Abstract
Both microbial iron reduction and microbial reduction of anodes in fuel cells can occur by way of soluble electron mediators. To test whether neutral red (NR) mediates iron reduction, as it does anode reduction, by Escherichia coli, ferrous iron levels were monitored in anaerobic cultures grown with amorphous iron oxide. Ferrous iron levels were 19.4 times higher in cultures fermenting pyruvate in the presence of NR than in the absence of NR. NR did not stimulate iron reduction in cultures respiring with nitrate. To explore the mechanism of NR-mediated iron reduction, cell extracts of E. coli were used. Cell extract-NADH-NR mixtures had an enzymatic iron reduction rate almost 15-fold higher than the chemical NR-mediated iron reduction rate observed in controls with no cell extract. Hydrogen was consumed during stationary phase (in which iron reduction was detectable) especially in cultures containing both NR and iron oxide. An E. coli hypE mutant, with no hydrogenase activity, was also impaired in NR-mediated iron reduction activity. NR-mediated iron reduction rates by cell extracts were 1.5 to 2 times higher with hydrogen or formate as the electron source than with NADH. Our findings suggest that hydrogenase donates electrons to NR for extracellular iron reduction. This process appears to be analogous to those of iron reduction by bacteria that use soluble electron mediators (e.g., humic acids and 2,6-anthraquinone disulfonate) and of anode reduction by bacteria using soluble mediators (e.g., NR and thionin) in microbial fuel cells.
Electrical applications and manipulations of bacterial metabolism have been examined periodically for over 30 years. In recent times, electrical applications of bacterial systems have received the most attention. Harvesting electrons from bacterial metabolism is being studied as a potential sustainable energy source, and electricity is being used to enhance fermentations of reduced organic chemicals (49). Electrodes, cation-exchange membranes, electron mediators, and overall system design have been modified, yet some of the underlying biological mechanisms of electron transfer between bacteria and electrodes remain unclear.
A persistent question in electrical applications of bacteria is how electrons enter and leave the cell. An answer may be approached from analogous systems in nature where extracellular insoluble electron acceptors (such as ferric iron) are reduced by bacteria. Dissimilatory iron-reducing bacteria reduce iron as a form of anaerobic respiration. In these systems, electron transfer is mediated by outer membrane cytochromes (15, 29) or by soluble electron shuttles such as 2,6-anthraquinone disulfonate (AQDS) (18). Evidence of common mechanisms for metal reduction and anode reduction is emerging. For example, AQDS, which can mediate electron transfer to ferric iron in some members of the family Geobacteraceae, was shown to stimulate electricity generation in microbial fuel cell cultures of one member of the family Geobacteraceae, Desulfuromonas acetoxidans (6). Iron reduction by way of soluble mediators clearly resembles electron transfer by artificial electron mediators in electrochemical bioreactors.
Fermentative bacteria are often used in microbial fuel cells, and some of these bacteria can reduce iron (24). Unlike dissimilatory iron reducers, fermentative bacteria do not derive significant energy from iron reduction (24). Electrical yields of bacterial fermentations are often enhanced by the addition of electron mediators, usually in the form of redox dyes (e.g., thionin [12, 21] and neutral red [NR] [34]). Humic acids allow some fermentative bacteria to reduce significant amounts of iron and to produce more oxidized end products (5). However, Escherichia coli is incapable of utilizing humic acids for iron reduction (5) and of reducing AQDS (43).
NR is water and lipid soluble, and it adheres to bacterial membranes (37). NR has a redox potential of −325 mV, similar to that of NADH (−320 mV), and a structure similar to that of flavins. Its redox potential suggests that NR could interact with metabolic steps prior to respiratory chains. NR has been shown to interact with bacterial metabolism in Clostridium acetobutylicum (20) and Butyribacterium methylotrophicum (42) by altering fermentative electron and carbon flows. NR might intercept electrons from or donate electrons to steps that involve NAD/NADH or perhaps directly exchange electrons with NADH itself. Park and Zeikus (34, 37) showed that NR chemically reduced and oxidized NAD/NADH in vitro. It has been demonstrated that NR is an effective electron mediator for electricity production by E. coli cultures in microbial fuel cells (34, 36). However, the exact mechanism by which NR mediates electron transfer from E. coli metabolism to the anode remains unanswered.
The purpose of this study is to examine the mechanism by which NR acts as an electron mediator to couple E. coli metabolism to the reduction of iron, an extracellular electron acceptor. This report also explores analogies between microbial anode and metal reduction. Experiments were conducted to compare ferrous iron generated in the presence and absence of NR in anaerobic cultures. Chemical and enzymatic NR-mediated iron reduction rates were compared by using cell extracts from fermenting E. coli. End product analysis and enzymatic assays with cell extracts and cell suspensions were used to determine the stationary-phase source of electrons. Finally, an E. coli hypE mutant unable to incorporate nickel into hydrogenases was used to assess the role of hydrogenase in NR-mediated iron reduction.
MATERIALS AND METHODS
Chemicals, bacteria, and growth conditions.
All chemicals were purchased from Sigma-Aldrich (St. Louis, Mo.). Escherichia coli K-12 (ATCC 10798) was obtained from the American Type Culture Collection. E. coli SE38 (hypE mutation, formerly called hydB103) (23, 38) is derived from a K-12 parent and was obtained from the E. coli Genetic Stock Center (MCD Biology Department, Yale University, New Haven, Conn.; http://cgsc.biology.yale.edu/cgsc.html). Bacteria were cultured at 37°C in 10.5-ml volumes in anaerobic modified M-9 medium (3) containing 3 g of Casamino Acids per liter, 105 mM pyruvate (unless otherwise stated), and vitamin (47) and mineral (25) solutions in rubber bung-sealed anaerobic test tubes. The pH was adjusted to 7.2, and the medium was made anaerobic by repeated evacuation and flushing with N2 before autoclaving was done.
Iron reduction by bacterial cultures.
Anaerobic modified M-9 medium was supplemented with ∼110 mM amorphous iron oxide [Fe(III) oxyhydroxide] (27) before autoclaving. Amorphous iron oxide was prepared as described previously (25). The total concentration of amorphous iron oxide was determined as that which was reducible by hydroxylamine (28). When appropriate, NR was supplied at 100 μM before autoclaving. Cultures were grown in the dark to prevent iron photoreduction and shaken at 250 rpm in horizontal tubes to evenly distribute the iron precipitate. In preparation for ferrous iron quantification, 100-μl samples were taken and immediately added to 900 μl of 0.5 N HCl. The iron was allowed to dissolve in the dark for at least 12 h before quantification.
Analytical techniques.
Ferrous iron was quantified by the ferrozine assay (26) with Fe(NH3)2(SO4)2 and FeSO4 in 0.5 N HCl as standards. Dissolved iron sample or standard (20 μl) was mixed with 980 μl of ferrozine reagent (26), and absorption was measured at 562 nm. Organic acids were quantified by high-performance liquid chromatography (HPLC) with a Waters/Breeze HPLC (Waters, Milford, Mass.) equipped with a refractive index detector and a UV detector set at 210 nm. A 300 by 7.8 mm Aminex HPX-87H column (Bio-Rad, Hercules, Calif.) was used at 40°C with 4 mM H2SO4 as the eluent. The hydrogen concentration was determined by gas chromatography (GC) using an Aerograph series 1400 chromatograph (Varian, Palo Alto, Calif.) equipped with a molecular sieve 5A column and a thermal conductivity detector. Bacterial protein was quantified by the bicinchoninic acid assay (BCA; Pierce, Rockford, Ill.) with bovine serum albumin as the standard. Cells were lysed by resuspending cell pellets in 0.25 N NaOH and freezing them on dry ice, and thawing them at 60°C five times. NaOH also promoted oxidation of any reduced iron that would otherwise contribute to false positives (data not shown), by interacting with the bidentate ligand of bicinchoninic acid (44). Iron oxide was spun down in a microcentrifuge (Eppendorf, Hamburg, Germany) at 5,000 rpm (2,040 × g) for 3 min before 50 μl of the supernatant sample was mixed with 950 μl of BCA reagent. BCA-protein mixtures were incubated for 30 min at 60°C before the absorption at 562 nm was recorded. Hydrogenase assays were performed as described in reference 45.
Cell lysate and cell suspension preparations.
Five hundred milliliters of modified M-9 medium was prepared anaerobically in 1-liter round flasks by repeated evacuation and flushing with N2 before autoclaving was performed. For growing nitrate-respiring cells, pyruvate was omitted, 15 mM NO3 was added, and the headspace was filled with 80% H2-20% CO2. E. coli starter culture (10 ml) was used to inoculate the flask. Cultures were grown at 37°C for 20 h into stationary phase. Cells were harvested by centrifugation for 15 min at 10,000 × g in N2-filled centrifuge bottles. Harvested cells were washed with anoxic 0.1 M Tris-HCl (pH 7.7). Cells were lysed by two passages through a French press at 1,200 to 1,400 lb/in2 under an N2 headspace. Cell lysates were stored between assays at −20°C. The cell extract protein concentration was quantified by the BCA assay. Cell suspensions were prepared in a similar manner except cells were not lysed and all assays were performed immediately following the preparation of cell suspensions to avoid lysis during storage.
Chemical and enzymatic iron reduction assays.
All assays were performed in rubber-stoppered cuvettes that were evacuated and flushed with N2 as described elsewhere (50). All additions were made by syringe. All reagents were dissolved in 0.1 M Tris-HCl (pH 7.7) and made anoxic by repeat evacuation and flushing with N2. Iron reduction was monitored by measuring the formation of Fe(II)-ferrozine over time at 562 nm (19) in a Cary 300 spectrophotometer (Varian). Ferric citrate was used instead of amorphous iron oxide because it is soluble and therefore uniformly distributed in reaction mixtures, allowing for the rate of iron reduction to be measured. The extinction coefficient used was 26 cm−1 mM−1, which was determined from the slopes of several standard curves. Reactions were performed at 37°C in mixtures that typically contained 0.23 mM ferric citrate, 0.43 g of ferrozine per liter, and 100 μM NR when appropriate.
RESULTS
(i) NR-mediated iron reduction by fermenting E. coli.
Unlike other fermentative bacteria (5) and dissimilatory iron-reducing bacteria (18), E. coli cannot use humic acids to reduce iron. Experiments were conducted to test whether NR can serve as an electron mediator for iron reduction by E. coli. Iron reduction by E. coli was examined in the presence and absence of NR during fermentation and anaerobic nitrate respiration. The results from stationary phase cultures, where iron reduction was detectable, are summarized in Table 1. Overall, NR greatly stimulated iron reduction in E. coli under fermentative conditions but not during respiration with nitrate. The same observations were made when 50 mM glucose was provided instead of pyruvate (data not shown).
TABLE 1.
Influence of NR on iron reduction by fermenting versus nitrate-respiring E. colia
| TEAb | Electron sourcec | Maximum growth (mg of cell protein/ml)d
|
Concn of Fe(II) produced (mM)
|
Standardized Fe(II) produced (μmol)e
|
|||
|---|---|---|---|---|---|---|---|
| −NR | +NR | −NR | +NR | −NR | +NR | ||
| None | Pyruvate | 0.44 ± 0.01 | 0.36 ± 0.01 | 1.9 ± 0.1 | 36.9 ± 2.0 | 0.39 ± 0.02 | 9.30 ± 0.37 |
| NO3 | Pyruvate | 0.23 ± 0.01 | 0.22 ± 0.01 | 1.0 ± 0.1 | 1.0 ± 0.1 | 1.42 ± 0.13 | 1.52 ± 0.12 |
| NO3 | Hydrogen | 0.06 ± 0.02 | 0.06 ± 0.01 | <0.1 | <0.1 | ND | ND |
| NO3 | Formate | 0.07 ± 0.01 | 0.06 ± 0.01 | <0.1 | <0.1 | ND | ND |
| NO3 | Lactate | 0.08 ± 0.00 | 0.12 ± 0.00 | <0.1 | <0.1 | ND | ND |
Averages±standard deviations for triplicate cultures in stationary phase (72 to 73 h). NR-mediated iron reduction was not detected during log phase. ND, not determined; −NR, with no NR added; +NR, with NR added.
TEA, terminal electron acceptor.
Primary electron sources and concentrations were pyruvate, ∼105 mM; formate, ∼100 mM; lactate, ∼35 mM. H2 (∼80%) in headspace and estimated at 0.6 mM dissolved according to the mole fraction solubility of hydrogen (1.35 × 10−5) at 308K.
Quantification of biomass at the end of log phase.
Amount of Fe(II) produced divided by milligrams of cell protein at the end of log phase and by the millimolar concentration of total electrons in the amount of electron source consumed for each individual culture before being averaged is given. Total electrons were predicted from the oxidation of the electron source to CO2 based on reference 17.
(ii) Comparison of chemical and enzymatic iron reductions.
NR can reduce and oxidize NAD/NADH in vitro when coupled to an electrode as an electron donor or acceptor (34, 37). To determine whether NR could oxidize NADH in vitro with soluble ferric citrate as the final electron acceptor, iron reduction rates were monitored spectrophotometrically in anoxic cuvettes. Iron reduction was not detectable in mixtures of NADH and ferric citrate but proceeded when oxidized NR was added to the reaction mixture (Table 2). NR did not reduce ferric citrate in the absence of NADH.
TABLE 2.
Comparison of enzymatic versus chemical iron reduction rates
| Conditiona | Reduction rate [μM Fe(II)/min]b |
|---|---|
| NADH + NR | 0.12 ± 0.03 |
| CE | 0.18 ± 0.13 |
| Heated CE | 0.04 ± 0.02 |
| CE + NR | 0.22 ± 0.08 |
| Heated CE + NR | 0.02 ± 0.01 |
| CE + NADH | 0.76 ± 0.04* |
| Heated CE + NADH | 0.11 ± 0.04 |
| CE + NADH + NR | 1.78 ± 0.02* |
| Heated CE + NADH + NR | 0.37 ± 0.05 |
| CE + For | 1.25 ± 0.04* |
| CE + For + NR | 3.01 ± 0.18* |
| CE + H2 | 0.87 ± 0.09* |
| CE + H2 + NR | 3.67 ± 0.61* |
| CE + Suc | 0.57 ± 0.01* |
| CE + Suc + NR | 0.78 ± 0.04* |
| CE + Lac | 1.08 ± 0.01* |
| CE + Lac + NR | 1.49 ± 0.01* |
NADH was supplied at 1.3 mM. CE, cell extract; lysate from stationary-phase pyruvate-grown culture of E. coli K-12 (20 mg/ml); supplied at 0.05 ml in a final assay volume of 1.05 ml. Heated CE, lysates were heated for 30 min at 80°C and allowed to cool before being added to the reaction mixture. For, formate, concentration of 4.8 mM. H2, ∼80% in headspace and estimated at 0.6 mM dissolved according to the mole fraction solubility of hydrogen (1.35 × 10−5) at 308K. Suc, succinate; final concentration of 6 mM. Lac, lactate; concentration of 4.8 mM.
The chemical reduction rate of 0.12 μM Fe(II)/min was subtracted from enzymatic values. Chemical reduction of iron by hydrogen, formate, succinate, and lactate was not observed in the presence or absence of NR. *, 99% confidence of a statistical difference from the cell suspension value, as determined by a single-tail, two-sample, unequal variance t test. Error values are standard deviations from triplicate assays.
Since ferric citrate reduction by NADH required NR, chemical NR-mediated iron reduction is thought to comprise two reactions, (i) NR reduction by NADH and (ii) iron reduction by NR. To determine what was limiting the rate of iron reduction, concentrations of ferric citrate, NADH, and NR were varied. The iron reduction rate did not vary at concentrations of ferric citrate between 50 and 410 μM. The iron reduction rate varied with NADH concentrations below 600 μM but not above. The iron reduction rate varied proportionately with NR concentrations between 50 and 200 μM. The rates of iron reduction were 70, 130, and 220 nM Fe(II)/min (standard deviation was 20 nM Fe(II)/min for all) at 50, 100, and 200 μM NR, respectively.
Previous work indicated that electrically reduced NR could provide electrons to enzymatic reactions (e.g., fumarate reductase) in the absence of NADH (37). To compare the chemical and enzymatic NR-mediated iron reduction rates, cell extracts from stationary-phase E. coli K-12 fermenting pyruvate were used to assay iron reduction. The results in Table 2 show that cell extracts could use NADH to reduce ferric citrate in the absence of NR. When NR is added to the cell extract-NADH mixtures, the rate of iron reduction increased almost 15-fold over the chemical NR-mediated iron reduction rate. Iron reduction activity was significantly decreased in controls by using heated cell extracts compared to untreated cell extracts.
(iii) Hydrogen-driven NR-mediated iron reduction.
NR can mediate electron transfer from NADH to iron or a graphite electrode, suggesting that NR could oxidize cytoplasmic NADH chemically and enzymatically in bacterial cultures (34). To test whether NADH is the primary source of electrons for NR-mediated iron reduction, fermentation end product concentrations were monitored in E. coli fermentations in the presence of amorphous iron oxide with or without NR. Since most of the iron reduction occurs during stationary phase, it was expected that fermentation end products would be oxidized during stationary phase to supply electrons for iron reduction. Formate and hydrogen oxidation both have suitable redox potentials (−420 and −410 mV, respectively) to reduce NR, and hydrogenase and formate dehydrogenase can reduce redox dyes, such as benzyl viologen (45). HPLC analysis of organic acid end products, including formate, from fermentations by E. coli K-12 showed no significant change over 6 days (data not shown). GC analysis of headspace hydrogen showed that the hydrogen concentration decreased during stationary phase. The results in Table 3 show that the hydrogen loss appears to be attributable to microbial consumption activity and that this activity is stimulated by the combined presence of iron oxide and NR. The amount of hydrogen consumed could account for 61 to 97% of the reduced iron, suggesting that hydrogen could be a major source of electrons for NR-mediated iron reduction.
TABLE 3.
Relationship between hydrogen consumption and iron reduction in stationary-phase E. coli culturesa
| Culture condition | H2 consumption (μmol)b | Iron reduced (μmol)c | Electrons consumed/ electrons reducedd |
|---|---|---|---|
| K-12 | 33 ± 6 | NA | NA |
| K-12 + Fe(III) | 48 ± 10 | 18 ± 2 | 0.38-2.96 |
| K-12 + NR | 18 ± 1 | NA | NA |
| K-12 + Fe(III) + NR | 169 ± 9* | 383 ± 83 | 0.61-0.97 |
Data are means ± standard deviations from triplicate cultures.
(H2 [mM] in the headspace × headspace volume after 11 h) − (H2 in the headspace × headspace volume after 84 h). Values take into account 10 ± 8 μmol lost from abiotic control of hydrogen and sterile water. *, 95% confidence of a statistical difference from K-12 hydrogen consumption value, as determined by a single-tail, two-sample, unequal variance t test.
[Fe(II) × liquid volume after 84 h] − [Fe(II) × liquid volume after 11 h].
([Hydrogen consumed × 2 electrons in culture with iron] − [hydrogen consumed × 2 electrons in culture without iron])/corresponding iron reduced.
To confirm the importance of hydrogen in NR-mediated iron reduction by E. coli, iron reduction by the hypE mutant, SE38, was compared with that by wild-type K-12. The hypE mutation prevents incorporation of nickel into hydrogenases, thereby eliminating the synthesis of active hydrogenases for hydrogen production and consumption (40). GC was used to confirm that SE38 did not produce detectable amounts of hydrogen and a hydrogenase assay confirmed that SE38 could not consume hydrogen. Interestingly, SE38 had less capacity for NR-mediated iron reduction (95% confidence; single-tail, two-sample, unequal variance t test), but still produced 65% of the amount of ferrous iron produced by wildtype K-12 (Table 4). HPLC analysis of organic acids during stationary phase did not indicate that SE38 consumed other fermentation end products, such as formate (data not shown).
TABLE 4.
Role of hydrogenase activity in NR-mediated iron reduction in E. coli fermentationsa
| E. coli strainb | Maximum growth (mg of cell protein/ml)c
|
Fe(II) produced (mM)
|
Standardized Fe(II) produced (μmol)d
|
|||
|---|---|---|---|---|---|---|
| −NR | +NR | −NR | +NR | −NR | +NR | |
| Wild-type K-12 | 0.44 ± 0.01 | 0.36 ± 0.01 | 1.9 ± 0.1 | 36.9 ± 2.0 | 0.39 ± 0.02 | 9.30 ± 0.37 |
| hypE mutant SE38 | 0.16 ± 0.00 | 0.12 ± 0.02 | 0.6 ± 0.0 | 6.7 ± 1.8 | 0.34 ± 0.02 | 6.11 ± 1.41 |
Data are means ± standard deviations from triplicate cultures samples.
SE38 values are from 84 h after inoculation to account for the slower growth rate such that SE38 and K-12 spent approximately the same amount of time in stationary phase.
Quantification of biomass at the end of log phase. −NR, with no added NR; +NR, with NR added.
Fe(II) produced divided by milligrams of cell protein at the end of log phase and by the millimolar concentration of total electrons in the amount of electron source consumed for each individual culture before being averaged. Total electrons was predicted from the oxidation of the electron source to CO2 based on reference 17.
Although hydrogen appeared to be a major source of electrons for NR-mediated iron reduction, it was not clear whether the electrons passed through NAD+ before reducing iron. To test the likelihood of NAD+ serving as an additional mediator in NR-mediated iron reduction, iron reduction rates with cell extracts and alternative electron sources were compared. The results in Table 2 show that the rate of NR-mediated iron reduction was significantly higher when formate or hydrogen was the electron source compared with NADH (95% confidence; single-tail, two-sample, unequal variance t test). However, NADH provided higher NR-mediated iron reduction rates than when lactate or succinate was the electron donor (95% confidence; single-tail, two-sample, unequal variance t test). NR also had less of a stimulatory effect on iron reduction rates in cell extracts with lactate or succinate. When ethanol was provided as the electron donor, iron reduction rates were similar to those of controls without added electron donor [e.g., 0.21 nM Fe(II)/min without NR and 0.25 nM Fe(II)/min with NR].
Although formate levels remained constant in stationary-phase cultures, kinetic assays with cell extracts showed that formate produces high NR-mediated iron reduction rates (Table 2). Cell suspensions were used in place of cell extracts to test whether the organized structure of the cell plays a role in determining which electron source can be used for NR-mediated iron reduction. Since formate and hydrogen were the most promising electron sources for NR-mediated iron reduction in cell extracts, other sources were not examined. The results in Table 5 show that both formate and hydrogen could serve as electron donors for NR-mediated iron reduction by cell suspensions of K-12. Formate, but not hydrogen, could serve as an electron donor for NR-mediated iron reduction in cell suspensions of SE38 (data not shown). The specific iron reduction rate by SE38 with formate was about half that of K-12 with formate (data not shown). Interestingly, when NR was omitted from the reaction mixture, the rates of iron reduction were statistically similar with and without supplied electron donor.
TABLE 5.
Effect of formate and hydrogen on iron reduction rates by E. coli cell suspensions
| Conditiona | Reduction rate [μM Fe(II)/min]b |
|---|---|
| CS | 0.06 ± 0.02 |
| CS + NR | 2.62 ± 0.05* |
| CS + For | 0.13 ± 0.09 |
| CS + For + NR | 5.67 ± 0.64* |
| CS + H2 | 0.03 ± 0.03 |
| CS + H2 + NR | 7.01 ± 0.97* |
CS, cell suspension from stationary-phase pyruvate-grown E. coli culture (13.9 mg/ml) was supplied at 50 μl in a final assay volume of 1.05 ml. For, formate (4.8 mM). H2, ∼80% in headspace and estimated at 0.6 mM dissolved according to the mole fraction solubility of hydrogen (1.35 × 10−5) at 308K.
The chemical reduction rate of 0.12 μM Fe(II)/min was subtracted from enzymatic values. Data are means ± standard deviations from triplicate assays. *, 99% confidence of a statistical difference from cell suspension value, as determined by a single-tail, two-sample, unequal variance t test. Chemical reduction of iron by hydrogen or formate was not observed in presence or absence of NR.
While hydrogenase played a role in NR-mediated iron reduction for fermentative cultures, the results in Table 1 show that hydrogen did not stimulate NR-mediated iron reduction in cultures respiring with nitrate. Cell suspensions of E. coli grown with hydrogen and nitrate were used to determine if respiring cells could carry out NR-mediated iron reduction with hydrogen in the absence of nitrate. Cell suspensions from nitrate-respiring cultures had about 16-fold-less hydrogenase activity than cells from fermenting cultures (data not shown). Initial iron reduction rates by H2-NO3-grown cell suspensions with hydrogen were statistically not different from the chemical iron reduction rate (data not shown).
DISCUSSION
Many of the trends with NR in this study, such as the reduction of an exogenous electron acceptor in bacterial fermentations and oxidation of NADH by NR, resemble trends seen in electrochemical bioreactors. Previous studies showed that NR allows bacteria to transfer electrons to a graphite electrode in microbial fuel cells and to oxidize NADH in the presence of an anode (32, 34-36). Additionally, NR-mediated iron reduction was detectable only during stationary phase. This observation is similar to previous observations in which growing E. coli produced 12-times-less electricity than stationary cells (34). We are testing the hypothesis that NR-mediated reduction of insoluble iron occurs by a physiochemical mechanism similar to that of anode reduction in microbial fuel cells. An excess precipitate of amorphous iron oxide can be viewed as an electron-accepting anode, allowing for NR-based electron harvesting studies to be performed in readily available test tubes without many of the physical variables of microbial fuel cells (e.g., electrode resistance and proton translocation rates). Harvested electrons can be quantified by measuring the amount of ferrous iron produced. The setup could also be used in preliminary studies of bacterial electrical applications by testing the ability of various microbes to perform electron transfer with NR, or potentially other electron mediators, based on ferrous iron generated.
NR-mediated iron reduction was observed only in fermenting E. coli and not in cultures respiring with nitrate. It is expected that nitrate would be thermodynamically favorable as an electron acceptor over NR since NR has a low redox potential. Cell suspensions from stationary-phase cultures grown with H2 and NO3 showed insignificant iron reduction activity in the presence of hydrogen. The low rate could be due in part to the lower hydrogenase activity in respiring cells or to a high overall cell redox potential.
Although E. coli cannot reduce iron with humic acids (5), it can reduce iron with NR. Compounds similar in structure to NR such as riboflavin, pyocyanin, quinones, and humic acids occur in natural environments, and they also reduce iron when coupled with bacterial metabolism (5, 11, 31, 48). Small aromatic compounds with a variety of proposed functions may have alternative roles, including iron reduction. For example, pyocyanin and AQDS can reduce iron but also have antibiotic activities (43). With the right electron mediator, it is possible that any bacteria could reduce iron and so fortuitous iron reduction may be widespread in nature. However, it should be noted that the amount of ferrous iron produced by bacteria respiring with iron greatly exceeds that produced by iron-reducing fermentative organisms and by NR-mediated iron reduction in this study. For example, Geobacter metallireducens completely oxidizes acetate to CO2 respiring with iron, resulting in 8 mol of Fe(II) per mol of acetate consumed (24). In comparison, the maximum iron reduced in this study was about 0.4 mol of Fe(II) per mol of pyruvate consumed. This amount is similar to the levels of ferrous iron produced by other fermentative bacteria by unknown mechanisms (24). Compared to humic acid-mediated iron reduction by fermentative bacteria, iron reduction by Propionibacterium freudenreichii produced 1 mol of Fe(II)/mol of lactate and Lactococcus lactis produced 2 mol of Fe(II)/mol of glucose (5).
Since aromatic compounds have a variety of roles in nature, bioelectrical devices using similar soluble mediators could potentially be used for purposes other than generating electricity and shifting end product distributions. For example, methanophenazine is of similar structure to NR and occurs in electron transport chains of Methanosarcina mazei (1, 9). Methanophenazine has been proposed as an electron mediator in a symbiotic relationship in consortia of methanogens and sulfate-reducing bacteria (30). Methanophenazine has been shown to couple to hydrogenase as an electron mediator. Perhaps this electron mediator is analogous to NR in allowing extracellular electron transport but between consortium members. Based on the ability to grow methanogens with electrically reduced NR (33), it may be worthwhile to try electrically reduced NR as a means to isolate and support individual members of this tight symbiosis and to prove or disprove the exciting prospect of an electron-based syntrophy.
Iron reduction proceeded when NR was added to NADH-ferric citrate mixtures. This result is not surprising since NR's structure is similar to that of pyocyanin and flavins, which also reduce iron (11, 48). Cell extracts stimulated NR-mediated iron reduction, suggesting that NR-mediated iron reduction in bacterial cultures is enzymatically coupled. In fact, a constant rate of chemical NR-mediated iron reduction would produce only ∼0.5 mM of ferrous iron after 72 h. Taking into account a lower concentration of NADH in cultures than in the kinetic assays and limitations such as NR diffusion or transport, it does not seem plausible for NR-mediated iron reduction in cultures to occur solely by the direct chemical oxidation of cytoplasmic NADH by NR. The reduction of NR is probably enzymatically facilitated, since NR reduction was rate limiting in chemical iron reduction assays with NADH, NR, and ferric citrate. It is unknown whether an enzyme or other cell component facilitates iron reduction by NR.
Hydrogen appears to be a source of electrons for NR-mediated iron reduction, based on decreasing hydrogen concentrations in stationary phase, lower iron reduction activity in cultures of the hypE mutant, and high iron reduction rates when hydrogen was the electron donor in cell extracts and cell suspensions. However, while formate concentrations remained constant during stationary phase, formate generated iron reduction rates in cell extracts and cell suspensions nearly as high as those with hydrogen. Both formate and hydrogen oxidations have redox potentials suitable for reducing NR (−420 and −410 mV, respectively). It is possible that formate turnover in cultures masked observations of formate oxidation that led to NR-mediated iron reduction. Formate turnover would help explain how SE38 reduced 65% of the amount of iron as did wildtype K-12. However, it is interesting that in K-12 cell extracts and cell suspensions, hydrogen always gave higher iron reduction rates than did formate. Since formate is soluble while hydrogen has a low solubility, one might expect formate to be more accessible and give higher reduction rates. The fact that hydrogen produced higher iron reduction rates than formate could suggest that formate first generates hydrogen via formate hydrogenlyase activity, which is then oxidized to reduce NR. Similarly, NR-mediated iron reduction rates by cell extracts with succinate and lactate were lower than those with NADH. Perhaps succinate and lactate must first be oxidized to generate NADH, which can then reduce NR in one or possibly several subsequent enzymatic steps. In any case, enzymatic reactions other than hydrogen oxidation must be involved in NR-mediated iron reduction since the hydrogen consumed during stationary phase could not account for all of the iron reduced and since SE38 still reduced a significant amount of iron, especially in the presence of NR. Formate oxidation remains a likely candidate for this role.
Our results implicate a role for hydrogenase in extracellular iron reduction and suggest a role for it in anode reduction in microbial fuel cells. In this regard, it would be of interest to evaluate the role of hydrogenase in extracellular iron reduction by dissimilatory iron-reducing bacteria and bacteria that can fortuitously reduce iron. A variety of dissimilatory iron-reducing bacteria can use hydrogen as an electron source during anaerobic respiration with ferric iron (25). However, hydrogen oxidation for iron reduction during iron respiration is probably different from the hydrogen oxidation leading to iron reduction observed in this study. The iron reduction observed in this study is suspected to be part of a secondary metabolism rather than directly involved in cell growth as is anaerobic respiration. Fortuitous iron reduction by fermentative bacteria could potentially involve hydrogen oxidation and an electron mediator in a manner similar to the phenomenon described in this study.
It is not entirely clear which of the four E. coli hydrogenases is involved in NR-mediated iron reduction. However, it is probably one or both of the hydrogen-oxidizing hydrogenases, hydrogenase-1 and hydrogenase-2 (40). Hydrogenase-2 activity primarily supplies electrons to electron transport chains (14), while a role for hydrogenase-1 remains ambiguous. Hydrogenase-1 could be involved in recycling hydrogen produced by hydrogenase-3 (41) or could act as a hydrogen-scavenging enzyme, primarily active at certain hydrogen concentrations (40). More recently, the hydrogen-oxidizing isoenzymes have been distinguished based on their activity at different redox potentials, hydrogenase-2 being active at lower redox potentials and hydrogenase-1 being active at higher redox potentials (22). Hydrogenase-2 therefore seems more likely to interact with NR than hydrogenase-1, since NR has a relatively low redox potential of −325 mV. Due to the involvement of hydrogenase-2 with respiratory electron transport chains, it is possible that a respiratory quinone would have a higher affinity for electrons from hydrogen oxidation than an artificial electron acceptor, NR. This may explain why no iron reduction was observed in cultures oxidizing hydrogen and respiring with nitrate.
In assays involving cell suspensions (Table 5), it is clear that NR is required to break down some part of organized cell processes to transfer electrons to iron. NR could interfere with the organization of an electron transfer network and/or it could be crossing the boundaries of the inner and outer membranes. It is important that for assays with cell suspensions, the oxidized iron source, ferric citrate, is soluble unlike the amorphous iron oxide used in cell cultures. E. coli is capable of using ferric citrate as an iron source for growth, and it has specific, regulated outer and inner membrane active transport mechanisms for its uptake (7). Since there are dense cell concentrations and high iron concentrations in the assay conditions, it is unlikely that there is rapid import of iron into the cytoplasm for cell growth. Therefore, it is also unlikely that NR-mediated iron reduction is occurring in the cytoplasm. Ferric citrate can form polynuclear complexes with an average molecular weight of 2 × 105 (4), which would probably not cross the outer membrane. However, it has also been reported that ferric citrate can freely diffuse into the periplasm (16). Taking into account that the active site of hydrogenase-2 is oriented into the periplasm (39), it is possible that NR-mediated reduction of ferric citrate occurs in the periplasm. However, reduction of iron itself in the periplasm does not explain reduction of extracellular insoluble iron and anode reduction.
This report provides insight into how an electron mediator can interact with bacterial metabolism to facilitate reduction of an extracellular insoluble acceptor. However, there is still much to understand, particularly in the transport of electron mediators themselves. Mechanisms for mediator transport could involve mechanisms for iron acquisition and antibiotic resistance. For example, some fermentative bacteria may reduce iron to facilitate its acquisition. E. coli produces extracellular iron reductases (10, 46) for iron acquisition, while other bacteria produce small molecules able to reduce iron, such as pyocyanin (11) and riboflavin (48), which resemble phenazine mediators (e.g., NR). Although repressed anaerobically (13), TonB-dependent transport of the siderophore ferrichrome by FhuABCD will also transport the antibiotic albomycin, a structural analogue of ferrichrome, and the structurally dissimilar rifamycin-derivative CGP 4832 (8). Active transport of CGP 4832 decreased the MIC by 200 to 400 fold (8). Based on this result, one can imagine how active transport could result in high iron reduction rates with electron mediators. Similarly, Shewanella oneidensis, which can reduce iron with the electron mediator AQDS, relies on an energy-dependent outer membrane active transport mechanism involving TolC for AQDS export as well as for export of the antibiotic pyocyanin (43). TolC is an outer membrane efflux pump that associates with periplasmic and cytoplasmic membrane proteins to actively expel antibiotics and secrete proteins as part of a type I secretion system (2). Based on the similarities of NR to pyocyanin, E. coli could require TolC for the efflux of reduced NR or of an NR-stimulated cellular factor capable of reducing iron. Such factors could be an iron reductase (described for E. coli in facilitating iron acquisition [10, 46]) or a small molecule such as riboflavin, which has been described in Helicobacter pylori and other bacteria but not in E. coli (48). Future studies involving electron mediators for microbial iron and anode reduction should concentrate not only on electron transfer reactions but also on energy-dependent mediator transport and not assume diffusion.
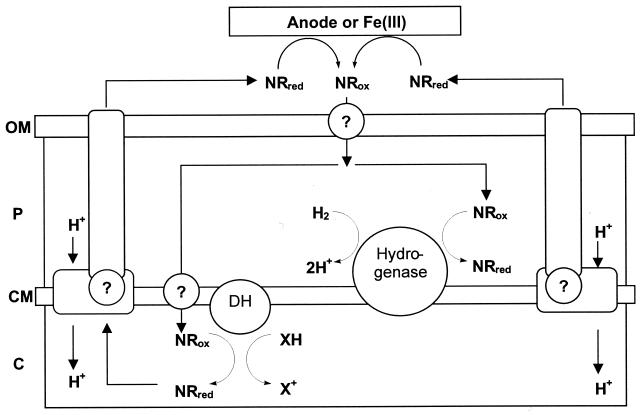
The exact mechanism for microbial reduction of extracellular electron acceptors by way of soluble mediators is not known and could employ several mechanisms. A hypothetical model for NR-mediated reduction of extracellular electron acceptors is proposed in Fig. 1. The model depicts events that may occur in fermentative E. coli cultures where hydrogen produced during fermentation is being reconsumed. First, oxidized NR enters the periplasm and cytoplasm by an unknown mechanism. NR is reduced in the periplasm by hydrogenase and also by unknown redox enzymes (perhaps formate dehydrogenase on the cytoplasmic face of the inner membrane). Reduced NR then exits the cell by an unknown process possibly involving energy-dependent transport. Once in the external environment NR is oxidized by ferric iron or an anode. Our data support this model but do not exclude alternative models. For example, an additional electron mediator capable of interacting with NR could be involved in the reduction of extracellular acceptors. Such an alternative model would help explain observations of anode reduction with bound NR (32, 35, 36).
FIG. 1.
Hypothetical model depicting the role of NR in electron transfer reactions and electron translocation from metabolizing E. coli cells to an extracellular electron acceptor. NR enters the periplasm and cytoplasm by an unknown mechanism. NR is enzymatically reduced by hydrogenase and by other enzymes, such as dehydrogenases. NR then leaves the cell by an unknown mechanism and reduces extracellular iron or an electrode. NRred, reduced NR; NRox, oxidized NR; OM, outermembrane; P, periplasm; CM, cytoplasmic membrane; C, cytoplasm; DH, dehydrogenase; XH, reduced cofactor; X+, oxidized cofactor; ?, possible transporter involved in NR transport.
In summary, NR allows fermentative, but not anaerobically respiring, E. coli to reduce iron. The mechanism of NR-mediated iron reduction does not rely solely on the direct oxidation of a cytoplasmic NADH pool, but rather it is enzymatically coupled to hydrogen oxidation and oxidation of other compounds, formate being a likely candidate.
Acknowledgments
We thank Pil Kim, Harini Krishnamurthy, David Finkelstein, Maris Laivenieks, and Claire Vieille for helpful discussions and technical assistance. We are grateful for the mutant SE38 supplied by the E. coli Genetic Stock Center. We are indebted to John Breznak for the use of GC equipment.
This work was supported by the Office of Naval Research, U.S. Navy grant no. N00014-01-1-0190, and National Science Foundation no. 0323966e. James McKinlay was supported by the Michigan State University Graduate School and the Michigan State University Andrew Rasmussen Fellowship.
REFERENCES
- 1.Abken, H. J., M. Tietze, J. Brodersen, S. Baumer, U. Beifuss, and U. Deppenmeier. 1998. Isolation and characterization of methanophenazine and function of phenazines in membrane-bound electron transport of Methanosarcina mazei Gol. J. Bacteriol. 180:2027-2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen, C., C. Hughes, and V. Koronakis. 2001. Protein export and drug efflux through bacterial channel-tunnels. Curr. Opin. Cell Biol. 13:412-416. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1993. Current protocols in molecular biology, vol. 1. Greene Publishing & Wiley-Interscience, New York, N.Y.
- 4.Bates, G. W., C. Billups, and P. Saltman. 1967. The kinetics and mechanism of iron(III) exchange between chelates and transferrin. J. Biol. Chem. 242:2810-2815. [PubMed] [Google Scholar]
- 5.Benz, M., B. Schink, and A. Brune. 1998. Humic acid reduction by Propionibacterium freudenreichii and other fermenting bacteria. Appl. Environ. Microbiol. 64:4507-4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bond, D. R., D. E. Holmes, L. M. Tender, and D. R. Lovley. 2002. Electrode-reducing microorganisms that harvest energy from marine sediments. Science 295:483-485. [DOI] [PubMed] [Google Scholar]
- 7.Braun, V. 2003. Iron uptake by Escherichia coli. Front. Biosci. 8:S1409-S1421. [DOI] [PubMed] [Google Scholar]
- 8.Braun, V., and M. Braun. 2002. Active transport of iron and siderophore antibiotics. Curr. Opin. Microbiol. 5:194-201. [DOI] [PubMed] [Google Scholar]
- 9.Brodersen, J., S. Baumer, H. J. Abken, G. Gottschalk, and U. Deppenmeier. 1999. Inhibition of membrane-bound electron transport of the methanogenic archaeon Methanosarcina mazei Gol by diphenyleneiodonium. Eur. J. Biochem. 259:218-224. [DOI] [PubMed] [Google Scholar]
- 10.Cowart, R. E. 2002. Reduction of iron by extracellular iron reductases: implications for microbial iron acquisition. Arch. Biochem. Biophys. 400:273-281. [DOI] [PubMed] [Google Scholar]
- 11.Cox, C. D. 1986. Role of pyocyanin in the acquisition of iron from transferrin. Infect. Immun. 52:263-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delaney, G. M., H. P. Bennetto, J. R. Mason, S. D. Roller, J. L. Stirling, and C. F. Thurston. 1984. Electron-transfer coupling in microbial fuel cells. 2. Performance of fuel cells containing selected microorganism-mediator-substrate combinations. J. Chem. Technol. Biotechnol. 34B:13-27. [Google Scholar]
- 13.Dorman, C. J., G. C. Barr, N. N. Bhriain, and C. F. Higgins. 1988. DNA supercoiling and the anaerobic and growth phase regulation of tonB gene expression. J. Bacteriol. 170:2816-2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dubini, A., R. L. Pye, R. L. Jack, T. Palmer, and F. Sargent. 2002. How bacteria get energy from hydrogen: a genetic analysis of periplasmic hydrogen oxidation in Escherichia coli. Int. J. Hydrogen Energ. 27:1413-1420. [Google Scholar]
- 15.Gaspard, S., F. Vazquez, and C. Holliger. 1998. Localization and solubilization of the iron(III) reductase of Geobacter sulfurreducens. Appl. Environ. Microbiol. 64:3188-3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harle, C., I. Kim, A. Angerer, and V. Braun. 1995. Signal transfer through three compartments: transcription initiation of the Escherichia coli ferric citrate transport system from the cell surface. EMBO J. 14:1430-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harris, R. F., and S. S. Adams. 1979. Determination of the carbon-bound electron composition of microbial cells and metabolites by dichromate oxidation. Appl. Environ. Microbiol. 37:237-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hernandez, M., and D. Newman. 2001. Extracellular electron transfer. Cell. Mol. Life Sci. 58:1562-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaufmann, F., and D. R. Lovley. 2001. Isolation and characterization of a soluble NADPH-dependent Fe(III) reductase from Geobacter sulfurreducens. J. Bacteriol. 183:4468-4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim, B. H., and J. G. Zeikus. 1992. Hydrogen metabolism in Clostridium acetobutylicum fermentation. J. Microbiol. Biotechnol. 2:248-254. [Google Scholar]
- 21.Kim, N., Y. Choi, S. Jung, and S. Kim. 2000. Effect of initial carbon sources on the performance of microbial fuel cells containing Proteus vulgaris. Biotechnol. Bioeng. 70:109-114. [DOI] [PubMed] [Google Scholar]
- 22.Laurinavichene, T. V., N. A. Zorin, and A. A. Tsygankov. 2002. Effect of redox potential on activity of hydrogenase 1 and hydrogenase 2 in Escherichia coli. Arch. Microbiol. 178:437-442. [DOI] [PubMed] [Google Scholar]
- 23.Lee, J. H., P. Patel, P. Sankar, and K. T. Shanmugam. 1985. Isolation and characterization of mutant strains of Escherichia coli altered in H2 metabolism. J. Bacteriol. 162:344-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lovely, D. 1991. Dissimilatory Fe(III) and Mn(IV) reduction. Microbiol. Rev. 55:259-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lovely, D. 2000. Dissimilatory Fe(III)- and Mn(IV)-reducing prokaryotes. In Martin Dworkin (ed.), The prokaryotes. Springer-Verlag [Online.] http://141.150.157.117:8080/prokPUB/index.htm. Accessed December 2003.
- 26.Lovely, D., and E. J. P. Phillips. 1986. Availability of ferric iron for microbial reduction in bottom sediments of the freshwater tidal Potomac River. Appl. Environ. Microbiol. 52:751-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lovely, D. R., and E. J. P. Phillips. 1986. Organic matter mineralization with reduction of ferric iron in anaerobic sediments. Appl. Environ. Microbiol. 51:683-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lovely, D. R., and E. J. P. Phillips. 1987. Rapid assay for microbially reducible ferric iron in aquatic sediments. Appl. Environ. Microbiol. 53:1536-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Myers, C., and J. Myers. 1992. Localization of cytochromes to the outer membrane of anaerobically grown Shewanella putrefaciens MR-1. J. Bacteriol. 174:3429-3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nauhaus, K., A. Boetius, M. Kruger, and F. Widdel. 2002. In vitro demonstration of anaerobic oxidation of methane coupled to sulphate reduction in sediment from a marine gas hydrate area. Environ. Microbiol. 4:296-305. [DOI] [PubMed] [Google Scholar]
- 31.Newman, D. K., and R. Kolter. 2000. A role for excreted quinones in extracellular electron transfer. Nature 405:94-97. [DOI] [PubMed] [Google Scholar]
- 32.Park, D. H. 2000. Electricity production in biofuel cell using modified graphite electrode with neutral red. Biotechnol. Lett. 22:1301-1304. [Google Scholar]
- 33.Park, D. H., M. Laivenieks, M. V. Guettler, M. K. Jain, and J. G. Zeikus. 1999. Microbial utilization of electrically reduced neutral red as the sole electron donor for growth and metabolite production. Appl. Environ. Microbiol. 65:2912-2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park, D. H., and J. G. Zeikus. 2000. Electricity generation in microbial fuel cells using neutral red as an electronophore. Appl. Environ. Microbiol. 66:1292-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park, D. H., and J. G. Zeikus. 2002. Impact of electrode composition on electricity generation in a single-compartment fuel cell using Shewanella putrefaciens. Appl. Microbiol. Biotechnol. 59:58-61. [DOI] [PubMed] [Google Scholar]
- 36.Park, D. H., and J. G. Zeikus. 2003. Improved fuel cell and electrode designs for producing electricity from microbial degradation. Biotechnol. Bioeng. 81:348-355. [DOI] [PubMed] [Google Scholar]
- 37.Park, D. H., and J. G. Zeikus. 1999. Utilization of electrically reduced neutral red by Actinobacillus succinogenes: physiological function of neutral red in membrane-driven fumarate reduction and energy conservation. J. Bacteriol. 181:2403-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sankar, P., J. H. Lee, and K. T. Shanmugam. 1985. Cloning of hydrogenase genes and fine structure analysis of an operon essential for H2 metabolism in Escherichia coli. J. Bacteriol. 162:353-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sargent, F., S. P. Ballantine, P. A. Rugman, T. Palmer, and D. Boxer. 1998. Reassignment of the gene encoding the Escherichia coli hydrogenase 2 small subunit. Eur. J. Biochem. 255:746-754. [DOI] [PubMed] [Google Scholar]
- 40.Sawers, G. 1994. The hydrogenases and formate dehydrogenases of Escherichia coli. Antonie Leeuwenhoek 66:57-88. [DOI] [PubMed] [Google Scholar]
- 41.Sawers, R. G., D. J. Jamieson, C. F. Higgins, and D. H. Boxer. 1986. Characterization and physiological roles of membrane-bound hydrogenase isoenzymes from Salmonella typhimurium. J. Bacteriol. 168:398-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shen, G.-J., B. A. Annous, R. W. Lovitt, M. K. Jain, and J. G. Zeikus. 1996. Biochemical route and control of butyrate synthesis in Butyribacterium methyltrophicum. Appl. Microbiol. Biotechnol. 45:355-362. [Google Scholar]
- 43.Shyu, J., D. Lies, and D. Newman. 2002. Protective role of tolC in efflux of the electron shuttle anthraquinone-2,6-disulfonate. J. Bacteriol. 184:1806-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stookey, L. 1970. Ferrozine new spectrophotometric reagent for iron. Anal. Chem. 42:779-782. [Google Scholar]
- 45.Van der Werf, M. J., M. V. Guettler, M. K. Jain, and J. G. Zeikus. 1997. Environmental and physiological factors affecting the succinate product ratio during carbohydrate fermentation by Actinobacillus sp. 130Z. Arch. Microbiol. 167:332-342. [DOI] [PubMed] [Google Scholar]
- 46.Vartivarian, S. E., and R. E. Cowart. 1999. Extracellular iron reductases: identification of a new class of enzymes by siderophore-producing microorganisms. Arch. Biochem. Biophys. 364:75-82. [DOI] [PubMed] [Google Scholar]
- 47.Wolin, E., M. Wolin, and R. Wolfe. 1963. Formation of methane by bacterial extracts. J. Biol. Chem. 238:2882-2886. [PubMed] [Google Scholar]
- 48.Worst, D. J., M. Gerrits, C. Vandenbroucke-Grauls, and J. Kusters. 1998. Helicobacter pylori ribBA-mediated riboflavin production is involved in iron acquisition. J. Bacteriol. 180:1473-1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zeikus, J. G. 2003. Bioelectrical synthesis of chemicals, fuels and drugs. In C. H. Hou (ed.), Handbook of industrial biocatalysis, in press. Marcel Decker, Inc., New York, N.Y.
- 50.Zeikus, J. G., G. Fuchs, W. Kenealy, and R. K. Thauer. 1977. Oxidoreductases involved in cell carbon synthesis of Methanobacterium thermoautotrophicum. J. Bacteriol. 132:604-613. [DOI] [PMC free article] [PubMed] [Google Scholar]