Abstract
Problematic fermentations are common in the wine industry. Assimilable nitrogen deficiency is the most prevalent cause of sluggish fermentations and can reduce fermentation rates significantly. A lack of nitrogen diminishes a yeast's metabolic activity, as well as the biomass yield, although it has not been clear which of these two interdependent factors is more significant in sluggish fermentations. Under winemaking conditions with different initial nitrogen concentrations, metabolic flux analysis was used to isolate the effects. We quantified yeast physiology and identified key metabolic fluxes. We also performed cell concentration experiments to establish how biomass yield affects the fermentation rate. Intracellular analysis showed that trehalose accumulation, which is highly correlated with ethanol production, could be responsible for sustaining cell viability in nitrogen-poor musts independent of the initial assimilable nitrogen content. Other than the higher initial maintenance costs in sluggish fermentations, the main difference between normal and sluggish fermentations was that the metabolic flux distributions in nitrogen-deficient cultures revealed that the specific sugar uptake rate was substantially lower. The results of cell concentration experiments, however, showed that in spite of lower sugar uptake, adding biomass from sluggish cultures not only reduced the time to finish a problematic fermentation but also was less likely to affect the quality of the resulting wine as it did not alter the chemistry of the must.
Sluggish and stuck fermentations are common in the wine industry. Factors that affect the yeast growth rate and that lead to problematic fermentations include limited nutrient contents, ethanol toxicity, fatty acid toxicity, temperature extremes (5), the ratio of nitrogen sources to carbon sources in the medium (21), and the initial quantity and quality of the amino acids (29). Sluggish cultures may prolong the process time from days to weeks. In stuck fermentations, high levels of residual sugar are left following the arrest of fermentation. Several difficulties occur at the cellular level; these include diminished sugar transport capacity, the inability of yeasts to produce compatible solutes in response to high osmolality (20, 28), and cell membrane integrity issues in the presence of high concentrations of ethanol (8). As such mechanisms may act simultaneously in cells and to various extents, it is difficult to isolate and reproduce the mechanisms in stuck and sluggish fermentations.
Quantitatively, nitrogen is the second most abundant nutrient in wine fermentations. It is essential for yeast metabolism and growth. Consequently, a lack of nitrogen triggers sluggish fermentations (1, 8, 13). In previous studies workers found differences in nitrogen-related curves (e.g., curves of biomass versus assimilable nitrogen) which indicated that the fermentation rate and biomass yield functions are distinct (9, 10; W. Agenbach, Proc. S. Afr. Soc. Enol. Vitic., p. 66-88, 1977). In other studies the workers associated nitrogen deficiency with a high sugar transporter turnover rate, which resulted in a loss of sugar uptake capacity in the cells (5, 32). The cellular membrane is the primary region for controlling sugar uptake and the subsequent ethanol production (34, 39).
Currently, the most common method for dealing with nitrogen-deficient fermentations is adding supplementary nitrogen (usually ammonium phosphate). The timing of the addition is key for ensuring a successful fermentation (3, 32). Early addition affects both the fermentation rate and the biomass yield. Late addition has a minimal effect on biomass formation, however, and only increases the fermentation rate (3). Unfortunately, it is not possible to distinguish clearly between the effect of nitrogen on the fermentation rate and the effect of nitrogen on the biomass yield as the two effects are interdependent.
We propose that the rate of fermentation is indeed a two-component function comprising an intracellular component (a property of the cell metabolism) and a cellular component (which is dependent on the mass of cells actively fermenting).
In this work, we used metabolic flux balancing and biomass concentration experiments to isolate the two components and to quantitatively evaluate how these components affect the rate of fermentation. The metabolic phenotype is best represented by the flux distribution throughout the network of intracellular pathways (26, 38, 40). Adding biomass with cells at the same metabolic state allowed us to evaluate the effects on the fermentation rate. We then assessed the relative significance of the two components in nitrogen-deficient sluggish fermentations.
MATERIALS AND METHODS
Yeast strain and growth conditions.
Saccharomyces cerevisiae Prise de Mousse EC1118 (Lalvin, Zug, Switzerland), a strain commercially available worldwide for the wine industry, was used throughout this study. Initial seed cultures were grown in YPD medium at 28°C in aerobic conditions. Defined artificial must, which simulated standard grape juice, was used in bioreactor fermentations. Modified MS300 medium (33) contained (per liter) 120 g of glucose, 120 g of fructose, 6 g of citric acid, 6 g of dl-malic acid, 750 mg of KH2PO4, 500 mg of KH2SO4, 250 mg of MgSO4 · 7H2O, 155 mg of CaCl2 · 2 H2O, 200 mg of NaCl, 4 mg of MnSO4 · H2O, 4 mg of ZnSO4, 1 mg of CuSO4 · 5H2O, 1 mg of KI, 0.4 mg of CoCl2 · 6H2O, 1 mg of H3BO3, 1 mg of NaMoO4 · 2H2O, 20 mg of myo-inositol, 2 mg of nicotinic acid, 1.5 mg of calcium panthothenate, 0.25 mg of thiamine hydrochloride, 0.25 mg of pyridoxine hydrochloride, and 0.003 mg of biotin. The nitrogen concentration for normal fermentations was 380 mg liter−1, and the nitrogen concentration for sluggish fermentations was 65 mg liter−1. The nitrogen sources used were 18.6% (wt/wt) ammoniacal nitrogen (NH4Cl), 20.5% (wt/wt) l-proline, 16.9% (wt/wt) l-glutamine, 1.25% (wt/wt) l-arginine, 6% (wt/wt) l-tryptophan, 4.9% (wt/wt) l-alanine, 4% (wt/wt) l-glutamic acid, 2.6% (wt/wt) l-serine, 2.6% (wt/wt) l-threonine, 1.6% (wt/wt) l-leucine, 1.5% (wt/wt) l-aspartic acid, 1.5% (wt/wt) l-valine, 1.3% (wt/wt) l-phenylalanine, 1.1% (wt/wt) l-isoleucine, 1.1% (wt/wt) l-histidine, 1.1% (wt/wt) l-methionine, 0.6% (wt/wt) l-tyrosine, 0.6% (wt/wt) l-glycine, 0.6% (wt/wt) l-lysine, and 0.4% (wt/wt). The following anaerobic factors were added to the medium: 15 mg of ergosterol per liter, 5 mg of sodium oleate per liter, and 0.5 ml of Tween 80 per liter. Since proline is not assimilable by yeast under anaerobic growth conditions (18), the ammonium salt and α-amino acids (all amino acids except proline) were considered assimilable sources of nitrogen. Therefore, for normal and sluggish fermentations, the concentrations of assimilable nitrogen were 300 and 50 mg liter−1, respectively.
Cultivation conditions.
The bioreactors, a 2-liter Bioflo IIc bioreactor (New Brunswick Scientific Co., Edison, N.J.) with a 1.5-liter working volume, a 3-liter Bioflo IIc bioreactor with a 2.5-liter working volume, and a 50-liter Bioengineering bioreactor (Bioengineering, Wald, Switzerland) with a 35-liter working volume, were inoculated to obtain an initial density of 106 cells ml−1. Cells were washed with 0.9% NaCl to eliminate any remaining nitrogen from rich media prior to inoculation. The temperature was maintained at 28°C, and the pH was maintained at 3.5. Nitrogen was sparged through the medium for 30 min at a rate of 250 ml min−1 before inoculation to eliminate any oxygen in the medium. The medium was agitated at 100 rpm to keep the cells in suspension. Carbon dioxide production in addition to nitrogen sparging and agitation ensured that that the conditions were anaerobic throughout the experiment.
Two batch cultures were compared in this work. In a normal fermentation, 300 mg of assimilable nitrogen (ammonia and amino acids) per liter was used, while a second culture contained 50 mg of assimilable nitrogen per liter to force sluggish fermentation. Three independent experiments were carried out for each type of batch culture.
Cell concentration experiments.
At the end of the exponential phase of one of the sluggish fermentations, an 11.5-liter sample containing 1.2 g (dry weight) of cells per liter was removed from the 35-liter batch culture. A 7.5-liter aliquot was centrifuged (3,000 × g at room temperature), and the pellet was resuspended in 1.5 liters of supernatant. Hence, the cell concentration was increased fivefold without altering the chemical composition of the must. In a similar fashion, biomass from the remaining 4 liters was centrifuged and resuspended in 2 liters of supernatant to obtain a twofold increase in the cell concentration. After resuspension, the concentrated cultures were loaded into sterile bioreactors and sparged with nitrogen before the two fermentations were allowed to continue.
Analytical techniques.
Carbon dioxide evolution in the bioreactor was determined with a Gallus 1000 volumetric flux transductor (Schlumberger, Buenos Aires, Argentina). Culture samples were taken periodically to establish the fermentation status. These samples were analyzed to determine the dry cell weight, the cell number, and the concentrations of glucose, fructose, intra- and extracellular organic acids and amino acids, ammonia, and free amino acid nitrogen. Dry cell weight was estimated by filtering cells and washing them twice with distilled water and then drying the preparation to a constant weight at 85°C. Cell numbers were estimated with a Neubauer chamber (Brand Biotech).
Glucose, fructose, and trehalose concentrations were measured by high-performance liquid chromatography (HPLC) by using a Waters high-performance carbohydrate cartridge as described by the manufacturer (Waters Corporation). Organic acids, glycerol, and ethanol concentrations were measured by HPLC by using a Bio-Rad HPX-87H column (40). Amino acids were derivatized with Waters AccQ · Fluor reagent, and then the concentrations were measured by HPLC by using an AccQ · Tag amino acid analysis column in accordance with the instructions of the manufacturer (Waters Corporation). The ammonia concentration was measured enzymatically by using glutamate dehydrogenase (Sigma Chemical Co., St. Louis, Mo.). The concentration of free amino acid nitrogen was determined by using the σ-phthaldehyde/N-acetyl-l-cysteine spectrophotometric assay (NOPA) procedure (12). Since the NOPA reagent reacts only with primary amino groups, the nitrogen in other groups in a molecule is unaffected. In arginine, for example, just one nitrogen molecule reacts.
Viability was measured by using a LIVE/DEAD yeast viability kit as described by the manufacturer (Molecular Probes, Eugene, Oreg.) and a fluorescence microscope (Leitz Laborlux S) equipped with an XF56 filter (Omega Optical). Metabolically active cells contained small red fluorescent intravacuolar structures, while dead cells emitted diffuse, green-yellow fluorescence.
Intracellular metabolites were extracted with boiling buffered ethanol (14), and then the concentrations were determined by HPLC. Glycogen and trehalose were extracted with 0.25 M Na2CO3 prior to enzymatic hydrolysis to glucose (27). DNA concentrations were determined by the Burton method (15). RNA concentrations were determined by the Schmidt-Tannhauser procedure (4). Protein concentrations were determined by the biuret method (41). The total cellular carbohydrate concentrations were determined by the phenol method (15). Lipid concentrations were determined gravimetrically after chloroform and methanol extraction (11).
Stoichiometric model.
The stoichiometric model used to represent the metabolic network of S. cerevisiae was adapted from the model of Nissen et al. (26). A few modifications were necessary to account for fructose uptake, transport reactions, and amino acid biosynthesis pathways. The pathway network consists of glycolysis, the pentose phosphate pathway, the pyruvate carboxylase reaction, the synthesis of ethanol, glycerol, and acetate, the tricarboxylic acid cycle, synthesis and transport reactions for the amino acids arginine, glutamine, tryptophan, alanine, glutamate, serine, threonine, leucine, aspartate, valine, phenylalanine, and isoleucine (16, 42, 43), transport reactions for incorporation and secretion of various metabolites, and the synthesis pathways for macromolecular components (26).
Metabolic flux analysis: consistency analysis.
The flow rates of material (i.e., the fluxes through the pathways of the bioreaction network) are estimated from measurements of substrate uptake and product formation rates. Measurements and fluxes are linked through metabolite (stoichiometric) balances (38). Intracellular metabolite pools are assumed to be at a steady state; however, it is necessary to establish the macromolecular composition in order to quantify the drain of metabolites in biomass synthesis (19).
An analysis of metabolite balancing was carried out to determine the flux distribution in each culture at three equivalent stages during fermentation. To compare normal and sluggish flux distributions, the metabolic fluxes for the cultures were compared at the same alcohol concentration. The substrate uptake rates and product and macromolecular component formation rates were calculated for batch cultures as previously described (37) and were used to estimate the metabolic fluxes. The relative standard deviations considered for the measured rates, based on the corresponding measurement variances, were as follows: carbon dioxide evolution rate, ammonia, and macromolecular components, 10%; and glucose, organic acids, and amino acids, 5%. The stoichiometric matrix derived from the model has a condition number of <100 and indicates that the model is numerically robust (38).
Prior to any flux analysis, we verified that the balance for biomass macromolecular components accounted for 95 to 100% of the cell weight in all of the samples analyzed and, after this, that carbon balances accounted for 95 to 100% in the three flux distributions obtained.
RESULTS
Two fermentations were performed under winemaking conditions. The cultures differed only in the initial nitrogen content. The first culture contained 300 mg of assimilable nitrogen per liter, which is sufficient to avoid sluggish fermentations (6). The second culture, on the other hand, contained 50 mg of assimilable nitrogen per liter, a concentration which is sufficiently low to cause a sluggish fermentation. Each type of fermentation was performed in triplicate.
Normal fermentations.
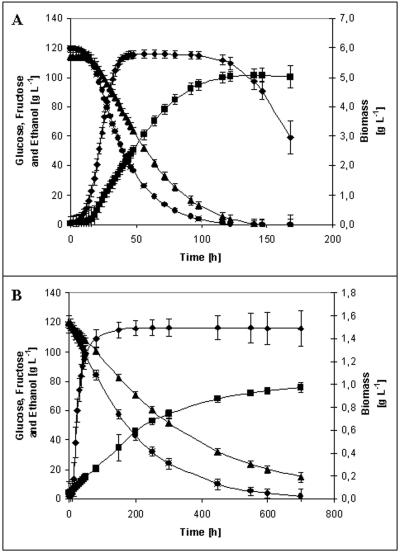
In normal fermentations, the average time to reach dryness (<4 g of sugar liter−1) was 6 days (170 h) (Fig. 1A). Glucose was consumed in preference to fructose (23). This was due to differences in the various transporters' affinities for these sugars (7, 30). The biomass growth curve revealed an exponential phase and a stationary phase that began at 48 h, at which point the biomass concentration was 5.8 g liter−1. Ethanol synthesis occurred mainly in the stationary phase and resulted in a final ethanol concentration of 12.7% (vol/vol) (Table 1). These values are typical of experiments reported for normal fermentations for several commercial strains grown under similar conditions (31).
FIG. 1.
(A) Normal fermentation profile (300 mg of N liter−1). (B) Sluggish fermentation profile (50 mg of N liter−1). Symbols: •, glucose consumption; ▴, fructose consumption; ▪, ethanol production; ♦, biomass production.
TABLE 1.
Final fermentation values
| Culturea | Fermentation time (h)b | Cell dry wt (g liter−1) | Sugar fermentation rate (g liter−1 day−1) | Ethanol concn (%, vol/vol) | Glycerol concn (g liter−1) | Succinic acid concn (g liter−1) | Acetic acid concn (g liter−1) |
|---|---|---|---|---|---|---|---|
| Normal (300 mg of N liter−1) | 170 ± 12 | 5.8 ± 0.1 | 33.5 ± 2.6 | 12.7 ± 0.9 | 7.8 ± 0.6 | 1.8 ± 0.2 | 1.0 ± 0.1 |
| Sluggish (50 mg of N liter−1) | 700 ± 10 | 1.5 ± 0.1 | 7.6 ± 0.3 | 9.5 ± 0.4 | 7.2 ± 0.3 | 1.0 ± 0.1 | 1.0 ± 0.1 |
| Concentrated twofold | 330 | 3.2 | 17.1 | 13.4 | 8.1 | 1.4 | 0.6 |
| Concentrated fivefold | 120 | 8.0 | 47.7 | 11.2 | 9.4 | 1.4 | 0.8 |
Data for the normal and sluggish fermentations were obtained from three independent experiments. Musts for the concentration experiments were obtained from a single sluggish fermentation.
Time for 93% of the sugar consumption for sluggish fermentations.
Assimilable nitrogen was depleted after 24 h of culture, and this coincided with the highest specific growth rate (0.2 h−1). Even though all assimilable nitrogen was consumed, the cell viability remained greater than 97% until all of the sugar was consumed.
Besides ethanol, the yeast produced a number of other products during the fermentation. The final concentration of glycerol, quantitatively the second most important product of wine fermentation, was 7.8 g liter−1, and glycerol was produced mainly during the exponential growth phase, in response to an imbalance in reduction equivalents triggered by protein synthesis (35). Other significant compounds produced by the yeast were succinic and acetic acids, whose final concentrations were 1.8 and 1.0 g liter−1, respectively.
Sluggish fermentations.
The average batch time in sluggish fermentations was 29 days (700 h), and these fermentations left 16 g of residual sugar liter−1 (Fig. 1B). As in the normal fermentations, glucose consumption was preferred over fructose consumption. The biomass entered the stationary phase after 72 h, when the biomass concentration was 1.5 g liter−1. The ethanol concentration, meanwhile, reached 9.5% (vol/vol) (Table 1).
Assimilable nitrogen was completely depleted from the medium in the first 35 h (data not shown). Nonetheless, the yeast continued to consume sugar for nearly 27 days in a nitrogen-free medium. Moreover, the cell viability remained high (97%) until the end of the fermentation (data not shown). The final concentrations of glycerol, succinic acid, and acetic acid were 7.2, 1.0, and 1.0 g liter−1, respectively.
Cellular composition.
The macromolecular composition of yeast was determined for both types of cultures. The relative sugar consumption was used as the basis for comparison as the time scales were different (2). Insufficient biomass made determination of macromolecular composition unreliable before 20 h of culture.
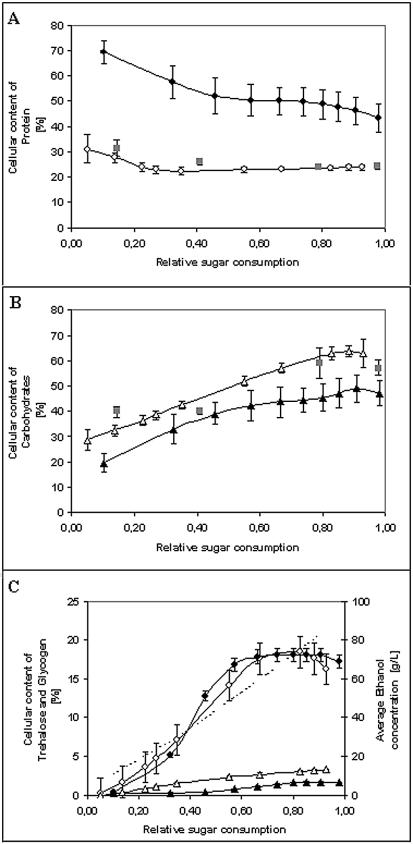
The protein content decreased from 70 to 45% of the cell weight for normal conditions and from 30 to 24% of the cell weight for conditions that led to sluggish fermentations (Fig. 2A). The RNA content also decreased throughout the fermentations (from 9 to 4% and from 24 to 7%, respectively) (data not shown). Conversely, the levels of the total carbohydrates increased from 20 to 50% in normal cultures and from 26 to 63% in sluggish fermentations (Fig. 2B). The decreases in the protein and RNA contents were due to the reduced requirements for these components when the cells entered the stationary phase and to increased cell weight resulting from the accumulation of carbohydrates (35). An interesting finding was the high initial RNA level in sluggish fermentations, which could have been the result of metabolic adjustments in response to lower nitrogen availability. In both normal and sluggish fermentations, the sum of the total carbohydrate, protein, and RNA contents accounted for 96% of the dry cell weight on average. This proportion was constant throughout the fermentations, and the remaining cell weight comprised less than 1% DNA and around 3% lipids.
FIG. 2.
Macromolecular composition for normal cultures (solid symbols), sluggish cultures (open symbols), and concentrated cultures (gray squares). (A) Protein. (B) Total carbohydrates. (C) Trehalose (circles), glycogen (triangles), and ethanol (dotted line).
Glycogen and trehalose accumulated when nitrogen was exhausted upon entry into the stationary phase for a normal fermentation and throughout the stationary phase for sluggish fermentations (Fig. 2C). Glycogen accounted for 1.6% of the dry cell weight in normal fermentations and 3% of the dry cell weight in sluggish fermentations. Trehalose, however, accounted for 18% of the dry cell weight at the end of the stationary phase, and the level was highly correlated with the increase in the amount of ethanol in both cultures (Fig. 2C). Regardless of the differences in the initial nitrogen concentrations, we found that the amounts and profiles of accumulated trehalose were similar in all types of cultures.
Metabolic fluxes.
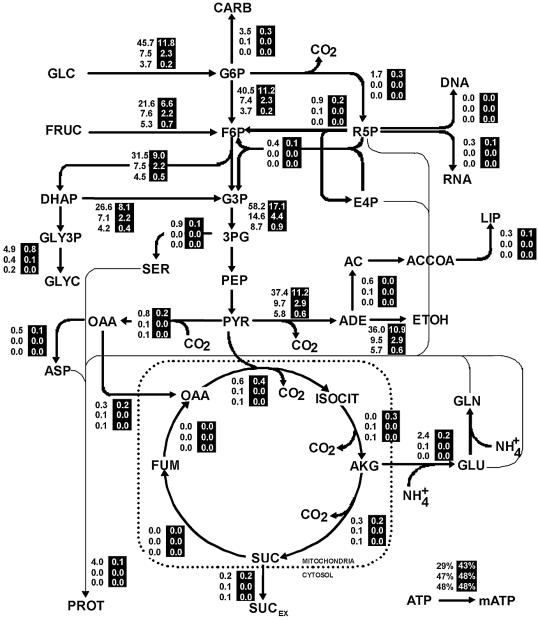
Metabolic carbon flux distributions were obtained for normal and sluggish fermentations in order to study the effect of assimilable nitrogen on the metabolism of the yeast. Three fermentation points were compared at specific concentrations of alcohol: in the exponential phase, at the onset of the stationary phase, and during the stationary phase. We decided to examine alcohol content rather than relative sugar consumption because although the amount of alcohol is related to sugar consumption and reflects the internal state of the cell, it also reveals environmental conditions caused by alcohol-induced stress. The flux distributions in the network at ethanol concentrations of 1.2, 6.4, and 9.5% (vol/vol) are shown in Fig. 3. The fluxes were expressed as specific fluxes (in millimoles of C per gram [dry weight] of cells per hour) to aid comparisons of fermentations with different biomass concentrations.
FIG. 3.
Metabolic fluxes in normal fermentations (black numbers) and sluggish fermentations (white numbers on a black background) at the following three alcohol concentrations (from top to bottom in each case): 1.2, 6.4, and 9.5% (vol/vol). The values are expressed in millimoles of C per gram (dry weight) of cells per hour. The flux used for maintenance is expressed as a percentage of the total ATP produced. CARB, carbohydrates; GLC, glucose; FRUC, fructose; F6P, fructose 6-phosphate; R5P, ribose 5-phosphate; DHAP, dihydroxyacetone phosphate; G3P, glyceraldehyde 3-phosphate; E4P, erythrose 4-phosphate; GLY3P, glycerol 3-phosphate; 3PG, 3-phosphoglycerate; GLYC, glycerol; SER, serine; PEP, phosphoenolpyruvate; AC, acetate; ACCOA, acetyl coenzyme A; LIP, lipids; OAA, oxaloacetate; PYR, pyruvate; ADE, acetaldehyde; ETOH, ethanol; ASP, aspartate; ISOCIT, isocitrate; GLN, glutamine; FUM, fumarate; AKG, α-ketoglutarate; GLU, glutamic acid; PROT, protein; SUC, succinate; SUCEX, extracellular succinate; mATP, maintenance ATP.
During the exponential phase, 65% of the carbon consumed came from glucose in both types of cultures. At the onset of the stationary phase carbon was consumed equally from fructose and glucose. In the late stationary phase fructose supplied 60% of the carbon in normal fermentations and 75% of the carbon in sluggish fermentations. In each phase, most of the carbon was used in energy production (ethanol synthesis). The carbon fraction used for ethanol production increased from the exponential phase to the late stationary phase, from 53 to 63% in a normal fermentation and from 59 to 67% in a sluggish fermentation (Fig. 3).
The decline in the flux towards glycerol (see Appendix, equation 13) was consistent with the decrease in reduction equivalents following the growth phase (35). In all three phases less than 4% of the carbon was used for the production of other metabolites (mainly succinic and acetic acids). The fraction of carbon directed to the pentose phosphate pathway was growth rate dependent. Thus, when the requirement for intermediates from this pathway diminished (at the start of the stationary phase), the flux through the pathway decreased (equation 14). The flux of carbon in the tricarboxylic acid cycle was kept very low under all conditions, mainly to supply the necessary metabolites for biosynthesis.
Normal fermentations had larger fluxes than sluggish fermentations at the three points compared. Only small differences in flux distributions were observed between the two types of cultures when the fluxes were normalized with respect to total sugar entry. Therefore, the flux distribution was largely unaffected by the initial nitrogen concentration within the metabolic network. In fact, the main difference between normal and sluggish fermentations was the kinetics of carbon entry into the cells.
Cell concentration experiments.
In a new approach, we isolated the effect of biomass from the effect of metabolic activity in low-assimilable-nitrogen musts by increasing the concentration of cells in the same metabolic state. At the beginning of the stationary phase of a sluggish fermentation, two samples were extracted, and the biomass was concentrated twofold for one sample and fivefold for the other sample. The two resulting fermentations were then continued with larger amounts of biomass without altering other fermentation parameters, and the results were compared with the results of three sluggish fermentations.
After concentration, the amounts of biomass obtained in the stationary phase in the two experiments were 3.2 g liter−1 for the culture concentrated twofold and 8.0 g liter−1 for the culture concentrated fivefold. The fermentation times were 53 and 86% shorter, respectively, than the average time taken for the sluggish fermentations to consume 93% of the sugar (Table 1). The rate of sugar fermentation increased linearly with the biomass concentration (two- and sixfold, respectively).
At the end of the fermentations of both concentrated cultures there was no residual sugar, which was reflected in significantly higher final concentrations of ethanol (Table 1). The profiles of proteins, total carbohydrates (Fig. 2A and B), trehalose, and glycogen (data not shown) were still similar to the profiles for a sluggish fermentation.
DISCUSSION
Nitrogen deficiency, the most common cause of sluggish fermentations, indirectly affects the fermentation rate through intracellular activity and biomass yield. To gain further insight into the phenomenon of problematic cultures caused by low levels of assimilable nitrogen, we studied fermentations using an artificial must under winemaking conditions (anaerobiosis, low pH, high initial sugar content).
Depletion of assimilable nitrogen occurred halfway through the exponential phase of both normal and sluggish fermentations, irrespective of the initial amount of nitrogen available, and this confirmed that nitrogen is the growth-limiting substrate. In the sluggish fermentations even the time required to consume lower levels of initial nitrogen was less than that for normal cultures. The biomass yield was significantly lower in sluggish cultures, and indeed, every stage of such fermentations occurred later than it occurred in normal cultures.
In low-assimilable-nitrogen sluggish fermentations the size of the protein fraction was consistently one-half the size in normal fermentations. Lower protein levels made the relative amounts of RNA and carbohydrates proportionally higher in sluggish fermentations. The protein, RNA, and carbohydrate profiles were similar for both culture types even though the quantities of these macromolecules were different in absolute terms.
Synthesis of trehalose and glycogen occurred mainly at the onset of the stationary phase in normal fermentations and mostly in the stationary phase in sluggish fermentations. Total carbohydrates accumulated throughout the fermentations not only due to the production of reserve carbohydrates but also due to the production of other carbohydrates (data not shown). Nissen et al. (26) described the accumulation of mannan, glucan, and chitin as an effect of the decline in the specific growth rate of S. cerevisiae.
Trehalose has been reported to accumulate in response to nutrient depletion (36), as well as in response to other types of stress, including heat shock and osmotic stress (17, 28). We found that there was a strong correlation between trehalose accumulation and ethanol production. Trehalose is directly related to the survival of a cell exposed to ethanol (24). Wine yeasts respond to an increase in the ethanol level by accumulating trehalose to mitigate damage to membranes and proteins. Trehalose manages to reduce ion leakage caused by ethanol (25). It also protects the cell from endocytosis inhibition due to high ethanol levels (22), which contributes to more efficient nitrogen utilization, and this may explain sustained cell viability in nitrogen-poor musts. As we found no significant change in medium osmolality (there was a change from 1,500 to 2,000 mOsm kg−1, calculated from the contributions of sugar, ethanol, and glycerol), in our experiments trehalose was not accumulated in response to higher osmotic pressure.
The principal difference in the flux distribution between a normal fermentation and a sluggish fermentation was the kinetics of sugar entry into the network. The specific carbon uptake rate was always higher in a normal culture (3.6-fold higher in the exponential phase and 10-fold higher in the late stationary phase) (Fig. 3). Thus, there was a net difference in the amount of sugar transported per gram of biomass. This could have been due either to weaker transporter activity in sluggish fermentation cells or to fewer transporters per cell. With regard to the latter possibility, it has been found that nitrogen deficiency has an impact on the transporter turnover rate and on the expression of at least one transporter, HXT1 (5). In sluggish fermentations there are lower protein levels and also a proportionally lower flux compared to the protein levels and flux in a normal fermentation. Normalized flux distributions are similar, so we wonder whether the lack of nitrogen affects the expression of specific proteins or diminishes the expression of the entire proteome. Expressing the fluxes in terms of total protein instead of dry cell weight reduced the differences observed in the sugar transport rate twofold. This suggests that there is a combination of specific and general protein regulation.
Inside the metabolic network, fluxes associated with nitrogen sources represent a fraction of the total flux; the most noticeable difference is the flux towards protein. Consequently, in both culture types, differences in nitrogen-related pathways had no effect on flux distribution inside the network.
The carbon fraction used for ethanol production (energy production) increased throughout the fermentation in both cultures, from 53 to 63% for normal cultures and from 59 to 64% for sluggish cultures. As there is no energy requirement for growth in the stationary phase, this increase in energy was used entirely for maintenance. In both musts, there were high energy costs in the late stationary phase in response to ion diffusion into the cytoplasm (due to ethanol concentration) and protein turnover (due to nitrogen depletion). In sluggish fermentations, only the initial maintenance requirements were higher than those in normal fermentations (Fig. 3). The higher initial energy cost may have occurred as the cells regulated their metabolism in response to the lower levels of assimilable nitrogen available. Afterwards, maintenance requirements were directly correlated to ethanol stress in both cultures.
Evaluating the impact of increasing biomass upon the rate of fermentation in cell concentration experiments revealed that (i) the higher the concentration of biomass, the quicker fermentation was completed, even when cells grown with a severe shortage of nitrogen were used; and (ii) that the rate of fermentation was a linear function of biomass, while fermentation time was an exponential function of biomass.
The reduced fermentation times observed in concentrated cultures established that the main problem with a limited initial nitrogen content is that it curbs biomass formation. Despite a lower specific instantaneous sugar uptake rate, the concentrated sluggish fermentations concluded rapidly. The lack of fermentative mass was responsible not only for lower fermentation rates but also for incomplete sugar consumption and a lower ethanol yield in wine obtained from nitrogen-deficient musts.
In summary, by isolating the effects of metabolic flux and the amount of biomass we found that the viable cell concentration governs the fermentation rates in nitrogen-poor musts. From an industrial perspective, these findings suggest two alternatives to deal with nitrogen-deficient musts. The first alternative involves early nitrogen supplementation, as the resulting increase in biomass should ensure a normal fermentation profile. The second alternative involves addition of viable biomass from other fermentation tanks (possibly by decantation or filtration) that would also result in the restoration of the normal fermentation profile. Not only does adding biomass from other fermentations reduce the time to completion, but it should also be less likely to affect negatively the resulting wine, as there should be no significant changes in the concentrations of the main components present in the must. Nevertheless, possible effects on aroma and flavor profiles warrant further investigation.
Acknowledgments
We thank Alex Crawford for a thorough revision of the manuscript.
This work was supported by Fondo Nacional para el Desarollo Científico y Tecnológico de Chile (FONDECYT) grant 2010087 and by Beca de Apoyo a la Realización de Tesis Doctoral (CONICYT). Cristian Varela and Francisco Pizarro were supported by a doctoral fellowship from Consejo Nacional de Investigación Científica y Tecnológica de Chile (CONICYT).
APPENDIX
In the following biochemical reactions used in the yeast model the subscripts CYT, MIT, and EX indicate cytosolic, mitochondrial, and extracellular metabolites, respectively; AICAR refers to 5′-phosphoriboxyl-5-aminoimidazole-4-carboxamida; THF refers to tetrahydrofolate.
Glycolysis
1. Glucose + 1/6 ATP → Glucose 6-phosphate + 1/6 ADP + 1/6 H+
2. Fructose + 1/6 ATP → Fructose 6-phosphate + 1/6 ADP + 1/6 H+
3. Glucose 6-phosphate → Fructose 6-phosphate
4. Fructose 6-phosphate + 1/6 ATP → 3/6 Glyceraldehyde 3-phosphate + 3/6 Dihydroxyacetone phosphate + 1/6 ADP + 1/6 H+
5. Dihydroxyacetone phosphate → Glyceraldehyde 3-phosphate
6. Glyceraldehyde 3-phosphate + 1/3 NAD+CYT + 1/3 ADP + 1/3 P + 1/3 H2O → 3-Phosphoglycerate + 1/3 ATP + 1/3 NADHCYT + 2/3 H+
7. 3-Phosphoglycerate → Phosphoenolpyruvate + 1/3 H2O
8. Phosphoenolpyruvate + 1/3 ADP + 1/3 H+ → Pyruvate + 1/3 ATP
Ethanol, glycerol, and acetate synthesis
9. Pyruvate + 1/3 H+ → 2/3 Acetaldehyde + 1/3 CO2
10. Acetaldehyde + 1/2 NADHCYT + 1/2 H+ → Ethanol + 1/2 NAD+CYT
11. Acetaldehyde + 1/2 NADP+CYT + 1/2 H2O → Acetate + 1/2 NADPHCYT + H+
12. Dihydroxyacetone phosphate + 1/3 NADHCYT + 1/3 H+ → Glycerol 3-phosphate + 1/3 NAD+CYT
13. Glycerol 3-phosphate + 1/3 H+ → Glycerol + 1/3 P
Pentose phosphate pathway
14. Glucose 6-phosphate + 2/6 NADP+CYT + 1/6 H2O → 5/6 Ribose 5-phosphate + 2/6 NADPHCYT + 2/6 H+ + 1/6 CO2
15. Ribose 5-phosphate → 2/5 Erythrose 4-phosphate + 3/5 Fructose 6-phosphate
16. 5/9 Ribose 5-phosphate + 4/9 Erythrose 4-phosphate → 6/9 Fructose 6-phosphate + 3/9 Glyceraldehyde 3-phosphate
Tricarboxylic acid cycle
17. 3/7 Pyruvate + 4/7 OxaloacetateMIT + 1/7 NAD+MIT + 1/7 H2O → 6/7 Isocitrate + 1/7 CO2 + 1/7 NADHMIT + 1/7 H+
18. Isocitrate + 1/6 NAD+MIT → 5/6 α-Ketoglutarate + 1/6 CO2 + 1/6 NADHMIT
19. α-Ketoglutarate + 1/5 NAD+MIT + 1/5 ADP + 1/5 P + 1/5 H2O → 4/5 Succinate + 1/5 ATP + 1/5 CO2 + 1/5 NADHMIT + 1/5 H+
20. Succinate + 1/4 Flavin adenine dinucleotide → Fumarate + 1/4 Reduced flavin adenine dinucleotide + 1/4 H+
21. Fumarate + 1/4 H2O + 1/4 NAD+MIT → OxaloacetateMIT + 1/4 NADHMIT + 1/4 H+
22. Acetate + 1/2 Coenzyme A + ATP + H+ → Acetyl coenzyme A + ADP + P + 1/2 H2O
23. Isocitrate + 1/6 NADP+MIT → 5/6 α-Ketoglutarate + 1/6 CO2 + 1/6 NADPHMIT
Anaplerotic reaction: pyruvate carboxylase
24. 3/4 Pyruvate + 1/4 CO2 + 1/4 ATP → OxaloacetateCYT + 1/4 ADP + 1/4 P + 1/4 H+
Carbohydrate synthesis
25. Glucose 6-phosphate + 1/6 ATP → Carbohydrates + 1/6 ADP + 2/6 P
Nitrogen metabolism and amino acid biosynthesis
26. α-Ketoglutarate + 1/5 NH4+ + 1/5 NADPHCYT + 1/5 H+ → Glutamate + 1/5 H2O + 1/5 NADP+CYT
27. Glutamate + 1/5 NH4+ + 1/5 ATP → Glutamine + 1/5 ADP + 1/5 P + 1/5 H2O
28. 4/9 OxaloacetateCYT + 5/9 Glutamate → 4/9 Aspartate + 5/9 α-Ketoglutarate
29. 3/8 3-Phosphoglycerate + 5/8 Glutamate + 1/8 NAD+CYT → 3/8 Serine + 5/8 α-Ketoglutarate + 3/8 P + 1/8 NADHCYT
30. 10/15 Glutamate + 4/15 Aspartate + 1/15 NH4+ + 1/15 CO2 + 5/15 ATP + 1/15 NADHCYT → 6/15 Arginine + 4/15 Fumarate + 5/15 α-Ketoglutarate + 5/15 ADP + 5/15 P + 1/15 NAD+CYT + 3/15 H2O + 4/15 H+
31. 3/8 Pyruvate + 5/8 Glutamate → 3/8 Alanine + 5/8 α-Ketoglutarate
32. Aspartate + 2/4 ATP + 2/4 NADPHCYT + 2/4 H+ → Threonine + 1/4 H2O + 2/4 ADP + 2/4 P + 2/4 NADP+CYT
33. 6/13 Pyruvate + 2/13 Acetyl coenzyme A + 5/13 Glutamate → 6/13 Leucine + 5/13 α-Ketoglutarate + 2/13 CO2 + 1/13 H2O + 1/13 Coenzyme A
34. 6/5 Pyruvate + Glutamate + 1/5 NADHCYT + 1/5 H+ → Valine + α-Ketoglutarate + 1/5 CO2 + 1/5 NAD+CYT + 1/5 H2O
35. 6/5 Phosphoenolpyruvate + 4/5 Erythrose 4-phosphate + Glutamate + 1/5 NADHCYT + 1/5 H+ + 1/5 ATP → 9/5 Phenylalanine + α-Ketoglutarate + 1/5 CO2 + 1/5 ADP + 1/5 P + 1/5 NAD+CYT + H2O
36. 4/6 Threonine + 3/6 Pyruvate + 5/6 Glutamate + 1/6 NADHCYT + 1/6 H+ → Isoleucine + 5/6 α-Ketoglutarate + 1/6 NH4+ + 1/6 CO2 + 1/6 H2O + 1/6 NAD+CYT
Synthesis of DNA, RNA, proteins, and lipids
37. 5/23 Ribose 5-phosphate + 10/23 Glutamine + 3/23 Serine + 4/23 Aspartate + 1/23 CO2 + 6/23 ATP + 1/23 NADP+CYT → 9/23 AICAR + 10/23 Glutamate + 4/23 Fumarate + 1/23 NADPHCYT + 6/23 ADP + 6/23 P + 5/23 H+ + 2/23 H2O
38. 0.4579 AICAR + 0.4371 Glutamine + 0.0842 THF + 0.3313 Aspartate + 0.2544 Ribose 5-phosphate + 0.4625 ATP + 0.0509 NADPHCYT + 0.1540 NAD+CYT + 0.3301 H2O → DNA + 0.4371 Glutamate + 0.1278 Fumarate + 0.0509 NADP+CYT + 0.1540 NADHCYT + 0.5166 H+ + 0.4625 ADP + 0.4625 P
39. 0.5112 AICAR + 0.5271 Glutamine + 0.0568 THF + 0.2993 Aspartate + 0.2400 Ribose 5-phosphate + 0.4890 ATP + 0.0568 NADP+CYT + 0.1348 NAD+CYT + 0.3427 H2O → RNA + 0.5271 Glutamate + 0.1073 Fumarate + 0.1348 NADHCYT + 0.0568 NADPHCYT + 0.7080 H+ + 0.4890 ADP + 0.4890 P
40. 0.0263 Ribose 5-phosphate + 0.7189 Glutamate + 0.1311 Glutamine + 0.2319 Aspartate + 0.3944 Pyruvate + 0.0869 Serine + 0.0597 Erythrose 4-phosphate + 0.0895 Phosphoenolpyruvate + 1.0084 ATP + 0.0522 NADPHMIT + 0.0742 NAD+MIT + 0.0201 NADHCYT + 0.0883 NADPHCYT + 0.0083 SO4−2 + 0.7685 H2O → Proteins + 0.5596 α-Ketoglutarate + 0.0518 Fumarate + 0.0044 Glyceraldehyde 3-phosphate + 0.1155 CO2 + 0.0110 NH4+ + 0.0124 THF + 0.0742 NADHMIT + 1.0617 H+ + 0.0522 NADP+MIT + 0.0201 NAD+CYT + 0.0883 NADP+CYT + 1.0084 ADP + 1.0084 P
41. 0.8326 Acetyl coenzyme A + 0.0662 Glyceraldehyde 3-phosphate + 0.1012 Serine + 0.4000 ATP + 0.7111 NADPHCYT + 0.0259 H+ → Lipids + 0.0258 H2O + 0.4000 ADP + 0.4000 P + 0.4163 Coenzyme A + 0.7111 NADP+CYT
Oxaloacetate shuttle
42. OxaloacetateCYT + 1/4 ATP → OxaloacetateMIT + 1/4 ADP + 1/4 P
Mitochondrial synthesis of ethanol
43. Acetaldehyde + 1/2 NADHMIT + 1/2 H → Ethanol + 1/2 NAD+MIT
Transport reactions
44. NH4+EX + ATP → NH4+ + ADP + P
45. Acetate → AcetateEX
46. Ethanol → EthanolEX
47. Glycerol → GlycerolEX
48. Succinate → SuccinateEX
49. AspartateEX + 1/4 ATP → Aspartate + 1/4 ADP + 1/4 P
50. GlutamateEX + 1/5 ATP → Glutamate + 1/5 ADP + 1/5 P
51. GlutamineEX + 1/5 ATP → Glutamine + 1/5 ADP + 1/5 P
52. SerineEX + 1/3 ATP → Serine + 1/3 ADP + 1/3 P
53. ArginineEX + 1/6 ATP → Arginine + 1/6 ADP + 1/6 P
54. TryptophanEX + 1/11 ATP → Tryptophan + 1/11 ADP + 1/11 P
55. AlanineEX + 1/3 ATP → Alanine + 1/3 ADP + 1/3 P
56. ThreonineEX + 1/4 ATP → Threonine + 1/4 ADP + 1/4 P
57. LeucineEX + 1/6 ATP → Leucine + 1/6 ADP + 1/6 P
58. ValineEX + 1/5 ATP → Valine + 1/5 ADP + 1/5 P
59. PhenylalanineEX + 1/9 ATP → Phenylalanine + 1/9 ADP + 1/9 P
60. IsoleucineEX + 1/6 ATP → Isoleucine + 1/6 ADP + 1/6 P
ATP dissipation reaction
61. ATP → ADP
REFERENCES
- 1.Alexandre, H., and C. Charpentier. 1998. Biochemical aspects of stuck and sluggish fermentation in grape must. J. Ind. Microbiol Biot. 20:20-27. [Google Scholar]
- 2.Bely, M., J. Sablayrolles, and P. Barre. 1990. Description of alcoholic fermentation kinetics—its variability and significance. Am. J. Enol. Vitic. 41:319-324. [Google Scholar]
- 3.Bely, M., J. M. Sablayrolles, and P. Barre. 1990. Automatic detection of assimilable nitrogen deficiencies during alcoholic fermentation in enological conditions. J. Ferment. Bioeng. 70:246-252. [Google Scholar]
- 4.Benthin, S., J. Nielsen, and J. Villadsen. 1991. A simple and reliable method for the determination of cellular RNA content. Biotechnol. Tech. 5:39-42. [Google Scholar]
- 5.Bisson, L. 1999. Stuck and sluggish fermentations. Am. J. Enol. Vitic. 50:107-119. [Google Scholar]
- 6.Bisson, L., and C. Butzke. 2000. Diagnosis and rectification of stuck and sluggish fermentations. Am. J. Enol. Vitic. 51:168-177. [Google Scholar]
- 7.Boles, E., and C. Hollenberg. 1997. The molecular genetics of hexose transport in yeasts. FEMS Microbiol. Rev. 21:85-111. [DOI] [PubMed] [Google Scholar]
- 8.Boulton, R. 1996. Principles and practices of winemaking. Chapman & Hall, New York, N.Y.
- 9.Cantarelli, C. 1957. On the activation of alcoholic fermentation in winemaking. Am. J. Enol. Vitic. 8:113-120. [Google Scholar]
- 10.Cantarelli, C. 1957. On the activation of alcoholic fermentation in winemaking, part II. Am. J. Enol. Vitic. 8:167-175. [Google Scholar]
- 11.Dillemans, M., L. Van Nedervelde, and A. Debourg. 2001. An approach to the mode of action of a novel yeast factor increasing yeast brewing performance. J. Am. Soc. Brew. Chem. 59:101-106. [Google Scholar]
- 12.Dukes, B., and C. Butzke. 1998. Rapid determination of primary amino acids in grape juice using an o-phthaldialdehyde/N-acetyl-l-cysteine spectrophotometric assay. Am. J. Enol. Vitic. 49:125-134. [Google Scholar]
- 13.Fleet, G., and G. Heard. 1992. Yeasts—growth during fermentation, p. 27-54. In G. H. Fleet (ed.), Wine microbiology and biotechnology. Harwood Academic Publishers, Camberwell, Australia.
- 14.Gonzalez, B., J. Francois, and M. Renaud. 1997. A rapid and reliable method for metabolite extraction in yeast using boiling buffered ethanol. Yeast 13:1347-1355. [DOI] [PubMed] [Google Scholar]
- 15.Herbert, D., P. Phipps, and R. Strange. 1971. Chemical analysis of microbial cells, p. 209-344. In J. Norris and D. Ribbons (ed.), Methods in Microbiology, vol. 5B. Academic Press, London, United Kingdom.
- 16.Horak, J. 1986. Amino acid transport in eukaryotic microorganisms. Biochim. Biophys. Acta 864:223-256. [DOI] [PubMed] [Google Scholar]
- 17.Hottiger, T., P. Schmutz, and A. Wiemken. 1987. Heat-induced accumulation and futile cycling of trehalose in Saccharomyces cerevisiae. J. Bacteriol. 169:5518-5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ingledew, W., C. Magnus, and F. Sosulski. 1987. Influence of oxygen on proline utilization during the wine fermentation. Am. J. Enol. Vitic. 38:246-248. [Google Scholar]
- 19.Jorgensen, H., J. Nielsen, J. Villadsen, and H. Mollgaard. 1995. Metabolic flux distributions in Penicillium chrysogenum during fed-batch cultivations. Biotechnol. Bioeng. 46:117-131. [DOI] [PubMed] [Google Scholar]
- 20.Kim, J., P. Alizadeh, T. Harding, A. Hefner-Gravink, and D. Klionsky. 1996. Disruption of the yeast ATH1 gene confers better survival after dehydration, freezing, and ethanol shock: potential commercial applications. Appl. Environ. Microbiol. 62:1563-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larsson, C., U. von Stockar, I. Marison, and L. Gustafsson. 1993. Growth and metabolism of Saccharomyces cerevisiae in chemostat cultures under carbon-, nitrogen-, or carbon- and nitrogen-limiting conditions. J. Bacteriol. 175:4809-4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lucero, P., E. Penalver, E. Moreno, and R. Lagunas. 2000. Internal trehalose protects endocytosis from inhibition by ethanol in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 66:4456-4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luyten, K., C. Riou, and B. Blondin. 2002. The hexose transporters of Saccharomyces cerevisiae play different roles during enological fermentation. Yeast 19:713-726. [DOI] [PubMed] [Google Scholar]
- 24.Mansure, J., A. Panek, L. Crowe, and J. Crowe. 1994. Trehalose inhibits ethanol effects on intact yeast cells and liposomes. Biochim. Biophys. Acta 1191:309-316. [DOI] [PubMed] [Google Scholar]
- 25.Mansure, J., R. Souza, and A. Panek. 1997. Trehalose metabolism in Saccharomyces cerevisiae during alcoholic fermentation. Biotechnol. Lett. 19:1201-1203. [Google Scholar]
- 26.Nissen, T., U. Schulze, J. Nielsen, and J. Villadsen. 1997. Flux distributions in anaerobic, glucose-limited continuous cultures of Saccharomyces cerevisiae. Microbiology 143:203-218. [DOI] [PubMed] [Google Scholar]
- 27.Parrou, J., and J. Francois. 1997. A simplified procedure for a rapid and reliable assay of both glycogen and trehalose in whole yeast cells. Anal. Biochem. 248:186-188. [DOI] [PubMed] [Google Scholar]
- 28.Parrou, J., M. Teste, and J. Francois. 1997. Effects of various types of stress on the metabolism of reserve carbohydrates in Saccharomyces cerevisiae: genetic evidence for a stress-induced recycling of glycogen and trehalose. Microbiology 143:1891-1900. [DOI] [PubMed] [Google Scholar]
- 29.Pretorius, I. S. 2000. Tailoring wine yeast for the new millennium: novel approaches to the ancient art of winemaking. Yeast 16:675-729. [DOI] [PubMed] [Google Scholar]
- 30.Reifenberger, E., E. Boles, and M. Ciriacy. 1997. Kinetic characterization of individual hexose transporters of Saccharomyces cerevisiae and their relation to the triggering mechanisms of glucose repression. Eur. J. Biochem. 245:324-333. [DOI] [PubMed] [Google Scholar]
- 31.Remize, F., J. Roustan, J. Sablayrolles, P. Barre, and S. Dequin. 1999. Glycerol overproduction by engineered Saccharomyces cerevisiae wine yeast strains leads to substantial changes in by-product formation and to a stimulation of fermentation rate in stationary phase. Appl. Environ. Microbiol. 65:143-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salmon, J. 1989. Effect of sugar transport inactivation in Saccharomyces cerevisiae on sluggish and stuck enological fermentations. Appl. Environ. Microbiol. 55:953-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salmon, J., and P. Barre. 1998. Improvement of nitrogen assimilation and fermentation kinetics under enological conditions by derepression of alternative nitrogen-assimilatory pathways in an industrial Saccharomyces cerevisiae strain. Appl. Environ. Microbiol. 64:3831-3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schaaff, I., J. Heinisch, and F. K. Zimmermann. 1989. Overproduction of glycolytic enzymes in yeast. Yeast 5:285-290. [DOI] [PubMed] [Google Scholar]
- 35.Schulze, U., G. Liden, J. Nielsen, and J. Villadsen. 1996. Physiological effects of nitrogen starvation in an anaerobic batch culture of Saccharomyces cerevisiae. Microbiology 142:2299-2310. [DOI] [PubMed] [Google Scholar]
- 36.Sillje, H., J. Paalman, E. ter Schure, S. Olsthoorn, A. Verkleij, J. Boonstra, and C. Verrips. 1999. Function of trehalose and glycogen in cell cycle progression and cell viability in Saccharomyces cerevisiae. J. Bacteriol. 181:396-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stephanopoulos, G., A. Aristidou, and J. Nielsen. 1998. Metabolic engineering: principles and methodologies. Academic Press, San Diego, Calif.
- 38.Vallino, J., and G. Stephanopoulos. 1993. Metabolic flux distributions in Corynebacterium glutamicum during growth and lysine overproduction. Biotechnol. Bioeng. 41:633-646. [DOI] [PubMed] [Google Scholar]
- 39.Vanderaar, P. C., T. S. Lopes, J. Klootwijk, P. Groeneveld, H. W. Vanverseveld, and A. H. Stouthamer. 1990. Consequences of phosphoglycerate kinase overproduction for the growth and physiology of Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 32:577-587. [Google Scholar]
- 40.Varela, C., E. Agosin, M. Baez, M. Klapa, and G. Stephanopoulos. 2003. Metabolic flux redistribution in Corynebacterium glutamicum in response to osmotic stress. Appl. Microbiol. Biotechnol. 60:547-555. [DOI] [PubMed] [Google Scholar]
- 41.Verduyn, C., E. Postma, W. Scheffers, and J. Vandijken. 1990. Physiology of Saccharomyces cerevisiae in anaerobic glucose-limited chemostat cultures. J. Gen. Microbiol. 136:395-403. [DOI] [PubMed] [Google Scholar]
- 42.Voet, D., and J. Voet. 1995. Biochemistry, 2nd ed. J. Wiley & Sons, New York, N.Y.
- 43.Walker, G. 1998. Yeast physiology and biotechnology. J. Wiley & Sons, Chichester, United Kingdom.