Abstract
A total of 30 Megasphaera elsdenii strains, selectively isolated from the feces of organically raised swine by using Me109 M medium, and one bovine strain were analyzed for tetracycline resistance genotypic and phenotypic traits. Tetracycline-resistant strains carried tet(O), tet(W), or a tet gene mosaic of tet(O) and tet(W). M. elsdenii strains carrying tet(OWO) genes exhibited the highest tetracycline MICs (128 to >256 μg/ml), suggesting that tet(O)-tet(W) mosaic genes provide the selective advantage of greater tetracycline resistance for this species. Seven tet genotypes are now known for M. elsdenii, an archetype commensal anaerobe and model for tet gene evolution in the mammalian intestinal tract.
Megasphaera elsdenii is a commensal (mutualist) species in the gastrointestinal tracts of ruminant and nonruminant mammals, including humans (7, 25, 26). This anaerobic bacterium contributes to the overall metabolism that takes place in those microbial ecosystems (2, 5, 13, 15). M. elsdenii has been the focus of both prebiotic and probiotic applications for improving animal health (10, 12, 19, 27).
In a recent study of intestinal bacteria resistant to tetracycline, we detected resistant M. elsdenii strains at high population levels (approximately 107 CFU/g) in cecal samples from healthy swine (24). Eight strains were isolated and characterized. The M. elsdenii strains are highly resistant to chlortetracycline (MIC = 256 to >256 μg/ml) and carry one of two “tet(OWO)” genes for tetracycline resistance. [Throughout the manuscript, “tet(OW)” and “tet(OWO)” are used as convenient, practical terms for describing M. elsdenii recombinant tet genes and genotypes. As noted previously (24), these designations are not recognized under present tet classification guidelines (14). It is our hope that future guidelines will be developed to accommodate these novel interclass hybrid genes.] These tet genes are interclass mosaic genes apparently formed by double-crossover recombinations between tet(O) and tet(W) genes.
Our previous study used a nutritionally complex medium with high concentrations of chlortetracycline and, thus, was biased to select tetracycline-resistant M. elsdenii strains. In this study, Me109M medium was developed and used to select M. elsdenii strains without using chlortetracycline. The goals were twofold: first, to obtain tetracycline-sensitive strains of M. elsdenii useful both for investigating tet gene transfer and for probiotic applications, and second, to discover whether or not M. elsdenii strains have additional tet genotypes.
Selective isolation of M. elsdenii—Me109M medium.
On the basis of previous studies (4-6, 9, 14, 16, 17, 19-21) and preliminary experiments in our laboratory, Me109M medium was developed to selectively culture M. elsdenii. Me109M contained (per liter): tryptone-peptone, 4 g; yeast extract, 2 g; salts solution A (6 g of K2HPO4/liter of water), 40 ml; salts solution B [6 g of KH2PO4, 12 g of (NH4)2SO4, 12 g of NaCl, 1.2 g of MgSO4-7H2O, 0.6 g of CaCl2/liter of water], 40 ml; Na-dl-lactate syrup (60% [wt/wt]), 12.7 ml; resazurin solution (0.1% [wt/vol]), 1 ml; l-cysteine-HCl, 0.5 g; distilled water, 900 ml; Difco Bacto agar, 12 g. The pH of the medium was adjusted to 5.0. A total of 10 ml of a monensin solution (5 mg/ml of ethanol) was added to the autoclaved medium. Agar plates of medium were prepared aerobically and, at least 2 days before inoculation, transferred into a Coy anaerobic chamber at room temperature.
M. elsdenii strains were isolated under anoxic conditions from fecal samples of 10 grower-phase (40- to 50-kg) swine purchased from two Iowa farms that have raised animals organically (National Organic Foods Production Act; Alternative Farming Systems Information Center [www.nalusda.gov/afsic/ofp]), without antibiotic feeding, for at least 4 years. The swine were housed at the National Animal Disease Center, with no exposure to other animals, in chemically decontaminated buildings with strict entry requirements for human personnel. The animals were supplied water ad libitum and fed their original organic diet, which was free of detectable tetracycline (3).
M. elsdenii population densities averaged 2.4 × 108 CFU/g of feces (wet weight), and there were no significant differences in counts between animals from the two farms. M. elsdenii colonies appeared as large (2- to 3-mm-diameter), yellow-white, dome-shaped colonies after 72 h of incubation at 38°C and contained large cocci in pairs or chains. These colonies represented approximately one-third of the colonies growing on Me109M agar and 0.4% of the total cultivable fecal bacteria (data not presented). Me109M is highly selective for M. elsdenii and has been used to detect population levels of this species as low as 104 CFU/g of feces (T. B. Stanton, unpublished data). A total of 30 strains (3 from each animal) were cloned by subculturing and confirmed to be M. elsdenii on the basis of physiological properties and 16S rRNA-V3 sequence analysis (24).
M. elsdenii tetracycline-resistant genotypes.
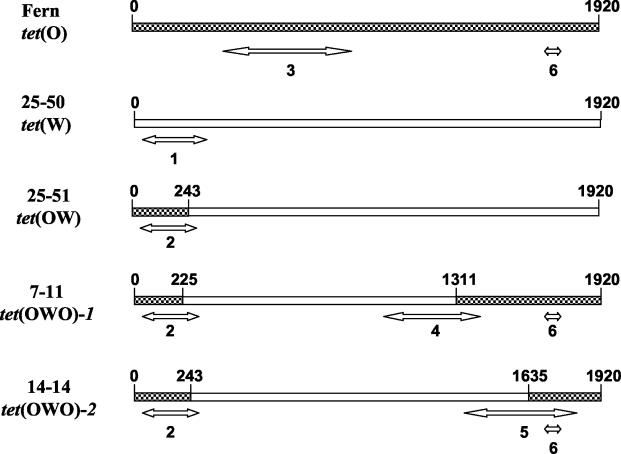
The 30 swine strains and bovine strain Fern, previously isolated during unrelated studies, were analyzed by PCR assays (24) to differentiate known M. elsdenii tetracycline resistance genes (Fig. 1; Table 1). Nine strains did not contain detectable tet genes. The tet(OWO)-1 gene identified previously in swine M. elsdenii strain 7-11 (Fig. 1) was not detected in any of the isolates. A total of 11 strains had the tet(OWO)-2 genotype reported previously for M. elsdenii strain 14-14 (Fig. 1; Table 1). Another 11 strains carried genes—tet(O), tet(W), and tet(OW)—not previously detected for M. elsdenii. The tet(OW) genes apparently originate from single-crossover recombinations between tet(O) and tet(W).
FIG. 1.
Schematic depiction of PCR assays for differentiating M. elsdenii tetracycline-resistant genotypes. Strain and tetracycline resistance genotype designations are given at the left of the figure. Checkered regions of genes have high-level sequence identity with tet(O); open regions have high-level sequence identity with tet(W). Products of PCR amplification are depicted by double-headed arrows. Crossover regions of the genes were established by sequence analysis.
TABLE 1.
M. elsdenii tetracycline resistance genotypes determined by PCR analysesa
| M. elsdenii strain(s) | PCRb
|
tet genotypec | |||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | ||
| 24-50; 26-50; 27-50; 28-50; 29-54; 30-54; 31-54; 32-54; 33-54 | − | − | − | − | − | − | No tet |
| Fern | − | − | + | − | − | + | tet(O) |
| 25-50; 29-55 | + | − | − | − | − | − | tet(W) |
| 24-51; 25-51; 27-51; 28-53; 29-57; 30-55; 31-55; 33-57 | − | + | − | − | − | − | tet(OW) |
| 24-53; 25-52; 26-51; 26-52; 27-52; 28-52; 30-56; 31-57; 32-56; 32-57; 33-56 | − | + | − | − | + | + | tet(OWO)-2 |
All M. elsdenii strains are swine fecal isolates except strain Fern, which was isolated from bovine rumen contents.
“+” indicates product and “−” indicates no product for PCR depicted in Fig. 1.
Additional tet genotypes were discovered upon sequencing tet(OW) and tet(W) genes, as described in the text.
Sequence comparisons revealed differences between the strain 27-51 and 25-51 tet(OW) genes;these have been designated tet(OW)-1 and tet(OW)-2, respectively. In the strain 25-51 tet(OW)-2 gene, the tet(O)-to-tet(W) crossover position occurs at base position 243, resembling that of strain 14-14 (Fig. 1). The corresponding crossover position of the strain 27-51 tet(OW)-1 gene at base position 225 is similar to that of strain 7-11 (Fig. 1).
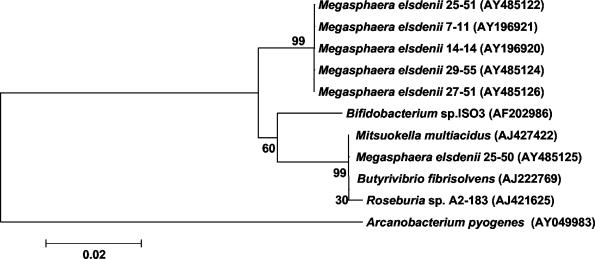
The tet(W) genes of M. elsdenii strains 25-50 and 29-55 likely have different origins and have been designated, respectively, tet(W)-1 and tet(W)-2. When the Tet(W) portions (356-amino-acid segments) common to M. elsdenii Tet(W), Tet(OW), and Tet(OWO) proteins are compared, the amino acid sequences fall into two main clusters marked by a bootstrap support value of 99 (Fig. 2). Sequences between the two clusters differ by 11 amino acids. One cluster of segments is comprised of M. elsdenii Tet(W), Tet(OW), and Tet(OWO) sequences. The sequences are identical, suggesting that recombination between a tet gene identical to tet(W)-2 of M. elsdenii strain 29-55 and a tet(O) gene produced the presently known mosaic genes of M. elsdenii. These sequences do not match any known Tet(W) sequences of other intestinal species. By contrast, not only the M. elsdenii 25-50 Tet(W)-1 segment (Fig. 2) but also the complete 25-50 Tet(W)-1 sequence is identical to those of Tet(W) proteins of Butyrivibrio fibrisolvens and Mitsuokella multiacidus, suggesting that these tet(W) genes share a common origin and may be communicable among these intestinal anaerobes. A larger sample of tet(W) sequences from diverse bacterial species will undoubtedly increase the robustness of the tree and could provide further insight into horizontal gene flow among species in the intestinal tract.
FIG. 2.
Phylogenetic analysis of a Tet(W) common region (365 amino acids) shared by various species. GenBank accession numbers for nucleotide sequences used to derive amino acid sequences are given in parentheses. The unrooted neighbor-joining tree was generated using Mega2 (11). A Poisson substitution model was applied to distance calculations. Branches are labeled by bootstrap values from 10,000 replicates. Bar, number of substitutions per amino acid.
Seven tetracycline resistance genotypes of M. elsdenii have now been detected in these and previous studies (24). M. elsdenii appears to be a commensal “warehouse” for tet(W), recombinant tet(O)-tet(W), and perhaps tet(O) genes in the mammalian intestinal tract. Whether M. elsdenii cells are the site for tet(O) and tet(W) recombination or are the recipients of mosaic tet genes remains unclear (24).
Tetracycline MICs of M. elsdenii strains.
Tetracycline MICs for M. elsdenii strains were determined by the agar dilution method according to NCCLS recommendations (18). All strains carrying tet genes exhibited tetracycline antimicrobial MICs that were 4- to >100-fold greater than those exhibited for strains lacking the genes (Table 2). Of the strains, 11 carrying mosaic tet(OWO) genes consistently exhibited the highest tetracycline MICs (128 to >256 μg/ml) (Table 2). This finding implies that recombinant tet(O)-tet(W) genes provide a selective advantage of increased tetracycline resistance to M. elsdenii cells. An important test of this hypothesis will be to examine the resistance properties of the various tet genes in an isogenic background in, for example, the same M. elsdenii strain.
TABLE 2.
Tetracycline MIC values for M. elsdenii strains
| M. elsdenii strain(s)a | tet genotypeb | MIC (μg/ml)c
|
||
|---|---|---|---|---|
| Tet | Oxytet | Chlortet | ||
| 24-50; 27-50; 33-54 (LC-1T; B159; T81) | No gene | 4-8 | 8-16 | 2-8 |
| Fern | tet(O) | 64 | 64 | 256 |
| 25-50; 29-55 | tet(W) | 32-64 | 64 | 64-128 |
| 24-51; 27-51; 25-51; 31-55 | tet(OW) | 64 | 64-128 | 128-256 |
| 7-11 (2-9) | tet(OWO)-1 | 128 | 256 | >256 |
| 24-53; 27-52; 33-56 (4-13; 7-12; 14-14; 15-5; 19-3; 20-11) | tet(OWO)-2 | 128 | 256->256 | 256->256 |
Strain 7-11 was used as a M. elsdenii tetracycline-resistant reference strain in the assays. Tetracycline MIC values for M. elsdenii strains enclosed by parentheses have been reported previously (26).
Genotypes are based on PCR assay results (Fig. 1; Table 1). Strains 27-51 and 25-51 had tet(OW)-1 and tet(OW)-2 genotypes, respectively, as identified by sequence analysis. Strains 25-50 and 29-55 had tet(W)-1 and tet(W)-2 genotypes, respectively.
Tetracycline (Tet), oxytetracycline (Oxytet), and chlortetracycline (Chlortet) MIC values for NCCLS reference strain Bacteroides fragilis ATCC 25285 (Tcs) were 2, 8, and 4 μg/ml, respectively. For B. thetaiotaomicron ATCC 29741 (Tcr), the MIC value of all three antibiotics was 64 μg/ml.
Both tetracycline-sensitive and tetracycline-resistant M. elsdenii strains were isolated from the same fecal dilutions from every organically raised animal, an indication that both types are present at population levels of 107 to 108 CFU/g of feces. Previously, M. elsdenii strains exhibiting high tetracycline MIC levels and carrying tet(OWO) genes were conservatively estimated at 107 CFU/g of cecal contents of conventionally raised swine regardless of whether or not the animals were fed chlortetracycline (24). Tetracycline-resistant M. elsdenii strains persist at high population levels in swine in the absence of antibiotic use.
Due to the large diversity of uncultivated or difficult-to-culture commensal anaerobes, it has been suggested that one or a few bacterial species might serve as useful indicators in the analysis of reservoir populations (7a). Escherichia coli and Bacteroides and Enterococcus spp. have been used to monitor the flow and persistence of antibiotic resistance determinants within and between intestinal ecosystems (1, 8, 22, 23, 28, 29). M. elsdenii is a common anaerobe in the intestinal tracts of both ruminant and nonruminant mammals, including humans. Currently known intestinal Megasphaera comprise only one species, M. elsdenii. M. elsdenii strains grow rapidly on simple anaerobic culture medium, can be selectively isolated on Me109M agar, and have easily recognizable cell and colony morphologies. M. elsdenii strains are among the most numerous tetracycline-resistant populations in swine intestinal tracts (24). On the basis of these considerations, M. elsdenii offers several attractive features as an indicator or archetype commensal species for monitoring the antibiotic resistance status and reservoir potential of the intestinal microbiota.
Nucleotide sequence accession numbers.
The tet gene sequences of five M. elsdenii strains were determined and have been deposited in GenBank under the following accession numbers: strain Fern, tet(O) (AY485123); strain 25-50, tet(W)-1 (AY485125); strain 29-55, tet(W)-2 (AY485124); strain 27-51, tet(OW)-1 (AY485126); strain 25-51, tet(OW)-2 (AY485122).
Acknowledgments
We acknowledge Sam Humphrey for consistently competent technical support. Brooke Peterson-Burch provided links to Mega2 software and insights regarding computer analysis of Tet(W) phylogeny for which we are grateful. We thank Tom Casey and Brooke Peterson-Burch for helpful, in-depth reviews of the manuscript.
Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
REFERENCES
- 1.Aarestrup, F. M., H. Hasman, L. B. Jensen, M. Moreno, I. A. Herrero, L. Dominguez, M. Finn, and A. Franklin. 2002. Antimicrobial resistance among enterococci from pigs in three European countries. Appl. Environ. Microbiol. 68:4127-4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allison, M. J. 1978. Production of branched-chain volatile fatty acids by certain anaerobic bacteria. Appl. Environ. Microbiol. 35:872-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anonymous. 1969. 5.2.02. AOAC official method 968.49. Chlortetracycline in feeds. J. Assoc. Off. Anal. Chem. 51:750. [Google Scholar]
- 4.Callaway, T. R., K. A. Adams, and J. B. Russell. 1999. The ability of “low G + C gram-positive” ruminal bacteria to resist monensin and counteract potassium depletion. Curr. Microbiol. 39:226-230. [DOI] [PubMed] [Google Scholar]
- 5.Counotte, G. H. M., R. A. Prins, R. H. A. M. Janssen, and M. J. A. deBie. 1981. Role of Megasphaera elsdenii in the fermentation of dl-[2-13C]lactate in the rumen of dairy cattle. Appl. Environ. Microbiol. 42:649-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dennis, S. M., T. G. Nagaraja, and E. E. Bartley. 1981. Effects of lasalocid or monensin on lactate-producing or -using rumen bacteria. J. Anim. Sci. 52:418-426. [DOI] [PubMed] [Google Scholar]
- 7.Hespell, R. B., D. E. Akin, and B. A. Dehority. 1997. Bacteria, fungi, and protozoa of the rumen, p. 59-141. In R. I. Mackie, B. A. White, and R. E. Isaacson (ed.), Gastrointestinal microbes and host interactions, vol. II. Chapman & Hall, New York, N.Y. [Google Scholar]
- 7a.Isaacson, R. E., and M. E. Torrence (ed.). 2002. The role of antibiotics in agriculture. [Online.] American Academy of Microbiology, Washington, D.C. http://www.asm.org/ASM/files/CCLIBRARYFILES/FILENAME/0000000487/AntibioticsAgReport%5BEnglish%5D.pdf. [PubMed]
- 8.Johnsen, P. J., G. S. Simonsen, O. Olsvik, T. Midtvedt, and A. Sundsfjord. 2002. Stability, persistence, and evolution of plasmid-encoded VanA glycopeptide resistance in enterococci in the absence of antibiotic selection in vitro and in gnotobiotic mice. Microb. Drug Resist. 8:161-170. [DOI] [PubMed] [Google Scholar]
- 9.Kim, Y. J., R. H. Liu, J. L. Rychlik, and J. B. Russell. 2002. The enrichment of a ruminal bacterium (Megasphaera elsdenii YJ-4) that produces the trans-10, cis-12 isomer of conjugated linoleic acid. J. Appl. Microbiol. 92:976-982. [DOI] [PubMed] [Google Scholar]
- 10.Klieve, A. V., D. Hennessy, D. Ouwerkerk, R. J. Forster, R. I. Mackie, and G. T. Attwood. 2003. Establishing populations of Megasphaera elsdenii YE 34 and Butyrivibrio fibrisolvens YE 44 in the rumen of cattle fed high grain diets. J. Appl. Microbiol. 95:621-630. [DOI] [PubMed] [Google Scholar]
- 11.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 12.Kung, L., Jr., and A. O. Hession. 1995. Preventing in vitro lactate accumulation in ruminal fermentations by inoculation with Megasphaera elsdenii. J. Anim. Sci. 73:250-256. [DOI] [PubMed] [Google Scholar]
- 13.Ladd, J. N., and D. J. Walker. 1959. The fermentation of lactate and acrylate by the rumen micro-organism LC. Biochem. J. 71:364-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mackie, R. I., F. M. C. Gilchrist, and S. Heath. 1984. An in vivo study of ruminal micro-organisms influencing lactate turnover and its contribution to volatile fatty acid production. J. Agric. Sci. 103:37-51. [Google Scholar]
- 15.Miura, H., M. Horiguchi, and T. Matsumoto. 1980. Nutritional interdependence among rumen bacteria, Bacteroides amylophilus, Megasphaera elsdenii, and Ruminococcus albus. Appl. Environ. Microbiol. 40:294-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miwa, T., H. Esaki, J. Umemori, and T. Hino. 1997. Activity of H+-ATPase in ruminal bacteria with special reference to acid tolerance. Appl. Environ. Microbiol. 63:2155-2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miyazaki, H., T. Hino, and H. Itabashi. 1991. Effects of extracellular pH on the intracellular pH, membrane potential, and growth yield of Megasphaera elsdenii in relation to the influence of monensin, ethanol, and acetate. J. Gen. Appl. Microbiol. 37:415-422. [Google Scholar]
- 18.National Committee for Clinical Laboratory Standards. 1997. Methods for antimicrobial susceptibility testing of anaerobic bacteria; approved standard, 4th ed., vol. 17, no. 22. M11-A4. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 19.Ouwerkerk, D., A. V. Klieve, and R. J. Forster. 2002. Enumeration of Megasphaeraelsdenii in rumen contents by real-time Taq nuclease assay. J. Appl. Microbiol. 92:753-758. [DOI] [PubMed] [Google Scholar]
- 20.Russell, J. B. 1991. Intracellular pH of acid-tolerant ruminal bacteria. Appl. Environ. Microbiol. 57:3383-3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rychlik, J. L., R. LaVera, and J. B. Russell. 2002. Amino acid deamination by ruminal Megasphaera elsdenii strains. Curr. Microbiol. 45:340-345. [DOI] [PubMed] [Google Scholar]
- 22.Shoemaker, N. B., H. Vlamakis, K. Hayes, and A. A. Salyers. 2001. Evidence for extensive resistance gene transfer among Bacteroides spp. and among Bacteroides and other genera in the human colon. Appl. Environ. Microbiol. 67:561-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith, H. W. 1975. Persistence of tetracycline resistance in pig E. coli. Nature 258:628-630. [DOI] [PubMed] [Google Scholar]
- 24.Stanton, T. B., and S. B. Humphrey. 2003. Isolation of tetracycline-resistant Megasphaera elsdenii strains with novel mosaic gene combinations of tet(O) and tet(W) from swine. Appl. Environ. Microbiol. 69:3874-3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stewart, C. S. 1997. Microorganisms in hindgut fermentors, p. 142-186. In R. I. Mackie, B. A. White, and R. E. Isaacson (ed.), Gastrointestinal microbiology II. Gastrointestinal microbes and host interactions, vol. 2. Chapman & Hall, New York, N.Y. [Google Scholar]
- 26.Sugihara, P. T., V. L. Sutter, H. R. Attebery, K. S. Bricknell, and S. M. Finegold. 1974. Isolation of Acidaminococcus fermentans and Megasphaera elsdenii from normal human feces. Appl. Microbiol. 27:274-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsukahara, T., H. Koyama, M. Okada, and K. Ushida. 2002. Stimulation of butyrate production by gluconic acid in batch culture of pig cecal digesta and identification of butyrate-producing bacteria. J. Nutr. 132:2229-2234. [DOI] [PubMed] [Google Scholar]
- 28.Werner, G., I. Klare, and W. Witte. 2002. Molecular analysis of streptogramin resistance in enterococci. Int. J. Med. Microbiol. 292:81-94. [DOI] [PubMed] [Google Scholar]
- 29.Witte, W., I. Klare, and G. Werner. 2002. Molecular ecological studies on spread of antibiotic resistance genes. Anim. Biotechnol. 13:57-70. [DOI] [PubMed] [Google Scholar]