Abstract
Toluene-o-xylene monooxygenase (ToMO) from Pseudomonas stutzeri OX1 oxidizes toluene to 3- and 4-methylcatechol and oxidizes benzene to form phenol; in this study ToMO was found to also form catechol and 1,2,3-trihydroxybenzene (1,2,3-THB) from phenol. To synthesize novel dihydroxy and trihydroxy derivatives of benzene and toluene, DNA shuffling of the alpha-hydroxylase fragment of ToMO (TouA) and saturation mutagenesis of the TouA active site residues I100, Q141, T201, and F205 were used to generate random mutants. The mutants were initially identified by screening with a rapid agar plate assay and then were examined further by high-performance liquid chromatography and gas chromatography. Several regiospecific mutants with high rates of activity were identified; for example, Escherichia coli TG1/pBS(Kan)ToMO expressing the F205G TouA saturation mutagenesis variant formed 4-methylresorcinol (0.78 nmol/min/mg of protein), 3-methylcatechol (0.25 nmol/min/mg of protein), and methylhydroquinone (0.088 nmol/min/mg of protein) from o-cresol, whereas wild-type ToMO formed only 3-methylcatechol (1.1 nmol/min/mg of protein). From o-cresol, the I100Q saturation mutagenesis mutant and the M180T/E284G DNA shuffling mutant formed methylhydroquinone (0.50 and 0.19 nmol/min/mg of protein, respectively) and 3-methylcatechol (0.49 and 1.5 nmol/min/mg of protein, respectively). The F205G mutant formed catechol (0.52 nmol/min/mg of protein), resorcinol (0.090 nmol/min/mg of protein), and hydroquinone (0.070 nmol/min/mg of protein) from phenol, whereas wild-type ToMO formed only catechol (1.5 nmol/min/mg of protein). Both the I100Q mutant and the M180T/E284G mutant formed hydroquinone (1.2 and 0.040 nmol/min/mg of protein, respectively) and catechol (0.28 and 2.0 nmol/min/mg of protein, respectively) from phenol. Dihydroxybenzenes were further oxidized to trihydroxybenzenes with different regiospecificities; for example, the I100Q mutant formed 1,2,4-THB from catechol, whereas wild-type ToMO formed 1,2,3-THB (pyrogallol). Regiospecific oxidation of the natural substrate toluene was also checked; for example, the I100Q mutant formed 22% o-cresol, 44% m-cresol, and 34% p-cresol, whereas wild-type ToMO formed 32% o-cresol, 21% m-cresol, and 47% p-cresol.
Di- and trihydroxy aromatic compounds are important industrial chemicals that have many applications, as shown by worldwide production of catechol, resorcinol, and hydroquinone at a level of 110,000 tons/year (22). Catechol is used as an intermediate in the food, pharmaceutical, and agrochemical industries (22), and hydroquinone is used in photography, in cosmetics, and in both medical and industrial X-ray films (11, 22). Resorcinol and its derivatives are used to inhibit rust in paints, to regulate plant growth, and as capacitor electrolytes (8, 15, 38). Production of 4-methylresorcinol is uncommon, and prices can exceed $200,000/kg (http://www.apinchemicals.com). Methylhydroquinone has recently been reported to be used in the synthesis of (±)-helibisabonol A and puraquinonic acid, which are precursors of agrochemical herbicides and antileukemia drugs, respectively (14, 16). 1,2,3-Trihydroxybenzene (1,2,3-THB) (pyrogallol), the first synthetic dye for hair, is primarily used as a modifier in oxidation dyes (36) and as a pharmaceutical intermediate, and it has also been used as a topical antipsoriatic (22). Hydroxyhydroquinone (1,2,4-THB) has been used in dyes and as a corrosion inhibitor (22).
Some of these compounds cannot be easily synthesized chemically, and the traditional chemical processes are often lengthy and require expensive starting materials (26). Direct microbial synthesis of such compounds from inexpensive substrates might provide a more cost-effective and more environmentally benign approach, and biocatalysis is likely to account for 30% of the chemical business by 2050 (37). In this paper, we report the formation of valuable hydroxylated benzenes from benzene, toluene, phenol, o-cresol, m-cresol, p-cresol, catechol, hydroquinone, and resorcinol with toluene-o-xylene monooxygenase (ToMO) from Pseudomonas stutzeri OX1.
ToMO hydroxylates toluene in the ortho, meta, and para positions, as well as o-xylene in both the 3 and 4 positions, and it oxidizes many substrates, including o-xylene, m-xylene, p-xylene, toluene, benzene, ethyl benzene, styrene, naphthalene (2), and trichloroethylene (TCE) (6). It is the only known oxygenase which attacks tetrachloroethylene (30). The six genes coding for ToMO are touABE (three-component hydroxylase with two catalytic oxygen-bridged dinuclear centers, A2B2E2), touC (ferredoxin), touD (mediating protein), and touF (NADH-ferredoxin oxidoreductase) (2, 4). ToMO TouA (499 amino acids) has the highest levels of amino acid identity to the hydroxylase (TbuA1) of toluene 3-monooxygenase (T3MO) of Pseudomonas pickettii PKO1 (68%) and the hydroxylase (TmoA) of toluene 4-monooxygenase (T4MO) of Pseudomonas mendocina KR1 (66.8%) (3), but these molecules are distinct enzymes based on differences in the regiospecific oxidation of toluene.
The importance of residue V106 as an active residue in toluene monooxygenases resulting from directed evolution of toluene ortho-monooxygenase (TOM) of Burkholderia cepacia G4 has been reported previously (5). This beneficial mutation resulted in a twofold increase in the initial TCE degradation rate and a sixfold increase in naphthalene oxidation. This residue corresponds to I100 of the alpha-subunit TouA of the hydroxylase in ToMO.
The methane monooxygenase (MMO) active site residues have been identified by X-ray crystallography (9, 27, 28), and by comparison to MMO, the active site residues for T4MO, T3MO, and toluene 2-monooxygenase from Pseudomonas sp. strain JS150 have been predicted by Pikus et al. (25). The T201 residue encoded by tmoA in T4MO has been studied by saturation mutagenesis, and T4MO Q141C, Q141V, I180F, and F205I mutants with mutations in tmoA have been studied previously by using site-directed mutagenesis (24, 25). For oxidation of m-xylene by the Q141C T4MO mutant, 3-methylbenzyl alcohol formation increased sixfold from 2.2% to 11.7%, and for p-xylene oxidation the product distribution completely switched to 2,5-dimethylphenol (78%) from 4-methylbenzyl alcohol (22%). For hydroxylation of toluene by the F205I T4MO mutant, the percentage of m-cresol formation increased fivefold from 2.8% to 14.5% (25). The T201F T4MO mutant with a mutation in tmoA exhibited a large shift in the product distribution and also formed 10-fold more benzyl alcohol from toluene (24). Catechol or THB formation by these mutants was not studied as these reactions were not known previously.
Here, DNA shuffling of touA and saturation mutagenesis of TouA at residues I100, Q141, T201, and F205 in ToMO were used to study the regiospecific hydroxylation of toluene and benzene by ToMO, as well as to enhance the synthesis of catechols, methylcatechols, and THB. It was discovered that ToMO forms catechol from phenol, as well as THB from catechol, hydroquinone, and resorcinol. Overall, five new reactions were found for wild-type ToMO, and protein engineering was used to construct mutants that synthesize eight novel, industrially significant products.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and SDS-PAGE.
Escherichia coli strain TG1 (10) was used as the host with pBS(Kan)ToMO and variants of this plasmid. Cells were initially streaked from −80°C glycerol stocks on Luria-Bertani (LB) agar plates (32) containing 100 μg of kanamycin per ml and incubated at 37°C. After growth on LB agar plates, cells were cultured from a fresh single colony in LB medium (32) supplemented with 100 μg of kanamycin per ml at 37°C with shaking at 250 rpm (New Brunswick Scientific Co., Edison, N.J.). The relative expression of the touA loci from E. coli TG1/pBS(Kan)ToMO was evaluated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (32) by using a 12% Tris-HCl gel both with and without 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) (Fisher Scientific. Co., Fairlawn, N.J.).
Chemicals.
Benzene, toluene, phenol, p-cresol, catechol, 3-methylcatechol, 4-methylcatechol, and methylhydroquinone were purchased from Fisher Scientific Co.; resorcinol, 1,2,3-THB, 1,2,4-THB, o-cresol, and m-cresol were purchased from Sigma Chemical Co. (St. Louis, Mo.); hydroquinone, benzyl alcohol, 2-hydroxybenzyl alcohol, 3-hydroxybenzyl alcohol, 4-hydroxybenzyl alcohol, 2-methylresorcinol, and 5-methylresorcinol were obtained from Acros Organics (Morris Plains, N.J.); and 4-methylresorcinol was obtained from Apin Chemicals (Abingdon, United Kingdom).
Construction of pBS(Kan)ToMO.
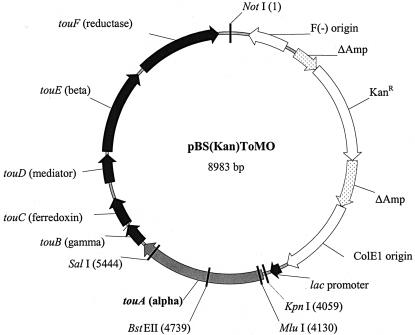
To create pBS(Kan)ToMO for constitutive expression of ToMO, the touABCDEF locus was PCR amplified from plasmid pBZ1260 (2) with a mixture of Taq and Pfu polymerases (1:1), primer ToMO-KpnI-KACFront (Table 1), which generated a unique KpnI site upstream of the touA start codon, and primer ToMO-NotI-KACRear, which generated a unique NotI site downstream of the touF stop codon (Fig. 1). The PCR product was then cloned into the multiple cloning site in pBS(Kan) (5) after double digestion with KpnI and NotI to create pBS(Kan)ToMO (8,983 bp). Colonies expressing active ToMO were distinguished based on their dark blue color on LB agar plates.
TABLE 1.
Primers used for constructing pBS(Kan)ToMO, for sequencing the ToMO touABCDEF locus of P. stutzeri OX1 in pBS(Kan)ToMO and pBZ1260, and for mutagenizing the ToMO touA locus via DNA shuffling and saturation mutagenesis at codons for I100, Q141, T201, and F205
| Primer | Nucleotide sequence |
|---|---|
| Construction primers | |
| ToMO-KpnI-KACFront | 5′-GGTGCGGTACCACCCATTAGCG-3′ |
| ToMO-NotI-KACRear | 5′-CTCCTGCAGCGGCCGCTGTTAATGC-3′ |
| Sequencing primers | |
| ToMO1 | 5′-GCCAAGCGCGCAATTAACCCTC-3′ |
| ToMO2 | 5′-CCGTATGCCAATGTCAAGCGGAGC-3′ |
| ToMO3 | 5′-CGTCAGTTGCTTGATCTAGGGCTCG-3′ |
| ToMO4 | 5′-GGGTGACGACGCTCATGATTATGAG-3′ |
| ToMO5 | 5′-GGGCGTTGAAGGCGGCGATATG-3′ |
| ToMO6 | 5′-GGGCGGGGGATGTGGCGATTGCG-3′ |
| ToMO7 | 5′-GCCCACTCAAACACGATGACTGG-3′ |
| ToMO8 | 5′-GGTAGTCTTGCCCAATCCGAAGG-3′ |
| ToMO9 | 5′-GCAACTCCGGCGGGTGTGGG-3′ |
| ToMO10 | 5′-CGGTCTCGCCTACCTTCATCCG-3′ |
| Mutagenesis primers | |
| I100-front | 5′-CACTATGCAACTTCACTTCGGAGCGNNNGCACTTG-3′ |
| I100-rear | 5′-GCGGCGTATTCTTCAAGTGCNNNCGCTCCGAAGTG-3′ |
| Q141-front | 5′-CCGCCATGGGCAAATCNNNCTTTAC-3′ |
| Q141-rear | 5′-GGCATACGGAAAGTAAAGNNNGATTTGCCC-3′ |
| T201-front | 5′-CGCATTCGAAACAGGCTTCNNNAATATGC-3′ |
| T201-rear | 5′-CCGAGAAACTGCATATTNNNGAAGCC-3′ |
| F205-front | 5′-GCTTCACCAATATGCAGNNNCTCGG-3′ |
| F205-rear | 5′-CAGCGGCCAAACCGAGNNNCTGC-3′ |
| ToMO-SalI-rear | 5′-CCCACTCATAATCATGAGCGTCG-3′ |
| ToMO-KpnI-front | 5′-CCGGCTCGTATGTTGTGTGGAATTGTGAGCGG-3′ |
| ToMO-BstEII-rear | 5′-CCAGGATCTTGAGCGACGGTCCACCTTGCTGTGCG-3′ |
FIG. 1.
Vector pBS(Kan)ToMO used for constitutive expression of wild-type ToMO and mutants. KanR, kanamycin resistance gene. The six genes coding for ToMO are touABE (three-component hydroxylase, A2B2E2), touC (ferredoxin), touD (mediating protein), and touF (NADH-ferredoxin oxidoreductase).
Saturation mutagenesis and DNA shuffling of ToMO.
Saturation mutagenesis at residues I100, Q141, T201, and F205 of the alpha subunit (touA) of ToMO (499 amino acids) was performed as described previously (20), and Table 1 shows the primers used for cloning. To substitute 19 other amino acids at residue I100 of TouA, primers I100-front and I100-rear with NNN at the codon encoding I100 were used, and primers ToMO-KpnI-front and ToMO-BstEII-rear were upstream and downstream of the unique KpnI and BstEII restriction sites (Fig. 1) (BstEII occurs naturally in touA, and KpnI occurs in the multiple cloning site). Pfu polymerase (Stratagene, La Jolla, Calif.) was used in the PCR to minimize random point mutations, and pBS(Kan)ToMO was used as the template (Fig. 1). A 364-bp DNA fragment was amplified by using primers ToMO-KpnI-front and I100-rear, and a 385-bp DNA fragment was amplified by using primers I100-front and ToMO-BstEII-rear. After purification from agarose gels, the two fragments were combined at a 1:1 ratio as templates and used with the ToMO-KpnI-front and ToMO-BstEII-rear primers to obtain the full-length, 701-bp product that had random mutations at residue I100 of TouA and had two unique restriction enzyme sites. A PCR program consisting of 30 cycles of 94°C for 45 s, 55°C for 45 s, and 72°C for 2.15 min with final extension for 72°C for 7 min was used. The resulting PCR product was cloned into pBS(Kan)ToMO after double digestion with KpnI and BstEII, which replaced the corresponding fragment in the original plasmid. The resulting plasmid library was electroporated into E. coli TG1 competent cells by using a GenePulser/Pulse Controller (Bio-Rad, Hercules, Calif.) at 15 kV/cm, 25 μF, and 200 Ω.
Similarly, to substitute 19 other amino acids at residue Q141 of TouA, a 594-bp DNA fragment was amplified by using primers ToMO-KpnI-front and Q141-rear (Table 1), and a 332-bp DNA fragment was amplified by using primers Q141-front and ToMO-BstEII-rear (Table 1). The two fragments were combined at a 1:1 ratio as templates to obtain the full-length product (890 bp) with the ToMO-KpnI-front and ToMO-BstEII-rear primers, and the product was cloned by using KpnI and BstEII (Fig. 1).
To substitute 19 other amino acids at residue T201 of TouA, a 773-bp DNA fragment was amplified by using primers ToMO-KpnI-front and T201-rear (Table 1), and an 881-bp DNA fragment was amplified by using primers T201-front and ToMO-SalI-rear (Table 1). The two fragments were combined at a 1:1 ratio as templates to obtain the full-length product (1,615 bp) with the ToMO-KpnI-front and ToMO-SalI-rear primers, and the product was cloned by using MluI and SalI (Fig. 1).
To substitute 19 other amino acids at residue F205 of TouA, a 784-bp DNA fragment was amplified by using primers ToMO-KpnI-front and F205-rear (Table 1), and an 867-bp DNA fragment was amplified by using primers F205-front and ToMO-SalI-rear (Table 1). The two fragments were combined at a 1:1 ratio as templates to obtain the full-length product (1,615 bp) with the ToMO-KpnI-front and ToMO-SalI-rear primers, and the product was cloned by using MluI and SalI (Fig. 1).
DNA shuffling of 90% of touA was performed as described previously for the TOM gene (5). ToMO-KpnI-front and ToMO-SalI-rear (Table 1) were designed to be 50 to 100 bp upstream and downstream of the MluI and SalI natural restriction sites, respectively, used for cloning; these two sites lie within the coding region of touA so that the promoter and the ribosome binding site regions were not shuffled. The shuffled product was then cloned into plasmid pBS(Kan)ToMO to replace the 1,314-bp region between the natural MluI and SalI sites of the gene encoding wild-type ToMO with shuffled DNA (90% of touA) (Fig. 1). Amplifications were carried out with the GeneAmp 2400 PCR system (Perkin-Elmer, Norwalk, Conn.). A plasmid midi kit and a QIAquick gel extraction kit (QIAGEN, Inc., Chatsworth, Calif.) were used to isolate plasmids and DNA fragments from agarose gels, respectively. A Bio-Rad GenePulser/Pulse Controller was used for electroporation at 15 kV/cm, 25 μF, and 200 Ω.
Colony screening.
The plate assay which we used was a variation of the method described previously by Meyer et al. (19). The mutant libraries were first streaked from transformant plates onto plates containing LB agar supplemented with 100 μg of kanamycin per ml and 1% (wt/vol) glucose to turn off the tou operon in order to increase stability. The glucose-grown colonies were then transferred to fresh plates containing LB agar supplemented with 100 μg of kanamycin per ml and 1 mM substrate (toluene or benzene) with a nylon membrane. After incubation for 24 h at room temperature in a chamber, the colonies were checked visually to search for colonies that developed a dark brown-red color around the cell mass, indicating the formation of derivatives of hydroquinone, resorcinol, or catechol from toluene or benzene. The control expressing wild-type ToMO remained yellow to light red on toluene- or benzene-containing agar plates (indicating the formation of catechol derivatives only). The negative control expressing no monooxygenase, TG1/pBS(Kan), remained colorless on toluene- or benzene-containing plates. At least three replicates were checked before high-performance liquid chromatography (HPLC) analysis was performed.
Product identification and rates of formation.
The possible mutants initially identified by screening by the agar plate assay were further examined by reverse-phase HPLC. Experiments were conducted with exponentially grown cells at an optical density at 600 nm (OD600) of 1.0. The cells were washed once by centrifugation at 6,000 × g for 5 min at 25°C (JA-17 rotor in a J2 series centrifuge [Beckman, Palo Alto, Calif.]) and were resuspended with 1 volume of 50 mM Tris-HNO3 buffer (pH 7.0) to an OD600 of 5 to 10. Fifteen-milliliter glass vials containing the cell suspensions (2.5 ml) were each sealed with a Teflon-coated septum and aluminum seal, and the substrates benzene, phenol, toluene, o-cresol, m-cresol, p-cresol, catechol, hydroquinone, and resorcinol were added from ethanol stock solutions at a concentration of 0.8 mM. After contact at room temperature with shaking at 250 rpm in a KS250 benchtop shaker (IKA Laboratories, Cincinnati, Ohio) for 15 to 240 min, 1 ml of each cell suspension was centrifuged at 13,000 × g for 2 to 3 min, and the supernatant (500 μl) was filtered with a 1-ml syringe (Becton Dickinson) coupled to a nylon membrane filter unit (Millex-HN; pore size, 0.45 μm; diameter, 4 mm). By using HPLC, the dihydroxy and trihydroxy derivatives of benzene and toluene were analyzed immediately, and the monohyroxy derivatives of benzene or toluene were either kept at −20°C (for not more than 24 h) or analyzed immediately. A Zorbax SB-C8 column (5 μm; 4.6 by 250 mm; Agilent Technologies) was used with a 515 solvent delivery system (Waters Corporation, Milford, Mass.) coupled to a Waters 996 photodiode array detector. To detect the methyl-substituted catechols, methyl-substituted resorcinols, and methylhydroquinone and to determine their rates of formation, gradient elution was performed with H2O (0.1% formic acid) and acetonitrile (70:30 for 0 to 8 min, 40:60 for 15 min, and 70:30 for 20 min) as the mobile phase at a flow rate of 1 ml/min. To detect catechol, resorcinol, and hydroquinone and to determine their rates of formation, an isocratic mobile phase consisting of H2O (0.1% formic acid) and acetonitrile (70:30) was used. To detect the THBs formed from catechol, resorcinol, and hydroquinone, an isocratic mobile phase consisting of H2O (0.1% formic acid) and acetonitrile (90:10) was used. To confirm product identification, the retention times and UV-visible spectra of standard chemicals were compared with those of the enzyme-derived samples (Table 2), and the enzyme products were coeluted with authentic standards. At least two independent cultures were analyzed for each substrate and strain tested, and at least five injections were made for each substrate.
TABLE 2.
Retention times and maximum wavelengths of substrates and products used in HPLC analysis
| Compound | Retention time (min) | λmax (nm) |
|---|---|---|
| Toluenea | 20.0 | 264 |
| Benzenea | 19.4 | 274 |
| Benzyl alcohola | 7.5 | 258 |
| o-Cresola | 14.2 | 272 |
| m-Cresola | 13.6 | 278 |
| p-Cresola | 13.6 | 278 |
| 3-Methylcatechola | 8.7 | 274 |
| 4-Methylcatechola | 7.6 | 282 |
| 2-Methylresorcinola | 5.9 | 272 |
| 4-Methylresorcinola | 6.0 | 280 |
| 5-Methylresorcinola | 6.1 | 274 |
| Methylhydroquinonea | 5.2 | 290 |
| 2-Hydroxybenzyl alcohola | 5.1 | 274 |
| 3-Hydroxybenzyl alcohola | 4.5 | 274 |
| 4-Hydroxybenzyl alcohola | 4.0 | 274 |
| Phenolb | 8.7 | 271 |
| Catecholb | 5.1 | 276 |
| Catecholc | 12.8 | |
| Hydroquinoneb | 3.7 | 290 |
| Hydroquinonec | 6.5 | |
| Resorcinolb | 4.3 | 274 |
| Resorcinolc | 10.0 | |
| 1,2,3-THBc | 6.0 | 267 |
| 1,2,4-THBc | 4.6 | 288 |
Standard used for product identification by the gradient method for methylcatechols, methylresorcinols, methylhydroquinone, and hydroxybenzyl alcohols.
Standard used for product identification by the isocratic method (70:30) for the formation of catechol, hydroquinone, and resorcinol from phenol.
Standard used for product identification by the isocratic method (90:10) for the formation of 1,2,3-THB and 1,2,4-THB from catechol, hydroquinone, or resorcinol. The ratio of H2O (0.1% formic acid) to acetonitrile was changed from 70:30 to 90:10 to aid identificaton of the more hydrophilic THBs and to distinguish these compounds from the solvent peak obtained with the 70:30 method.
The initial product formation rates when a substrate concentration of 0.8 mM was used were determined by sampling at 15-min intervals for 2 h and were expressed in nanomoles per minute per milligram of protein by converting product peak areas to concentrations by using standard curves prepared at the specific absorbance wavelength (Table 2) for each product formed. The protein content was 0.22 mg of protein/ml/OD600 unit for recombinant E. coli TG1, as determined with a protein assay kit (Sigma Diagnostics Inc., St. Louis, Mo.). The whole-cell catalytic parameters Vmax and Km were determined for formation of catechol from phenol (25, 50, 100, 200, and 400 μM) for wild-type ToMO and the M180T/E284G variant, as well as for formation of 4-methylcatechol from p-cresol (25, 50, 100, 200, and 400 μM) for wild-type ToMO and the I100Q variant. The experiments involved HPLC measurement of the initial rates of formation of catechol from 25, 50, 100, 200, and 400 μM phenol.
Toluene oxidation and regiospecificity.
To determine the toluene oxidation products, experiments were conducted with exponentially grown cells at an OD600 of 1.0. The cells were washed once by centrifugation at 6,000 × g for 5 min at 25°C (JA-17 rotor in a J2 series centrifuge [Beckman]), resuspended with 1 volume of 50 mM Tris-HNO3 (pH 7.0) to an OD600 of 10, and then incubated with 91 μM toluene (based on Henry's law [7]) at room temperature and harvested every 5 min. Hexadecane (0.5 mM) was used as an internal standard for calculations (retention time, 17.8 min) and was added prior to extraction. Ethyl acetate-hexadecane (0.5 mM) was used to extract the toluene by adding 1 ml to 2 ml of a cell suspension. The suspension was centrifuged for 1 to 2 min, and the ethyl acetate phase (upper phase) was analyzed by gas chromatography by using a 6890N gas chromatograph (Hewlett-Packard, Wilmington, Del.) equipped with an EC-WAX capillary column (30 m by 0.25 mm; thickness, 0.25 μm; Alltech Associates, Inc., Deerfield, Ill.) and a flame ionization detector. The injector and detector temperatures were maintained at 250 and 275°C, respectively, and a split ratio of 3:1 was used. The He carrier gas flow rate was maintained at 0.8 ml/min. The temperature program was as follows: 80°C for 5 min, increase from 80 to 205°C at a rate of 5°C/min, increase from 205 to 280°C at a rate of 15°C/min, and 280°C for 5 min. Under these conditions, p- and m-cresol are separated, and the retention times for toluene and o-, p-, and m-cresol were 4.2, 27.6, 29.3, and 29.5 min, respectively. The experiments were performed at least two times for each strain tested. The molar amount of toluene degraded and the molar amounts of o-, m-, and p-cresol formed were calculated by constructing a calibration curve, and the retention times were compared with the retention times of standards.
DNA sequencing.
A dideoxy chain termination procedure (33) with an ABI Prism BigDye terminator cycle sequencing Ready Reaction kit (Perkin-Elmer, Wellesley, Mass.) and a PE Biosystems ABI 373 DNA sequencer (Perkin-Elmer) was used to determine the ToMO nucleotide sequence. Ten primers (Table 1) that were 20 to 25 bp long were generated from the wild-type ToMO sequence (GenBank accession number AJ005663) (3) for sequencing of the ToMO touABCDEF locus in one direction for pBS(Kan)ToMO (Fig. 1) and pBZ1260. The sequence data generated were analyzed by using the Vector NTI software (InfoxMax, Inc., Bethesda, Md.).
Modeling of ToMO TouA.
Part of the wild-type ToMO TouA alpha subunit (amino acid residues P65 to E284) was modeled using SWISS-MODEL Server (13, 23, 34) and was based on the soluble MMO (sMMO) MmoX alpha subunit (polymer chain D) from Methylococcus capsulatus (Bath) (27). The I100Q, F205G, and M180T mutations were modeled from the wild-type TouA ToMO model by use of the Swiss-Pdb Viewer program (DeepView) (13, 23, 34). The program Swiss-Pdb Viewer made the amino acid substitutions isosterically for ToMO TouA based on residue interactions, steric hindrance, and energy minimization.
RESULTS
Screening.
DNA shuffling of 90% of touA and saturation mutagenesis at the positions encoding four different amino acids (I100, Q141, T201, and F205) in touA were performed to create mutations in ToMO in order to synthesize novel di- and trihydroxy aromatic compounds from benzene, phenol, toluene, o-cresol, m-cresol, p-cresol, catechol, hydroquinone, and resorcinol. A library of 8,000 mutants was generated for saturation mutagenesis. The number of colonies in saturation mutagenesis that had to be screened in order to check all possible substitutions with a 99% probability was calculated to be 293 (29); hence, around 500 colonies for each residue (a total of 4,000 colonies) were screened for toluene and benzene by using the agar plate assay. Formation of a dark red-brown halo indicated that catechol, hydroquinone, or resorcinol or methyl derivatives of these compounds were formed. For DNA shuffling of touA, an additional 2,000 colonies were screened.
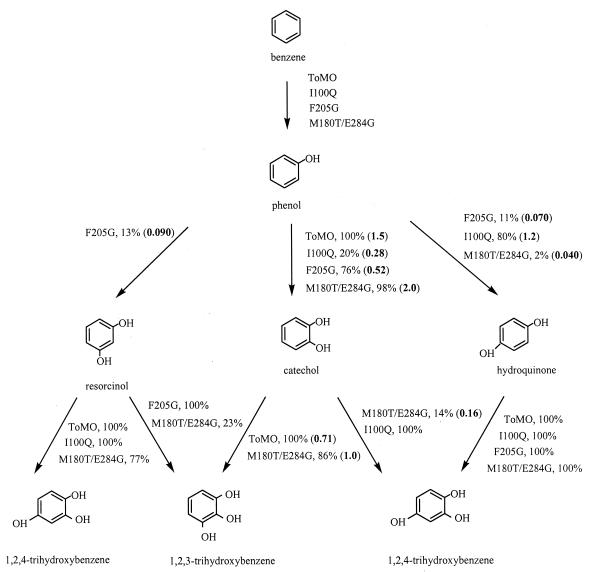
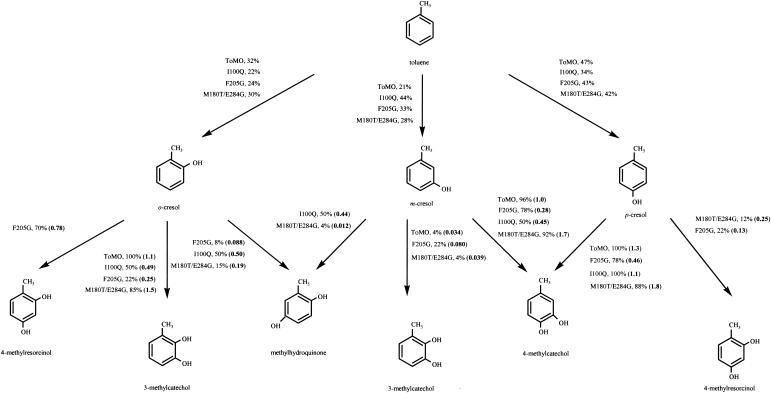
We identified mutants that formed dark red-brown halos with amino acid changes at TouA residues I100 and F205 (by saturation mutagenesis) and at residues M180 and E284 (by DNA shuffling). No benzene or toluene mutants that formed dark red-brown halos were found after saturation mutagenesis for TouA residue T201 or Q141. Wild-type ToMO and mutant cultures were further examined by HPLC, and Fig. 2 and 3 show both the products and the initial reaction rates for the oxidation of benzene, phenol, toluene, o-cresol, m-cresol, p-cresol, catechol, hydroquinone, and resorcinol. The best mutants were sequenced to identify the beneficial amino acid changes (Table 3).
FIG. 2.
Pathways for the oxidation of benzene (0.8 mM) to phenol, for the oxidation of phenol (0.8 mM) to dihydroxybenzenes, and for the oxidation of dihydroxybenzenes (0.8 mM) to trihydroxybenzenes by E. coli TG1/pBS(Kan)ToMO expressing wild-type ToMO and the I100Q, F205G, and M180T/E284G TouA variants. The molar product percentages are indicated; the values in boldface type in parentheses indicate the product formation rates (in nanomoles per minute per milligram of protein).
FIG. 3.
Pathways for the oxidation of toluene (0.8 mM) to o-cresol, m-cresol, and p-cresol and for the oxidation of o-cresol (0.8 mM), m-cresol (0.8 mM), and p-cresol (0.8 mM) to methylcatechols, methyl-resorcinols, and methylhydroquinone by E. coli TG1/pBS(Kan)ToMO expressing wild-type ToMO and the I100Q, F205G, and M180T/E284G TouA variants. The molar product percentages are indicated; the values in boldface type in parentheses indicate the product formation rates (in nanomoles per minute per milligram of protein).
TABLE 3.
Rates of substrate utilization by TG1/pBS(Kan)ToMO expressing wild-type ToMO and TouA variants I100Q, F205G, and M180T/E284G
| Enzyme | Substrate utilization rates (nmol consumed/min/mg of protein)
|
||||
|---|---|---|---|---|---|
| Phenol | o-Cresol | m-Cresol | p-Cresol | Catechol | |
| Wild type | 1.8 | 1.1 | 1.0 | 1.4 | 0.75 |
| I100Q | 1.5 | 1.0 | 0.86 | 1.1 | NDa |
| F205G | 1.1 | 1.0 | 0.34 | 0.52 | ND |
| M180T/E284G | 2.8 | 2.2 | 1.5 | 2.2 | 1.1 |
ND, not determined.
Oxidation of benzene by wild-type ToMO and TouA variants.
The pathways for oxidation of benzene to phenol, dihydroxybenzene derivatives, and trihydroxybenzene derivatives with wild-type ToMO and the I100Q, F205G, and M180T/E284G TouA variants are shown in Fig. 2. Previously, it was reported that ToMO hydroxylates benzene to phenol (2). Here, we discovered that E. coli TG1/pBS(Kan)ToMO expressing wild-type ToMO performs a second hydroxylation and forms catechol at an initial rate of 1.5 nmol/min/mg of protein from 0.8 mM phenol (Fig. 2). We also discovered that ToMO further hydroxylates catechol to form 1,2,3-THB (0.71 nmol/min/mg of protein). The negative control, TG1 expressing pBS(Kan), did not form catechol or 1,2,3-THB. Similarly, we found that 1,2,3-THB is formed from catechol with other wild-type aromatic monooxygenases, such as T3MO of P. pickettii PKO1, T4MO of P. mendocina KR1, and TOM of B. cepacia G4 (35).
For both wild-type ToMO and the TouA variants, there was good agreement between the rates of disappearance of all of the substrates (phenol, o-cresol, m-cresol, p-cresol, and catechol) and the overall rates of appearance of the products (THB, catechol, hydroquinone, resorcinol, 3-methylcatechol, 4-methylcatechol, methylhydroquinone, and 4-methylresorcinol) (Table 3). For example, with wild-type ToMO, the rate of formation of 1,2,3-THB (0.71 nmol/min/mg of protein) and the rate of disappearance of catechol (0.75 nmol/min/mg of protein) were consistent.
The I100Q TouA variant hydroxylated benzene and formed phenol like wild-type ToMO (Fig. 2), and then it hydroxylated phenol at positions 2 and 4 and formed primarily hydroquinone (80%), as well as catechol (20%), whereas wild-type ToMO performed the second hydroxylation only at position 2 and formed catechol. The rate of hydroquinone formation by the I100Q variant was comparable to the rate of catechol formation by the wild type. The I100Q variant further hydroxylated catechol to form 1,2,4-THB instead of the 1,2,3-THB formed by wild-type ToMO. Both the I100Q variant and wild-type ToMO formed 1,2,4-THB from hydroquinone and resorcinol (Fig. 2); these reactions are two previously unknown reactions for wild-type ToMO.
The F205G TouA variant hydroxylated benzene and formed phenol (like both wild-type ToMO and the I100Q variant) (Fig. 2); however, it hydroxylated phenol at positions 2, 3, and 4, and it formed significant amounts of resorcinol (13%), catechol (76%), and hydroquinone (11%), whereas wild-type ToMO formed only catechol. The F205G variant further hydroxylated hydroquinone and formed 1,2,4-THB like wild-type ToMO, but it formed 1,2,3-THB (100%) from resorcinol, which is different from the wild-type ToMO and I100Q variant reactions. No product peak was observed when we used 0.8 mM catechol with TG1 expressing variant F205G.
The M180T/E284G TouA variant hydroxylated benzene and formed phenol like wild-type ToMO and the I100Q and F205G variants. The M180T/E284G variant performed the second hydroxylation at positions 2 and 4 and formed catechol (98%), as well as hydroquinone (2%); the rate of catechol formation was 33% higher than the wild-type rate at a concentration of 0.8 mM. The M180T/E284G variant further hydroxylated hydroquinone and formed 1,2,4-THB, like wild-type ToMO. In contrast to wild-type ToMO, the M180T/E284G variant formed both 1,2,3-THB (86%) and 1,2,4-THB (14%) from catechol. The M180T/E284G variant also formed both 1,2,3-THB and 1,2,4-THB from resorcinol, unlike wild-type ToMO (Fig. 2).
To investigate the enhanced rate of catechol formation by the M180T/E284G variant compared to the rate of formation by wild-type ToMO, the apparent Vmax was measured for whole cells, and the values were found to be 5.7 and 6.1 nmol/min/mg of protein for the M180T/E284G variant and the wild type, respectively, whereas the apparent Km was 7.5 μM for the M180T/E284G variant and 25 μM for wild-type ToMO. Hence, Vmax/Km was threefold greater for the M180T/E284G variant. The rates of formation of 3-methylcatechol and 4-methylcatechol for the M180T/E284G variant were also greater than the rates of formation for wild-type ToMO (1.4- to 1.7-fold) (Fig. 3).
Oxidation of toluene by wild-type ToMO and TouA variants.
Oxidation of toluene as a substrate was also evaluated with the enzyme variants to see whether addition of methyl groups to the benzene ring affected the regiospecificity and to determine if the oxidation rate of the natural substrate was altered significantly. Table 4 summarizes the products obtained from whole-cell oxidation of toluene with wild-type ToMO and with the I100Q, F205G, and M180T/E284G TouA variants. It was shown previously that wild-type ToMO hydroxylates toluene to o-cresol, m-cresol, and p-cresol and 3- and 4-methylcatechol (2), and the same results were obtained in this study, except that slight 3-methylcatechol formation (4%) from m-cresol was found in addition to formation of 4-methylcatechol (96%) (Fig. 3).
TABLE 4.
Toluene oxidation rates and regiospecificity for TG1/pBS(Kan)ToMO expressing wild-type ToMO and TouA variants I100Q, F205G, and M180T/E284G as determined by gas chromatographya
| Enzyme | Oxidation rate (nmol/min/mg of protein) | Regiospecificity (%)
|
||
|---|---|---|---|---|
| o-Cresol | m-Cresol | p-Cresol | ||
| Wild type | 2.63 ± 0.15 | 32 | 21 | 47 |
| I100Q | 1.53 ± 0.08 | 22 | 44 | 34 |
| F205G | 1.07 ± 0.03 | 24 | 33 | 43 |
| M180T/E284G | 3.12 ± 0.05 | 32 | 26 | 42 |
The initial toluene concentration was 91 μM based on Henry's law (250 μM if all of the volatile organic compound was in the liquid phase).
To determine the toluene activity and regiospecificity, gas chromatography was used rather than HPLC since m-cresol and p-cresol were not separated under the HPLC conditions used. The wild-type product distribution agrees well with that reported previously by Bertoni et al. (2). Both the I100Q and F205G TouA mutations caused a shift in product distribution for the first hydroxylation of toluene and resulted in elevated m-cresol formation at the expense of o-cresol formation (Table 4). The M180T/E284G mutation caused no substantial shift in the product distribution for the first hydroxylation (32% o-cresol, 26% m-cresol, and 42% p-cresol). The oxidation rates for toluene are also shown in Table 4; the M180T/E284G variant oxidized toluene (91 μM according to Henry's law) slightly faster than the wild type oxidized toluene (20%).
The I100Q and F205G variants had a different regiospecificity for the second hydroxylation of toluene. Figure 3 shows the oxidation of o-cresol with wild-type ToMO and with the I100Q, F205G, and M180T/E284G TouA variants. Wild-type ToMO formed only 3-methylcatechol; however, the I100Q variant formed methylhydroquinone (50%) and 3-methylcatechol (50%). Unlike wild-type ToMO, the F205G variant formed 4-methylresorcinol (70%), 3-methylcatechol (22%), and methylhydroquinone (8%) from o-cresol. The M180T/E284G variant formed 3-methylcatechol (85%) at slightly elevated rates compared with the wild-type isoform rate, along with methylhydroquinone (15%).
Figure 3 shows the pathways for oxidation of m-cresol. Wild-type ToMO formed 4-methylcatechol (96%) and 3-methylcatechol (4%). Unlike wild-type ToMO, the I100Q variant formed methylhydroquinone (50%) and 4-methylcatechol (50%). Like wild-type ToMO, the F205G variant formed 4-methylcatechol (78%) and 3-methylcatechol (22%); however, the F205G variant formed 3-methylcatechol 2.4-fold faster from 0.8 mM m-cresol. Unlike wild-type ToMO and the I100Q and F205G variants, the M180T/E284G variant formed 4-methylcatechol (92%; 1.7-fold faster than the wild type), 3-methylcatechol (4%), and methylhydroquinone (4%).
Figure 3 also shows the pathways for oxidation of p-cresol by ToMO and the variants. ToMO and the I100Q variant formed only 4-methylcatechol at comparable rates; however, the F205G and M180T/E284G variants formed both 4-methylcatechol (78 and 88%, respectively) and 4-methylresorcinol (22 and 12%, respectively).
To characterize the conversion of p-cresol to 4-methylcatechol more fully (Fig. 3), the apparent Vmax values for whole cells were found to be 3.3 nmol/min/mg of protein for the I100Q variant and 2.7 nmol/min/mg of protein for the wild type. The apparent Km values were 111 μM for the I100Q variant and 58 μM for the wild type; hence, Vmax/Km was reduced 63% for the I100Q variant.
All of the methyl-substituted catechol, methylhydroquinone, and methyl-substituted resorcinol samples were relatively unstable due to nonenzymatic reactions since standards incubated with Tris-HNO3 buffer (no cells) overnight formed unknown compounds (as observed by HPLC). However, this did not affect our results because the analyses were done in the first 4 h.
DNA sequence.
By using primers (Table 1) generated from the ToMO sequence in the GenBank database (accession number AJ005663) (3), the entire wild-type ToMO locus from plasmid pBS(Kan)ToMO (touA region sequenced at least 10 times in the same direction) (Fig. 1) and the ToMO locus of pBZ1260 (2) were sequenced. pBZ1260 was the source of our ToMO tou locus and the original source of the GenBank DNA sequence (3). Both touABCDEF in pBS(Kan)ToMO and touABCDEF in pBZ1260 have the S269C codon change in touF [TCT for the GenBank sequence and TGT for the pBS(Kan)ToMO and pBZ1260 sequences]. In addition, pBS(Kan)ToMO has the K445E codon change in touA [AAA for the GenBank sequence and GAA for the pBS(Kan)ToMO sequence] and the N283S codon change in touF [AAT for the GenBank sequence and AGT for the pBS(Kan)ToMO sequence], which do not affect activity (there is no change in the regiospecific oxidation of toluene).
Enzyme expression level.
The I100Q and F205G TouA variants were expression down mutants, as shown by SDS-PAGE; a single nucleotide change in one codon led to a less elevated protein expression level (1.5- to 2-fold). Hence, the enzymes were even more active than the rates shown in Fig. 2 and 3 indicate. Similar changes in expression levels due to mutations have been observed with naphthalene dioxygenase and para-nitrobenzyl esterase (21, 31). The variation in the protein expression level could have been due to modification of the primary amino acid sequence, which led to an increase in protein lability, or the single nucleotide change may have led to increased lability of the transcript (the ribosome binding site and promoter were not altered). The expression level of the M180T/E284G variant was approximately the same as that of wild-type ToMO.
ToMO TouA modeling.
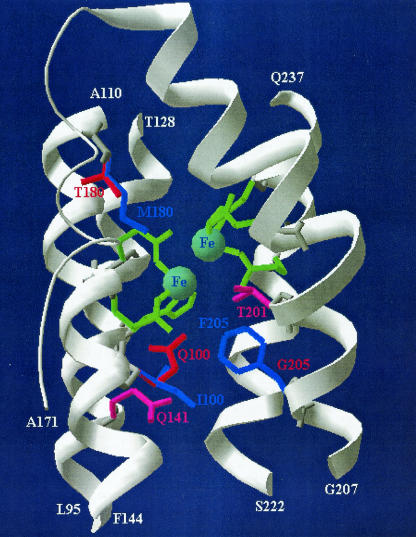
The approximate three-dimensional coordinates for the TouA four-helix bundle anchoring the active site are shown in Fig. 4 (the model was based on the crystal structure of sMMO). The accuracy of the wild-type ToMO TouA alpha-subunit model was judged by conservation of the spatial positions of the diiron coordinating residues in ToMO (E104, E134, H137, E197, E231, and H234) compared to the residues of sMMO (E114, E144, H147, E209, E243, and H246) (27).
FIG. 4.
Active site of ToMO TouA alpha-subunit three-dimensional structure model constructed by using the crystal structure of sMMO from M. capsulatus (Bath) as the template. Residues in green (E110, E140, H143, E201, E235, and H238) are metal binding sites that form the diiron center (light blue). The I100Q, F205G, and M180T mutations are shown in red, and the wild-type I100, F205, and M180 residues are shown in blue. Saturation mutagenesis at Q141 and T201 resulted in no regiospecific mutants and is indicated by pink. Portions of the four-helix bundle of TouA (helix B, P87 to F117; helix C, P121 to K150; helix E, S185 to E214; and helix F, T219 to Q237) anchoring the diiron active site are shown in white; these regions end with L95 and A110 (helix B), T128 and F144 (helix C), A171 and G207 (loop plus helix E), and S222 and Q237 (helix F).
Although homology modeling has limitations (12, 34) (only 30% sequence identity for the modeled part in this study), the model did help us visualize the positions of the side chains for the I100Q, M180T, and F205G variants (Fig. 4). The E284G amino acid substitution in the M180T/E284G variant is not near the active site (data not shown). If the proximal location of the mutations with respect to the active site (Fig. 4) is considered, it appears that the I100Q, M180T, and F205G mutations might lead to variations in the active site shape as the volume occupied by the side chains is altered. The substantial differences in the regiospecific oxidation of phenol, o-cresol, m-cresol, p-cresol, catechol, and resorcinol by the I100Q, M180T/E284G, and F205G variants suggest that these substrates dock in the active site in an altered manner when these residues are changed.
DISCUSSION
We show clearly in this paper that in addition to forming 3- and 4-methylcatechol from o-, m-, and p-cresol, wild-type ToMO is capable of two additional, successive oxidations of phenol to form catechol and 1,2,3-THB (Fig. 2), and wild-type ToMO forms 1,2,4-THB from either resorcinol or hydroquinone and generates a small amount of 3-methylcatechol from m-cresol. Furthermore, the regiospecific hydroxylation of the substrates phenol, o-cresol, m- cresol, p-cresol, catechol, and resorcinol may be changed by introducing the I100Q, F205G, and M180T/E284G mutations into ToMO TouA. These mutations did not alter the first hydroxylation of toluene significantly (Table 4), since the mutants and the wild type are nonspecific, but they caused different second regiospecific hydroxylations of toluene or benzene to occur (Fig. 2 and 3). These mutations facilitate the synthesis of either resorcinol or hydroquinone from phenol, the synthesis of 1,2,3-THB from resorcinol, the synthesis of 1,2,4-THB from catechol, the synthesis of 4-methylresorcinol from either o-cresol or p-cresol, and the synthesis of methylhydroquinone from either o-cresol or m-cresol. Hence, by using ToMO and the I100Q, F205G, and M180T/E284G TouA mutants, it is now possible to form all the dihydroxy derivatives of the benzene ring (except 2-methylresorcinol and 5-methylresorcinol) from benzene, o-cresol, m-cresol, and p-cresol with a single enzyme and its variants.
Previously, it was shown that a phenol-acclimated activated sludge formed 3-methylcatechol (the main product), 4-methylresorcinol, and methylhydroquinone from o-cresol (17, 18), but the microorganisms and the enzymes responsible were not elucidated. A pathway for the degradation of o-cresol by Pseudomonas sp. strain CP4 has been proposed (but not confirmed); in this pathway 4-methylresorcinol is a possible product and 3-methlycatechol is a confirmed product (1).
Pikus et al. studied one substitution in the related enzyme T4MO, I180F, but this change resulted in no change in the product distribution for the oxidation of toluene, m-xylene, and p-xylene (25). Here, we report that M180 of ToMO TouA, which corresponds to I180 of T4MO TmoA and which was identified by DNA shuffling, might play an important role in the regiospecificity of the oxidation of phenol, o-cresol, m-cresol, p-cresol, catechol, and resorcinol. The M180T/E284G variant obtained from DNA shuffling has a second amino acid change at TouA residue E284, which may also help change the regiospecificity of oxidation by this mutant. The significance of residue E284 has not been described previously. The M180T/E284G variant is also an up-mutant for nitrobenzene and nitrophenol oxidation (data not shown). Thus, in the future these two residues (M180 and E284) will be studied by using saturation mutagenesis.
The whole-cell rate of formation of catechol from phenol (1.5 nmol/min/mg of protein) and the whole-cell rate of formation of 1,2,3-THB from catechol (0.71 nmol/min/mg of protein) were found to be comparable to the oxidation rate of the natural substrate toluene (2.63 ± 0.15 nmol/min/mg of protein) by whole cells expressing wild-type ToMO (Fig. 2 and Tables 3 and 4). Hence, the rates of these two newly discovered reactions are relatively high, although the reactions do not appear to be physiologically relevant. The initial rates of formation of the uncommon chemical 4-methylresorcinol by the F205G and M180T/E284G variants and the rates of methylhydroquinone formation by the I100Q variant from o- and m-cresol were also high. The overall rates of product formation from phenol, o-cresol, m-cresol, and p-cresol with the I100Q and F205G variants were as high as the wild-type ToMO rates of product formation, whereas with the M180T/E284G variant the product formation rates were slightly greater (1.3- to 1.6-fold); hence, regiospecificity was changed for these variants without reducing the rates. In contrast, some of the recently described P. mendocina KR1 T4MO regiospecific mutants (24, 25) have lower activities; for example, the rate of toluene oxidation (kcat) was around 10-fold lower with a Q141C T4MO mutant than with the wild-type T4MO (25); the rates of toluene oxidation were also lower with other T201 T4MO mutants that were selected to be active (24).
Methyl substitution on the benzene ring did not affect the regiospecificity of oxidation by wild-type ToMO and TouA variants. For example, wild-type ToMO formed catechol from benzene and methylcatechols from toluene. The I100Q and M180T/E284G variants formed catechol and hydroquinone from benzene and methylcatechols and methylhydroquinone from toluene. The F205G variant, on the other hand, formed catechol, hydroquinone, and resorcinol from benzene, as well as methylcatechols, methylhydroquinone, and 4-methylresorcinol from toluene.
The regiospecificities of hydroxylation by the I100Q, F205G, and M180T/E284G TouA variants were different from that of the wild-type ToMO; hence, residues I100, F205, and M180/E284 of the alpha subunit TouA of ToMO (499 amino acids) govern phenol, o-cresol, m-cresol, p-cresol, catechol, and resorcinol hydroxylation. These results also confirm that the ToMO hydroxylase subunit influences hydroxylation regiospecificity. The I100Q mutation also increased TCE degradation with ToMO (data not shown), as observed for variant Tom-Green of B. cepacia G4, which has an amino acid change at residue 106 (V106A) that corresponds to the change at residue I100 in ToMO (5). Hence, the results here support the hypothesis that residue I100 is a key amino acid residue in the alpha subunit of toluene monooxygenases.
Acknowledgments
This study was supported by the U.S. Army Research (grant DAAD19-00-1-0568).
We thank Ayelet Fishman for her help with the HPLC analysis and acknowledge that K. A. Canada of the Wood lab constructed strain E. coli TG1/pBS(Kan)ToMO.
REFERENCES
- 1.Ahamad, P. Y. A., A. A. M. Kunhi, and S. Divakar. 2001. New metabolic pathway for o-cresol degradation by Pseudomonas sp. CP4 as evidenced by H NMR spectroscopic studies. World J. Microbiol. Biotechnol. 17:371-377. [Google Scholar]
- 2.Bertoni, G., F. Bolognese, E. Galli, and P. Barbieri. 1996. Cloning of the genes for and characterization of the early stages of toluene and o-xylene catabolism in Pseudomonas stutzeri OX1. Appl. Environ. Microbiol. 62:3704-3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bertoni, G., M. Martino, E. Galli, and P. Barbieri. 1998. Analysis of the gene cluster encoding toluene/o-xylene monooxygenase from Pseudomonas stutzeri OX1. Appl. Environ. Microbiol. 64:3626-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cafaro, V., R. Scognamiglio, A. Viggiani, V. Izzo, I. Passaro, E. Notomista, F. D. Piaz, A. Amoresano, A. Casbarra, P. Pucci, and A. D. Donato. 2002. Expression and purification of the recombinant subunits of toluene/o-xylene monooxygenase and reconstitution of the active complex. Eur. J. Biochem. 269:5689-5699. [DOI] [PubMed] [Google Scholar]
- 5.Canada, K. A., S. Iwashita, H. Shim, and T. K. Wood. 2002. Directed evolution of toluene ortho-monooxygenase for enhanced 1-naphthol synthesis and chlorinated ethene degradation. J. Bacteriol. 184:344-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chauhan, S., P. Barbieri, and T. K. Wood. 1998. Oxidation of trichloroethylene, 1,1-dichloroethylene, and chloroform by toluene/o-xylene monooxygenase from Pseudomonas stutzeri OX1. Appl. Environ. Microbiol. 64:3023-3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dolfing, J., A. J. V. D. Wijngaard, and D. B. Janssen. 1993. Microbiological aspects of the removal of chlorinated hydrocarbons from air. Biodegradation 4:261-282. [DOI] [PubMed] [Google Scholar]
- 8.Dressler, H. 1994. Resorcinol. Its uses and derivatives, 1st ed. Plenum Press, New York, N.Y.
- 9.Elango, N., R. Radhakrishnan, W. A. Froland, B. J. Wallar, C. A. Earhart, J. D. Lipscomb, and D. H. Ohlendorf. 1997. Crystal structure of the hydroxylase component of methane monooxygenase from Methylosinus trichosporium OB3b. Protein Sci. 6:556-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gibson, T. J. 1984. Studies on the Epstein-Barr virus genome. Ph.D. thesis. Cambridge University, Cambridge, England.
- 11.Gillner, M., G. S. Moore, H. Cederberg, and K. Gustafsson. 1994. Hydroquinone, environmental health criteria 157. International Programme on Chemical Safety, Geneva Switzerland.
- 12.Guex, N., A. Diemand, and M. C. Peitsch. 1999. Protein modeling for all. Trends Biotechnol. 24:364-367. [DOI] [PubMed] [Google Scholar]
- 13.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modelling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 14.Hisaindee, S., and D. L. J. Clive. 2001. A synthesis of puraquinonic acid. Tetrahedron Lett. 42:2253-2255. [Google Scholar]
- 15.Krolikowska, A., W. Bokszczanin, A. Kozlowski, and T. Dzikowicz. April. 1991. Mixtures of dihydroxybenzene derivatives and alkylphenols as rust inhibitors for paints. Poland patent 153,464.
- 16.Macias, F. A., D. Marin, D. Chinchilla, and J. M. G. Molinillo. 2002. First total synthesis of (+/-)-helibisabonol A. Tetrahedron Lett. 43:6417-6420. [Google Scholar]
- 17.Masunaga, S., Y. Urushigawa, and Y. Yonezawa. 1986. Biodegradation pathway of o-cresol by heterogeneous culture. Phenol acclimated activated sludge. Water Res. 20:477-484. [Google Scholar]
- 18.Masunaga, S., Y. Urushigawa, and Y. Yonezawa. 1983. Microbial transformation of o-cresol to dihydroxytoluenes by phenol acclimated activated sludge. Chemosphere 12:1075-1082. [Google Scholar]
- 19.Meyer, A., A. Schmid, M. Held, A. H. Westphal, M. Röthlisberger, H. E. Kohler, W. J. H. V. Berkel, and B. Witholt. 2002. Changing the substrate reactivity of 2-hydroxybiphenyl 3-monooxygenase from Pseudomonas azelaica HBP1 by directed evolution. J. Biol. Chem. 277:5575-5582. [DOI] [PubMed] [Google Scholar]
- 20.Miyazaki, K., and F. H. Arnold. 1999. Exploring nonnatural evolutionary pathways by saturation mutagenesis: rapid improvement of protein function. J. Mol. Evol. 49:716-720. [DOI] [PubMed] [Google Scholar]
- 21.Moore, J. C., and F. H. Arnold. 1996. Directed evolution of a para-nitrobenzyl esterase for aqueous-organic solvents. Nat. Biotechnol. 14:458-467. [DOI] [PubMed] [Google Scholar]
- 22.Othmer, K. 1991. Kirk-Othmer encyclopedia of chemical technology, 4th ed. Wiley-Interscience Publishers, New York, N.Y.
- 23.Peitsch, M. C. 1995. Protein modeling by E-mail. Bio/Technology 13:658-660. [Google Scholar]
- 24.Pikus, J. D., K. H. Mitchell, J. M. Studts, K. McClay, R. J. Steffan, and B. G. Fox. 2000. Threonine 201 in the diiron enzyme toluene 4-monooxygenase is not required for catalysis. Biochemistry 39:791-799. [DOI] [PubMed] [Google Scholar]
- 25.Pikus, J. D., J. M. Studts, K. McClay, R. J. Steffan, and B. G. Fox. 1997. Changes in the regiospecificity of aromatic hydroxylation produced by active site engineering in the diiron enzyme toluene 4-monooxygenase. Biochemistry 36:9283-9289. [DOI] [PubMed] [Google Scholar]
- 26.Robinson, G. K., G. M. Stephens, H. Dalton, and P. J. Geary. 1992. The production of catechols from benzene and toluene by Pseudomonas putida in glucose fed-batch culture. Biocatalysis 6:81-100. [Google Scholar]
- 27.Rosenzweig, A. C., H. Brandstetter, D. A. Whittington, P. Nordlund, S. J. Lippard, and C. A. Frederick. 1997. Crystal structures of the methane monooxygenase hydroxylase from Methylococcus capsulatus (Bath): implications for substrate gating and component interactions. Proteins Struct. Funct. Genet. 29:141-152. [PubMed] [Google Scholar]
- 28.Rosenzweig, A. C., P. Nordlund, P. M. Takahara, C. A. Frederick, and S. J. Lippard. 1995. Geometry of the soluble methane monooxygenase catalytic diiron center in two oxidation states. Chem. Biol. 2:409-418. [PubMed] [Google Scholar]
- 29.Rui, L., Y. M. Kwon, A. Fishman, K. F. Reardon, and T. K. Wood. 2004. Saturation mutagenesis of toluene ortho-monooxygenase from Burkholderia cepacia G4 for enhanced 1-naphthol synthesis and chloroform degradation. Appl. Environ. Microbiol. 70:3246-3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ryoo, D., H. Shim, K. Canada, P. Barbieri, and T. K. Wood. 2000. Aerobic degradation of tetrachloroethylene by toluene-o-xylene monooxygenase of Pseudomonas stutzeri OX1. Nat. Biotechnol. 18:775-778. [DOI] [PubMed] [Google Scholar]
- 31.Sakamoto, T., J. M. Joern, A. Arisawa, and F. H. Arnold. 2001. Laboratory evolution of toluene dioxygenase to accept 4-picoline as a substrate. Appl. Environ. Microbiol. 67:3882-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwede, T., J. Kopp, N. Guex, and M. C. Peitsch. 2003. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 31:3381-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tao, Y., A. Fishman, W. E. Bentley, and T. K. Wood. Oxidation of benzene to phenol, catechol, and 1,2,3-trihydroxybenzene by toluene 4-monooxygenase of Pseudomonas mendocina KR1 and toluene 3-monooxygenase of Ralstonia pickettii PKO1. Appl. Environ. Microbiol., in press. [DOI] [PMC free article] [PubMed]
- 36.Tice, R. 1998. Review of toxicological literature, pyrogallol 87-66-1. National Toxicology Program, Durham, N.C.
- 37.van Beilen, J. B., W. A. Duetz, S. A., and B. Witholt. 2003. Practical issues in the application of oxygenases. Trends Biotechnol. 21:170-177. [DOI] [PubMed] [Google Scholar]
- 38.Yonezawa, T. March. 2003. Capacitor electrolytes containing dihydroxytoluenes for low specific resistance. Japan patent 2,003,068,585.