Abstract
Mussel adhesive proteins have been suggested as a basis for environmentally friendly adhesives for use in aqueous conditions and in medicine. However, attempts to produce functional and economical recombinant mussel adhesive proteins (mainly foot protein type 1) in several systems have failed. Here, the cDNA coding for Mytilus galloprovincialis foot protein type 5 (Mgfp-5) was isolated for the first time. Using this cDNA, we produced a recombinant Mgfp-5 fused with a hexahistidine affinity ligand, which was expressed in a soluble form in Escherichia coli and was highly purified using affinity chromatography. The adhesive properties of purified recombinant Mgfp-5 were compared with the commercial extracted mussel adhesive Cell-Tak by investigating adhesion force using atomic force microscopy, material surface coating, and quartz crystal microbalance. Even though further macroscale assays are needed, these microscale assays showed that recombinant Mgfp-5 has significant adhesive ability and may be useful as a bioadhesive in medical or underwater environments.
Mussels, which are a common food throughout the world, have been studied as a potential source of a water-resistant bioadhesive (25, 27). Mussels produce and secrete specialized adhesives that function in water, allowing them to attach themselves in marine environments, which are characterized by salinity, humidity, tides, turbulence, and waves (25). They adhere tightly to surfaces underwater by using the byssus secreted from their foot. The byssus consists of a bundle of threads; at the end of each thread is an adhesive plaque containing a water-resistant glue that enables the plaque to anchor to wet solid surfaces (21). This strong and water-insoluble adhesion has attracted interest for potential use in biotechnological applications. Mussel adhesive proteins are able to form not only permanent and strong but also flexible underwater bonds to substrates such as glass, Teflon, metal, and plastic. Moreover, their biodegradable properties make them environmentally friendly. Mussel adhesive proteins can also be used as medical adhesives since they are nontoxic to the human body and do not impose immunogenicity (6, 11, 31).
The byssus and the adhesive plaque of mussels are composed of several different proteins. Studies of mussel adhesive proteins and their characteristics over 20 years have identified three distinct types of collagenous proteins from the byssus (4, 20, 29) and five distinct types of adhesive plaque proteins (8, 14, 18, 21, 26, 30). The adhesive proteins are involved in adhesion of mussels to wet surfaces (5). Interestingly, every type of mussel adhesive protein contains a high ratio of 3,4-dihydroxyphenyl-l-alanine (DOPA), which is derived from hydroxylation of tyrosine residues (5, 26, 33). Moreover, adhesive proteins that are close to the adhesion interface have a high proportion of DOPA residues (14, 18, 26, 30). It was also reported that mussel adhesive protein analogs without DOPA showed greatly reduced ability for adhesion. DOPA residues also enable mussel adhesive protein molecules to cross-link each other by oxidative conversion to o-quinone (32, 33).
Although natural extraction was initially used to isolate mussel adhesive protein for commercial purposes, this process is labor-intensive and inefficient, requiring around 10,000 mussels for 1 mg of protein (17). Mussel foot protein type 1 (fp-1) is considered a key protein for adhesion of mussels in wet environments (8, 26); it contains ∼13 mol% DOPA and consists of about 80 repeats of a decapeptide consensus sequence. Production of recombinant fp-1 protein has been attempted in several expression systems, including Escherichia coli and yeast (8, 22). However, these attempts have failed to express functional and economical mussel adhesive proteins for several reasons, including a highly biased amino acid composition (5 amino acid types comprised ∼89% of the total amino acids), different codon usage preference between mussel and other expression systems (tRNA utilization problem), and small amount of adhesive produced (16, 17, 22). Thus, successful mass production technologies have not yet been developed for mussel adhesive proteins. However, profitable genetic production of mussel glue might be possible in the near future due to findings over the last two decades, such as identification of several kinds of proteins in mussel adhesion pads, location of each type of mussel adhesive protein, and identification of the most important amino acid residue(s) for adhesion (31, 32, 33).
A recent study reported the identification of mussel foot protein type 5 (fp-5) from the mussel adhesive pad of Mytilus edulis (Mefp-5), and it appears the protein may have a use as a bioadhesive since it has the highest known DOPA content among mussel adhesive proteins (25 to 30 mol%) (30). However, that study did not investigate Mefp-5 adhesion characteristics since natural extraction resulted in very little purified protein. In the present study, we isolated the cDNA of the novel mussel adhesive protein fp-5 from M. galloprovincialis (Mgfp-5) and expressed the protein in E. coli. We found that recombinant Mgfp-5 had significant adhesive properties, suggesting its potential use in aqueous and medical environments.
MATERIALS AND METHODS
Isolation of mgfp-5 cDNA.
Due to the similarity between the two mussel species, we used previously reported sequence information for the M. edulis mefp-5 gene (30) in order to isolate the mgfp-5 sequence from an M. galloprovincialis foot cDNA library (a generous gift from J. Herbert Waite) that was constructed using a ZAP expression vector (Stratagene). To acquire the mgfp-5 sequence minus the signal sequence, PCR was performed with a mixture of 5 μl of M. galloprovincialis foot cDNA library, two mefp-5 primers (forward, 5′-GGCCTGCAGCAGTTCTGAAGAATACAAGGG-3′; reverse, 5′-GTAGATCTATACGCCGGACCAGTGAACAG-3′), and 2.5 U of Taq DNA polymerase (MBI-fermentas) for 30 cycles. The 243-bp partial product was obtained and cloned into a pGEM-T vector (Promega) for amplification and sequencing (Fig. 1A). To obtain the upstream signal sequence region of mgfp-5, nested PCR was executed using M. galloprovincialis foot cDNA library, a T3 promoter primer of the ZAP vector, and an mgfp-5 antisense primer (5′-CTTGTATTTTCCGCTGTTTTT-3′) based on the mgfp-5 partial sequence. About 300 bp of signal was obtained and cloned into a pGEM-T vector for sequencing. To obtain the C-terminal poly(A) tail region, another nested PCR was performed using mgfp-5 sense (5′-AAAAACAGCGGAAAATACAAG-3′) and T7 promoter primers. About 350 bp of signal was obtained and cloned into a pGEM-T vector for sequencing. To confirm the sequence of cloned mgfp-5 cDNA from the M. galloprovincialis foot cDNA library, reverse transcriptase PCR using purified total RNA from M. galloprovincialis foot and subsequent sequencing were performed. M. galloprovincialis mussels approximately 8 cm in shell length were obtained from Juk-do market in Pohang, Korea. After the mussel foot was dissected, total RNA purification was performed using a modified Chomczynski and Sacchi method (3).
FIG. 1.
Sequence alignments for nucleotides (A) and amino acids (B) of Mgfp-5 and Mefp-5. (A) Lowercase letters on the upper line show the mefp-5 nucleotide sequence; capital letters on the lower line indicate a mismatch of the mgfp-5 nucleotide sequence compared to the mefp-5 sequence. Dots indicate an identical mgfp-5 nucleotide compared with mefp-5. Dashes indicate a gap introduced to preserve alignment. (B) Capital letters on the upper line show the Mefp-5 amino acid sequence; capital letters on the lower line indicate a mismatch in the Mgfp-5 amino acid sequence compared to Mefp-5. Dots indicate an identical Mgfp-5 amino acid compared to Mefp-5. Dashes indicate a gap introduced to preserve alignment.
Strains.
E. coli TOP10 [F− mcrA Δ(mrr-hsdRMS-mcrBC) Φ801acZΔM15 Δ lacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG] (Invitrogen) was used for recombinant plasmid construction. E. coli BL21 [F− ompT hsdSB (rB− mB−) gal dcm] (10) was used as a host strain for expressing recombinant Mgfp-5.
Construction of expression vector.
The 243-bp mature mgfp-5 cDNA PCR fragment was inserted into the PstI and EcoRI sites of the pTrcHisA vector (Invitrogen) that contains a hexahistidine (His6) tag at the N terminus, to simplify protein purification, and a trc promoter that is inducible by isopropyl-β-d-thiogalactopyranoside (IPTG). Due to the presence of original sequences from the pTrcHisA vector such as the gene 10 leader, anti-Xpress antibiotic epitope, and enterokinase recognition sequence, the final total length of coding sequence for recombinant protein was 351 bp. The constructed vector was denoted pMDG05.
Cell culture and analytical methods.
For strain construction and protein expression, E. coli cells were grown in Luria-Bertani (LB) medium. The constructed transformant harboring the plasmid was stored at −80°C. Culture experiments were performed with 3 liters LB medium supplemented with 50 μg of ampicillin (Sigma) per ml in a 7-liter bioreactor (BioTron) at 37°C, with shaking at 250 rpm. Cell growth was monitored by measuring the optical density at 600 nm (OD600) using a UV-visible spectrophotometer (UV-1601PC; Shimadzu). When cultures reached an OD600 of 0.7 to 0.8, 1 mM (final concentration) IPTG was added to the culture broth for induction of recombinant Mgfp-5. The samples were centrifuged at a 10,000 × g for 10 min at 4°C, and cell pellets were stored at −80°C for further analyses. Total protein concentration was determined using the Bio-Rad Bradford assay with bovine serum albumin (BSA) as a protein standard.
Purification of recombinant Mgfp-5.
Immobilized-metal affinity chromatography purification was performed using the Acta Prime purification system (Amersham Biosciences) at room temperature at 1 ml per min. Ni-nitrilotriacatate NTA agarose (Qiagen) (10 ml) charged with 10 ml of 0.1 M NiSO4 (Samchun Chemicals) was used as the affinity purification resin. Affinity purification was performed under denaturing conditions. Harvested cell pellets were resuspended in 5 ml of buffer B (8 M urea, 10 mM Tris-HCl, 100 mM sodium phosphate [pH 8.0]) per g (wet weight). Samples were lysed by gentle shaking for 1 h at room temperature, lysates were centrifuged at 14,000 rpm for 20 min at room temperature, and the supernatant was collected for purification. The column was equilibrated with 5 resin volumes of buffer B, after which 10 ml of denatured cell lysate was loaded, and then the column washed with buffer C (8 M urea, 10 mM Tris-HCl, 100 mM sodium phosphate, [pH 6.3]) and buffer D (8 M urea, 10 mM Tris-HCl, 100 mM sodium phosphate [pH 5.9]). Target recombinant Mgfp-5 was eluted with buffer E (8 M urea, 10 mM Tris-HCl, 100 mM sodium phosphate [pH 4.5]). Eluted recombinant Mgfp-5 samples were dialyzed in 5% acetic acid overnight at 4°C, using Spectra/Por molecular porous membrane tubing (Spectrum Laboratories).
SDS-PAGE and Western blot analysis.
Samples were resuspended in protein sample buffer (0.5 M Tris-HCl [pH 6.8], 10% glycerol, 5% sodium dodecyl sulfate [SDS], 5% β-mercaptoethanol, 0.25 % bromophenol blue) and heated to 100°C for 5 min. After centrifugation for 1 min, proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE) (15% [wt/vol] polyacrylamide) and then detected using Coomassie blue staining (Bio-Rad) or silver staining (Bio-Rad) or were Western blotted. For Western blot analysis, proteins were transferred onto a nitrocellulose membrane (Amersham Pharmacia) using a Mini-Trans blot cell (Bio-Rad) and Bjerrum and Schafer-Nielsen transfer buffer (48 mM Tris, 39 mM glycine, 20% methanol) for 30 min at 15 V. Proteins of interest were detected using a monoclonal anti-hexahistidine antibody (1:1,000 [vol/vol]; R&D Systems) and an anti-mouse immunoglobulin G conjugated with alkaline phosphatase (1:1,000 [vol/vol]; Sigma). The membrane was then washed and developed colorimetrically with FAST Red TR/Naphthol AS-MX (4-chloro-2-methylbenzene diazonium/3-hydroxy-2-naphthoic acid 2,4-dimethylanilide phosphate; Sigma). The membrane was scanned, and its image was analyzed using Gel-Pro Analyzer software (Media Cybernetics).
MALDI-TOF mass spectroscopy analysis.
Matrix-assisted laser desorption ionization (MALDI) time-of-flight (TOF) mass spectrometry analysis was performed on a PerSeptive Voyager DE instrument (Perkin-Elmer). Sinapinic acid in 30% acetonitrile-0.1% trifluoroacetic acid was used as the matrix solution. Samples were diluted 1:25 with matrix solution, and 1 μl per plate was spotted onto gold-plated sample plates and evaporated using a vacuum pump. Mass spectra were acquired in positive-ion mode using an accelerating voltage of 25,000 V, grid voltage at 70 to 80%, guidewire voltage at 0.3%, delay time of 200 to 500 ns, and N2 laser power at 1,600 to 1,900 (arbitrary units). Internal calibration was performed using BSA with [M + H]+ at 66.431 and [M + 2H]2+ at 33.216.
Modification of tyrosine residues.
To investigate adhesion, purified recombinant Mgfp-5 that was resolved in 5% acetic acid buffer to prevent auto-oxidation of DOPA residues to o-quinone under basic pH conditions was modified with tyrosinase to convert tyrosine residues into DOPA. Prior to adhesion assays, 1.44 mg of either recombinant Mgfp-5, BSA, or Cell-Tak per ml was modified with 10 Unit of tyrosinase (Sigma) at room temperature for 6 h with shaking. BSA in 5% acetic acid buffer was used as a nonadhesive protein control (2, 16). As a positive control, Cell-Tak is a commercially available naturally extracted M. edulis mussel adhesive protein mixture of fp-1 and fp-2 that already contains DOPA residues in 5% acetic acid buffer.
Investigation of coating of various surfaces.
The ability of recombinant Mgfp-5 to coat the following surfaces was investigated: glass slide, poly(methyl methacrylate) plate, polystyrene plate, commercial silicone-based antifouling agent (SigmaGlide; Sigma Coatings)-coated slide, and aluminum plate. Each material surface was cleaned by washing with distilled water several times and drying with nitrogen gas. A 10-μl drop of 1.44-mg/ml protein solution was added to each material surface and incubated in a humid environment for 12 h at 25°C. After being dried, each surface was washed thoroughly with deionized water for 2 h with shaking. The coating by each protein was visualized using Coomassie blue staining.
Measurement of protein adsorption on a gold surface.
The quartz crystal (Seiko EG & G) used for the quartz crystal microbalance (QCM) experiment was an AT-cut piece of quartz 5 mm in diameter with a basic resonant frequency of 9 MHz. A 5-μl drop of 1.44-mg/ml protein sample was placed onto the gold surface of the quartz crystal and incubated in a humid environment for 12 h at 25°C. After being dried, the gold surface was rinsed thoroughly in deionized water for 2 h with shaking and the remaining deionized water was evaporated using a vacuum chamber. Dried quartz crystal was connected to a quartz crystal analyzer (QCA917; Seiko EG & G), and variations in resonance frequency were measured and converted to mass using the following equation, on the basis that resonant frequency decreases as a function of increasing mass (23, 24):
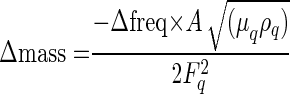 |
where Δmass is mass change, Δfreq is resonant-frequency change, μq is the AT-cut quartz crystal constant (2.947 × 1.011 g/cm/s2), ρq is the quartz crystal density (2.648 g/cm2), Fq is the reference frequency (9.00 MHz), and A is the quartz crystal surface area (0.196 cm2).
Measurement of adhesion force.
The force-distance curve was obtained using atomic force microscopy (AFM) (SPA400; Seiko Instruments) (1, 7, 9). AFM cantilevers were modified by the technique of Ducker et al. (7). The cantilevers used for the present experiments were Olympus oxide-sharpened silicon nitrate probes (Veeco & Seiko Instruments), and the spring constant supplied by the manufacturer was 0.57 or 11 N/m. A glass sphere (Park Science) with a radius of 20 μm was attached under microscopic observation to the tip of the cantilever by using an epoxy resin (Vantico), and the modified cantilever was cured at room temperature for 24 h. It has been shown that intermolecular force measurements by the modified cantilever are not distorted by cured epoxy resin (19). The modified AFM cantilevers were mounted into cells and placed in contact with 10-μl sample solutions (1.44 mg/ml; BSA, Cell-Tak, and recombinant Mgfp-5) on glass slides for 30 min in order to absorb proteins onto the glass bead. After 30 min contact, each modified probe was withdrawn from the protein solution and brought into contact on a new clean glass surface to measure the adhesion force. A force-distance curve was obtained by separation of the modified cantilever from the glass surface.
RESULTS
Elucidation of the mgfp-5 sequence.
Based on the M. edulis mefp-5 sequence (30), we designed primers and performed PCR in order to isolate the mgfp-5 gene from the M. galloprovincialis foot cDNA library. A positive signal of about 230 bp was obtained after PCR, and sequencing and alignment analysis indicated that the PCR fragment had almost the same sequence as mefp-5. To obtain the complete mgfp-5 sequence, including the signal sequence and C-terminal region, separate nested PCRs were performed using T3 and mgfp-5 antisense primers and mgfp-5 sense and T7 primers. Almost the full sequence of cloned mgfp-5 cDNA was elucidated, except for a few base pairs of signal sequence. We confirmed the cloned mgfp-5 sequence from the M. galloprovincialis foot cDNA library by using reverse transcriptase PCR on isolated total RNA from the M. galloprovincialis foot. Sequence alignment analysis showed that the cloned mgfp-5 was 94% homologous with mefp-5 (minus the signal sequence) (Fig. 1A). When we converted the DNA sequence to the amino acid sequence, we found that Mgfp-5 almost perfectly matched Mefp-5, with the exceptions being a substitution of alanine for threonine at residue 14 and two inserted amino acid residues (lysine at 68 and glycine at 73) (Fig. 1B). Interestingly, among the total of 76 Mgfp-5 residues, 5 were histidine, and each was adjacent to a tyrosine residue.
Expression and purification of recombinant Mgfp-5.
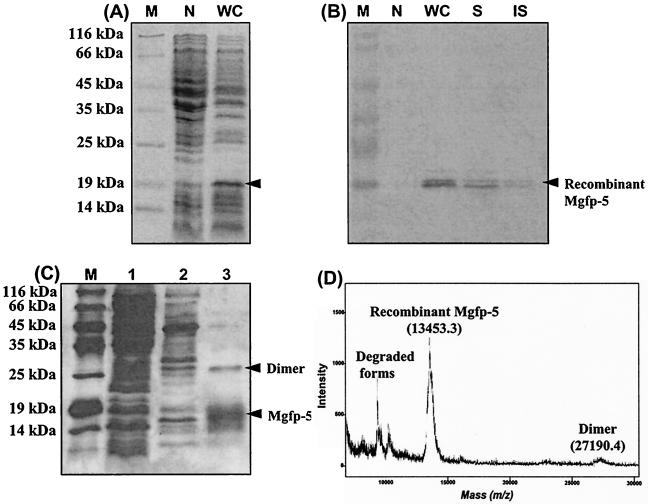
To express recombinant Mgfp-5 in E. coli, mgfp-5 cDNA was cloned into an expression vector containing a His6 tag sequence and an IPTG-inducible trc promoter. Recombinant Mgfp-5 was successfully expressed (∼40 mg/liter; ∼10% of total soluble protein) in E. coli culture by using a 7-liter bioreactor with a 3-liter working volume. The apparent molecular mass of expressed recombinant Mgfp-5 was about 18 kDa from SDS-PAGE (Fig. 2A), which is larger than the 13.5 kDa calculated from the amino acid sequence. Recombinant Mgfp-5 was separated into soluble and insoluble fractions, and Western blot analysis showed that the majority of the recombinant protein was soluble (Fig. 2B).
FIG. 2.
(A and B) SDS-PAGE with Coomassia blue staining (A) and Western blot analysis (B) of recombinant Mgfp-5 from E. coli BL21. Lanes: M, protein molecular mass marker; N, whole-cell sample of the BL21 strain that harbors the parent vector pTrcHisA (negative control); WC, whole-cell sample; S, soluble supernatant fraction; IS, insoluble cell debris fraction. Recombinant cells were cultured in LB medium at 37°C with shaking at 250 rpm. (C) SDS-PAGE analysis, with silver staining, of immobilized metal affinity purification fractions. Lanes: M, protein molecular mass marker; 1, flowthrough fraction; 2, washout fraction; 3, eluted fraction. (D) MALDI-TOF mass spectrometry analysis of purified recombinant Mgfp-5. We used 15% polyacrylamide gels for all analyses. Monoclonal anti-hexahistidine antibody was used for the Western blot analysis.
It is essential that recombinant Mgfp-5 be purified to investigate its adhesive ability. Since expressed recombinant Mgfp-5 had a His6 tag, purification was performed by immobilized-metal affinity chromatography. Recombinant Mgfp-5 was purified to above 95% by one-step affinity purification (Fig. 2C). However, due to the intrinsically adhesive properties of Mgfp-5, the majority of protein was attached to the chromatographic resin, resulting in a final purification yield of ∼7%. Amino acid composition analysis of purified recombinant Mgfp-5 showed that it almost matched the predicted composition of Mgfp-5 (data not shown). MALDI-TOF mass spectrometry analysis showed that the actual molecular mass of recombinant Mgfp-5 was identical to the calculated mass of ∼13.5 kDa (Fig. 2D). This analysis also showed that some recombinant Mgfp-5 existed as a dimer and that some had been degraded. In summary, Western blot, amino acid composition, and MALDI-TOF MS analyses confirmed that the purified protein was recombinant Mgfp-5.
Adhesive ability of recombinant Mgfp-5.
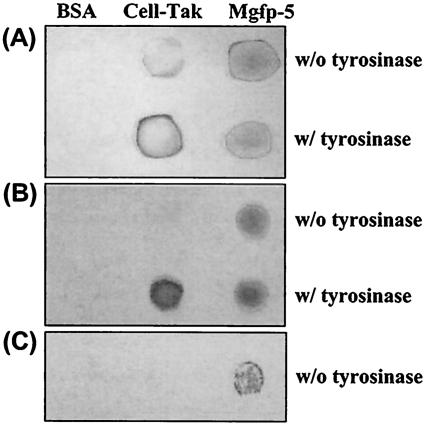
To investigate adhesion of recombinant Mgfp-5, we performed a comparative study with BSA as a negative control and Cell-Tak as a positive control, with or without modification of tyrosines to DOPA residues and finally to o-quinone residues by tyrosinase treatment (tyrosinase catalyzes two steps: the conversion of tyrosine to DOPA, followed by the conversion of DOPA to o-quinone). To prevent auto-oxidation of DOPA residues to o-quinone and formation of intramolecular cross-linking in basic-pH environments, purified recombinant Mgfp-5 proteins were freeze-dried and resolved in 5% acetic acid buffer before being subjected to tyrosinase treatment. Cell-Tak is a commercially available naturally extracted M. edulis adhesive-protein mixture of fp-1 and fp-2 that contains DOPA residues in 5% acetic acid buffer. We examined adsorption and coating by placing drops of protein solutions (10 μl of 1.44 mg/ml) on several material surfaces, namely, a glass slide, a poly(methyl methacrylate) plate, and a commercial silicone-based antifouling agent-coated slide (Fig. 3). We also examined coating to a polystyrene plate and an aluminum plate (data not shown). Slides were incubated for 12 h at 25°C, washed with deionized water for 2 h, and Coomassie blue-stained to visualize the remaining protein. We found that recombinant Mgfp-5 generally remained on all tested substrates while BSA was totally washed away. Moreover, recombinant Mgfp-5 showed remarkable coating ability even when left unmodified (no tyrosinase treatment). Interestingly, recombinant Mgfp-5 coated the commercial antifouling agent-coated surface while Cell-Tak did not (Fig. 3C). These coating experiments indicate that recombinant Mgfp-5 has high adsorption or adhesive ability on many material surfaces.
FIG. 3.
Recombinant Mgfp-5 coating of a glass slide (A), a poly(methyl methacrylate) plate (B), and a commercial silicone-based antifouling agent SigmaGlide-coated slide (C). BSA was used as a negative control, and commercial Cell-Tak was used as a positive control. Modification of each protein sample was performed using 10 U of tyrosinase per μl for 6 h at room temperature. A 10-μl drop of 1.44-mg/ml protein sample was placed on each surface, the surfaces were incubated for 12 h at 25 °C, and the slides were washed with deionized water for 2 h. Coated proteins were visualized using Coomassie blue staining.
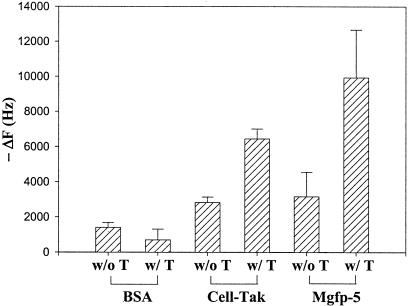
A QCM was employed to quantify recombinant Mgfp-5 adsorption. On a QCM, the frequency of the quartz crystal resonance varies with mass changes on the gold surface of the crystal. Drops of protein solution (5 μl of 1.44 mg/ml) were placed on the gold surface, which was incubated for 12 h at 25 °C and washed with deionized water for 2 h, and the weights were then measured. We found that recombinant Mgfp-5 and Cell-Tak showed high-frequency changes compared to BSA, indicating that the former two remained on the gold surface of the quartz crystal (Fig. 4). Both tyrosinase-treated recombinant Mgfp-5 and Cell-Tak showed much higher-frequency changes than did nontreated samples. Since a decrease in the resonance frequency by 1 kHz is proportional to an increase in mass of 1.068 ng (23, 24), we found that the modified recombinant Mgfp-5 mass was measured as 10.533 μg, which was greater than the initial mass of modified recombinant Mgfp-5 placed on the slide (∼7.2 μg by the Bradford assay).
FIG. 4.
QCM analysis of absorption of recombinant Mgfp-5 to a gold surface. BSA was used as a negative control, and commercial Cell-Tak was used as a positive control. T in the sample legends indicates that protein was modified by treatment with 10 U of tyrosinase per μl for 6 h at room temperature. A 5-μl drop of 1.44-mg/ml protein sample was placed on the gold surface of a quartz crystal, and the crystal was incubated for 12 h at 25°C and washed with deionized water for 2 h. Variations in quartz crystal resonance frequency were measured using a quartz crystal analyzer. Each value and error bar represents the mean of two independent experiments and its standard deviation.
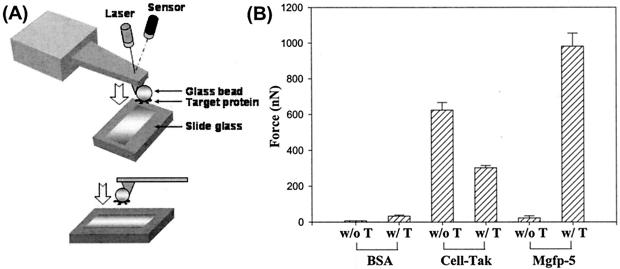
The above method measures adsorption to one surface only; therefore, we employed AFM to measure the adhesion force between two surfaces. We modified the AFM cantilever by attaching a bead (∼20-μm radius and of the same material as the contact surface, i.e., glass) to the tip (Fig. 5A) (1, 7, 9). We used a low-spring-constant cantilever (0.57 N/m) at first, but since the results were out of the deflection range due to the high binding affinity of recombinant Mgfp-5 protein for the substrate (data not shown), we then used a stiffer cantilever (11 N/m). These studies showed the adhesion force of tyrosinase-treated recombinant Mgfp-5 was much higher (∼981 nN) than those of modified BSA (∼32 nN) and non-treated Cell-Tak (∼624 nN) (Fig. 5B). Interestingly, unmodified recombinant Mgfp-5 showed a very low adhesion force (∼22 nN) and tyrosinase-treated Cell-Tak revealed a decrease in adhesion force (∼302 nN).
FIG. 5.
Measurement of the adhesion force of recombinant Mgfp-5 by using modified AFM analysis. (A) Modification of the AFM cantilever. A glass sphere with a radius of 20 μm was attached under microscopic observation to the cantilever tip with epoxy resin, and the modified cantilevers were cured at room temperature for 24 h. (B) AFM analysis. BSA was used as a negative control, and commercial Cell-Tak was used as a positive control. T in the sample legends indicates that the protein was modified by treatment with 10 U of tyrosinase per μl for 6 h at room temperature. The spring constant of the AFM cantilever supplied by the manufacturer was 11 N/m. The modified AFM cantilevers were placed in contact with 10-μl sample solutions (1.44 mg/ml) on a slide glass for 30 min, and the force-distance curve was obtained by separation of the cantilever from the glass surface. Each value and error bar represents the mean of measurements from five different pulls and its standard deviation.
DISCUSSION
We cloned and expressed the novel mussel adhesive protein Mgfp-5 in a soluble form in E. coli. Interestingly, growth of E. coli expressing recombinant Mgfp-5 almost stopped for several hours after IPTG induction, after which the cells started growing again, with a slight proteolytic degradation of expressed recombinant Mgfp-5 (data not shown), suggesting that recombinant Mgfp-5 might be harmful to E. coli. Expressed recombinant Mgfp-5 showed somewhat similar properties reported previously for other mussel adhesive proteins, such as a higher apparent molecular mass on SDS-PAGE analysis and strong binding to chromatographic resin. Expressed recombinant Mgfp-5 showed an apparent molecular mass of ∼18 kDa on SDS-PAGE, while MALDI-TOF mass spectrometry analysis of purified recombinant Mgfp-5 showed that it had exactly the same mass as predicted from the sequence (∼13.5 kDa) (Fig. 2B). Similar findings were reported for Mefp-3 (18) and barnacle cement proteins (15). The difference in apparent and actual mass of recombinant Mgfp-5 might result from the protein being basic (the calculated pI value was 9.3), since proteins with higher pI values (i.e., basic proteins) tend to bind more SDS molecules, which would increase their molecular mass.
We found that yields of purified recombinant Mgfp-5 from soluble supernatants were poor under native conditions when using IMAC (∼1.5%) and that this was due to strong binding or adsorption to the chromatographic resin (data not shown). Therefore, we investigated the purification of recombinant Mgfp-5 under denaturing conditions. Surprisingly, we found that denatured purified recombinant Mgfp-5 showed similar adhesion to its nondenatured counterpart (data not shown). This might be due to the peculiar linear structure of fp-5 (unpublished data). The relationship between structure and adhesion characteristics of the fp-5 adhesive protein is still unknown, and further investigation is needed. Even though chromatographic purification of recombinant Mgfp-5 was somewhat enhanced under denaturing conditions, most recombinant Mgfp-5 proteins strongly bound or adsorbed to the resin. Thus, despite its high purity (above 95%), we could not purify more than 7% of the total target protein (which represented approximately 2.6 mg per liter of fermentation volume). It appears that nonchromatographic separation and purification methods will be needed for recombinant Mgfp-5, and these are currently under development in our laboratory.
Due to insufficient amounts of purified protein, macroscale adhesion measurements for recombinant Mgfp-5 could not be attempted. Therefore, we used several adhesion test methods that are suitable for small quantities of sample protein, such as AFM, QCM, and surface-coating tests (31). These microscale adhesion measurements (Fig. 3 to 5) showed that recombinant Mgfp-5 had better binding or adsorption properties than did the natural adhesive protein mixture Cell-Tak. Even though the affinities of binding or adsorption to several kinds of surfaces might not show a direct correlation to adhesive ability, they can surely demonstrate the potential of recombinant Mgfp-5 as an adhesive. AFM with modified cantilever might be the best microscale method to characterize adhesion strength, and so further modifications of the AFM adhesion testing method are under development to obtain more precise and reproducible results.
The adhesive bonding strength of DOPA-containing synthetic polymers is reported to be almost linearly correlated with the content of DOPA (32, 33). Moreover, DOPA in mussel adhesive proteins serves plays key roles in adhesion: formation of interfacial complexes with substratum and cross-linking among proteins (28, 31). Posttranslational modification of tyrosine to DOPA occurs at the highest levels in proteins located near the adhesion plaque-substratum interface. Adhesion mechanisms involving DOPA are not yet fully understood in relation to mussel adhesive protein (5). Adhesive protein fp-5 is located at the interface between the mussel adhesive plaque and the substratum and also has the highest DOPA content known for fp molecules (25 to 30 mol%, compared to ∼13 mol% for fp-1) (30, 31). Consistent with these observations, the recombinant Mgfp-5 in the present study (which had a tyrosine content of ∼17 mol%, lower than that of ∼26 mol% for natural Mgfp-5) showed high adsorption ability in adhesion test experiments (note that the DOPA percentage is usually lower than the tyrosine percentage due to incomplete modification efficiency). We found that unmodified recombinant Mgfp-5 showed very low adhesion ability in the AFM experiment (Fig. 5). Lack of DOPA residues might cause this result, which has consistency with the previous researches for the role of DOPA in the adhesion mechanism (32, 33). QCM tests for recombinant Mgfp-5 also showed increased adsorption ability by tyrosinase treatment (Fig. 4). We used another method to show tyrosinase activity through nitroblue tetrazolium staining for DOPA residues, and we found that recombinant Mgfp-5 was not stained before tyrosine treatment but was stained after tyrosinase treatment (data not shown). Therefore, we confirmed that tyrosinase treatment improved the adhesion or adsorption ability of recombinant Mgfp-5 by modification of tyrosine to DOPA.
In QCM analysis, resonance frequency changes reflect mass changes, and in the present study the final mass of recombinant Mgfp-5 was higher than the mass initially placed on the slide. This increase in mass might be due to trapping of water molecules in the protein and on the gold surface (13) and/or to the different measurement methods used to measure mass before and after application, i.e., the Bradford assay and QCM, respectively.
The tip of the AFM cantilever can be easily deformed on contact with the protein or a surface, and its area of contact is unknown and varies with each trial. Moreover, it is difficult to determine whether the force being measured derives from interaction of the protein with the AFM tip or with the material surface due to the difference in composition between the cantilever (silicon nitrate) and the contact surface (glass slide in this work). Therefore, by attaching a bead of the same material as the contact surface (glass in this case) to the AFM tip, we can define the contact area and prevent problems associated with deformation of the tip. Also, we can measure the binding force that is generated solely by interaction between the protein and the target surface.
Cell-Tak can be used without modification since it is a naturally extracted adhesive protein mixture from mussel and contains modified DOPA residues. However, tyrosinase-treated Cell-Tak showed better adsorption ability than did nontreated Cell-Tak in coating (Fig. 3) and QCM (Fig. 4) experiments. This observation might indicate that not all tyrosine residues of mussel Cell-Tak are naturally modified to DOPA residues (12). Even though tyrosinase treatment improved the adsorption ability of Cell-Tak, it caused a decrease in adhesion ability in AFM tests (Fig. 5). This might be due to oxidation of DOPA to o-quinone residues and formation of intramolecular cross-linking in Cell-Tak (12). Since Cell-Tak already contains enough DOPA residues and since tyrosinase catalyzes the oxidation of DOPA, the o-quinone residues that are formed may cause stronger adsorption on the surface by formation of intramolecular cross-linking (Fig. 3 and 4). In contrast, this intramolecular cross-linking may impose weaker adhesion bonding of Cell-Tak on the modified AFM cantilever with the glass slide by formation of a rigid protein surface (Fig. 5). BSA was expected to have no adhesive properties and was therefore used as a negative control in the present study. However, BSA also showed very low-level adsorption (Fig. 4 and 5B), which may be because it contains 3.5% tyrosine residues (among 607 total amino acids) and/or was tyrosinase treated.
The existence of several phosphorylations to serine residues in natural Mefp-5 has been reported, and it was discussed that the negative charges of phosphate may allow it to interact with various kinds of metal ions on the substratum (30). Therefore, modification of recombinant Mgfp-5 with phosphatase might improve the adhesion or adsorption ability. This is under investigation.
In conclusion, we cloned and expressed soluble recombinant Mgfp-5 and found that this novel recombinant mussel adhesive protein had significant adhesive ability. These findings suggest that recombinant Mgfp-5 could be used as a commercial bioadhesive for medical purposes or in underwater environments.
Acknowledgments
This work was supported by National R&D Project for Useful Materials from Marine Organisms from the Ministry of Maritime Affairs and Fisheries, National R&D Project for Nano Science and Technology from the Ministry of Science and Technology, the Brain Korea 21 Program from the Ministry of Education, and POSTECH Biotech Center.
We thank J. Herbert Waite (UCSB) for providing the M. galloprovincialis foot cDNA library and for critical reviews and comments. We also thank Jong Hoon Hahn (POSTECH), Kil Won Cho (POSTECH), Hyun Jin Park (POSTECH), and Young Soo Kim (POSTECH) for help with AFM and QCM analyses, and we thank Namjun Cho (KUT) and Yong Hwan Kim (KRICT) for helpful comments about adhesion.
REFERENCES
- 1.Biggs, S. 1995. Steric and bridging forces between surfaces bearing adsorbed polymer: an atomic force microscopy study. Langmuir 11:156-162. [Google Scholar]
- 2.Bowen, W. R., N. Hilal, R. W. Lovitt, and C. J. Wright. 1998. Direct measurement of interactions between adsorbed protein layers using an atomic force microscope. J. Colloid Interf. Sci. 197:348-352. [DOI] [PubMed] [Google Scholar]
- 3.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid quanidium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 4.Corne, K. J., X. Qin, and J. H. Waite. 1997. Extensible collagen in mussel byssus: a natural block polymer. Science 277:1830-1832. [DOI] [PubMed] [Google Scholar]
- 5.Deming, T. J. 1999. Mussel byssus and biomolecular materials. Curr. Opin. Chem. Biol. 3:100-105. [DOI] [PubMed] [Google Scholar]
- 6.Dove, J., and P. Sheridan. 1986. Adhesive protein from mussels: possibilities for dentistry, medicine, and industry. J. Am. Dent. Assoc. 112:879. [DOI] [PubMed] [Google Scholar]
- 7.Ducker, W. A., T. J. Senden, and R. M. Pashley. 1991. Direct measurement of colloidal forces using an atomic force microscope. Nature 353:239-241. [Google Scholar]
- 8.Filpula, D. R., S. M. Lee, R. P. Link, S. L. Strausberg, and R. L. Strausberg. 1990. Structural and functional repetition in a marine mussel adhesive protein. Biotechnol. Prog. 6:171-177. [DOI] [PubMed] [Google Scholar]
- 9.Frank, B. P., and G. Belfort. 2002. Adhesion of Mytilus edulis foot protein 1 on silica: ion effects on biofouling. Biotechnol. Prog. 18:580-586. [DOI] [PubMed] [Google Scholar]
- 10.Grodberg, J., and J. J. Dunn. 1988. ompT encodes the Escherichia coli outer membrane protease that cleaves T7 RNA polymerase during purification. J. Bacteriol. 170:1245-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Green, K. 1995. Mussel adhesive protein, p. 19-27. In D. H. Sierra and R. Saltz (ed.), Surgical adhesives and sealants: current technology and applications. Technomic, Lancaster, Pa.
- 12.Hansen, D. C., S. G. Corcoran, and J. H. Waite. 1998. Enzymatic tempering of a mussel adhesive protein film. Langmuir 14:1139-1147. [Google Scholar]
- 13.Höök, F., B. Kaeemo, T. Nylander, C. Fant, K. Sott, and H. Elwing. 2001. Variation in coupled water, viscoelastic properties and film thickness of a mefp-1 protein film during adsorption and cross-linking: a quartz crystal microbalance with dissipation monitoring, ellipsometry and surface plasmon resonance study. Anal. Chem. 74:5796-5804. [DOI] [PubMed] [Google Scholar]
- 14.Inoue, K., Y. Takeuchi, D. Miki, and S. Odo. 1995. Mussel adhesive plaque proteins is a novel member of epidermal growth factor-like gene family. J. Biol. Chem. 270:6698-6701. [DOI] [PubMed] [Google Scholar]
- 15.Kamino, K., K. Inoue, T. Maruyama, N. Takamatsu, S. Harayama, and Y. Shizuri. 2000. Barnacle cement proteins: importance of disulfide bonds in their insolubility. J. Biol. Chem. 275:27360-27365. [DOI] [PubMed] [Google Scholar]
- 16.Kitamura, M., K. Kawakami, N. Nakamuram, K. Tsumoto, H. Uchiyama, Y. Ueda, I. Kumagai, and T. Nakaya. 1999. Expression of a model peptide of a marine mussel adhesive protein in Escherichia coli and characterization of its structural and functional properties. J. Polym. Sci. Ser. A 37:729-736. [Google Scholar]
- 17.Morgan, D. 1990. Two firms race to derive profits from mussels' glue. Scientist 4:1-6. [Google Scholar]
- 18.Papov, V., T. Diamond, K. Biemann, and J. H. Waite. 1995. Hydroxyarginine-containing polyphenolic proteins in the adhesive plaques of the marine mussel Mytilus edulis. J. Biol. Chem. 270:20183-20192. [DOI] [PubMed] [Google Scholar]
- 19.Pincet, F., E. Perez, and J. Wolfe. 1995. Does glue contaminate the surface force apparatus? Langmuir 11:373-374. [Google Scholar]
- 20.Qin, X., K. J. Corne, and J. H. Waite. 1997. Tough tendons. Mussel byssus has collagen with silk-like domains. J. Biol. Chem. 272:32623-32627. [DOI] [PubMed] [Google Scholar]
- 21.Rzepecki, L. M., K. M. Hansen, and J. H. Waite. 1992. Characterization of a cystein-rich polyphenolic protein family from Mytilus edulis. Biol. Bull. 183:123-137. [DOI] [PubMed] [Google Scholar]
- 22.Salerno, A. J., and I. Goldberg. 1993. Cloning, expression, and characterization of a synthetic analog to the biadhesive precursor protein of the sea Mussel Mytilus edulis. Appl. Microbiol. Biotechnol. 58:209-214. [DOI] [PubMed] [Google Scholar]
- 23.Sauerbrey, G. 1959. Verwendung von Schwingquarzen zur Wägung dünner Schichten und zur Mikrowägung. Z. Phys. 155:206. [Google Scholar]
- 24.Thomson, M., A. L. Kipling, W. C. Duncan-Hewitt, L. V. G. Rajaković, and B. A. Čavic- Vlasak. 1991. Thickness- shear-mode acoustic wave sensors in the liquid phase: a review. Analyst 116:881-889. [Google Scholar]
- 25.Waite, J. H. 1983. Adhesion in byssally attached bivalves. Biol. Rev. 58:209-231. [Google Scholar]
- 26.Waite, J. H. 1983. Evidence for a repeating 3,4-dihydroxyphenylalanine and hydroxyproline- containing decapeptide in the adhesive protein of the mussel, Mytilus edulis. Biol. Chem. 258:2911-2915. [PubMed] [Google Scholar]
- 27.Waite, J. H. 1987. Nature's underwater adhesive specialist. Int. J. Adhesion Adhesives 7:9-14. [Google Scholar]
- 28.Waite, J. H. 1992. Formation of mussel byssus: anatomy of a natural manufacturing process. Results Probl. Cell Differ. 19:27-54. [DOI] [PubMed] [Google Scholar]
- 29.Waite, J. H., X. Qin, and K. J. Corne. 1998. The peculiar collagens of mussel byssus. Matrix Biol. 17:93-106. [DOI] [PubMed] [Google Scholar]
- 30.Waite, J. H., and X. Qin. 2001. Polyphosphoprotein from the adhesive pads of Mytilus edulis. Biochemistry 40:2887-2893. [DOI] [PubMed] [Google Scholar]
- 31.Waite, J. H. 2002. Adhesion á la moule. Integr. Comp. Biol. 42:1172-1180. [DOI] [PubMed] [Google Scholar]
- 32.Yu, M., and T. J. Deming. 1998. Synthetic polypeptide mimics of marine adhesive. Macromolecules 31:4739-4745. [DOI] [PubMed] [Google Scholar]
- 33.Yu, M., J. Hwang, and T. J. Deming. 1999. Role of l-3,4-dihydroxylphenylalanine in mussel adhesive proteins. J. Am. Chem. Soc. 121:5825-5826. [Google Scholar]