22.Vandecasteele, S. J., W. E. Peetermans, R. Merckx, and J. Van Eldere. 2001. Quantification of expression of Staphylococcus epidermidis housekeeping genes with Taqman quantitative PCR during in vitro growth and under different conditions. J. Bacteriol. 183:7094-7101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vandecasteele, S. J., W. E. Peetermans, R. Merckx, M. Van Ranst, and J. Van Eldere. 2002. Use of gDNA as internal standard for gene expression in staphylococci in vitro and in vivo. Biochem. Biophys. Res. Commun. 291:528-534. [DOI] [PubMed] [Google Scholar]
- 24.Walker, N. J. 2002. A technique whose time has come. Science 296:557-559. [DOI] [PubMed] [Google Scholar]
- 25.Wawrik, B., J. H. Paul, and F. R. Tabita. 2002. Real-time PCR quantification of rbcL (ribulose-1,5-bisphosphate carboxylase/oxygenase) mRNA in diatoms and pelagophytes. Appl. Environ. Microbiol. 68:3771-3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weinbauer, M. G., I. Fritz, D. F. Wenderoth, and M. G. Höfle. 2002. Simultaneous extraction from bacterioplankton of total RNA and DNA suitable for quantitative structure and function analyses. Appl. Environ. Microbiol. 68:1082-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weinbauer, M. G., and M. G. Höfle. 2001. Quantification of nucleic acids from aquatic environments by using green-fluorescent dyes and microtiter plates, p. 2.1.3-2.1.10. In A. Akkermans, J. van Elsas, and F. de Bruijn (ed.), Molecular microbial ecology manual, 5th suppl. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 28.World Health Organization. 2002. The world health report. Reducing risks, promoting healthy life. World Health Organization, Geneva, Switzerland.
- 29.Zhang, J., and C. D. Byrne. 1999. Differential priming of RNA templates during cDNA synthesis markedly affects both accuracy and reproducibility of quantitative competitive reverse-transcriptase PCR. Biochem. J. 337:231-241. [PMC free article] [PubMed] [Google Scholar]